Abstract
Summary: Often in human genetic analysis, multiple tables of single nucleotide polymorphism (SNP) statistics are shown alongside a Haploview style correlation plot. Readers are then asked to make inferences that incorporate knowledge across these multiple sets of results. To better facilitate a collective understanding of all available data, we developed a Ruby-based web application, LD-Plus, to generate figures that simultaneously display physical location of SNPs, binary SNP attributes (such as coding/non-coding or presence on genotyping platforms), common haplotypes and their frequencies and continuously scaled values (such as Fst, minor allele frequency, genotyping efficiency or P-values), all in the context of the D′ and r2 linkage disequilibrium structures. Combining these results into one comprehensive figure reduces dereferencing between figures and tables, and can provide unique insights into genetic features that are not clearly seen when results are partitioned across multiple figures and tables.
Availability: LD-Plus is freely available for non-commercial research institutions. For full details see http://chgr.mc.vanderbilt.edu/ritchielab/ldplus.
Contact: ritchie@chgr.mc.vanderbilt.edu
1 INTRODUCTION
After completing the first phase of the International HapMap Project, triangular correlation plots implemented in Haploview software (Barrett et al., 2005) have become a ubiquitous component of population-based genetic analysis reports. These plots provide a nice visualization of how single nucleotide polymorphisms (SNPs) travel together in human subpopulations and samples due to linkage disequilibrium (LD). Often, haplotype block structure and haplotype frequency information can be helpful in interpreting other statistical results, such as Hardy–Weinberg statistics, Wright's Fst, population-specific allele frequency differences or association P-values. The need for a visualization tool that can simultaneously display all these types of data can be readily seen in various human genetics publications.
When characterizing population-based genetic differences in genes of phenotypic interest, a bevy of analyses are often conducted to explore potential differences in allele frequency and haplotype structure (and associated ancestral recombination events), and examine evidence of population divergence. These statistics are often reported separately, indexed by reference SNP (RS) number, as seen in (Sile et al., 2008). Determining if significant allele frequency differences occur within a haplotype block, or due to haplotype block structure, requires visually dereferencing SNPs by RS number, and finding the haplotype structure on a separate figure.
Genome-wide association studies and fine-mapping studies following linkage analysis also often display regional association statistics in the context of LD. In some cases, this is achieved by aligning two disjoint figures (see Fig. 1 from Gudmundsson et al., 2009), or by displaying data in terms of physical position, which does not always allow easy interpretation. Also, association P-values are rarely presented in the context of other quality control data, such as Hardy–Weinberg statistics, genotyping efficiency, minor allele frequency, quality scores and other relevant data that can aid the interpretation of association test statistics.
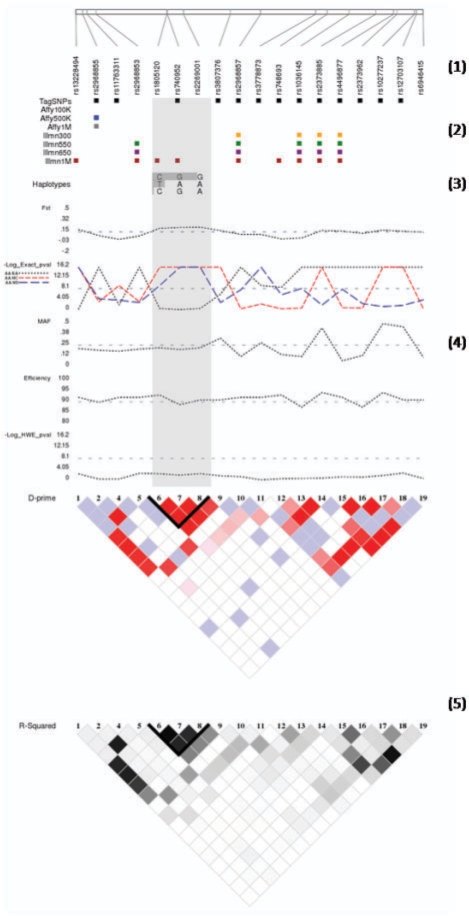
Fig. 1.
