Abstract
Summary: Recent parallel pyrosequencing methods and the increasing number of finished genomes encourage the sequencing and investigation of closely related strains. Although the sequencing itself becomes easier and cheaper with each machine generation, the finishing of the genomes remains difficult. Instead of the desired whole genomic sequence, a set of contigs is the result of the assembly. In this applications note, we present the tool r2cat (related reference contig arrangement tool) that helps in the task of comparative assembly and also provides an interactive visualization for synteny inspection.
Availability: http://bibiserv.techfak.uni-bielefeld.de/r2cat
Contact: peter.husemann@cebitec.uni-bielefeld.de
1 INTRODUCTION
With the advent of high-throughput sequencing machines, it has become easier and cheaper to sequence a genome. A decade ago, a sequencing project lasted for years and required a million-dollar budget, whereas today the sequencing itself takes days and costs only a few thousand dollars. Nevertheless, the effort to close a genome completely is still non-negligible, and thus one very important step in genome finishing remains the closure of gaps between contigs. This task becomes easier if the order and the relative orientation of the contigs is known. Mapping the contigs on a closely related genome provides this kind of information. Consequently, a program that orders contigs regarding their matches and visualizes the synteny of contigs and a reference genome can be helpful to close the gaps.
A number of tools have been developed to aid in this task such as Projector2 (van Hijum et al., 2005), a web service that maps contigs on a template genome and visualizes the result, OSLay (Richter et al., 2007) which computes an optimal syntenic layout for a set of contigs, or ABACAS (Assefa et al., 2009) that orders contigs using several external programs for matching, primer design and visualization.
Our program r2cat (related reference contig arrangement tool) is able to quickly match a set of contigs onto a related genome, order the contigs according to their matches and display the result in an interactive synteny plot. The matching, however, is not restricted to contigs, such that the program can also be used to visualize the synteny of two finished genomes. The software is open source and available within the Comparative Genomics – Contig Arrangement Toolsuite (cg-cat; http://bibiserv.techfak.uni-bielefeld.de/cg-cat) on the Bielefeld Bioinformatics Server (BiBiServ).
2 METHODS
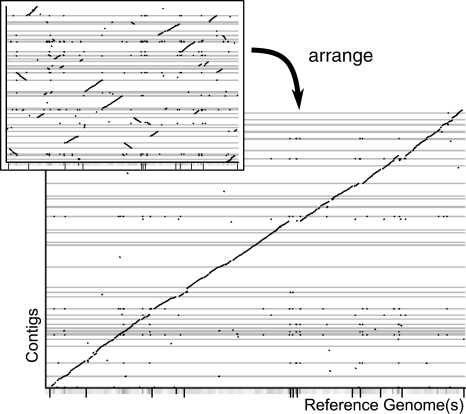
In a first step, similar regions between the contigs and a related reference genome have to be determined. For this task, a q-gram filter (Rasmussen et al., 2006) is used. Regions of up to 8% difference are found that have at least 44 exact matches of possibly overlapping 11mers, which are each not further apart than 64 bases. All these matching regions are displayed in an interactive synteny plot, as shown in Figure 1. The contigs can then in a second step be ordered and oriented automatically according to their matches. To this end, a sliding window approach determines that position of a contig on the reference sequence, where it gains the most matches. A manual correction, however, is easily possible.
Fig. 1.
Synteny plots produced by r2cat. The contigs of C.urealyticum (NCBI number: NC_010545) are mapped onto the reference sequence of C.jeikeium (NC_007164).
The resulting order can then be helpful for gap closing purposes in the finishing phase of a sequencing project, assuming that the corresponding genomes have a high degree of synteny.
3 IMPLEMENTATION
The tool r2cat that implements the matching, ordering and visualization is written in Java and can be started from the Internet without installation using the Java WebStart Framework. The sources are licensed under GPL and available from the author.
Matching and ordering: the fast built-in matching runs well for prokaryotic genomes up to 12 MB. The matching routine is capable of handling multichromosomal genomes, provided in multi-FASTA files, and also finds matches for the reverse complement of each contig. After the matching, the contigs can be arranged automatically. The matches, as well as the inferred order and orientation, can be stored in and retrieved from human readable text files. These can be parsed from other programs as well or modified by hand if necessary.
