Abstract
Summary: We have developed an algorithm to detect copy number variants (CNVs) in homozygous organisms, such as inbred laboratory strains of mice, from short read sequence data. Our novel approach exploits the fact that inbred mice are homozygous at virtually every position in the genome to detect CNVs using a hidden Markov model (HMM). This HMM uses both the density of sequence reads mapped to the genome, and the rate of apparent heterozygous single nucleotide polymorphisms, to determine genomic copy number. We tested our algorithm on short read sequence data generated from re-sequencing chromosome 17 of the mouse strains A/J and CAST/EiJ with the Illumina platform. In total, we identified 118 copy number variants (43 for A/J and 75 for CAST/EiJ). We investigated the performance of our algorithm through comparison to CNVs previously identified by array-comparative genomic hybridization (array CGH). We performed quantitative-PCR validation on a subset of the calls that differed from the array CGH data sets.
Availability: The software described in this manuscript, named cnD for copy number detector, is free and released under the GPL. The program is implemented in the D programming language using the Tango library. Source code and pre-compiled binaries are available at http://www.sanger.ac.uk/resources/software/cnd.html
Contact: rd@sanger.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Copy number variants (CNVs) are segments of DNA that have been duplicated, or lost, in the genome of one individual or strain with respect to another. CNVs are thought to contribute significantly to phenotypic differences between mouse strains. In humans, CNVs have been causally linked to a range of disorders including schizophrenia (Moon et al., 2006), autism (Sebat et al., 2007) and birth defect syndromes (Lu et al., 2008). High-resolution surveys for CNVs have been performed in common laboratory strains of mice using array-comparative genomic hybridization (array CGH) (Cahan et al., 2009; Cutler et al., 2007; Graubert et al., 2007; Henrichsen et al., 2009; She et al., 2008). These studies have found a significant level of variation between strains, such that as much as 15% of the reference C57BL/6J mouse genome may be found as CNVs in another strain. While array CGH can be an effective way of identifying CNVs, aCGH studies are limited in resolution by the number of probes that can be placed on a microarray. The widespread adoption of short read sequencing platforms has led to a rapid decrease in the cost of whole-genome re-sequencing making it a viable alternative to array CGH (Xie and Tammi, 2009). Hidden Markov Models (HMM) have previously been used to detect copy number variation from array CGH data (Cahan et al., 2008; Fridlyand et al., 2004). We have developed a HMM to detect CNVs in inbred strains from the alignments of short read sequences to a reference genome.
2 DESCRIPTION
The central idea behind our model is that the alignment of reads from regions with copy number gains (with respect to a reference genome) will be ‘collapsed’ to a single location on the reference genome. The effect of this will be 2-fold. First, the sequence depth of this location on the reference genome will be increased by an integral amount corresponding to the relative number of copies that exist in the sequenced strain. Second, any base-pair differences between the copied regions will appear to be heterozygous single nucleotide polymorphisms (SNPs) with respect to the reference. This fact is crucial to our model as laboratory strains of mice are inbred to be effectively homozygous at every position in the genome, hence any apparent heterozygous SNPs that are not sequencing errors are actually paralogous sequence variants and therefore define regions collapsed in the reference genome. Conversely, the alignment of reads from regions with copy number losses in the sequenced genome will be distributed over the corresponding copies in the reference genome and hence the reference regions will have lower sequence depth, with the important distinction that there will not be a heterozygous SNP signal. Our HMM exploits these factors to detect regions of copy number gain and loss.
Our algorithm proceeds in three stages. First, the sequence reads are aligned to the mouse reference genome (build NCBI 37, Mouse Genome Sequencing Consortium, Waterston et al., 2002) using the MAQ aligner (Li et al., 2008). MAQ calls SNPs and classifies them as homozygous or heterozygous. Summary statistics are computed for the sequence read depth, the number of heterozygous SNPs and the average number of hits per read over 1 kb windows of the reference genome sequence. This triplet of data for each 1 kb region of the reference genome is input to the HMM which classifies each region as corresponding to a gain, loss or no change in copy number.
2.1 The HMM
We developed a 10-state HMM of the copy number structure of the genome being sequenced. There are five major states of the model, representing normal sequence, a 2-fold increase in copy number, a 3-fold increase in copy number, a 2-fold decrease in copy number and zero copy number. In addition, each major state of the model has a sub-state corresponding to highly repetitive sequence, allowing the model to accommodate the frequent high-copy repeat elements dispersed throughout mammalian genomes. In all states expect for the repeat states the depth distribution is modeled by a normal distribution with the mean and variance reflecting the copy number of the state. For states representing a copy number gain, the heterozygous SNP rate is modeled by a negative binomial distribution. The heterozygous SNP rate is modeled by a Poisson distribution in all other states. More information about the HMM and emission distributions is given in the supplemental material.
The parameters of the model are learned for each chromosome in the input data set by Viterbi training for both the transition probabilities and emission distribution parameters (Durbin et al., 1998). After the model parameters have been determined, the sequence of states is computed by a final application of the Viterbi algorithm. The output of the Viterbi algorithm is processed to extract contiguous regions of gain or loss. The minimum threshold for detection is the input window size, typically one kilobase. There is a final optional filtering step to remove calls below a minimum size threshold.