LD-Plus figure of SNPs in KCNH2 of an African–American sample. Physical position of SNPs in the genome is displayed (1). Annotation of binary SNP characteristics is shown (in this example, inclusion in HapMap data and various genotyping platforms) (2). Haplotypes and haplotype frequencies are shown (3) for each defined haplotype block, and block boundaries are shaded throughout the figure to allow easy reference. Continuous SNP characteristics such as Fst, exact test statistics, genotyping efficiency, etc. are shown as line graphs (4). Haploview style LD plots in both D′ and r2 are shown, with defined haplotype blocks outlined in black (5). These data are based on 98 Coriell DNA samples, genotyped at 18 SNPs across the KCNH2 gene region (Bush et al., 2009).
To address these issues, we developed LD-Plus as a simple visualization tool for displaying and relating discrete and continuous SNP attributes to haplotype blocks, haplotypes and genomic features simultaneously.
2 DESIGN AND IMPLEMENTATION
LD-Plus requires a minimum of two files: a Haploview formatted PED file and the marker information file (or map file). Running LD-Plus with this minimal set of input files will generate a Haploview style correlation plot. This LD plot contains numerous tracks as described below. These additional annotation files can be readily created using a spreadsheet application or basic text editor.
Physical genome track. SNPs identified by RS number or other identifier are shown in relative physical distance. Regions of the genome, such as exons or functional elements, can be annotated and highlighted in physical distance. Features are simply input as a text file with feature labels and base-pair ranges.
SNP attributes track. Binary SNP (yes/no) attributes are highlighted, such as inclusion on a genotyping platform or non-synonymous SNPs. SNP attributes are input as a text file with 0/1 values for each attribute column.
Haplotypes track. Within each specified haplotype block, haplotype frequencies are shown as horizontal bar graphs, overlaid with the alleles that constitute the haplotype. Haplotypes are input with the starting SNP ID, haplotype base-pairs and frequency.
Statistics track. Continuously scaled variables, such as log P-values, minor allele frequencies, or genotyping efficiency are plotted as line graphs. SNP statistics are input as a text file with SNP ID and continuous values for each statistic. Axis ranges are specified in the table header.
LD track. Haploview style LD plots showing both D′ and r2 are shown with defined haplotype blocks. SNPs within the same LD block are shaded in gray throughout the plot to provide the context of LD for other tracks.
Documentation on how to properly format input files is available at the web address. LD-Plus was developed using the Ruby programming language and the RMagick Ruby Vector Graphics library, and uses Haploview procedures to generate LD statistics.
3 CONCLUSION
LD-Plus complements the common Haploview style plot by providing additional statistical context to LD statistics. Multiple dimensions of genomic data are displayed in a manner allowing easy comparison and evaluation of SNP relationships. Haplotype block boundaries are carried throughout the figure, providing haplotype context to other SNP statistics. This figure design can effectively represent nearly all of the information contained in the 10 figures and tables in (Sile et al., 2008) using only four figures (one for each ethnicity).
LD-Plus nicely complements other visualization tools such as the UCSC browser (Karolchik et al., 2008) and SNAP (Johnson et al., 2008) that relate LD statistics to physical distance. Displaying LD and SNP-based statistics in terms of physical position can at times be difficult, as the close proximity of some SNPs may require zooming to resolve statistical tracks. LD-Plus provides an equidistant display of SNP statistics and LD values and overlays haplotype block and frequency information.
Using LD-Plus, an investigator could recognize that a strongly associated SNP occurs in an LD block with haplotypes that contain a common coding variant or that a significant SNP is in an LD block that spans a regulatory element. The comprehensive figures generated by LD-Plus provide a unique perspective for multiple SNP statistics often collected in numerous genetic studies, and help to condense the wealth of data generated into a familiar, interpretable format.
Funding: National Institutes of Health (HL65962 and LM010040).
Conflict of Interest: none declared.
REFERENCES
- Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bush W, et al. Genetic variation in the rhythmonome: ethnic variation and haplotype structure in candidate genes for arrhythmias. Pharmacogenomics. 2009;10:1043–1053. doi: 10.2217/pgs.09.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat. Genet. 2009;41:460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sile S, et al. Haplotype diversity in four genes (CLCNKA, CLCNKB, BSND, NEDD4L) involved in renal salt reabsorption. Hum. Hered. 2008;65:33–46. doi: 10.1159/000106060. [DOI] [PMC free article] [PubMed] [Google Scholar]