Abstract
Motivation: The sequencing of the human genome has made it possible to identify an informative set of >1 million single nucleotide polymorphisms (SNPs) across the genome that can be used to carry out genome-wide association studies (GWASs). The availability of massive amounts of GWAS data has necessitated the development of new biostatistical methods for quality control, imputation and analysis issues including multiple testing. This work has been successful and has enabled the discovery of new associations that have been replicated in multiple studies. However, it is now recognized that most SNPs discovered via GWAS have small effects on disease susceptibility and thus may not be suitable for improving health care through genetic testing. One likely explanation for the mixed results of GWAS is that the current biostatistical analysis paradigm is by design agnostic or unbiased in that it ignores all prior knowledge about disease pathobiology. Further, the linear modeling framework that is employed in GWAS often considers only one SNP at a time thus ignoring their genomic and environmental context. There is now a shift away from the biostatistical approach toward a more holistic approach that recognizes the complexity of the genotype–phenotype relationship that is characterized by significant heterogeneity and gene–gene and gene–environment interaction. We argue here that bioinformatics has an important role to play in addressing the complexity of the underlying genetic basis of common human diseases. The goal of this review is to identify and discuss those GWAS challenges that will require computational methods.
Contact: jason.h.moore@dartmouth.edu
1 INTRODUCTION
The current strategy for revealing the genetic basis of disease susceptibility is to carry out a genome-wide association study (GWAS) with a million or more single nucleotide polymorphisms (SNPs) that capture much of the common variation in the human genome (Hirscchorn et al., 2005; Wang et al., 2005). This approach is based on the idea that genetic variations with alleles that are common in the population will explain much of the heritability of common diseases (Reich and Lander, 2001). This is referred to as the common disease common variant hypothesis and has been recently reviewed by Schork et al. (2009). These studies were made possible by the sequencing of the human genome (International Human Genome Sequencing Consortium, 2004) and the completion of the subsequent human haplotype mapping (HapMap) project that discovered millions of common SNPs and documented the correlation structure or linkage disequilibrium of the alleles at those loci (The International HapMap Consortium, 2005). This knowledge about genome variation in combination with novel bioengineering methods made it possible to design chips for measuring more than a million SNPs for several hundred dollars or less per sample. As the price of genome-wide genotyping has dropped, the number of studies utilizing GWAS has increased dramatically and this approach is now relatively common. As Manolio et al. (2008) and Donnelly (2008) have recently reviewed, hundreds of replicated susceptibility loci for as many as 70 diseases and traits have been reported from GWAS. Unfortunately, despite high expectations, few of the loci identified via GWAS are associated with a moderate or large increase in disease risk and some well-known genetic risk factors have been missed (Williams et al., 2007). In fact, the relative risks of most new loci are on the order of 1.1 to 1.2 at best which suggests that these individual SNPs may not be useful for genetic testing. This limitation has been pointed out in a recent study by Jakobsdottir et al. (2009) that showed SNPs identified by GWAS for a variety of diseases make very poor classifiers of disease, calling into question their usefulness for risk assessment in personal genetics (Moore and Williams, 2009).
To illustrate successes and failures of GWAS, consider the following review of breast, prostate, colorectal, lung and skin cancer that shows that a number of new susceptibility loci have been identified using the GWAS approach (Easton and Eeles, 2008). As mentioned above, the loci identified by GWAS typically have very small effect sizes. This is true for cancer where the increase in risk for the susceptibility alleles at each of the loci discovered by GWAS is generally 1.3-fold or less. Let us first consider familial breast cancer as a rare disease that has a very high heritability and is thus believed to have a relatively simple etiology. Easton et al. (2007) reported five significant, replicated associations that were identified by GWAS and replicated in several independent samples of subjects. Four of the discovered variants were in known genes and one was located in a hypothetical gene. Assuming a multiplicative model, these five loci together explain only 3.6% of the excess familial risk of breast cancer. Due to these small effect sizes, Ripperger et al. (2009) concluded that these loci are not suitable for use in genetic testing. In a follow-up study with two additional stages of testing and replication, two additional susceptibility loci were identified with odds ratios of 1.11 and 0.95, respectively. These two loci account for much <1% of the familial risk of breast cancer (Ahmed et al., 2009). When combined with the previous known genetic risk factors for familial breast cancer, the estimated fraction of risk explained is ∼5.9%. This is in stark contrast to BRCA1 and BRCA2 mutations that together account for between 20 and 40% of familial breast cancer. While the application of GWAS to familial breast cancer has generated new knowledge and perhaps new biology, it has not resulted in new genetic tests that can be used to predict and prevent familial breast cancer. The results for common diseases such as sporadic breast cancer and type II diabetes that have a much more complex underlying genetic architecture are similarly discouraging.
As another example, consider a recent GWAS applied to pancreatic cancer. Amundadottir et al. (2009) measured >500 000 SNPs in a detection sample of 1896 patients with pancreatic cancer and 1939 controls ascertained from the same population as the cases. The authors also used a replication sample of 2457 cases and 2654 controls. A logistic regression analysis of both samples identified a single SNP with an odds ratio of 1.2. This single SNP was located in an intron of the ABO blood group gene. This result confirmed previous epidemiological studies showing that the O blood group is associated with a lower risk of pancreatic cancer. Interestingly, this association was first reported >50 years ago and thus does not represent a novel finding. The failure to identify new susceptibility loci for some diseases using GWAS in relatively large sample sizes highlights some of the limitations of this approach.
These studies and many others highlight the positive and negative aspects of GWAS for common diseases and complex traits. One explanation for the mixed results of GWAS is that the current biostatistical analysis paradigm is, by design, agnostic or unbiased in that it ignores what is known about disease pathobiology. Further, the linear modeling framework often used for GWAS analysis usually considers only one SNP at a time thus ignoring the genomic and environmental context of each SNP (Moore and Williams, 2009). As Clark et al. (2004) predicted, our success with GWAS depends critically on the assumptions we make about disease complexity. Recently, there has been a shift away from the one SNP at a time approach toward a more holistic approach that recognizes the complexity of the genotype–phenotype relationship that is likely characterized by significant genetic heterogeneity and gene–gene and gene–environment interaction. We argue here that bioinformatics can play an important role in addressing the complexity of the underlying genetic basis of many common human diseases. The goal of this review is to identify and discuss those GWAS challenges that require computational rather than, or in addition to, biostatistical methods. We focus on computational methods for data mining and machine learning and bioinformatics methods for incorporating prior biological knowledge into data analysis algorithms. We conclude with a discussion about maximizing the utility of bioinformatics software for GWAS analysis. Readers are directed elsewhere for recent reviews of GWAS study design, quality control, imputation and biostatistical analysis issues (e.g. Amos, 2007; Chanock et al., 2007; Kraft and Cox, 2008; Spencer et al., 2009; Ziegler et al., 2008).
2 DATA MINING AND MACHINE LEARNING
2.1 Why are data mining and machine learning methods needed?
An important goal of human genetics and genetic epidemiology is to understand the mapping relationship between interindividual variation in DNA sequences, variation in environmental exposure and variation in disease susceptibility. Stated another way, how do one or more changes in an individual's DNA sequence increase or decrease their risk of developing disease through complex networks of biomolecules that are hierarchically organized, highly interactive and dependent on environmental exposures? Understanding the role of genomic variation and environmental context in disease susceptibility is likely to improve diagnosis, prevention and treatment. Success in this important public health endeavor will depend critically on the amount of non-linearity in the mapping of genotype to phenotype and our ability to address it. Here, we define as non-linear an outcome that cannot be easily predicted by the sum of the individual genetic markers. Non-linearities can arise from phenomena such as locus heterogeneity (i.e. different DNA sequence variations leading to the same phenotype), phenocopy (i.e. environmentally determined phenotypes that do not have a genetic basis) and the dependence of genotypic effects on environmental exposure (i.e. gene–environment interactions or plastic reaction norms) and genotypes at other loci (i.e. gene–gene interactions or epistasis). Each of these phenomena have been recently reviewed and discussed by Thornton-Wells et al. (2004) who call for an analytical retooling to address these complexities. Combining the complexities summarized here with GWAS data yields significant computational challenges.
