Abstract
Background
Treatment decisions and prognosis assessment for liposarcoma is based on a classification that depends on morphological and genetic features. Revisions by experienced referral pathologists are often advocated.
Methods
The process of histopathological classification in referring hospitals and subsequently in a referral center in relation to molecular biological information is evaluated. A total of 331 consecutive liposarcoma patients were evaluated for the added value of histological review at time of referral. Subsequently, cases were reclassified with implementation of present-day molecular information. For all patients, complete data on staging, treatment, and follow-up were available.
Results
Upon histological revision, 15/54 (28%) diagnoses were reclassified in the first decade, 14/65 (22%) in the second, and 14/53 (26%) in the last decade. Molecular biological analysis enabled well-differentiated liposarcoma with or without dedifferentiated component to be better recognized as such and distinguished from myxoid liposarcoma and pleomorphic liposarcoma. Inclusion of cytogenetic information resulted in reclassification after revision in 4/18 (22%) cases in the first decade, 10/38 (26%) cases in the second decade, and 19/75 (25%) cases in the last decade.
Conclusions
This study indicates that liposarcomas are heterogeneous tumors. Expert assessment and implementation of molecular biological analysis are valuable for adequate classification as a basis for treatment decisions.
Liposarcomas are a rare and heterogeneous group of malignant neoplasms. Treatment decisions and prognosis assessment for patients with liposarcomas is based on a classification that depends on morphological and genetic features of the tumor. According to the World Health Organization (WHO) classification, liposarcomas can be divided into morphological subtypes that are characterized by a distinct morphological spectrum and clear molecular features. The first category is well-differentiated liposarcoma and includes a variety of histological subtypes: lipoma-like, sclerosing, inflammatory, and spindle cell subtypes. The second category is dedifferentiated liposarcoma and can be found in up to 10% of well-differentiated liposarcoma of any type and has a more aggressive course. The third category consists of myxoid liposarcoma, of which a proportion of cases progress to round cell liposarcoma. The last category consists of pleomorphic liposarcoma.1 At clinical presentation these tumors range from small superficial low-grade tumors to large infiltrating poorly differentiated retroperitoneal tumors.2
Differentiation between lipomas and liposarcomas and the classification into subtypes is essential in tailoring treatment and for prognostic information. In contrast to lipomas, most liposarcomas should be widely excised to avoid recurrence. Myxoid/round cell liposarcoma is a highly radiosensitive tumor.3–5 Chemotherapy sensitivity varies considerably between liposarcoma subtypes, with a significant higher response rate to first-line chemotherapy in myxoid/round cell liposarcoma as compared with well-/dedifferentiated liposarcoma (48% versus 11%).6 Moreover, favorable response to specific agents such as trabectedin have been described for myxoid liposarcoma.7 Furthermore, in contrast to other types of liposarcoma, myxoid/round cell liposarcomas is specifically characterized by its multifocal extrapulmonary presentation. Therefore, particular attention to this characteristic extrapulmonary metastasizing pattern is advocated in staging myxoid and round cell liposarcoma.
Over the last few years, several studies focusing on lipomatous tumors have led to the delineation of new variants as well as to the introduction of new concepts, mainly as a result of fruitful interactions between molecular genetics and pathology. As a result, genetic alterations are now considered as an integral part of the WHO classification. Characteristic genetic alteration for well- and dedifferentiated liposarcoma is the amplification of the 12q13-15 region, which amongst other genes, includes MDM2 and CDK4.8,9 The class of myxoid/round cell liposarcoma is characterized by a classical t(12;16) (q13;p11) or t(12;22) (q13;q12) translocation, with a FUS–CHOP or EWSR1–CHOP gene fusion respectively.10–14 These alterations are mutually exclusive and distinctive for their disease class.
For the classification of lipomatous tumors, revision of slides by an experienced referral pathologist or a reference panel is frequently performed. Analysis of molecular biological changes in lipomatous tumors has provided new expert information in liposarcoma management. Here, we compared pathologic liposarcoma reports and evaluated diagnostic discrepancies of histopathological liposarcoma classification in referring hospitals and our referral center in relation to additional available molecular biological classifications.
