Abstract
By directly monitoring stirred protein solutions with Raman spectroscopy, the reversible unfolding of proteins caused by fluid shear is examined for several natural proteins with varying structural properties and molecular weight. While complete denaturation is not observed, a wide range of spectral variances occur for the different proteins, indicating subtle conformational changes that appear to be protein-specific. A number of significant overall trends are apparent from the study. For globular proteins, the overall extent of spectral variance increases with protein size and the proportion of β-structure. For two less structured proteins, fetuin and α-casein, the observed changes are of relatively low magnitude, despite the greater molecular structural mobility of these proteins. This implies that other protein-specific factors, such as posttranslational modifications, may also be significant. Individual band changes occurring in the spectral profiles of each individual protein are also discussed in detail.
Introduction
Protein unfolding has been, and continues to be, extensively studied due to its relevance not only to protein engineering but also to biological processes including misfolding-related pathologies such as Alzheimer's and Parkinson's diseases. Spectroscopic techniques such as circular dichroism, infrared, and Raman spectroscopy are increasingly being applied to investigations of conformational transitions in proteins due to their ability to follow perturbation-induced unfolding and folding processes (1,2). Several different physicochemical parameters, including pH, temperature, and mechanical force, have been shown to induce conformational changes including unfolding, molten globule formation, and fibril formation (3–7). However, the effect of fluid flow on protein structure has not been examined in detail, despite the fact that protein dynamics research and biotechnology applications often involve dynamic processes such as pumping, filtration, and mixing, that expose protein solutions to complex shear fields. Previous studies have produced strong evidence that fluid shear can unfold proteins; however, this is not true in all cases. For example, flow-induced unfolding of β-lactoglobulin (8,9), glycoproteins IB (10), and von Willebrand Factor (11,12) have been reported, whereas alternative studies of horse cytochrome c were used to suggest that proteins may not unfold under high shear rates (13). The use of different proteins, fluidic devices, and measurement techniques renders it difficult to compare results across different studies in the literature, and has possibly contributed to the lack of a clear understanding of shear-induced unfolding. Given the variations in protein stability as a function of structure (14–17), it is reasonable to suggest that the likelihood of a protein experiencing conformational variation under a certain set of flow conditions will be related to its structure and, therefore, the properties of flow-induced unfolding may be protein-specific.
To test this hypothesis, a series of experiments have been undertaken whereby six different natural proteins with varying structural properties have been separately exposed in solution to controlled fluid dynamic conditions and monitored during this process using Raman spectroscopy. The method by which these experiments have been conducted is similar to that of a previous investigation of flow-induced unfolding in lysozyme, which revealed that shearing of lysozyme in water altered the protein's backbone structure, whereas similar shear rates in 95% glycerol solutions affected the solvent exposure of side-chain residues located toward the exterior of the protein (18). In this procedure, a variable-speed, closed Taylor-Couette flow cell was designed to subject the protein solutions to shearing and to allow accurate placement within the spectrometer, thus enabling spectra of proteins in flow to be recorded in situ under controlled conditions. As the observed changes were reversible, the ability to record the protein structure during shearing was important. For the current experiments, the six proteins of varying structure and function have each been studied under the same flow conditions, and with their solutions prepared in a manner as closely matched as possible. This approach provides the opportunity to verify that flow-induced conformational changes are related to the structure and probably the function of the protein, and to determine the structural features most associated with funfolding.
Materials and Methods
Six different protein samples were studied: lysozyme (MW14,300), β-lactoglobulin (MW 18,350), insulin (MW 5808), concanavalin A (MW 26,500), α-casein (MW 23,600), and fetuin (MW 48,000). Samples were purchased from Sigma (St. Louis, MO) and used without further purification. The dry material was dissolved in distilled deionized H2O at a concentration of 10 mg/mL with the exception of insulin. As insulin is only sparingly soluble at neutral pH, and requires an acidic pH to be dissolved without aggregation, it was dissolved at a concentration of 5 mg/mL in distilled deionized H2O at a pH of 2.0. This resulted in a pH of ∼1.6 for the insulin solution, measured using a pH209 meter (HANNA Instruments, Woonsocket, RI). Raman measurements were performed using a ChiralRAMAN spectrometer (Biotools, Jupiter, FL). All spectra were acquired with a total exposure time of 1 min, a laser wavelength of 532.5 nm, a laser power of 0.60W at the sample, and a spectral resolution of ∼7 cm−1. The protein solutions were placed in a specially designed Taylor-Couette flow cell, which was positioned in the Raman instrument. The cell, which is characterized by a rotating inner cylinder of radius 20.6 mm mounted inside a stationary glass cylinder of radius 25.5 mm, has been previously described in detail (18). For each protein solution, a reference spectrum was first collected under stationary conditions, and then further spectra were acquired with the flow cell inner cylinder driven at rotational speeds of 30, 60, 90, 120, and 150 rpm, before a final spectrum was collected as soon as the cylinder stopped rotating. All experiments were conducted at room temperature, so that the measured flow cell temperature remained between 21.5°C and 24.5°C. The flow field corresponding to these conditions has been previously characterized for this apparatus using particle image velocimetry and is known to vary between time-invariant Taylor vortex flow and wavy vortex flow (18). The time-averaged topology of the flow in the Raman interrogation region does not vary significantly over this range of rotational speeds, whereas the stresses are expected to change almost linearly with cylinder velocity. Finally, it should be noted that the incident light is unpolarized, so any preferential alignment of protein molecules in the flow would not be detected by this experimental design.