To illustrate non-linear mapping from genotype to phenotype, consider the following example from sporadic Alzheimer disease (AD). In 2004, Infante et al. (2004) reported that polymorphisms in the interleukin-6 (IL-6) and interleukin-10 (IL-10) genes had an interaction effect on the risk of AD. This study of 232 AD patients and 191 controls reported that patients with the IL-6 C/C and IL-10 A/A genotypes had a five times lower risk of AD than control subjects (P=0.005). What makes this association interesting is the absence of a statistically significant association for the IL-10 A/A genotype (P=0.102). Further, there is significant biological plausibility for this interaction given the importance of inflammation in AD and the significant role of Il-6 as a pro-inflammatory molecule and IL-10 as an anti-inflammatory molecule. Of course, the gold standard in genetic association studies is replication. In 2009, Combarros et al. (2009) replicated the interaction of the IL-6 and IL-10 genes in a collaboration of seven AD studies with a total of 1757 AD cases and 6295 controls. The statistical replication of the non-linear interaction and the biological plausibility of the finding strongly suggest that these two genetic markers or nearby markers contribute to the development of AD.
Moore and Ritchie (2004) have provided an overview of three significant challenges that must be overcome if we are to successfully identify those genetic variations that are associated with health and disease using a genome-wide approach. First, powerful data mining and machine learning methods will need to be developed to computationally model the relationship between combinations of SNPs, other genetic variations and environmental exposure with disease susceptibility. This is because traditional parametric statistical approaches such as logistic regression have limited power for modeling high-order non-linear interactions that are likely important in the etiology of complex diseases (Moore and Williams, 2002). A second challenge is the selection of SNPs that should be included in the analysis. If non-linear interactions between genes explain a significant proportion of the heritability of common diseases, then combinations of SNPs will need to be evaluated from a list of thousands or millions of candidates. Filtering algorithms and/or stochastic search or wrapper algorithms will play an important role in GWAS because there are more combinations of SNPs to examine than can be exhaustively evaluated using modern computational horsepower. A third challenge is the biological interpretation of non-linear genetic models. Even when a computational model can be used to identify SNPs with genotypes that increase susceptibility to disease, the specifics of the mathematical relationships cannot be translated into prevention and treatment strategies without interpreting the results in the context of human biology. Making etiological inferences from computational models may be the most important and the most difficult challenge of all (Moore and Ritchie, 2004).
2.2 The modeling challenge
The parametric linear statistical model plays a very important role in modern genetic epidemiology because it has solid theoretical foundation, is easy to implement using a wide range of different software packages such as SAS and R and is easy to interpret. Despite these good reasons to use linear models, they do have limitations for detecting non-linear patterns of interaction (Moore and Williams, 2002). In addition, they are not likely to explain a large part of the variance of any given trait (Moore, 2003) which may explain some of the missing heritability that has not been accounted for by GWAS (Manolio et al., 2009). The first issue is that statistical or computational modeling of non-linear interactions and other complex phenomena such as locus heterogeneity inherently requires looking at combinations of SNPs. Considering multiple SNPs simultaneously is analytically challenging because the study samples or instances get spread thinly across multiple combinations of genotypes. This is because the number of genotype combinations goes up exponentially as each SNP is added to the model. Estimation of parameters in a linear model can be problematic where no data are observed for individual genotype combinations (i.e. empty cells). The second issue is that parametric linear models are generally implemented such that interaction effects are only modeled using factors that exhibit independent marginal effects. This makes model fitting easier but implicitly assumes that genetic architecture is simple and that important predictors will have detectable marginal effects. Further, it is well documented that linear models have greater power to detect marginal effects than interactions (Lewontin, 1974; Wahlsten, 1990). For example, the focused interaction testing framework (FITF) approach of Millstein et al. (2006) provides a powerful logistic regression approach to detecting interactions but conditions on marginal effects. Interactions in the absence of significant marginal effects are missed by FITF and other similar methods.
How common are gene–gene interactions in the absence of significant marginal effects likely to be? Moore (2003) argues that a simple genetic architecture characterized by SNPs with large marginal effects is an unrealistic assumption for many common human diseases. Rather, it is likely that complex phenomena such as epistasis or gene–gene interactions will make up much of the genetic architecture. As Moore (2003) summarizes, there are several reasons for this. First, epistasis is on old idea that has been around since the early 1900s. Early geneticists such as Bateson (1909) recognized the importance of gene–gene interactions for explaining deviations from Mendelian patterns of inheritance. More important than this historical note is the fact that epistasis is still discussed today as a key component of genetic architecture. Second, biological systems are driven by complex biomolecular interactions. As such, it makes sense that gene–gene interactions would play an important role in the genotype to phenotype mapping relationship. Studies emerging from the genetic analysis of bacteria and yeast document widespread epistasis at the biological level. Third, single SNP results do not always replicate even if cases where there is a general consensus that the association signal is real. This has been the norm for GWAS where very few SNPs with significant marginal effects replicate in multiple independent samples. As Marchini et al. (2005) and Greene et al. (2009c) suggest, this may be partly due to underlying patterns of epistasis. Finally, epistasis is commonly found when properly investigated (Templeton, 2000). Studies that look for gene–gene interactions in a manner that does not condition on marginal effects commonly find such non-linear effects. We predict that the data mining and machine learning methods reviewed here will reveal numerous significant interactions and other complex genotype–phenotype relationships when they are widely applied to GWAS data.
The limitations of the linear model and other parametric statistical approaches have motivated the development of data mining and machine learning methods (e.g. Hastie et al., 2009; Mitchell, 1997). The advantage of these computational approaches is that they make fewer assumptions about the functional form of the model and the effects being modeled (McKinney et al., 2006). In other words, data mining and machine learning methods are much more consistent with the idea of letting the data tell us what the model is rather than forcing the data to fit a preconceived notion of what a good model is. Several recent reviews highlight the need for new methods (Thornton-Wells et al., 2004) and discuss and compare different strategies for detecting statistical epistasis (Cordell, 2009; Motsinger et al., 2007). The methods reviewed by Cordell (2009) include novel approaches such as combinatorial partitioning (Culverhouse et al., 2004; Nelson et al., 2001) and logic regression (Kooperberg et al., 2001; Kooperberg and Ruczinski, 2005) and machine learning approaches such as random forests (RFs). Below, we briefly review two of these methods, RFs and multifactor dimensionality reduction (MDR) that have been developed to address these issues. Importantly, as Marchini et al. (2005) demonstrated, interaction analysis can have more power than traditional approaches despite issues such as multiple testing. This study provides an important, but often overlooked, foundation for the methods described below.
2.3 Computational modeling using decision trees and RFs
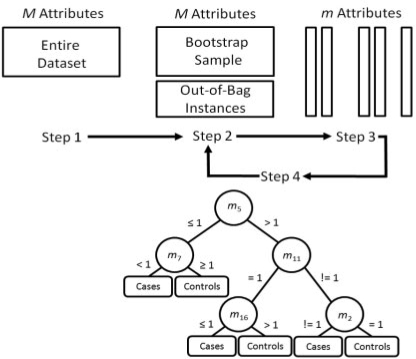
Classification or decision trees are a staple in the data mining and machine learning community due to their algorithmic simplicity and ease of interpretation. Decision trees are widely used for modeling the relationship between one or more attributes and a discrete end point such as case–control status (Mitchell, 1997). Here, we use the word attribute to mean a variable such as a SNP or a demographic variable such as gender that is used to make a prediction. Attribute is commonly used this way in data mining and machine learning. In statistics, an attribute is an independent variable, explanatory variable or predictor. A decision tree classifies subjects as case or control by sorting them through a tree from node to node where each node is an attribute with a decision rule that guides that subject through different branches of the tree to a leaf that provides its classification. The primary advantage of this approach is that it is simple and the resulting tree can be interpreted as a series of IF-THEN rules that are easy to understand. For example, a genetic model of heterozygote effects with genotype data coded {AA = 0, Aa = 1, aa = 2} might look like IF genotype at SNP1=1 THEN case ELSE control. In this simple model, the root node of the tree would be SNP1 with decision rule ‘=1’ and leafs equal to case and control (e.g. see tree in Fig. 1). Additional nodes or attributes below the root node allows hierarchical dependencies (i.e. interactions) to be modeled. Here, we review RFs that extend decision trees for the analysis of more complex data.