Methods
Patient Cohort
All liposarcoma patients treated at The Netherlands Cancer Institute between 1977 and 2006 were evaluated (n = 331). Original diagnosis by a referring pathologist and diagnosis by a pathologist at The Netherlands Cancer Institute at revision that were available were recorded (n = 246). In 85 cases the patient was initially admitted to our tertiary hospital.
At referral, immunohistochemical analysis was used for exclusion of other sarcoma classes of other histogenetic backgrounds. Histological evaluation by a referral pathologist was performed without knowledge of molecular biological analysis. The additional value of molecular biological analysis was then evaluated.
Molecular Biological Analysis of Region 12q13-15 by MDM2 and CDK4 Analysis with Multiplex Ligation-Dependent Probe Amplification
Multiplex ligation-dependant probe amplification technique is a high-throughput polymerase chain reaction (PCR)-based method to determine the relative copy number of various DNA sequences in small samples of human DNA. It is based on the annealing of a mixture of hemiprobes on their cognate DNA sequences. One of the hemiprobes contains stuffer DNA of variable length (19–370 bp), which allows multiplex detection using capillary sequencer.
The Multiplex Ligation-Dependent Probe Amplification Kit P172 test (MRC-Holland, Amsterdam, The Netherlands) was used.15 Target DNA (200 ng) in 5 μl 10 mM Tris (pH8)/0.1 mM ethylenediamine tetraacetic acid (EDTA) was denatured for 5 min at 98°C, after which 3 μl probe mix was added. The mixture was heated at 95°C for 1 min and incubated at 60°C overnight (16 h). Ligation was performed with the temperature-stable Ligase-65 enzyme (MRC-Holland, Amsterdam, The Netherlands) for 15 min at 54°C. Then, the ligase was inactivated by incubation for 5 min at 98°C. Ten microliters of this ligation mix was premixed with 30 μl PCR buffer and put in a PCR machine at 60°C. Subsequently, 10 μl mix was added, containing deoxynucleoside triphosphate, Taq polymerase, and one unlabeled and one carboxyfluorescein-labeled PCR primer, being complementary to the universal primer sequences. PCR was carried out for 35 cycles (30 s at 95°C, 30 s at 60°C, and 60 s at 72°C). The fragments were analyzed on a ABI model 310 or 3700 capillary sequencer (Applied Biosystems) using Genescan-ROX 500 size standards (Applied Biosystems). Fragment analysis was performed using Genescan and Genotyper software, amplification or deletions were calculated and evaluated while compared with 15 samples of DNA from normal tissue. After normalization, each probe set was scored as: 0.01–1.00, deleted; 1.01–2.99, normal copy number; 3.00–3.99, copy number gain; ≥4.00, amplification.
Detection of FUS–CHOP and EWSR1–CHOP Gene Fusion
RNA was extracted from representative sections of the formalin-fixed paraffin-embedded specimens according to the manufacturer’s method using high-purity RNA paraffin kit (Roche, Basel, Switzerland). Microdissection to optimize tumor cell content was performed if necessary. RNA concentration was measured using a nanodrop spectrophotometer. Reverse-transcription (RT)-PCR amplification was done using one-step RT-PCR. Due to the limiting quality of RNA from formalin-fixed paraffin-embedded material, different breakpoints were detected, using a combination of FUS–CHOP- and EWSR1–CHOP-specific internal primers, resulting in small PCR products.2 PCR cycles were operated on a regimen of 30 s of denaturation at 95°C, 30 s of primer annealing at 68°C, and 30 s extension/synthesis at 72°C, for 30 cycles. PCR products were separated by electrophoresis on 2.5% agarose gels containing ethidium bromide, visualized by ultraviolet (UV) light, and photographed. To confirm the type of fusion gene, PCR products were compared with positive controls of the different gene fusion combinations as confirmed by sequencing.
Treatment for Primary Liposarcomas
Lipomatous tumor larger than 3 cm were analyzed by imaging techniques. Biopsy was performed on all lipomatous tumors that the images suggested to be malignant. Sites of potential metastasis were evaluated by computed tomography (CT). Until 2000, most liposarcomas were treated with surgery and radiotherapy, irrespective of subtype or grade. After 2000, tailored treatment was introduced for subgroups. Surgery was the standard therapy for liposarcoma; (neo)adjuvant radiotherapy was considered for all intermediate- and high-grade liposarcomas with tumor-free margin of less than 10 mm in the absence of an intact fascial layer and for most recurrent cases not irradiated at primary presentation. Preoperative biopsies in radiological low-grade lipoma-like liposarcomas were considered redundant, as histologic evaluation of biopsies in these cases will not differentiate between benign deep-seated intramuscular lipomas and atypical lipomatous tumors/well-differentiated liposarcomas, and in all these patients the surgical procedure would aim for complete resection with narrow but free margins.