Data pretreatments
All spectral sets exhibited large solvent background that increased with rotational speed; Fig. 1 displays typical raw data for one protein, β-lactoglobulin. To directly compare the spectra, the appropriate water spectrum recorded in the flow cell for each of the rotational speeds was first subtracted from the raw spectra. Baseline subtraction was performed individually for each spectrum to remove baseline drift caused by flow rate-dependent backscatter noise and instrument instabilities. Electronic smoothing using a fast Fourier transform at two fast Fourier transform points was also applied to reduce noise without affecting the underlying bands. All data pretreatments were applied using ORIGIN 7.5 software (OriginLab, Northampton, MA).
Figure 1.
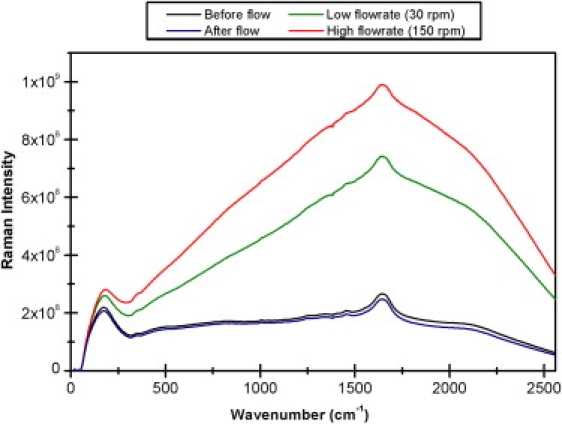
Raw Raman spectra of β-lactoglobulin recorded without flow, at low flow rate (30 rpm), high flow rate (150 rpm), and without flow immediately after flow was stopped.
After water subtraction was carried out, a spurious signal at ∼1380 cm−1 was observed in all spectra, most likely due to an inactive pixel in the charge-coupled device camera. This resulted in a negative peak which increased in intensity with increasing flow rates. Data in the region of ∼1370–1390 cm−1 were therefore omitted for clarity. Autocorrelations or variance plots were calculated from the final pretreated data using 2DCos software freely available on the internet (19), and plotted using ORIGIN 7.5.
Results and Discussion
Globular proteins
To analyze the way in which protein structural differences may affect protein susceptibility to shear-induced unfolding, it is necessary to first describe the secondary and tertiary structures of the proteins investigated. Four of these six proteins (concanavalin A, β-lactoglobulin, lysozyme, and insulin) have known globular structures, as shown in Fig. 2. Concanavalin A (Fig. 2 a), a saccharide-binding lectin from the jack bean (Canavalia ensiformis) involved in storage and defense (20,21), has an overall β-structure with loops and turns. Beta-lactoglobulin (Fig. 2 b), a major whey protein of cow's milk whose function is suggested to be involved in binding and transport of nonpolar molecules (4), is also mainly β-sheet with eight antiparallel β-strands forming an up-and-down β-barrel that forms a central cavity where binding is thought to take place (22). A three-turn helix is attached to the central cavity along with a further ninth β-strand (22,23). Hen egg-white lysozyme (Fig. 2 c) is an enzyme effective against bacteria and yeast and consists of mainly helical structure with some β-sheet in two domains, α and β. The α-domain is rich in α-helical structure whereas the β-domain contains β-sheet, several loops, and a further 310 helix. Insulin is a polypeptide hormone that is stored in pancreatic β-cells as a Zn2+-stabilized hexamer but functions as a Zn2+ free monomer. It has extensive effects on metabolism of carbohydrates and fats, especially the conversion of glucose to glycogen which lowers the blood glucose level (24). Its structure (Fig. 2 d) is mainly helical with its two polypeptide chains linked by one intrachain and two interchain disulfide bonds.
Figure 2.
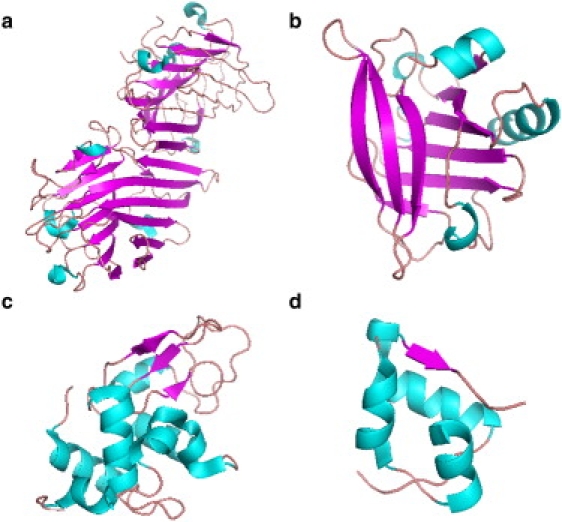
Cartoon representations of the structure of (a) concanavalin A (PDB 1GKB), (b) β-lactoglobulin (PDB 1BEB), (c) hen egg white lysozyme (PDB 1LSE), and (d) insulin (PDB 2A3G) drawn from atomic coordinates in the PDB using PyMOL (Delano Scientific, Palo Alto, CA).