Fig. 1.
Overview of the RF algorithm summarized in Section 2.3. Adapted from Reif et al. (2006).
A RF is a collection of individual decision tree classifiers, where each tree in the forest has been trained using a bootstrap sample of instances (i.e. subjects) from the data, and each attribute in the tree is chosen from among a random subset of attributes (Breiman, 2001). Classification of instances is based upon aggregate voting over all trees in the forest.
Individual trees are constructed as follows from data having N samples and M attributes:
Choose a training set by selecting N samples, with replacement, from the data.
At each node in the tree, randomly select m attributes from the entire set of M attributes in the data (the magnitude of m is constant throughout the forest building).
Choose the best split at that node from among the m attributes.
Iterate the second and third steps until the tree is fully grown (no pruning).
Repetition of this algorithm yields a forest of trees, each of which have been trained on bootstrap samples of instances. Thus, for a given tree, certain samples or instances will have been left out during training. Prediction error is estimated from these ‘out-of-bag’ instances. The out-of-bag instances are also used to estimate the importance of particular attributes via permutation testing. If randomly permuting values of a particular attribute does not affect the predictive ability of trees on out-of-bag samples, that attribute is assigned a low importance score (Bureau et al., 2005).
RFs are often used initially for selecting the subset of attributes that can then be modeled using a decision tree. We discuss using algorithms such as RF for filtering subsets of SNPs later in Section 2.6. An advantage of the RF approach is that the final decision tree models may uncover interactions among genes and/or environmental factors that do not exhibit strong marginal effects (Cook et al., 2004), especially when combined with methods such as ReliefF (see Section 2.6) for choosing the attributes to be used as nodes. Additionally, tree methods are suited to dealing with certain types of genetic heterogeneity, since early splits in the tree define separate model subsets in the data (Lunetta et al., 2004). RFs capitalize on the benefits of decision trees and have demonstrated excellent predictive performance when the forest is diverse (i.e. trees are not highly correlated with each other) and composed of individually strong classifier trees (Breiman, 2001). The RF method is a useful approach for studying gene–gene or gene–environment interactions because importance scores for particular attributes take interactions into account without demanding a prespecified model (Lunetta et al., 2004). However, most current implementations of the importance score are calculated in the context of all other attributes in the model. Therefore, assessing the interactions between particular sets of attributes must be done through careful model interpretation, although there has been preliminary success in jointly permuting explicit sets of attributes to capture their interactive effects (Bureau et al., 2005).
Lunetta et al. (2004) have previously shown that RFs outperform traditional methods such as the Fisher's exact test when the ‘risk’ SNPs interact. This study revealed that the relative superiority of the RF method increases as more interacting SNPs are added to the model. In addition, Bureau et al. (2005) have shown that RFs are robust in the presence of noisy or potential false positive SNPs relative to methods that rely on independent marginal effects. Initial results of RF applications to genetic data in studies of asthma (Bureau et al., 2005), rheumatoid arthritis (Sun et al., 2007) and glioblastoma (Chang et al., 2008), age-related macular degeneration (Jiang et al., 2009) and vaccination response (McKinney et al., 2009) are encouraging and it is anticipated that RF will prove a useful tool for detecting gene–gene interactions. They may also be useful when multiple different data types (e.g. proteomic biomarkers) are present (Reif et al., 2006, 2009) or for inferring gene networks (McKinney et al., 2009). The primary limitation of tree-based methods is that the standard implementations condition on marginal effects. That is, the algorithm finds the best single variable for the root node before adding additional variables as nodes in the model. Combining RF with ReliefF methods (described below) shows potential for overcoming this limitation (McKinney et al., 2009). Advantages of this approach include its basis on decision trees and the availability of the algorithm in many different open source software packages including R. In fact, the Willows package was designed specifically for tree-based analysis of SNP data (Zhang et al., 2009).
2.4 Computational modeling using MDR
Thornton-Wells et al. (2004) review the complex nature of the genotype–phenotype relationship and suggest that we need new statistical and computational tools to address these complexities. As a result, there is growing trend toward the development and evaluation of new and novel approaches that have more power for modeling non-linearity than parametric statistical approaches. As Cordell (2009) recently summarized, MDR is an example of one novel computational strategy for detecting and characterizing non-linear patterns of gene–gene interactions in genetic association studies. MDR was developed as a non-parametric (i.e. no parameters are estimated) and genetic model-free (i.e. no genetic model is assumed) data mining and machine learning strategy for identifying combinations of discrete genetic and environmental factors that are predictive of a discrete clinical end point (Hahn et al., 2003; Moore, 2004, 2007b; Moore and Hahn, 2004; Moore and White, 2006; Ritchie et al., 2001, 2003). Unlike most other methods, MDR was designed to detect interactions in the absence of detectable marginal effects and thus complements statistical approaches such as logistic regression and machine learning methods such as RFs and neural networks.
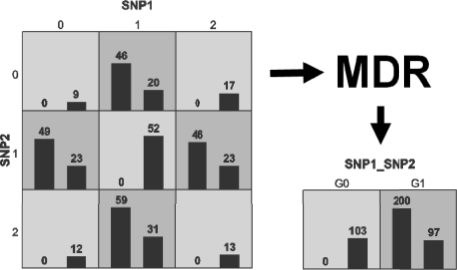
At the heart of the MDR approach is a feature or attribute construction algorithm that creates a new variable or attribute by pooling genotypes from multiple SNPs (Moore and White, 2006). The general process of defining a new attribute as a function of two or more other attributes is referred to as constructive induction, or attribute construction, and was first described by Michalski (1983). Constructive induction, using the MDR kernel, is accomplished in the following way (Fig. 2). Given a threshold T, a multilocus genotype combination is considered high risk if the ratio of cases (subjects with disease) to controls (healthy subjects) exceeds T, otherwise it is considered low risk. Genotype combinations considered to be high risk are labeled G1 while those considered low risk are labeled G0. This process constructs a new one-dimensional attribute with values of G0 and G1. It is this new single variable that is assessed, using any classification method. The MDR method is based on the idea that changing the representation space of the data will make it easier for methods such as logistic regression, classification trees or a naive Bayes classifier to detect attribute dependencies. As such, MDR significantly complements other classification method such as those reviewed by Hastie et al. (2001). This method has been confirmed in numerous simulation studies and a user-friendly open source MDR software package written in Java is freely available from www.epistasis.org.
Fig. 2.
Summary of the constructive induction process for MDR. The left bars within each cell represent the number of cases while the right bars represent the number of controls. Dark-shaded cells are high risk while the light-shaded cells are low risk. Prediction using any classifier can be carried out using the final constructed attribute.