3 RESULTS
We tested our model on Illumina short read sequence data from chromosome 17 for the A/J and CAST/EiJ strains of mouse that were sequenced to 22- and 34-fold, respectively (ERA accession number ERA000077). The data sets were generated using 36-bp paired-end reads of 200-bp insert libraries. For this experiment, we set a minimum call size threshold of 10 kb (see Supplementary data). We evaluated our calls against a collection of previously published aCGH copy number variation data (Cahan et al., 2009; Cutler et al., 2007; Henrichsen et al., 2009; She et al., 2008).
Our algorithm called 22 copy number gains (1.38 Mb of sequence) and 21 losses (0.49 Mb) for the A/J data set (see Fig. 1 and Supplementary Fig. 6 for example regions). The gain regions overlap 38% of the regions identified by aCGH (36% by sequence, 1.1 Mb). Seventy-seven percent of the gains cnD found were previously seen by aCGH. For CAST/EiJ, 45 gains (2.44 Mb of sequence) and 30 losses (1.16 Mb) were called. The gain regions overlap 76% of the gains called by aCGH (79% by sequence, 1.3 Mb). Thirty-six percent of the gains found by cnD were previously seen in the array CGH data set. This figure is much lower than that of A/J due to the fact that the CAST/EiJ strain was not used in the highest coverage aCGH study (Cahan et al., 2009). In both strains the regions of copy number loss called by our algorithm and aCGH differed widely (11% concordance by region for A/J and 32% for CAST/EiJ) owing to the relative difficulty of calling CNV losses compared to gains. We performed qPCR validation on a subset of both the gain calls that were novel to our algorithm (those not found by aCGH) and the novel gain calls found by aCGH. In total we attempted validation on 20 novel cnD gains, of which five were confirmed to be amplified relative to C57BL/6J. Of the 14 novel aCGH gains that we attempted to validate, one was confirmed to be a gain relative to C57BL/6J. Our concordance with array CGH and initial confirmation rates are similar to previously published copy number variation studies (Conrad et al., 2009; Redon et al., 2006; Scherer et al., 2007). Full details of the experimental validation are provided in the Supplementary data.
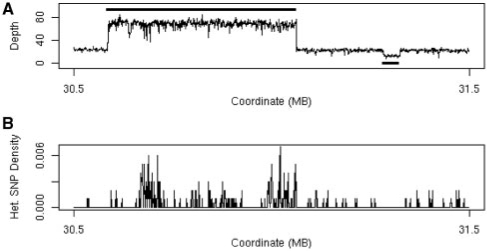
Fig. 1.
(A) Plot of sequencing depth across a one megabase region of A/J chromosome 17 clearly shows both a region of 3-fold increased copy number (30.6–31.1 Mb) and a region of decreased copy number (at 31.3 Mb). The solid black line above the depth plot indicates the called copy number gain and the solid black line below the plot indicates the called copy number loss. (B) Plot of the heterozygous SNP rate for the same region showing the high number of apparent heterozygous SNPs associated with the copy number gain.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Thomas Keane and Jim Stalker for implementing the initial data processing pipeline and Ian Sudbery for generating the chromosome 17 sequencing data.
Funding: Medical Research Council-UK and the Wellcome Trust (077192/Z/05/Z); Cancer-Research-UK to D.J.A.
Conflict of Interest: none declared.
REFERENCES
- Cahan P, et al. wuHMM: a robust algorithm to detect DNA copy number variation using long oligonucleotide microarray data. Nucleic Acids Res. 2008;36:e41. doi: 10.1093/nar/gkn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, et al. The impact of copy number variation on local gene expression in mouse hematopoietic stem and progenitor cells. Nat. Genet. 2009;41:430–437. doi: 10.1038/ng.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. [Epub ahead of print, doi: 1038/nature08516, October 7, 2009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler G, et al. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome Res. 2007;17:1743–1754. doi: 10.1101/gr.6754607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R, et al. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge, UK; New York: Cambridge University Press; 1998. Markov chains and hidden Markov models; p. 356. [Google Scholar]
- Fridlyand J, et al. Hidden Markov models approach to the analysis of array CGH data. J. Multivar. Anal. 2004;90:132–153. [Google Scholar]
- Graubert TA, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen CN, et al. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- Li H, et al. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XY, et al. Genomic imbalances in neonates with birth defects: high detection rates by using chromosomal microarray analysis. Pediatrics. 2008;122:1310–1318. doi: 10.1542/peds.2008-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HJ, et al. Identification of DNA copy-number aberrations by array-comparative genomic hybridization in patients with schizophrenia. Biochem. Biophys. Res. Commun. 2006;344:531–539. doi: 10.1016/j.bbrc.2006.03.156. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, et al. Challenges and standards in integrating surveys of structural variation. Nat. Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, et al. Mouse segmental duplication and copy number variation. Nat. Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.