Fig. 3 contains the pretreated spectra of these four proteins acquired with the inner cylinder rotating at 30 rpm (low flow rate) and at 150 rpm (high flow rate), as well as before and after the cylinder was rotated. A key finding which is immediately noticeable is that band intensity changes vary widely between the different proteins. Concanavalin A (Fig. 3 a) exhibits only very slight changes in some bands and negligible changes in most of the plotted frequency domain. For β-lactoglobulin (Fig. 3 b), minor changes in intensity can be observed. The lysozyme spectral set (Fig. 3 c) is associated with larger changes across numerous bands whereas the insulin spectral set (Fig. 3 d) exhibits the greatest changes of all the proteins studied. All of the changes observed are reversible, as illustrated by the similarity between the spectra acquired before and after flow for each case. For β-lactoglobulin and lysozyme, the changes occur at both low and high flow rates; however, insulin appears to experience little change at the low flow rate and significant changes at the higher flow rates. These results in insulin appear to support the theory that there is a threshold value of shear above which globules unfold (12).
Figure 3.
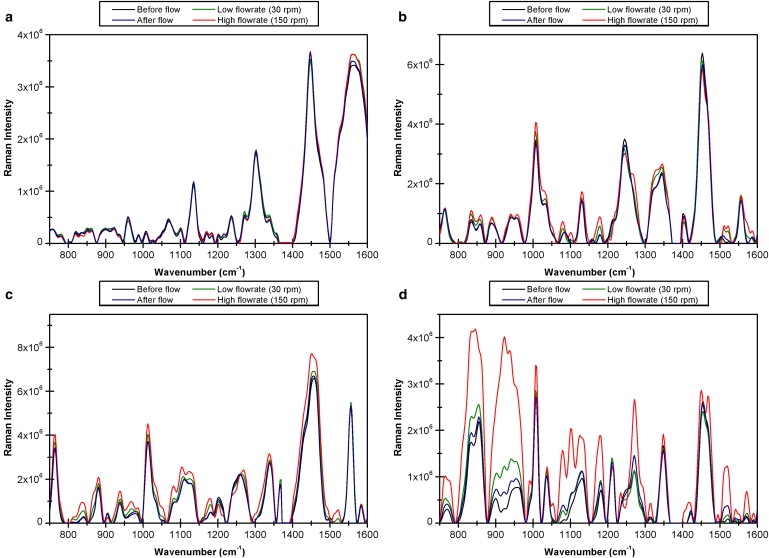
Pretreated Raman spectra of (a) Concanavalin A, (b) β-lactoglobulin, (c) hen egg white lysozyme, and (d) insulin recorded without flow, at low flow rate (30 rpm), high flow rate (150 rpm), and immediately after flow was stopped.
By applying the two-dimensional correlation analysis technique commonly referred to in the spectroscopy community as autocorrelation, differences in the extent of intensity variations can be directly compared. Two-dimensional correlation analysis is a cross-correlation technique, which can be applied to a set of perturbation-induced spectra as a function of two independent wavenumber positions (25). The output from two-dimensional correlation analysis can be represented in the form of synchronous plots, which identify similarities in behavior between data points at two independent wavenumbers, and asynchronous plots, which indicate overall differences in behavior between data points at the two independent wavenumbers. Such plots can be highly complex for the conformational changes exhibited by proteins and therefore further processing by means of the autocorrelation have proven useful (26). The autocorrelation refers to the plot of the relative intensities of the autopeaks in the synchronous contour maps, which display the overall extent of intensity changes of individual bands. As the two-dimensional correlation synchronous maps are a covariance or cross-product matrix (calculated by multiplying a two-dimensional data matrix by its transpose), the autocorrelation plot can also be referred to as the variance plot, which is appropriate here because the largest values correspond to the largest changes (27). Fig. 4 represents the variance in the different protein spectra collected across all five flow rates (30 rpm, 60 rpm, 90 rpm, 120 rpm, and 150 rpm) and before and after flow. The variances of all four proteins are plotted together for comparison purposes. The figure reinforces the initial interpretation that for the majority of spectral variations, with the exception of the methylene stretch-assigned bands occurring at ∼1446 and 1471 cm−1 (28,29), the largest intensity changes are occurring in insulin; this is followed in decreasing order by lysozyme and then β-lactoglobulin, with negligible changes being observed for concanavalin A.
Figure 4.
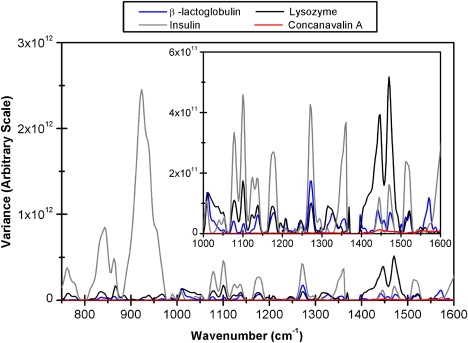
Variance plots generated from the pretreated flow-induced spectral intensity variations of concanavalin A, β-lactoglobulin, lysozyme, and insulin.