Palliative local treatment or chemotherapy was offered to patients with inoperable or nearly inoperable disease or with metastases according to the prevailing guidelines.6
Patient Follow-Up
In general, patient follow-up consisted of visits at 3-month intervals during the first year after treatment for the primary tumor, at 6-month intervals during the next 4 years, and at 1-year intervals thereafter. Magnetic resonance imaging of primary tumor site was obtained 3 months after resection. Follow-up visits consisted of physical examination and chest radiography. Magnetic resonance imaging or CT was performed on specific indications. Patients were discharged from follow-up after 15 years or more without evidence of disease. Overall survival was defined as time between first presentation to date of last follow-up or death.
Statistical Analysis
Survival of revised liposarcomas patients was visualized by a Kaplan–Meier method, and an estimation of disease-specific survival of the various subclasses was made. The pairwise log-rank test over the different strata was used to calculate significance among the survival curves. Statistical analysis was performed using Statistical Package for the Social Sciences software version 15.0.1 (SPSS, Chicago, IL). The primary end-point of this analysis was disease-specific survival. Ten-year estimates of disease-specific survival and the corresponding 95% confidence intervals are reported.
Results
A total of 331 patients (188 males, 143 females) were analyzed. Eighty-five patients were diagnosed in our institute. Histological evaluation in the remaining 246 patients was initially performed at the referring hospital. Additional molecular biological analysis was performed in 131 patients, including 80 referred patients. Additionally 34 lipomas were randomly retrieved from the hospital files to complete the lipomatous tumor spectrum. Molecular analyses were also performed on these cases.
Evaluation of Referring Classification by Histologic Revision in a Referral Center
Detailed information on the original and revised histological diagnoses is listed in Table 1.
Table 1.
Histologic classification in referring hospital compared with histologic classification in a referral hospital by decades
| 1977–1986 | 1987–1996 | 1997–2006 | Changed diagnosis | |||
|---|---|---|---|---|---|---|
| Referring Hospital | Referral hospital | Referring Hospital | Referral hospital | Referring Hospital | Referral hospital | |
| Lipoma (8) | Lipoma (1) | Lipoma (10) | Lipoma (5) | |||
| WLS (6) | WLS (9) | WLS (5) | 22/23 (96%) | |||
| MLS (1) | MLS (1) | |||||
| WLS (6) | WLS (3) | WLS (10) | WLS (9) | WLS (9) | WLS (6) | |
| DLS (1) | DLS (1) | DLS (2) | 7/25 (28%) | |||
| MLS (2) | MLS (1) | |||||
| DLS (0) | DLS (1) | DLS (1) | DLS (4) | DLS (3) | 1/5 (20%) | |
| PLS (1) | ||||||
| WLS (2) | WLS (2) | DLS (2) | ||||
| MLS (32) | MLS (29) | MLS (38) | MLS (= 36) | MLS (29) | MLS (27) | 7/99 (7%) |
| PLS (1) | ||||||
| WLS (1) | WLS (1) | 2/7 (29%) | ||||
| RLS (4) | RLS (3) | RLS (2) | RLS (1) | RLS (1) | RLS (1) | |
| MLS (1) | DLS (3) | 4/13 (31%) | ||||
| PLS (4) | PLS (3) | PLS (4) | PLS (4) | PLS (5) | PLS (2) | |
| WLS (3) | WLS (11) | WLS (3) | ||||
| DLS (1) | DLS (2) | DLS (4) | n.a. | |||
| MLS (22) | MLS (6) | MLS (2) | ||||
| RLS (0) | RLS (3) | RLS(0) | ||||
| PLS (8) | PLS (3) | PLS (3) | ||||
| (Lipo)sarcoma N.O.S. (35) | Liposarcoma N.O.S. (1) | (Lipo)sarcoma N.O.S. (25) | (Lipo)sarcoma N.O.S. (14) | (Lipo)sarcoma N.O.S. (2) | ||
| 43/172 (25%) | ||||||
| n.a = 74 | ||||||
| N = 246 | ||||||
Original diagnosis by a referring pathologist and diagnosis by a pathologist at The Netherlands Cancer Institute at revision that were available were recorded (n = 246). In 85 cases of liposarcoma the patient was initially admitted to our tertiary hospital
WLS, well-differentiated liposarcoma, DLS dedifferentiated liposarcoma, MLS myxoid liposarcoma, RLS round cell liposarcoma, PLS pleomorphic liposarcoma, N.O.S. not otherwise specified, n.a. not applicable