Visualization: the implemented visualization displays all matches in a dotplot thus providing a quick overview of the synteny. A horizontal bar at the bottom helps to assess the coverage of the matches: maximum coverage is displayed in black and fades to light grey with less coverage. Uncovered regions are marked explicitly. The implementation features an export of the synteny plot to either bitmap or vector-based graphics formats. Some of the latter are editable and are thus excellently suited for high-quality synteny plots to be used in publications and other print media. The view area itself is zoomable and panable. Contigs as well as single matches can be selected and displayed in separate table views. The contig table allows to reorder the contigs manually, if necessary, using drag and drop. The contigs can consequently be saved in the displayed order in FASTA format for further processing.
While the main focus of this tool is to order a set of contigs, the synteny visualization can also be used to investigate the relationship between two species if, instead of the contigs, the genomic sequence of a related genome is chosen for matching.
4 RESULTS
To show that the matching implemented in r2cat is competitive, we compared it with the three well-known matching programs BLAST, BLAT and MUMer. Each program was used on two prokaryotic datasets to match a set of contigs onto a reference genome. The first dataset “S.suis”, taken from Assefa et al. (2009), consists of 281 contigs (2.1 Mb) of a Streptococcus suis strain that were matched on the genome of another strain SC84 (2.1Mb, NCBI number: NC_012924). The second dataset “S.meliloti” consists of 446 contigs in 7.2 Mb of a Sinorhizobium meliloti strain that were matched on a reference genome with three replicons: one chromosome (3.65 Mb, NC_003047) and two megaplasmids (1.68 Mb, NC_003078; 1.35 Mb, NC_003037). Table 1 shows for each program and dataset the time that was needed for matching and additionally the number of contigs that could not be matched and thus could not be ordered.
Table 1.
Times for matching a set of contigs on a reference genome
|
S.suis |
S.meliloti |
|||
|---|---|---|---|---|
| Time (s) | Unmatched | Time (s) | Unmatched | |
| blast | 20.0 | 0 | 162.1 | 0 |
| blat | 46.9 | 94 | 700.8 | 84 |
| nucmer | 9.8 | 109 | 45.6 | 92 |
| r2cat | 6.2 | 102 | 45.4 | 75 |
Additionally, the number of contigs is given that could not be matched. The employed programs are BLAST (Altschul et al., 1990, blastall v. 2.2.19), BLAT (Kent, 2002, blat v. 15), MUMmer (Kurtz et al., 2004, nucmer v. 3.06), and our matching routine implemented within r2cat. The experiments were performed on a sparcv9 processor operating at 1593 MHz.
5 CONCLUSION
Our software r2cat is suited for a quick synteny visualization as well as contig ordering using a single reference genome. The speed of our matching is competitive to other established programs, and the automated contig arrangement is helpful in the finishing phase of a sequencing project by giving valuable hints on the order and orientation of the contigs. The vector graphics export of the visualization provides a handy way to generate publication quality graphics. Matching, ordering and the visualization are combined in a single application that can easily be used with Java WebStart. The program was already applied in the sequencing project of Rhizobium lupini (now Agrobacterium sp. H13.3).
A next step could be to extend the comparative assembly to employ several references and their phylogenetic relationships, as explored e.g. in Husemann and Stoye (2010).
ACKNOWLEDGEMENTS
The authors would like to thank J. Blom for the sorting idea, D. Wibberg and S. Jaenicke for feedback, and S. Schneiker-Bekel, A. Tauch and E. Trost for providing the sequence data.
Conflict of Interest: none declared.
REFERENCES
- Altschul S, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Assefa S, et al. ABACAS: algorithm based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husemann P, Stoye J. Phylogenetic comparative assembly. Algorithms Mol. Biol. 2010;5:3. doi: 10.1186/1748-7188-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KR, et al. Efficient q-gram filters for finding all epsilon-matches over a given length. J. Comp. Biol. 2006;13:296–308. doi: 10.1089/cmb.2006.13.296. [DOI] [PubMed] [Google Scholar]
- Richter DC, et al. OSLay: optimal syntenic layout of unfinished assemblies. Bioinformatics. 2007;23:1573–1579. doi: 10.1093/bioinformatics/btm153. [DOI] [PubMed] [Google Scholar]
- van Hijum S.A.FT, et al. Projector 2: contig mapping for efficient gap-closure of prokaryotic genome sequence assemblies. Nucleic Acids Res. 2005;33:W560–W566. doi: 10.1093/nar/gki356. [DOI] [PMC free article] [PubMed] [Google Scholar]