Since its initial description by Ritchie et al. (2001), many modifications and extensions to MDR have been proposed. These include entropy-based interpretation methods (Moore and White, 2006), the use of odds ratios (Chung et al., 2007), log-linear methods (Lee et al., 2007), generalized linear models (Lou et al., 2007), methods for imbalanced data (Velez et al., 2007), permutation testing methods (Greene et al., 2010a; Pattin et al., 2009), methods for dealing with missing data (Namkung et al., 2009a), model-based methods (Calle et al., 2008), parallel implementations (Bush et al., 2006; Sinnott-Arnstrong et al., 2009) and different evaluation metrics (Bush et al., 2008; Mei et al., 2007; Namkung et al., 2009b). These extensions have addressed many of the previous limitations of the MDR method. The MDR approach has also been successfully applied to a wide range of different genetic association studies. For example, Andrew et al. (2006) used MDR to model the relationship between polymorphisms in DNA repair enzyme genes and susceptibility to bladder cancer. A highly significant non-additive interaction was found between two SNPs in the Xeroderma pigmentosum group D (XPD) gene that was a better predictor of bladder cancer than smoking. Importantly, these results have been independently replicated (International Consortium of Bladder Cancer, 2009).
2.5 The attribute selection challenge
Combining the complexity of the genotype–phenotype relationship described above with the challenge of attribute (e.g. SNP) selection yields a needle-in-a-haystack problem. That is, there may be a particular combination of SNPs or SNPs and environmental factors that together with the right non-linear function are a significant predictor of disease susceptibility. However, individually each factor may not appear different than thousands of other SNPs that are not involved in the disease process and are thus noisy. Therefore, the learning algorithm is truly looking for a genetic needle in a genomic haystack. It is now commonly assumed that at least 106 carefully selected SNPs are necessary to capture much of the relevant variation across the human genome. With this many attributes, the number of higher order combinations is astronomical. These large datasets beg the question, what is the optimal computational approach to this problem?
There are two general approaches to selecting attributes for predictive models. The filter approach preprocesses the data by algorithmically assessing the quality or relevance of each variable and then using that information to select a subset for analysis. The wrapper approach iteratively selects subsets of attributes for classification using either a deterministic or stochastic algorithm. The key difference between the two approaches is that the learning algorithm plays no role in selecting those attributes to consider in the filter approach. As Freitas (2002) reviews, the advantage of the filter is speed while the wrapper approach has the potential to do a better job classifying subjects as sick or healthy. We first discuss several learning algorithms that have been applied to classifying healthy and disease subjects using their DNA sequence information and then discuss filter and wrapper approaches for the specific problem of detecting epistasis or non-linear patterns of gene–gene interactions on a genome-wide scale.
2.6 Attribute selection using filter algorithms
As discussed above, it is computationally infeasible to combinatorially explore all high-order interactions among the SNPs in a genome-wide association study. One approach is to filter out a subset of genetic variations with high quality (i.e. likely to be associated) that can then be efficiently analyzed using a method such as RFs or MDR (Moore and White, 2006; Wilke et al., 2005). There are many different statistical and computational methods for determining the quality of attributes. A standard statistical strategy in human genetics and genetic epidemiology is to assess the quality of each SNP using a chi-square test of independence followed by a correction of the significance level that takes into account an increased false positive (i.e. type I error) rate due to multiple tests. This is a very efficient filtering method for assessing the independent effects of SNPs on disease susceptibility but it ignores the dependencies or interactions between genes.
Kira and Rendell (1992) developed an algorithm called Relief that is capable of detecting complex attribute dependencies even in the absence of marginal effects. Relief estimates the quality of attributes through a type of nearest neighbor algorithm that selects neighbors (instances) from the same class and from the different class based on the vector of values across attributes. For the purposes of this description, we assume the class is dichotomous. Weights (W) or quality estimates for each attribute (A) are estimated based on whether the nearest neighbor (nearest hit, H) of a randomly selected instance (R) from the same class and the nearest neighbor from the other class (nearest miss, M) have the same or different values. This process of adjusting weights is repeated for m instances. The algorithm produces weights for each attribute ranging from −1 (worst) to +1 (best). The time complexity of Relief is O(m*n*a) where m is the number of instances randomly sampled from a dataset with n total instances and a attributes. Kononenko (1994) improved upon Relief by choosing n (usually set to 10) nearest neighbors instead of just one. This new ReliefF algorithm has been shown to be more robust to noisy attributes (Kononenko, 1994; Robnik-Šikonja and Kononenko, 2003) and is widely used in data mining applications.
ReliefF is able to capture attribute interactions because it selects nearest neighbors using the entire vector of values across all attributes. However, this advantage is also a disadvantage because the presence of many noisy or potential false positive attributes can reduce the signal the algorithm is trying to capture. Moore and White (2007b) proposed a ‘tuned’ ReliefF algorithm (TuRF) that systematically removes attributes that have low-quality estimates so that the ReliefF values if the remaining attributes can be reestimated. The motivation behind this algorithm is that the ReliefF estimates of the true functional attributes will improve as the noisy attributes are removed from the dataset. Moore and White (2007b) carried out a simulation study using previously published epistasis models (Velez et al., 2007) to evaluate the power of ReliefF, TuRF and a naïve chi-square test of independence for selecting functional attributes in a filtered subset. Moore and White (2007b) found that the power of ReliefF to pick (filter) the correct interacting attributes was consistently better (P≤0.05) than a naïve chi-square test of independence and that the TuRF algorithm was consistently better (P≤0.05) than ReliefF across all models studied. More recent extensions of the ReliefF algorithm have shown that using higher numbers of nearest neighbors greatly improve the power of ReliefF. For example, Greene et al. (2009b) showed that a spatially uniform ReliefF (SURF) that picks all neighbors within a predefined epsilon (i.e. distance or radius) greatly improves the power to detect interacting SNPs over that of ReliefF. McKinney et al. (2007) have combined ReliefF with measures of entropy to yield evaporative cooling ReliefF. This approach was highly successful when used to select SNPs for a RFs analysis (McKinney et al., 2009). The differences between how Relief, ReliefF and SURF select nearest neighbors are summarized in Figure 3. When combined with permutation tests designed to assess interactions, P-values can also be used to select SNPs with significant ReliefF scores (Greene et al., 2010b; Wongseree et al., 2009). These results suggest that algorithms based on ReliefF show promise for filtering interacting SNPs. The disadvantage of the filter approach is that important attributes might be discarded prior to analysis. Stochastic wrapper methods provide a flexible alternative and may be more powerful when the assumptions of the filter approach are not valid (Greene et al., 2009a).
Fig. 3.
Summary of how Relief, ReliefF and SURF select neighbors. Each panel in this figure shows the genotypes at two markers for a dataset of cases and controls. For the purpose of this example only these two markers will be considered and both are continuous. When analyzing real data, the process of selecting neighbors is the same, however, but there will be thousands of discrete valued markers (SNPs) each of which would be represented by one of thousands of dimensions. The individual for whom neighbors are being found is shown by the filled red circle. The neighbors that each approach uses for weighting are highlighted in blue. (A–C) Represent how Relief, ReliefF and SURF would select neighbors to be used in weighting. Relief selects the nearest individual of the same dichotomous class (blue circle) and the nearest individual of the other class (blue cross). ReliefF selects some user specified number of individuals (two in this example) to be used for weighting. SURF, instead of using a fixed number of neighbors, uses all individuals within a distance threshold. The dotted line shows a hypothetical distance threshold.