To provide a possible physical interpretation to this pattern, the secondary structures of the four proteins need to be compared. As illustrated in Fig. 2, a key difference in the structures of insulin and lysozyme compared to those of β-lactoglobulin and concanavalin A, is the proportion of β-structure. At one extreme, concanavalin A contains mainly β-structure and shows negligible changes in intensity. Conversely, insulin and lysozyme are mainly α-helical, with little β-structure, and are associated with significant intensity changes, with insulin showing the greatest changes. Some band intensity changes occur for β-lactoglobulin, which is mostly β-sheet with one α-helix, but these variances are not as strong as those observed for lysozyme or insulin. It is therefore reasonable to suggest that the extent of β-sheet structure within a protein determines the susceptibility of the protein to shear-induced conformational changes and the degree to which these changes may occur. In a recent time-of-flight neutron scattering study of well-folded and natively disordered proteins it was found that β-sheet structures appeared to be more rigid than a cluster of α-helices (14). Furthermore, several mechanical unfolding studies of various proteins also report a relationship between the secondary structure of the native topology and mechanical resistance to unfolding, suggesting that β- and mixed α/β proteins can withstand higher forces than α-helical proteins (15–17). Recent experiments and experimentally based simulations by Gabovich and Li (30), using a simple elastic theory, also suggested that unfolding of β-proteins requires notably higher forces as compared to the stretching of α-proteins. The presence of large amounts of β-sheet in concanavalin A, and to some extent β-lactoglobulin, may therefore provide the proteins with a rigidity which prevents them from changing their conformation when subjected to fluid shear.
As a possible alternative to this explanation, it should be noted that the susceptibility to flow-induced changes may also be related to protein size. Insulin (MW 5808) is much smaller than lysozyme (MW 14,300) which in turn is smaller than β-lactoglobulin (MW 18,350) and concanavalin A (MW 26,500). The relative length of each protein follows a similar trend. Therefore, the variances observed in Fig. 4 appear to directly correlate with protein size, which suggests that the smaller a protein molecule is, the more susceptible it is to shear-induced changes. Earlier studies have suggested that larger globular proteins are more resistant to mechanical forces. For example, it has been suggested that the critical shear rate for unfolding of a polymeric globule in flow increases as a function of globule radius (12), whereas the hydrostatic pressure required to denature proteins also increases with the size of the proteins due to the increased strength of the hydrophobic interactions in the interior of the protein (31). The spectral changes shown in Figs. 3 and 4 do not represent complete denaturation; however, it is feasible that the mechanisms responsible for the higher stress threshold for unfolding of larger proteins are the same as those that lead to the increased conformational changes reported here.
To better understand the nature of the changes, several individual bands are worth considering in detail. For insulin, the most pronounced spectral changes occur in bands located at ∼840, 863, 922, and 956 cm−1. In the Raman spectral set for this protein (Fig. 3 d), no definitive peaks occur at ∼840 and 863 cm−1; however, several peaks can be observed to change profile. It should be recalled that the strongest peaks in the variance plot represent the wavenumber regions associated with maximum intensity change, which do not necessarily correspond to the positions of maximum intensity found in the Raman spectra. In the spectra shown in Fig. 3 d, two bands can be observed at ∼834 and 856 cm−1 for the case where the protein solution is stationary. For the higher flow-rate case, the profile in this region appears to transform to one, containing three possible bands positioned at ∼832, 845, and 860 cm−1. The bands at ∼834 and 856 cm−1 are assigned to tyrosine residues (32). The ratio between these two bands has long been associated with the state of hydrogen bonding of the tyrosine phenoxyl group. If the ratio (I860/I832) is ∼2.5, then the phenolic oxygen is the acceptor of a strong hydrogen bond; if the ratio is ∼1.25, then the phenolic oxygen will be both an acceptor and a donor of moderate to weak hydrogen bond; and if the ratio is ∼0.3, then the phenolic hydroxyl is the proton donor in a strong hydrogen bond (33,34). A solvent-exposed tyrosine would be expected to have a ratio of ∼1.25, with the phenolic oxygen being an acceptor and a donator of strong hydrogen bonds. For the insulin spectra, there is a decrease in this ratio from ∼1.1 when there is no flow, to ∼0.9 at the highest flow rate (150 rpm). This ratio decrease suggests a possible decrease in solvent exposure of the tyrosine residues. There are two tyrosine residues (Tyr14 and Tyr19) that are both positioned in one of the α-helices, suggesting that changes are occurring in the α-helical structure in the presence of flow, although they may only be small alterations in geometry or hydration resulting in a change in the extent to which the phenolic oxygen is an acceptor of hydrogen bonding.
In the variance plots for lysozyme and β-lactoglobulin, much weaker peaks can be observed at ∼840 and 863 cm−1, indicating that these bands are also changing in intensity with flow, albeit to a lesser extent than for insulin. For lysozyme, the peak intensity ratio decreases from 0.8 to 0.7 when the rotational rate increases from 0 to 150 rpm, and for β-lactoglobulin the ratio fluctuates at ∼0.7. This suggests that there is no change in solvent exposure of tyrosine residues in β-lactoglobulin and only a possible small decrease in exposure of lysozyme tyrosine residues as the proteins are exposed to shear flow. There are three tyrosine residues in both β-lactoglobulin and hen egg white lysozyme. In lysozyme, two tyrosine residues are situated in the more interior loops and turns of the α-domain (Tyr20 and Tyr23) and a third in the loops of the β-domain (Tyr53). In β-lactoglobulin, two tyrosine residues are situated in elements of β-structure (Tyr42 and Tyr102) and one in a loop between two β-strands (Tyr99). Consequently, for both lysozyme and β-lactoglobulin there are no tyrosine residues located in α-helical structure, which may explain why smaller intensity variations are observed at ∼830 and 860 cm−1 for these two proteins than for insulin. The band at 1175 cm−1 has also been assigned to tyrosine and, as shown by the variance plot intensities, spectral variations occur to the greatest extent in insulin, followed by lysozyme and then β-lactoglobulin.