Twenty of 23 lipomatous tumors diagnosed by a referring pathologist as lipoma were reclassified as well-differentiated liposarcoma (87%). These patients had all been referred for local recurrence. The initial diagnosis was changed to a grade 1 sarcoma in 22/23 (96%) cases and to grade 2 myxoid liposarcoma in 1/23 (4%) case. Most tumors were located on the extremities 15/23 (65%), others being in the head and neck region 4/23 (17%) or the trunk 4/23 (17%). The median frequency of local recurrence before referral for lipomas was 1 (range 0–5); 16/23 (70%) recurred at least once after initial treatment. These clinical findings underline the correctness of the referral reclassification.
Pathologists in the referring hospital and our tertiary referral center concurred on the diagnosis of well-differentiated liposarcoma in 18/25 (72%) patients. The others were revised to dedifferentiated liposarcoma 4/25 (16%) or myxoid liposarcoma 3/25 (12%). The majority of these cases (12/25, 48%) were situated on the extremities, whereas 10/25 (40%) had a primary location in the retroperitoneum. There was consensus on the diagnosis of retroperitoneal tumors in 9/10 (90%), and in 7/12 (58%) of the referred extremity well-differentiated liposarcomas there was agreement. In this tumor group disagreement was predominantly found in extremity lesions.
In the group of lipomatous tumors that were classified as myxoid/ round cell liposarcoma by a referring pathologist, there was agreement in 92/99 (93%) patients between histopathological analysis of referring hospital and revision classification. The reclassifications (4/7, 57%) were predominantly retroperitoneal liposarcomas. A similar high degree of agreement was found for round cell tumors. Only 1/7 (14%) was reclassified after revision to well-differentiated liposarcoma.
Reclassification of pleomorphic liposarcoma occurred in 4/13 (31%). Three of the four were changed to dedifferentiated liposarcoma.
Reassessment of Referral Classification by Molecular Biological Information
Detailed information on revised histological diagnoses and molecular biological analysis is listed in Table 2. Molecular biological analysis confirmed the histological reclassification at revision to well-differentiated liposarcoma in all seven referred lipomas that had amplification on region 12q13-15. The randomly selected and reviewed lipomas showed an amplification of region 12q13-15 in 1/34 (3%) of cases.
Table 2.
Changes of revised histologic classification after molecular biological analysis, which was performed in 131 patients, and the additional 34 lipomas randomly retrieved from The Netherlands Cancer Institute files to complete the lipomatous tumor spectrum
| Revision classification | Molecular biological analysis | Definitive diagnosis | Change diagnosis | |
|---|---|---|---|---|
| Lipoma (34) | Amplification + | 1 | WLS | 1/34 (3%) |
| Amplification – | 33 | Lipoma | ||
| WLS (63) | Amplification + | 47 | WLS | |
| Amplification – | 13 | Lipoma | 15/63 (23%) | |
| Translocation – | 1 | WLS | ||
| Both amplification and translocation – | 2 | Lipoma | ||
| DLS (14) | Amplification + | 9 | DLS | |
| Amplification – | 4 | PLS | 4/14 (29%) | |
| Translocation – | 1 | DLS | ||
| MLS (32) | Amplification + translocation – | 11 | WLS/DLS | |
| Amplification – | 2 | MLS | ||
| Translocation + | 18 | MLS | 12/32 (38%) | |
| Translocation – | 1 | WLS/myxofibrosarcoma | ||
| RLS (9) | Translocation + | 9 | RLS | 0/9 (0%) |
| PLS (11) | Amplification + | 5 | DLS | 5/11 (45%) |
| Amplification – | 6 | PLS | ||
| Sarcoma n.o.s. (2) | Amplification + | 1 | DLS | n.a. |
| Translocation – | 1 | WLS | ||
| 165 | 165 | 37/ 163 (23%) | ||
| n.a = 2 | ||||
| N = 165 |
WLS, well-differentiated liposarcoma, DLS dedifferentiated liposarcoma, MLS myxoid liposarcoma, RLS round cell liposarcoma, PLS pleomorphic liposarcoma, N.O.S. not otherwise specified, Amplification amplification of region 12q13-15, Translocation detection of FUS–CHOP or EWS–CHOP gene fusion
+ present, – absent
Molecular analysis of lipomatous tumors that were reclassified as well-differentiated liposarcomas showed that in 47/63 (75%) of cases an amplification of region 12q13-15 was found, supporting a diagnosis of well-differentiated liposarcoma. In the remaining lipomatous tumors that were considered as well-differentiated liposarcomas 15/63 (23%) no amplification of region 12q13-15 was found (Fig. 1c).