2.7 Attribute selection using wrapper algorithms
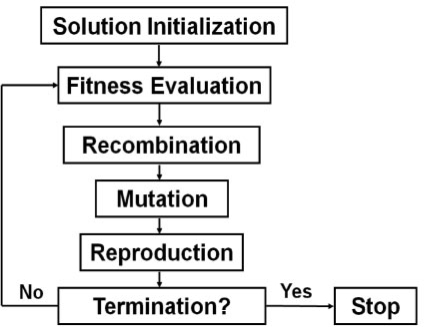
Wrapper methods may be more powerful than filter approaches because no attributes are discarded in the process. As a result, every attribute retains some probability of being selected for evaluation by the classifier. There are many different stochastic wrapper algorithms that can be applied to this problem (e.g. Michalewicz and Fogel, 2004). We review here evolutionary computing algorithms as an example stochastic search algorithm that has been developed for genetic association studies. Ritchie et al. (2003) and Moore et al. (2007), for example, have explored the use of a type of evolutionary computing algorithm called genetic programming (GP) for modeling and attribute selection in genetic association studies. GP is an automated computational discovery tool that is inspired by Darwinian evolution by natural selection (Banzhaf et al., 1998; Koza, 1992). The goal of GP is to ‘evolve’ computer programs to solve complex problems. This is accomplished by first generating or initializing a population of random computer programs that are composed of the basic building blocks needed to solve or approximate a solution to the problem. For genetic association studies this might be a list of SNPs, other important attributes such as age and gender along with a list of mathematical functions. Each randomly generated program is evaluated and the good programs are selected and recombined and mutated to form new computer programs. This process of selection based on fitness and recombination to generate variability is repeated until a best program or set of programs is identified. A flowchart for a simple GP is illustrated in Figure 4. GP and its many variations have been applied successfully in a wide range of different problem domains including bioinformatics (e.g. Fogel and Corne, 2003) and the genetic analysis of epistasis (Moore et al., 2007; Ritchie et al., 2003). It is important to note that GP differs considerably from genetic algorithms (GAs) in that the solution to a problem in GP is represented by a computer program rather than a linear bit string. This provides GP a great deal more flexibility than GA for both attribute selection and modeling.
Fig. 4.
Flowchart for a simple GP. The goal is to randomly generate an initial population of computer programs or solutions (e.g. genetic models), determine their fitness, select the best models, introduce variability and then iterate until the termination criteria are satisfied. This executes a parallel stochastic search using the principles of evolution by natural selection.
Consider, for example, the use of GP for attribute selection. Although we focused on GP here, a GA would likely work just as well if attribute selection is the only task. Moore and White (2007a) developed and evaluated a simple GP wrapper for attribute selection in the context of an MDR analysis. The goal of this study was to develop a stochastic wrapper method that is able to select attributes that interact in the absence of independent marginal effects. At face value, there is no reason to expect that a GP or any other wrapper method would perform better than a random attribute selector because there are no ‘building blocks’ for gene–gene interactions in the absence of marginal effects when accuracy is used as the fitness measure. That is, the fitness of any given classifier would look no better than any other when just one of the correct SNPs is in the MDR model. Preliminary studies by White et al. (2005) support this idea. For GP or any other wrapper to work there need to be recognizable building blocks. Moore and White (2007a) specifically evaluated whether including preprocessed attribute quality estimates using TuRF (see above) in a multiobjective fitness function improved attribute selection over a random search that just uses accuracy as the fitness of the models. Using a wide variety of simulated data, Moore and White (2007a) demonstrated that including TuRF scores in addition to accuracy in the fitness function significantly improved the power of GP to pick the correct two functional SNPs out of 1000 total SNPs. The use of expert knowledge to guide GP population initialization (Greene et al., 2009d), mutation (Greene et al., 2007), recombination (Moore and White, 2006) and a computational evolution system (Greene et al., 2010b; Moore et al., 2008) have also all shown promise. There may also be an important role for linkage disequilibrium in providing missing building blocks (Bush et al., 2009).
3 BIOLOGICAL KNOWLEDGE DATABASES FOR ANALYSIS AND INTERPRETATION
ReliefF and other measures such as interaction information (Moore and White, 2006) are likely to be very useful for providing analytical means for filtering genetic variations prior to epistasis analysis using either RFs or MDR, for example. However, there is growing recognition that we should also use the wealth of accumulated knowledge about gene function to prioritize which genetic variations are analyzed for gene–gene interactions and other complex effects. For example, for any given disease there are often multiple biochemical pathways that have been experimentally confirmed to play an important role. Genes in these pathways can be selected for gene–gene interaction analysis thus significantly reducing the number of gene–gene interaction tests that need to be performed. Gene Ontology (GO), chromosomal location and protein–protein interactions are all example sources of expert knowledge that can be used in a similar manner.
Consider for example, the recent studies by Pattin et al. (2008, 2009) who have specifically reviewed protein–protein interaction databases as a source of expert knowledge that can be used to guide GWASs of epistasis. Here, you might expect that a gene coding for a protein that interacts with many other proteins might be a good candidate for interaction with one or more other genes. You could use this information in several different ways. First, you could employ a biological filter and only test for interactions among SNPs in those genes with many protein–protein interactions. Alternatively, you could weight each gene by its degree of protein–protein interaction and then use this expert knowledge in a stochastic wrapper algorithm. Of course, the most interesting genes might be those with fewer connections. In this case, you could use the inverse of the connectedness as your expert knowledge. Some might find it more useful to use the strength of the protein–protein interaction evidence rather than the number of connections. Here, the genes could be prioritized or weighted by their confidence score that reflects the quality of the experimental and computational evidence for their biochemical interaction with other protein products. The important consideration when using expert knowledge from biological databases is to harness this information in a way that makes sense to the biologist. Emily et al. (2009) demonstrate how protein–protein interactions were used to reduce the search for two-locus interactions using GWAS data from the Wellcome Trust Case-Control Consortium.
The use of expert knowledge from GO and biochemical pathways in GWAS has been recently investigated by a number of groups. For example, Baranzini et al. (2009), Bush et al. (2009), Elbers et al. (2009), Emily et al. (2009), Herold et al. (2009), Holmans et al. (2009), Medina et al. (2009), O'Dushlaine et al. (2009), Pan (2008), Peng et al. (2009), Saccone et al. (2008) and Torkamani et al. (2008) have all shown that using biological knowledge to guide genetic association studies may provide more meaningful results. Yu et al. (2009) provide a hypothesis testing framework for combining multiple SNPs from the same gene or from multiple genes in a pathway-based manner. Askland et al. (2009) recently showed that patterns of SNPs in biological pathways are more likely to replicate than individual SNPs in GWAS. This is highly consistent with the idea that interactions may be more important that marginal effects. Zamar et al. (2009) have provided a software tool called Path to assist with pathway-based analysis of SNPs. Wilke et al. (2008) have suggested that we should not even begin to analyze a GWAS until we have exhaustively studied each candidate gene and each pathway. The use of pathways and other biological knowledge to guide GWAS is an important emerging area and is gaining more attention at international conferences (Moore, 2009).
Perhaps the greatest challenge of any data mining exercise is interpreting the results. This is especially important in GWAS where biological plausibility helps give increased credibility to results (Greene et al., 2009c; Moore and Williams, 2005). Assessing biological plausibility, however, is difficult without software that can only be generated through collaboration among geneticists and bioinformaticists. Fortunately, there are a number of emerging software packages that are designed with this in mind. GenePattern, for example, provides an integrated set of analysis tools and knowledge sources that facilitates this process (Reich et al., 2006). Other tools such as the exploratory visual analysis database and software are designed specifically for integrating research results with biological knowledge from public databases (Reif et al., 2005) and have been applied to GWAS data (Askland et al., 2009). Several commercial packages for typing bioinformatics results to pathways and gene function include Pathway Studio from Ariadne and Ingenuity Pathway Analysis from Ingenuity Systems. Ontology-based methods (Tsoi et al., 2009) and literature-based systems (Yu et al., 2008) have also been recently proposed for aiding with interpretation.
The use of biological knowledge in genetic association studies does have some important limitations. First, success is highly dependent on the quality of the information in the databases. In assessing quality, we need to consider both accuracy of the information included and the completeness of the information. Our expectation is that the quality and completeness of the databases will continue to improve and the amount of good information will soon outweigh the bad. This of course depends critically on the both the throughput and quality of experimental methods used to reveal molecular details and their relationships. Second, it is important to keep in mind the disconnect between the biology that happens at the cellular level and the statistical patterns of genetic variation that we observe at the population level (Moore and Williams, 2005). It is very difficult to make inference about population-level risk from the knowledge of cellular function and vice versa. This will always to some degree limit our ability to use biological knowledge to assist with association studies in human populations.