The band at ∼956 cm−1 has previously been assigned to less-ordered structure (29,35) and therefore the increase in the intensity of this band for insulin may suggest an increase in less-ordered structure. The band at 922 cm−1 has not been conclusively assigned in the literature, but bands in the vicinity of 930 cm−1 are assigned to α-helix (36,37), and it may be that the changes occurring in this region are as a result of changes in α-helical structure.
Proteins with less defined structure
To further explore the links between protein structure and fluid shear-induced unfolding behavior, the study was extended to two proteins with less defined structure, namely α-casein and fetuin. There are four types of caseins (i.e., αs1-, αs2-, β-, and κ-casein) (38,39), which are the phosphoproteins that form the major component of milk. In bovine milk 78% of the protein content is casein, of which 65% is α-casein composed of the αs1- and αs2-subunits, whose functions include transport of calcium and other metal ions to neonates, protection against heat coagulation, binding to membrane receptors, and acting as a signal transducer (40). Several structural studies suggested that α-casein has an open tertiary structure with no prevailing periodic secondary structure (40,41). However, other studies have suggested some secondary structure including regions of α-helix, β-structure, including turns and loops, and PPII (38,41,42). Fetuins are glycoproteins secreted in the liver and detected in the blood plasma of all mammalian species (43). They are similar carrier proteins to serum albumins but found in abundance in fetal blood with diverse physiological roles including mineralization, brain development, and innate immunity (43). Fetuin is composed of ∼24% carbohydrate with three branched heteropolysaccharide units with similar monosaccaride composition (7). As with the majority of glycoproteins, the precise structure of the protein portion has yet to be conclusively determined. Human fetuin-A is reported as having a two-chain peptide structure, whereas the bovine fetuin has been traditionally reported as having only one chain; however, more recent studies suggest that bovine fetuin exists as a disulfide-bridged two-chain form, with a long N-terminal domain and a short C-terminal domain (43,44).
Fig. 5, a and b, displays, respectively, the spectra of α-casein and fetuin before flow, at low and high rates, and after flow. When these spectra sets are compared to the previous sets shown in Fig. 3, fewer intensity changes can be observed for α-casein and fetuin than for lysozyme, insulin, and β-lactoglobulin, although there are more changes in these unstructured proteins than occurred for concanavalin A. This is further supported by Fig. 6, which compares the variance plots of α-casein, fetuin, and insulin, and illustrates that the majority of spectral variations occurring in insulin are far larger than the changes observed in both α-casein and fetuin. The work by Gaspar et al. (14) involving time-of-flight neutron scattering suggested that the unstructured proteins, including α-casein, were more flexible than the globular proteins. However, our results indicate that both α-casein and fetuin are less susceptible to flow-induced conformational changes than the globular proteins lysozyme, insulin, and β-lactoglobulin. This implies that the tendency for a protein to unfold is not fully determined by its flexibility, and that other factors, such as the size and/or structure of the protein, may be more important. With regard to the possible effect of protein size, α-casein has only a slightly lower molecular weight (MW 23,600) than the largest, most stable protein, concanavalin A (MW 26,500), and fetuin has a larger molecular weight (MW 48,000) than both. This contradicts the clear trend, reported above for globular proteins, that mechanically induced conformational changes are directly related to protein size, and may suggest instead that structural type is a more effective control parameter. However, when considering this problem with relation to unstructured proteins, other factors specific to the studied proteins should be considered. For instance, both fetuin and α-casein have posttranslational modifications. Fetuin has the addition of three branched heteropolysaccharide units (7), and α-casein has eight-to-nine phosphate groups attached to serine residues (38,45); it may be that these modifications act to reduce the susceptibility of these proteins to flow-induced conformational changes, although this requires further investigation.
Figure 5.
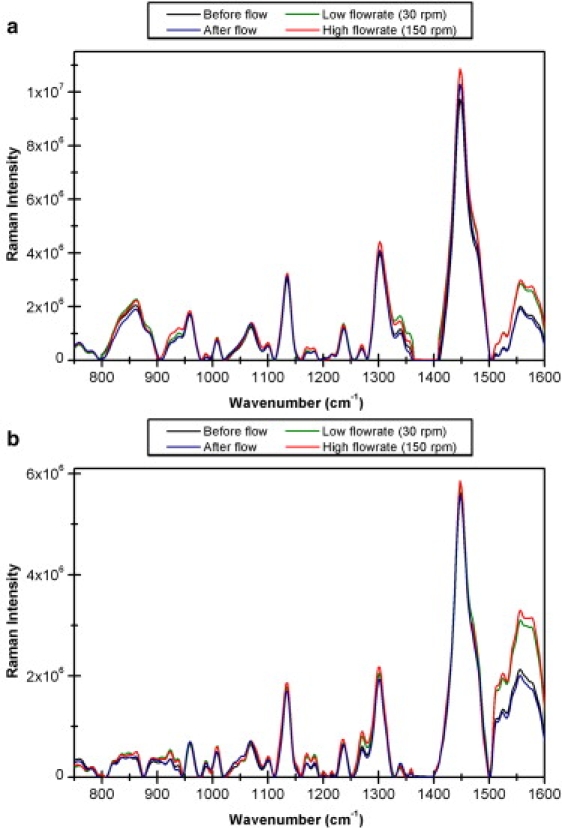
Pretreated Raman spectra of (a) α-casein and (b) fetuin recorded without flow, at low flow rate (30 rpm), high flow rate (150 rpm), and immediately after flow was stopped.