Fig. 1.
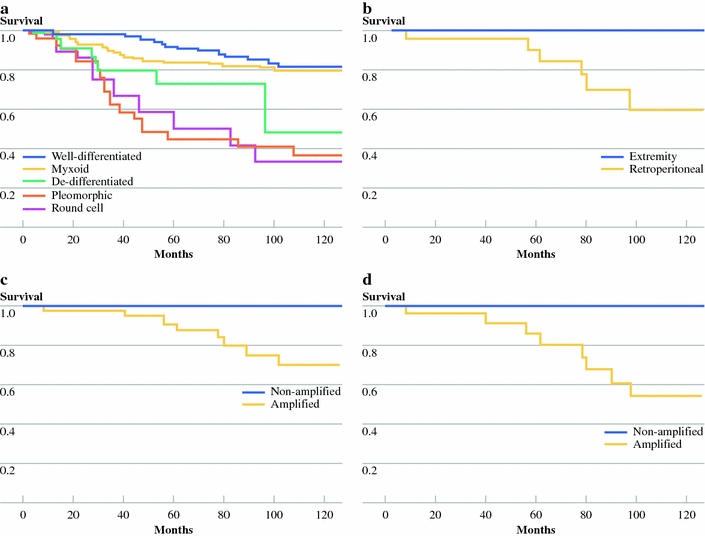
a Survival curves of primary liposarcoma patients based on revised histological classification. b Ten-year survival of revised extremity well-differentiated liposarcoma and survival of retroperitoneal well-differentiated liposarcoma. c Survival of all revised well-differentiated liposarcomas that were molecular biologically analyzed. The lower line indicates the amplified lipomatous tumors; the upper line indicates the nonamplified patients (P ≤ 0.066). d Ten-year survival of revised well-differentiated retroperitoneal liposarcoma, amplified versus nonamplified
Molecular biological analysis to demonstrate the classic translocation was performed in 30/92 (33%) lipomatous tumors revised as myxoid liposarcoma and confirmed the diagnosis in 18 (60%) of these cases, supporting the classification of myxoid liposarcoma. All retroperitoneal liposarcomas that were histologically classified as myxoid liposarcoma at revision (n = 11) were reclassified as well-differentiated liposarcoma in view of the absence of the classic translocation and because of presence of amplification of region 12q13-15 on molecular biological analysis. One extremity sarcoma was revised to myxoid liposarcoma, but none of the various fusion transcripts could be demonstrated, while histologically the sarcoma was revised to myxoid liposarcoma. Furthermore, a classic translocation was identified in all revised round cell liposarcomas that were analyzed.
Molecular biological analysis reclassified 5/11 that had been determined to be pleomorphic at revision. In five cases an amplification of region 12q13-15 was demonstrated.
Inclusion of molecular biological information as collected for this study resulted in reclassification in 4/18 (22%) cases in the time period 1977–1986. Molecular biological analysis refuted the revised histological diagnosis in 10/38 (26%) of cases between 1987 and 1996 and in 19/75 (25%) cases between 1997 and 2006.