4 SOFTWARE CHALLENGES
Perhaps the biggest challenge moving forward with GWAS analysis is to facilitate communication at all levels among biomedical researchers, biostatisticians and computer scientists. The key to successful bioinformatics is close face-to-face collaboration between the biologist, biostatistician and bioinformaticist. This is not always possible due to distances between institutions but is critical for moving forward with a systems-based research agenda where large volumes of data and information are the norm. The best bioinformatics tools will be those that experts in each area can use jointly.
We propose that one way to facilitate the close collaboration between biologists, biostatisticians and bioinformaticists is to make available user-friendly software packages that can be used jointly by researchers with expertise in experimental biology and researchers with expertise in statistics and computer science (Moore, 2007a). This will require software that is intuitive enough for a biologist and powerful enough for an analyst. To be intuitive to a biologist, the software needs to be easy to use and needs to provide output that is visual and easy to navigate. To be powerful, the software needs to provide the functionality that would allow a biostatistician and a bioinformaticist the flexibility to explore the more theoretical aspects of the algorithm. The key, however, to the success of any such software package is the ability of the biologist and the analysts to sit down together at the computer and jointly carry out an analysis. This is important for several reasons. First, the biologist can help answer questions the analyst might have that are related to domain-specific knowledge. Such questions might only arise at the time of the analysis and might otherwise be ignored. Similarly, the biologist might have ideas about specific questions to address with the software that might only be feasible with the help of the analyst. For example, a question that requires multiple processors to answer might need the assistance of someone with expertise in parallel computing.
The idea that biologists and analysts should work together is not new. Langley (2002) has suggested five lessons for the computational discovery process. First, traditional computational notations are not easily communicated to biologists. This is important because a computational model may not be interpretable by a biologist. Second, biologists often have initial models that should influence the discovery process. Domain-specific knowledge such as details about enzymatic reactions in a biochemical pathway can be critical to the discovery process. Third, biological data are often rare and difficult to obtain. It often takes years to collect and process the data to be analyzed. As such, it is important that the analysis is carefully planned and executed. Fourth, biologists want models that move beyond description to provide explanation of data. Explanation and interpretation are paramount to the biologist. Finally, biologists want computational assistance rather than automated discovery systems. Langley (2002) suggests that practitioners want interactive discovery environments that help them understand their data while at the same time giving them or their collaborating analyst control over the modeling process. Collectively, these five lessons suggest that synergy between biologists and bioinformaticists is critical. This is because each has important insights that may not get expressed or incorporated into the discovery process if either carries out the analysis in isolation. Future bioinformatics databases and analysis tools that successfully integrate these lessons will prove to be the most useful for GWAS and other high-throughput approaches.
5 SUMMARY AND CONCLUSIONS
Figure 5 summarizes a bioinformatics analysis strategy for GWAS based on the challenges outlined here. As discussed, it is important that biological knowledge available in public databases be integrated with GWAS data and clinical data. Following quality control procedures, available biological knowledge can be used to help guide both statistical and computational analyses as reviewed in Section 3. Given the complexity of the genotype–phenotype mapping relationship, we recommend two complementary strategies for computational analysis. The first strategy uses filter algorithms (see Section 2.6) such as those based on ReliefF or prior statistical results to select more manageable subsets of SNPs that can then be more efficiently analyzed using computational methods. The second uses wrapper algorithms (see Section 2.7) based on stochastic search methods such as GP for identifying optimal combinations of SNPs that are associated with disease end points. We suggest that computational modeling methods such as RF (see Section 2.3) and MDR (see Section 2.4) are needed to complement parametric statistical methods such as logistic regression for identifying non-linear patterns in GWAS data. The challenge of any statistical or computational analysis is the biological interpretation of a set of results. Use of prior knowledge about biological systems can facilitate this process. Once an inference is made and validated, new knowledge can be contributed to the public knowledge databases thus enhancing future iterations of this flowchart. We anticipate that this entire process will be greatly aided by the development of powerful and user-friendly software that can be optimally used by biologists, biostatisticians and bioinformaticists (see Section 4).
Fig. 5.
Flowchart for bioinformatics analyses of GWAS data. The use of filter and wrapper algorithms along with computational modeling approaches is recommended in addition to parametric statistical methods. Biological knowledge in public databases has a very important role to play at all levels of the analysis and interpretation.
GWAS have generated a number of important bioinformatics challenges including the modeling of complex genotype–phenotype relationships using data mining and machine learning methods, the use of biological knowledge databases to help guide and interpret genetic association studies and the development of powerful and user-friendly software that can facilitate interaction and collaboration among biologists and bioinformaticists. The flowchart shown in Figure 5 provides a starting point for the development of comprehensive and fully informative analyses of GWAS. This will become especially necessary as we generate more and more data and discover new complexities in the genome that make powerful bioinformatics research strategies even more critical for identifying genetic risk factors for common human diseases. With whole-genome sequencing on the horizon, we need to recruit the next generation of bioinformaticists to tackle these and other important computational challenges in genetic analysis because agnostic biostatistical approaches will only get us so far.
ACKNOWLEDGEMENTS
We would like to thank the anonymous reviewers for their very helpful comments and suggestions.
Funding: National Institutes of Health (LM010098, LM009012 and AI59694).
Conflict of Interest: none declared.
REFERENCES
- Ahmed S, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundadottir L, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI. Successful design and conduct of genome-wide association studies. Hum. Mol. Genet. 2007;16:R220–R225. doi: 10.1093/hmg/ddm161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AS, et al. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–1037. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- Askland K, et al. Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum. Genet. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- Banzhaf W, et al. Genetic Programming – An Introduction; On the Automatic Evolutionof Computer Programs and its Applications. San Francisco, CA, USA: Morgan Kaufmann; 1998. [Google Scholar]
- Baranzini SE, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum. Mol. Genet. 2009;18:2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W. Mendel's Principles of Heredity. Cambridge, UK: Cambridge University Press; 1909. [Google Scholar]
- Breiman L. Random Forests. Machine Learn. 2001;45:5–32. [Google Scholar]
- Bureau A, et al. Identifying SNPs predictive of phenotype using random forests. Genet. Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- Bush WS, et al. Parallel multifactor dimensionality reduction: a tool for the large-scale analysis of gene-gene interactions. Bioinformatics. 2006;22:2173–2174. doi: 10.1093/bioinformatics/btl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, et al. Alternative contingency table measures improve the power and detection of multifactor dimensionality reduction. BMC Bioinformatics. 2008;9:238. doi: 10.1186/1471-2105-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, et al. Biofilter: a knowledge-integration system for the multi-locus analysis of genome-wide association studies. Pac. Symp. Biocomput. 2009:368–379. [PMC free article] [PubMed] [Google Scholar]
- Calle ML, et al. Improving strategies for detecting genetic patterns of susceptibility in association studies. Stat. Med. 2008;27:6532–6546. doi: 10.1002/sim.3431. [DOI] [PubMed] [Google Scholar]