Figure 6.
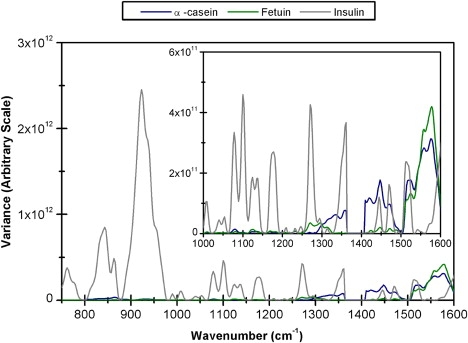
Comparison of variance plots generated from the pretreated flow-induced spectral intensity variations of insulin, fetuin, and α-casein.
Although the majority of spectral changes are greater in insulin than α-casein and fetuin, an exception occurs in the region ∼1530–1660 cm−1. Bands in this region are assigned to tryptophan residues (29,46) and the increase in intensity of these bands indicates a change in solvent exposure of tryptophan residues. No tryptophan residues are present in bovine insulin; however, there are six tryptophan residues present in lysozyme and no spectral variations occur at either ∼1556 or 1581 cm−1 in Fig. 4. Beta-lactoglobulin also has two tryptophan residues, and in Fig. 4 a small change can be observed in the band at ∼1570 cm−1. As shown in Figs. 5 and 6, the largest intensity variations for α-casein and fetuin occur in bands at ∼1556 and 1581 cm−1, indicating a possible increase in solvent exposure of tryptophan residues. Consequently, although α-casein and fetuin do not undergo flow-induced conformational changes to the same extent as insulin, lysozyme, and β-lactoglobulin, changes in side-chain exposure are detectable when these less structured proteins are in flow.
Conclusions
By acquiring Raman spectra of protein solutions within a specially designed Taylor-Couette flow cell, the conformational changes in proteins of varying structure have been monitored and compared. In all cases, the recorded changes were subtle and reversible, rather than providing evidence of complete unfolding. However, the extent of the conformational variations differed markedly for the six different proteins used for this study, despite the fact that each protein was exposed to the same flow conditions. The degree of conformational change in flow appears to be related to structure, and possibly the size of the protein.
For globular proteins there appears to be a correlation between the amount of β-structure and the extent of unfolding. Proteins with the greater proportion of β-structure (concanavalin A and β-lactoglobulin) exhibited less spectral variation than those with a larger percentage of α-helix (lysozyme and insulin). Additionally, there is a clear inverse relationship between the degree of conformational change and the size (molecular weight and radius) of the globule proteins. However, the investigations of two proteins with less defined structure, α-casein and fetuin, suggest that, in general, it is structure rather than molecular weight that determines the extent of flow-induced conformational changes. Furthermore, the relatively low degree of unfolding of these proteins compared to globular proteins β-lactoglobulin, lysozyme, and insulin, implies that flexibility is not necessarily the property that always determines protein stability in mechanical shear environments. Susceptibly to flow-induced changes in these two proteins may be reduced due to the posttranslational modifications of phosphorylation and glycosylation.
The most significant flow-induced changes occurred in the insulin spectra, which revealed decreased exposure of the tyrosine residues situated in α-helical structure. This suggests conformational changes are most likely occurring in α-helical structure with possible fluctuations in geometry or hydration. The insulin spectra also imply that there is a threshold value of shear above which the protein unfolds: only minor changes were observed at an inner cylinder rotational speed of 30 rpm. However, there were significant changes that occurred at the higher flow rates. This implies that the flow-induced changes are not only specific to protein structure but may also be highly shear-stress-dependent. These initial investigations into flow-induced conformational changes have demonstrated the vast potential of using Raman spectroscopy with specially designed flow cells to monitor flow-induced structural behavior of proteins. By adapting the flow cell to have more precise control over flow conditions, it should be possible to further investigate and determine relationships among fluid stresses, unfolding behavior and structural differences, and therefore develop a greater understanding of flow-induced conformational transitions and protein unfolding processes.