Discussion
This study indicates that expert revision is helpful in firmly delineating the tumor classification, especially when supported by molecular biological analysis.
Due to their heterogeneity, the frequency of discordant histopathological classification is higher in liposarcoma than in many other sarcomas. Diagnosis discrepancies in soft tissue sarcoma are reported by others.16–19 However, these reports described sarcomas in general without molecular biological analysis.
Considering the present treatment protocols, a therapy change would have been indicated in 35/172 (20%) cases based on histological revision alone and in 28/163 (17%) cases based on molecular biological analysis. Most changes at histologic revision were referred lipomas that were revised to well-differentiated liposarcoma, for which a more complete local resection is mandatory and follow-up should be prolonged. For myxoid liposarcoma, concordance was high with the exception of retroperitoneal tumors with morphological features mimicking myxoid liposarcoma. In pleomorphic liposarcoma, well-differentiated components were often not recognized in otherwise high-grade tumors.
The subgroup of well-differentiated liposarcoma needs further illustration. This histologic classification provided by referring hospital showed a significant lower 10-year survival (58%) when compared with patients with that same diagnosis after histological revision in our referral center (80%; P < 0.004). In histologically revised well-differentiated liposarcomas, 10-year survival tended to be better in nonamplified lipomatous tumors (100%) than in amplified liposarcomas (P = 0.066) (Fig. 1c). Moreover no disease-specific events were observed in the three retroperitoneal lesions that were histologically classified as well-differentiated liposarcoma but in which no amplification of region 12q13-15 was found (Fig. 1d). One of the explanations for these results may be that a (lipo)sarcomatous transformed component was missed by the referring pathologist in 4/25 lipomatous tumor cases. Secondly, survival differences between all revised well-differentiated liposarcomas that underwent molecular biological analysis were remarkable. The insights in the molecular pathogenesis of lipomatous tumors show a morphological overlap between ordinary lipomas and well-differentiated liposarcomas in 16/97 (16%) cases. The present study indicates that their position in the spectrum can be determined by means of molecular biological analysis. It should be noted that molecular analysis for liposarcoma in general may not be necessary in any case, and results should interpreted with caution and by experienced pathologists.
Classification of pleomorphic liposarcoma in literature is predominantly performed on the basis of histopathological analysis only, occasionally in combination with indistinct immunohistochemical markers.20–25 However, it appears that MDM2 and CDK4, are targeted most frequently in well-differentiated and dedifferentiated liposarcomas.8,9,26–29 One may therefore assume that pleomorphic liposarcoma expressing MDM2 or CDK4 amplification represents dedifferentiated liposarcoma without well-differentiated component. The histologic classification of pleomorphic liposarcoma is frequently questionable (Table 2). The molecular biological MDM2 and CDK4 analysis indicates that the diagnosis was disputable in 5/11 (45%) pleomorphic liposarcoma patients. It is possible that the incidence of the histologic diagnosis pleomorphic liposarcoma will continue to decrease due to the introduction of additional molecular biological analysis or immunohistochemical staining with MDM2 and CDK4 for all liposarcomas.
A few comments on our study are warranted. Firstly, in this study we predominantly used either multiplex ligation-dependent probe amplification or RT-PCR for analysis. These analyses were used in combination with histologic information to define a classification. In case of negative translocation or amplification analysis it must be noted that, in order to positively establish a well-/dedifferentiated liposarcoma character or myxoid/round cell liposarcoma, sometimes a combination of methods can lead to an undisputable diagnosis. Secondly, it should be realized that survival of liposarcoma patients probably also depends on tumor percentage dedifferentiation or percentage round cell component. Liposarcoma survival curves including molecular biologically defined classifications could improve accuracy. Finally, 10-year survival ranges from around 20% for high-grade dedifferentiated retroperitoneal liposarcoma up to 100% for low-grade well-differentiated liposarcomas on the extremity. Treatment for the former patients is complex and requires a multidisciplinary sarcoma team.30–34 Although in high-grade retroperitoneal cases referral to a sarcoma center is common, referral is sometimes omitted in more unclear cases. Due to a morphological continuum of lipomas and liposarcomas, exact recommendations to refer liposarcomas or to perform molecular biological analysis, in apparent straightforward cases, are difficult to make.