- Chang JS, et al. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol. Biomarkers Prev. 2008;17:1368–1373. doi: 10.1158/1055-9965.EPI-07-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chung Y, et al. Odds ratio based multifactor-dimensionality reduction method for detecting gene-gene interactions. Bioinformatics. 2007;23:71–76. doi: 10.1093/bioinformatics/btl557. [DOI] [PubMed] [Google Scholar]
- Clark AG, et al. Determinants of the success of whole-genome association testing. Genome Res. 2004;15:1463–1467. doi: 10.1101/gr.4244005. [DOI] [PubMed] [Google Scholar]
- Cook NR, et al. Tree and spline based association analysis of gene-gene interaction models for ischemic stroke. Stat. Med. 2004;23:1439–1453. doi: 10.1002/sim.1749. [DOI] [PubMed] [Google Scholar]
- Combarros O, et al. Replication by the Epistasis Project of the interaction between the genes for IL-6 and IL-10 in the risk of Alzheimer's disease. J. Neuroinflam. 2009;6:22. doi: 10.1186/1742-2094-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ. Genome-wide association studies: Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 2009 doi: 10.1038/nrg2579. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse R, et al. Detecting epistatic interactions contributing to quantitative traits. Genet. Epidemiol. 2004;27:141–152. doi: 10.1002/gepi.20006. [DOI] [PubMed] [Google Scholar]
- Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum. Mol. Genet. 2008;17:R109–R115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbers CC, et al. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet. Epidemiol. 2009;33:419–431. doi: 10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- Emily M, et al. Using biological networks to search for interacting loci in genome-wide association studies. Eur. J. Hum. Genet. 2009;17:1231–1240. doi: 10.1038/ejhg.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel GB, Corne DW. Evolutionary Computation in Bioinformatics. Boston: Morgan Kaufmann Publishers; 2003. [Google Scholar]
- Freitas A. Data Mining and Knowledge Discovery with Evolutionary Algorithms. New York, NY: Springer; 2002. [Google Scholar]
- Greene CS, et al. An expert knowledge-guided mutation operator for genome-wide genetic analysis using genetic programming. Lect. Notes Bioinformatics. 2007;4774:30–40. [Google Scholar]
- Greene CS, et al. Nature-inspired algorithms for the genetic analysis of epistasis in common human diseases: a theoretical assessment of wrapper vs. filter approaches. Proc. IEEE Cong. Evol. Comput. 2009a:800–807. [Google Scholar]
- Greene CS, et al. Spatially uniform relieff (SURF) for computationally-efficient filtering of gene-gene interactions. BioData Mining. 2009b;2:5. doi: 10.1186/1756-0381-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, et al. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS ONE. 2009c;4:e5639. doi: 10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, et al. Sensible initialization using expert knowledge for genome-wide analysis of epistasis using genetic programming. Proc. IEEE Cong. Evol. Comp. 2009d:1289–1296. doi: 10.1109/CEC.2009.4983093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, et al. Environmental sensing using expert knowledge in a computational evolution system for complex problem solving in human genetics. In: Riolo RL, et al., editors. Genetic Programming Theory and Practice VII. Ann Arbor: Springer; 2010a. in press. [Google Scholar]
- Greene CS, et al. Enabling personal genomics with an explicit test of epistasis. Pac. Symp. Biocomput. 2010b:327–336. doi: 10.1142/9789814295291_0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn LW, Moore JH. Ideal discrimination of discrete clinical endpoints using multilocus genotypes. In Silico Biol. 2004;4:183–194. [PubMed] [Google Scholar]
- Hahn LW, et al. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- Hastie T, et al. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York: Springer; 2001. [Google Scholar]
- Hastie T, et al. The Elements of Statistical Learning. 2. New York: Springer; 2009. [Google Scholar]
- Herold C, et al. INTERSNP: Genome-wide interaction analysis guided by a priori information. Bioinformatics. 2009;25:3275–3281. doi: 10.1093/bioinformatics/btp596. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Holmans P, et al. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante J, et al. Gene-gene interaction between interleukin-1A and interleukin-8 increases Alzheimer's disease risk. J. Neurol. 2004;251:482–483. doi: 10.1007/s00415-004-0375-6. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J, et al. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5:e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, et al. A random forest approach to the detection of epistatic interactions in case-control studies. BMC Bioinformatics. 2009;10:S65. doi: 10.1186/1471-2105-10-S1-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kira K, Rendell LA. Machine Learning: Proceedings of the AAAI'92. San Francisco: 1992. A practical approach to feature selection. [Google Scholar]
- Kononenko I. Machine Learning: ECML-94. New York: 1994. Estimating attributes: analysis and extension of relief; pp. 171–182. [Google Scholar]
- Kooperberg C, Ruczinski I. Identifying interacting SNPs using Monte Carlo logic regression. Genet. Epidemiol. 2005;28:157–170. doi: 10.1002/gepi.20042. [DOI] [PubMed] [Google Scholar]
- Kooperberg C, et al. Sequence analysis using logic regression. Genet. Epidemiol. 2001;21(Suppl. 1):S626–S631. doi: 10.1002/gepi.2001.21.s1.s626. [DOI] [PubMed] [Google Scholar]
- Koza JR. Genetic Programming: On the Programming of Computers by Means of Natural Selection. Cambridge, MA, USA: MIT Press; 1992. [Google Scholar]
- Kraft P, Cox DG. Study designs for genome-wide association studies. Adv. Genet. 2008;60:465–504. doi: 10.1016/S0065-2660(07)00417-8. [DOI] [PubMed] [Google Scholar]
- Langley P. Proceedings of the First International Workshop on Data Mining Lessons Learned. Sydney: 2002. Lessons for the computational discovery of scientific knowledge; pp. 9–12. [Google Scholar]
- Lee SY, et al. Log-linear model-based multifactor dimensionality reduction method to detect gene-gene interactions. Bioinformatics. 2007;23:2589–2595. doi: 10.1093/bioinformatics/btm396. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. The analysis of variance and the analysis of causes. Am. J. Hum. Genet. 1974;26:400–411. [PMC free article] [PubMed] [Google Scholar]
- Lou XY, et al. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am. J. Hum. Genet. 2007;80:1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetta KL, et al. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, et al. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, et al. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- McKinney BA, et al. Machine learning for detecting gene-gene interactions: a review. Appl. Bioinformatics. 2006;5:77–88. doi: 10.2165/00822942-200605020-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BA, et al. Evaporative cooling feature selection for genotypic data involving interactions. Bioinformatics. 2007;23:2113–2120. doi: 10.1093/bioinformatics/btm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BA, et al. Capturing the spectrum of interaction effects in genetic association studies by simulated evaporative cooling network analysis. PLoS Genet. 2009;5:e1000432. doi: 10.1371/journal.pgen.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina I, et al. Gene set-based analysis of polymorphisms: finding pathways or biological processes associated to traits in genome-wide association studies. Nucleic Acids Res. 2009;37:W340–W344. doi: 10.1093/nar/gkp481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H, et al. Multifactor dimensionality reduction-phenomics: a novel method to capture genetic heterogeneity with use of phenotypic variables. Am. J. Hum. Genet. 2007;81:1251–1261. doi: 10.1086/522307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalewicz Z, Fogel DB. How to Solve It: Modern Heuristics. New York: Springer; 2004. [Google Scholar]
- Michalski RS. A theory and methodology of inductive learning. Artif. Intell. 1983;20:111–161. [Google Scholar]
- Millstein J, et al. A testing framework for identifying susceptibility genes in the presence of epistasis. Am. J. Hum. Genet. 2006;78:15–27. doi: 10.1086/498850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T. Machine Learning. New York: McGraw-Hill; 1997. [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum. Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Moore JH. Computational analysis of gene-gene interactions in common human diseases using multifactor dimensionality reduction. Expert Rev. Mol. Diagn. 2004;4:795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- Moore JH. Bioinformatics. J. Cell Physiol. 2007a;213:365–369. doi: 10.1002/jcp.21218. [DOI] [PubMed] [Google Scholar]
- Moore JH. Genome-wide analysis of epistasis using multifactor dimensionality reduction: feature selection and construction in the domain of human genetics. In: Zhu X, Davidson I, editors. Knowledge Discovery and Data Mining: Challenges and Realities with Real World Data. IGI Global Hershey; 2007b. pp. 17–30. [Google Scholar]
- Moore JH. From genotypes to genometypes: putting the genome back in genome-wide association studies. Eur. J. Hum. Genet. 2009;17:1205–1206. doi: 10.1038/ejhg.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Williams SW. New strategies for identifying gene-gene interactions in hypertension. Ann. Med. 2002;34:88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- Moore JH, Ritchie MD. The challenges of whole-genome approaches to common diseases. JAMA. 2004;291:1642–1643. doi: 10.1001/jama.291.13.1642. [DOI] [PubMed] [Google Scholar]