References
- 1.Greenfield N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schweitzer-Stenner R. Advances in spectroscopy as a sensitive probe of peptide and protein structure. Vib. Spectrosc. 2006;42:98–117. [Google Scholar]
- 3.Su T., Purohit P.K. Mechanics of forced unfolding of proteins. Acta Biomater. 2009;5:1855–1863. doi: 10.1016/j.actbio.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 4.Taulier N., Chalikian T.V. Characterization of pH-induced transitions of β-lactoglobulin: ultrasonic, densimetric, and spectroscopic studies. J. Mol. Biol. 2001;314:873–889. doi: 10.1006/jmbi.2001.5188. [DOI] [PubMed] [Google Scholar]
- 5.Dobson C.M. Unfolded proteins, compact states and molten globules. Curr. Opin. Struct. Biol. 1992;2:6–12. [Google Scholar]
- 6.Lednev I.K., Ermolenkov V.V., Xu M. Deep-UV Raman spectrometer tunable between 193 and 205 nm for structural characterization of proteins. Anal. Bioanal. Chem. 2005;381:431–437. doi: 10.1007/s00216-004-2991-5. [DOI] [PubMed] [Google Scholar]
- 7.Naseem F., Khan R.H., Naeem A. Characterization of molten globule state of fetuin at low pH. Biochim. Biophys. Acta. 2003;1649:164–170. doi: 10.1016/s1570-9639(03)00169-9. [DOI] [PubMed] [Google Scholar]
- 8.Castelletto V., Hamley I.W. Beta-lactoglobulin fibers under capillary flow. Biomacromolecules. 2007;8:77–83. doi: 10.1021/bm0605917. [DOI] [PubMed] [Google Scholar]
- 9.Hill E.K., Krebs B., Dunstan D.E. Shear flow induces amyloid fibril formation. Biomacromolecules. 2006;7:10–13. doi: 10.1021/bm0505078. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z., Lou J., Schulten K. Flow-induced structural transition in the β-switch region of glycoprotein Ib. Biophys. J. 2008;95:1303–1313. doi: 10.1529/biophysj.108.132324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider S.W., Nuschele S., Schneider M.F. Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc. Natl. Acad. Sci. USA. 2007;104:7899–7903. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander-Katz A., Schneider M.F., Netz R.R. Shear-flow-induced unfolding of polymeric globules. Phys. Rev. Lett. 2006;97:138101. doi: 10.1103/PhysRevLett.97.138101. [DOI] [PubMed] [Google Scholar]
- 13.Jaspe J., Hagen S.J. Do protein molecules unfold in a simple shear flow? Biophys. J. 2006;91:3415–3424. doi: 10.1529/biophysj.106.089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaspar A.M., Appavou M.S., Doster W. Dynamics of well-folded and natively disordered proteins in solution: a time-of-flight neutron scattering study. Eur. Biophys. J. 2008;37:573–582. doi: 10.1007/s00249-008-0266-3. [DOI] [PubMed] [Google Scholar]
- 15.Li M.S. Secondary structure, mechanical stability, and location of transition state of proteins. Biophys. J. 2007;93:2644–2654. doi: 10.1529/biophysj.107.106138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockwell D.J., Beddard G.S., Radford S.E. Mechanically unfolding the small, topologically simple protein L. Biophys. J. 2005;89:506–519. doi: 10.1529/biophysj.105.061465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackbarow T., Chen X., Buehler M.J. Hierarchies, multiple energy barriers, and robustness govern the fracture mechanics of α-helical and β-sheet protein domains. Proc. Natl. Acad. Sci. USA. 2007;104:16410–16415. doi: 10.1073/pnas.0705759104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashton L., Dusting J., Blanch E.W. Shear-induced unfolding of lysozyme monitored in situ. Biophys. J. 2009;96:4231–4236. doi: 10.1016/j.bpj.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Two-dimensional correlation spectroscopy. Ozaji Lab. Webpages of Prof. Ozaki. http://sci-tech.ksc.kwansei.ac.jp/∼ozaki/e_2D.htm
- 20.Kantardjieff K.A., Hochtl P., Rupp B. Concanavalin A in a dimeric crystal form: revisiting structural accuracy and molecular flexibility. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:735–743. doi: 10.1107/s0907444901019588. [DOI] [PubMed] [Google Scholar]
- 21.Kadirvelraj R., Foley B.L., Woods R.J. Involvement of water in carbohydrate-protein binding: concanavalin A revisited. J. Am. Chem. Soc. 2008;130:16933–16942. doi: 10.1021/ja8039663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavel L., Andriot I., Guichard E. Interactions between β-lactoglobulin and aroma compounds: different binding behaviors as a function of ligand structure. J. Agric. Food Chem. 2008;56:10208–10217. doi: 10.1021/jf801841u. [DOI] [PubMed] [Google Scholar]
- 23.Kontopidis G., Holt C., Sawyer L. Invited review: β-lactoglobulin: binding properties, structure, and function. J. Dairy Sci. 2004;87:785–796. doi: 10.3168/jds.S0022-0302(04)73222-1. [DOI] [PubMed] [Google Scholar]
- 24.Dong J., Wan Z., Weiss M.A. Insulin assembly damps conformational fluctuations: Raman analysis of amide I linewidths in native states and fibrils. J. Mol. Biol. 2003;330:431–442. doi: 10.1016/s0022-2836(03)00536-9. [DOI] [PubMed] [Google Scholar]
- 25.Noda I., Ozaki Y. John Wiley; Chichester, UK: 2004. Two-Dimensional Correlation Spectroscopy: Applications in Vibrational and Optical Spectroscopy. [Google Scholar]