Acknowledgments
This manuscript is dedicated to the memory of our colleague J.L. (Hans) Peterse M.D. who tragically died suddenly during this project. We would like to thank H.J. Ruijter, L. Boerrigter, and D. Veldhuizen for their expert technical support. We would also like to thank O. E. Nieweg M.D. Ph.D. for his dedicated work in revising this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Kindblom LG. Lipomatous tumors-how we have reached our present views, what controversies remain and why we still face diagnostic problems: a tribute to Dr Franz Enzinger. Adv Anat Pathol. 2006;13:279–285. doi: 10.1097/01.pap.0000213053.00060.5a. [DOI] [PubMed] [Google Scholar]
- 2.de Vreeze RS, de Jong D, Tielen IH, et al. Primary retroperitoneal myxoid/round cell liposarcoma is a nonexisting disease: an immunohistochemical and molecular biological analysis. Mod Pathol. 2009;22(2):223–31. doi: 10.1038/modpathol.2008.164. [DOI] [PubMed] [Google Scholar]
- 3.de Vreeze RS, de Jong D, Haas RL. Effectiveness of radiotherapy in myxoid sarcomas is associated with a dense vascular pattern. Int J Radiat Oncol Biol Phys. 2008;72(5):1480–7. doi: 10.1016/j.ijrobp.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Pitson G, Robinson P, Wilke D, et al. Radiation response: an additional unique signature of myxoid liposarcoma. Int J Radiat Oncol Biol Phys. 2004;60:522–526. doi: 10.1016/j.ijrobp.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Engstrom K, Bergh P, Cederlund CG, et al. Irradiation of myxoid/round cell liposarcoma induces volume reduction and lipoma-like morphology. Acta Oncol. 2007;46:838–845. doi: 10.1080/02841860601080415. [DOI] [PubMed] [Google Scholar]
- 6.Jones RL, Fisher C, Al-Muderis O, et al. Differential sensitivity of liposarcoma subtypes to chemotherapy. Eur J Cancer. 2005;41:2853–2860. doi: 10.1016/j.ejca.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 7.Grosso F, Jones RL, Demetri GD, et al. Efficacy of trabectedin (ecteinascidin-743) in advanced pretreated myxoid liposarcomas: a retrospective study. Lancet Oncol. 2007;8:595–602. doi: 10.1016/S1470-2045(07)70175-4. [DOI] [PubMed] [Google Scholar]
- 8.Dei Tos AP, Doglioni C, Piccinin S, et al. Coordinated expression and amplification of the MDM2, CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol. 2000;190:531–536. doi: 10.1002/(SICI)1096-9896(200004)190:5<531::AID-PATH579>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 9.Hostein I, Pelmus M, Aurias A, et al. Evaluation of MDM2 and CDK4 amplification by real-time PCR on paraffin wax-embedded material: a potential tool for the diagnosis of atypical lipomatous tumours/well-differentiated liposarcomas. J Pathol. 2004;202:95–102. doi: 10.1002/path.1495. [DOI] [PubMed] [Google Scholar]
- 10.Panagopoulos I, Mertens F, Isaksson M, et al. A novel FUS/CHOP chimera in myxoid liposarcoma. Biochem Biophys Res Commun. 2000;279:838–845. doi: 10.1006/bbrc.2000.4026. [DOI] [PubMed] [Google Scholar]
- 11.Panagopoulos I, Hoglund M, Mertens F, et al. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12:489–494. [PubMed] [Google Scholar]
- 12.Hosaka T, Nakashima Y, Kusuzaki K, et al. A novel type of EWS-CHOP fusion gene in two cases of myxoid liposarcoma. J Mol Diagn. 2002;4:164–171. doi: 10.1016/S1525-1578(10)60698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singer S, Socci ND, Ambrosini G, et al. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Res. 2007;67:6626–6636. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- 14.Italiano A, Cardot N, Dupre F, et al. Gains and complex rearrangements of the 12q13–15 chromosomal region in ordinary lipomas: the “missing link” between lipomas and liposarcomas? Int J Cancer. 2007;121:308–315. doi: 10.1002/ijc.22685. [DOI] [PubMed] [Google Scholar]