- Moore JH, Williams SM. Traversing the conceptual divide between biological and statistical epistasis: systems biology and a more modern synthesis. Bioessays. 2005;27:637–646. doi: 10.1002/bies.20236. [DOI] [PubMed] [Google Scholar]
- Moore JH, White BC. Exploiting expert knowledge in genetic programming for genome-wide genetic analysis. Lect. Notes Comp. Sci. 2006;4193:969–977. [Google Scholar]
- Moore JH, White BC. Genome-wide genetic analysis using genetic programming: The critical need for expert knowledge. In: Riolo R, et al., editors. Genetic Programming Theory and Practice IV. New York: Springer; 2007a. pp. 11–28. [Google Scholar]
- Moore JH, White BC. Tuning ReliefF for genome-wide genetic analysis. Lect. Notes Comp. Sci. 2007b;4447:166–175. [Google Scholar]
- Moore JH, Williams SM. Epistasis and its implications for personal genetics. Am. J. Hum. Genet. 2009;85:309–320. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, et al. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J. Theor. Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Moore JH, et al. Symbolic modeling of epistasis. Hum. Hered. 2007;63:120–133. doi: 10.1159/000099184. [DOI] [PubMed] [Google Scholar]
- Moore JH, et al. Development and evaluation of an open-ended computational evolution system for the genetic analysis of susceptibility to common human diseases. Lect. Notes Comp. Sci. 2008a;4973:129–140. [Google Scholar]
- Moore JH, et al. Does complexity matter? Artificial evolution, computational evolution and the genetic analysis of epistasis in common human diseases. In: Riolo RL, editor. Genetic Programming Theory and Practice VI. Ann Arbor: Springer; 2008b. pp. 125–145. [Google Scholar]
- Motsinger AA, et al. Novel methods for detecting epistasis in pharmacogenomics studies. Pharmacogenomics. 2007;8:1229–1241. doi: 10.2217/14622416.8.9.1229. [DOI] [PubMed] [Google Scholar]
- Namkung J, et al. Identification of gene-gene interactions in the presence of missing data using the multifactor dimensionality reduction method. Genet. Epidemiol. 2009a;33:646–656. doi: 10.1002/gepi.20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkung J, et al. New evaluation measures for multifactor dimensionality reduction classifiers in gene-gene interaction analysis. Bioinformatics. 2009b;25:338–345. doi: 10.1093/bioinformatics/btn629. [DOI] [PubMed] [Google Scholar]
- Nelson MR, et al. A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Res. 2001;11:458–470. doi: 10.1101/gr.172901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dushlaine C, et al. The SNP ratio test: pathway analysis of genome-wide association datasets. Bioinformatics. 2009;25:2762–2763. doi: 10.1093/bioinformatics/btp448. [DOI] [PubMed] [Google Scholar]
- Pan W. Network-based model weighting to detect multiple loci influencing complex diseases. Hum Genet. 2008;124:225–234. doi: 10.1007/s00439-008-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattin KA, Moore JH. Exploiting the proteome to improve the genome-wide genetic analysis of epistasis in common human diseases. Hum. Genet. 2008;124:19–29. doi: 10.1007/s00439-008-0522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattin KA, Moore JH. Role for protein-protein interaction databases in human genetics. Exp. Rev. Proteomics. 2009;6:647–659. doi: 10.1586/epr.09.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattin KA, et al. A computationally efficient hypothesis testing method for epistasis analysis using multifactor dimensionality reduction. Genet. Epidemiol. 2009;33:87–94. doi: 10.1002/gepi.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, et al. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur. J. Hum. Genet. 2009;18:111–117. doi: 10.1038/ejhg.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Reif DM, et al. Exploratory visual analysis of pharmacogenomic results. Pac. Symp. Biocomput. 2005;2005:296–307. [PubMed] [Google Scholar]
- Reif DM, et al. Proceedings of the IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology. Washington D.C.: 2006. Feature selection using a random forests classifier for the integrated analysis of multiple data types; pp. 171–178. [Google Scholar]
- Reif DM, et al. Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun. 2009;10:112–119. doi: 10.1038/gene.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger T, et al. Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur. J. Hum. Genet. 2009;17:722–731. doi: 10.1038/ejhg.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, et al. Multifactor dimensionality reduction reveals high-order interactions among estrogen metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, et al. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, phenocopy, and genetic heterogeneity. Genet. Epidemiol. 2003a;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, et al. Optimization of neural network architecture using genetic programming improves detection and modeling of gene-gene interactions in studies of human diseases. BMC Bioinformatics. 2003b;4:28. doi: 10.1186/1471-2105-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robnik-Siknja M, Kononenko I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach. Learn. 2003;53:23–69. [Google Scholar]
- Saccone SF, et al. Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics. 2008;24:1805–1811. doi: 10.1093/bioinformatics/btn315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork NJ, et al. Common vs. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott-Armstrong NA, et al. Accelerating epistasis analysis in human genetics with consumer graphics hardware. BMC Res. Notes. 2009;2:149. doi: 10.1186/1756-0500-2-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CA, et al. Designing genome-wide association studies: Sample size, power imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MC, et al. International Consortium of Bladder Cancer. Polymorphisms in DNA repair genes, smoking and bladder cancer risk: findings from the international consortium of bladder cancer. Cancer Res. 2009;69:6857–6864. doi: 10.1158/0008-5472.CAN-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV, et al. Classification of rheumatoid arthritis status with candidate gene and genome-wide single-nucleotide polymorphisms using random forests. BMC Proc. 2007;1:S62. doi: 10.1186/1753-6561-1-s1-s62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Epistasis and complex traits. In: Wolf J, et al., editors. Epistasis and the Evolutionary Process. New York: Oxford University Press; 2000. pp. 41–57. [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton-Wells TA, et al. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet. 2004;20:640–647. doi: 10.1016/j.tig.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Torkamani A, et al. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, et al. Evaluation of genome-wide association study results through development of ontology fingerprints. Bioinformatics. 2009;25:1314–1320. doi: 10.1093/bioinformatics/btp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez DR, et al. A balanced accuracy metric for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet. Epidemiol. 2007;31:306–315. doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]
- Wahlsten D. Insensitivity of the analysis of variance to heredity-environment interactions. Behav. Brain Sci. 1990;13:109–161. [Google Scholar]
- Wang WY, et al. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Wilke R, et al. Combinatorial pharmacogenetics. Nat. Rev. Drug Disc. 2005;4:911–918. doi: 10.1038/nrd1874. [DOI] [PubMed] [Google Scholar]
- Wilke RA, et al. The pathway less traveled: Moving from candidate genes to candidate pathways in the analysis of genome-wide data from large scale pharmacogenetic association studies. Curr. Pharmacogenomics Personalized Med. 2008;6:150–159. doi: 10.2174/1875692110806030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SM, et al. Problems with genome-wide association studies. Science. 2007;316:1840–1842. [PubMed] [Google Scholar]
- Wongseree W, et al. Detecting purely epistatic multi-locus interactions by an omnibus permutation test on ensembles of two-locus analyses. BMC Bioinformatics. 2009;10:294. doi: 10.1186/1471-2105-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, et al. Pathway analysis by adaptive combination of P-values. Genet. Epidemiol. 2009;33:700–709. doi: 10.1002/gepi.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, et al. Gene Prospector: an evidence gateway for evaluating potential susceptibility genes and interacting risk factors for human diseases. BMC Bioinformatics. 2008;9:528. doi: 10.1186/1471-2105-9-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamar D, et al. Path: a tool to facilitate pathway-based genetic association analysis. Bioinformatics. 2009;25:2444–2446. doi: 10.1093/bioinformatics/btp431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. Willows: a memory efficient tree and forest construction package. BMC Bioinformatics. 2009;10:130. doi: 10.1186/1471-2105-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, et al. Biostatistical aspects of genome-wide association studies. Biometric. J. 2008;50:1–21. doi: 10.1002/bimj.200710398. [DOI] [PubMed] [Google Scholar]