- 26.Ashton L., Blanch E.W. Investigation of polypeptide conformational transitions with two-dimensional Raman optical activity correlation analysis, applying autocorrelation and moving window approaches. Appl. Spectrosc. 2008;5:469–475. doi: 10.1366/000370208784344433. [DOI] [PubMed] [Google Scholar]
- 27.Sasic S., Muszynski A., Ozaki Y. New insight into the mathematical background of generalized two-dimensional correlation spectroscopy and the influence of mean normalization pretreatment on two-dimensional correlation spectra. Appl. Spectrosc. 2001;55:343–349. [Google Scholar]
- 28.Vohník S., Hanson C., Thomas G.J. Conformation, stability, and active-site cysteine titrations of Escherichia coli D26A thioredoxin probed by Raman spectroscopy. Protein Sci. 1998;7:193–200. doi: 10.1002/pro.5560070120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howell N., Li-Chan E. Elucidation of interactions of lysozyme with whey proteins by Raman spectroscopy. Int. J. Food Sci. Technol. 1996;31:439–451. [Google Scholar]
- 30.Gabovich A.M., Li M.S. Mechanical stability of proteins. J. Chem. Phys. 2009;131:024121. doi: 10.1063/1.3170940. [DOI] [PubMed] [Google Scholar]
- 31.Chodankar S., Aswal V.K., Wagh A.G. Structural evolution during protein denaturation as induced by different methods. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2008;77:031901. doi: 10.1103/PhysRevE.77.031901. [DOI] [PubMed] [Google Scholar]
- 32.Siamwiza M.N., Lord R.C., Shimanouchi T. Interpretation of the doublet at 850 and 830 cm−1 in the Raman spectra of tyrosyl residues in proteins and certain model compounds. Biochemistry. 1975;14:4870–4876. doi: 10.1021/bi00693a014. [DOI] [PubMed] [Google Scholar]
- 33.Liang M., Chen V.Y.T., Chen W. A simple and direct isolation of whey components from raw milk by gel filtration chromatography and structural characterization by Fourier transform Raman spectroscopy. Talanta. 2006;69:1269–1277. doi: 10.1016/j.talanta.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Arp Z., Autrey D., Thomas G.J. Tyrosine Raman signatures of the filamentous virus Ff are diagnostic of non-hydrogen-bonded phenoxyls: demonstration by Raman and infrared spectroscopy of p-cresol vapor. Biochemistry. 2001;40:2522–2529. doi: 10.1021/bi0023753. [DOI] [PubMed] [Google Scholar]
- 35.Ashton L., Barron L.D., Blanch E.W. Two-dimensional Raman and Raman optical activity correlation analysis of the α-helix-disordered transition in poly(L-glutamic acid) Analyst. 2007;132:468–479. doi: 10.1039/b700421d. [DOI] [PubMed] [Google Scholar]
- 36.Miura T., Thomas G.J. Proteins: structure, function and engineering. In: Biswas B.B., Roy S., editors. Subcellular Biochemistry. Vol. 4. Plenum Press; New York: 1995. [Google Scholar]
- 37.Tsuboi M., Suzuki M., Thomas G.J. Intensity of the polarized Raman band at 1340–1345 cm−1 as an indicator of protein α-helix orientation: application to Pf1 filamentous virus. Biochemistry. 2000;39:2677–2684. doi: 10.1021/bi9918846. [DOI] [PubMed] [Google Scholar]
- 38.Kumosinski T.F., Brown E.M., Farrell H.M. Three-dimensional molecular modeling of bovine caseins: αs1-casein. J. Dairy Sci. 1991;74:2889–2895. doi: 10.3168/jds.S0022-0302(91)78470-1. [DOI] [PubMed] [Google Scholar]
- 39.Ferranti P., Scaloni A., Addeo F. The primary structure of water buffalo α (s1)- and β-casein identification of phosphorylation sites and characterization of a novel β-casein variant. J. Protein Chem. 1998;17:835–844. doi: 10.1023/a:1020786503978. [DOI] [PubMed] [Google Scholar]
- 40.Alaimo M.H., Farrell H.M., Germann M.W. Conformational analysis of the hydrophobic peptide αs1-casein (136–196) Biochim. Biophys. Acta. 1999;1431:410–420. doi: 10.1016/s0167-4838(99)00061-8. [DOI] [PubMed] [Google Scholar]
- 41.Creamer L.K., Richardson T., Parry D.A.D. Secondary structure of bovine αs1- and β-casein in solution. Arch. Biochem. Biophys. 1981;211:689–696. doi: 10.1016/0003-9861(81)90505-1. [DOI] [PubMed] [Google Scholar]
- 42.Jarvis R.M., Blanch E.W., Goodacre R. Quantification of casein phosphorylation with conformational interpretation using Raman spectroscopy. Analyst (Lond.) 2007;132:1053–1060. doi: 10.1039/b702944f. [DOI] [PubMed] [Google Scholar]
- 43.Kübler D., Gosenca D., Lehmann W.D. Proteolytic processing by matrix metalloproteinases and phosphorylation by protein kinase CK2 of fetuin-A, the major globulin of fetal calf serum. Biochimie. 2007;89:410–418. doi: 10.1016/j.biochi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Wind M., Gosenca D., Lehmann W.D. Stable isotope phospho-profiling of fibrinogen and fetuin subunits by element mass spectrometry coupled to capillary liquid chromatography. Anal. Biochem. 2003;317:26–33. doi: 10.1016/s0003-2697(03)00083-6. [DOI] [PubMed] [Google Scholar]
- 45.Mercier J.-C., Grosclaude F., Ribadeau-Dumas B. Structure primaire de la caseine s1-bovine [Primary structure of bovine αs1 casein. Complete sequence.] Eur. J. Biochem. 1971;23:41–51. doi: 10.1111/j.1432-1033.1971.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 46.Miura T., Takeuchi H., Harada I. Tryptophan Raman bands sensitive to hydrogen bonding and side chain conformation. J. Raman Spectrosc. 1989;20:667–671. [Google Scholar]