- 15.Schouten JP, McElgunn CJ, Waaijer R, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thway K, Fisher C. Histopathological diagnostic discrepancies in soft tissue tumours referred to a specialist centre. Sarcoma. 2009;2009:741975. doi: 10.1155/2009/741975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Presant CA, Russell WO, Alexander RW, et al. Soft-tissue and bone sarcoma histopathology peer review: the frequency of disagreement in diagnosis and the need for second pathology opinions. The Southeastern Cancer Study Group experience. J Clin Oncol. 1986;4:1658–1661. doi: 10.1200/JCO.1986.4.11.1658. [DOI] [PubMed] [Google Scholar]
- 18.Alvegard TA, Berg NO. Histopathology peer review of high-grade soft tissue sarcoma: the Scandinavian Sarcoma Group experience. J Clin Oncol. 1989;7:1845–1851. doi: 10.1200/JCO.1989.7.12.1845. [DOI] [PubMed] [Google Scholar]
- 19.Massi D, Beltrami G, Capanna R, et al. Histopathological re-classification of extremity pleomorphic soft tissue sarcoma has clinical relevance. Eur J Surg Oncol. 2004;30:1131–1136. doi: 10.1016/j.ejso.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Downes KA, Goldblum JR, Montgomery EA, et al. Pleomorphic liposarcoma: a clinicopathologic analysis of 19 cases. Mod Pathol. 2001;14:179–184. doi: 10.1038/modpathol.3880280. [DOI] [PubMed] [Google Scholar]
- 21.Nascimento AF, Raut CP. Diagnosis and management of pleomorphic sarcomas (so-called “MFH”) in adults. J Surg Oncol. 2008;97:330–339. doi: 10.1002/jso.20972. [DOI] [PubMed] [Google Scholar]
- 22.Hornick JL, Bosenberg MW, Mentzel T, et al. Pleomorphic liposarcoma: clinicopathologic analysis of 57 cases. Am J Surg Pathol. 2004;28:1257–1267. doi: 10.1097/01.pas.0000135524.73447.4a. [DOI] [PubMed] [Google Scholar]
- 23.Fritz B, Schubert F, Wrobel G, et al. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res. 2002;62:2993–2998. [PubMed] [Google Scholar]
- 24.Gebhard S, Coindre JM, Michels JJ, et al. Pleomorphic liposarcoma: clinicopathologic, immunohistochemical, and follow-up analysis of 63 cases: a study from the French Federation of Cancer Centers Sarcoma Group. Am J Surg Pathol. 2002;26:601–616. doi: 10.1097/00000478-200205000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Idbaih A, Coindre JM, Derre J, et al. Myxoid malignant fibrous histiocytoma and pleomorphic liposarcoma share very similar genomic imbalances. Lab Invest. 2005;85:176–181. doi: 10.1038/labinvest.3700202. [DOI] [PubMed] [Google Scholar]
- 26.Pilotti S, Della TG, Lavarino C, et al. Distinct mdm2/p53 expression patterns in liposarcoma subgroups: implications for different pathogenetic mechanisms. J Pathol. 1997;181:14–24. doi: 10.1002/(SICI)1096-9896(199701)181:1<14::AID-PATH730>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 27.Berner JM, Forus A, Elkahloun A, et al. Separate amplified regions encompassing CDK4 and MDM2 in human sarcomas. Genes Chromosomes Cancer. 1996;17:254–259. doi: 10.1002/(SICI)1098-2264(199612)17:4<254::AID-GCC7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Sirvent N, Coindre JM, Maire G, et al. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am J Surg Pathol. 2007;31:1476–1489. doi: 10.1097/PAS.0b013e3180581fff. [DOI] [PubMed] [Google Scholar]
- 29.Dei Tos AP, Doglioni C, Piccinin S, et al. Molecular abnormalities of the p53 pathway in dedifferentiated liposarcoma. J Pathol. 1997;181:8–13. doi: 10.1002/(SICI)1096-9896(199701)181:1<8::AID-PATH700>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 30.Mankin HJ, Mankin CJ, Simon MA. The hazards of the biopsy, revisited. Members of the Musculoskeletal Tumor Society. J Bone Joint Surg Am. 1996;78:656–663. doi: 10.2106/00004623-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 31.Ray-Coquard I, Thiesse P, Ranchere-Vince D, et al. Conformity to clinical practice guidelines, multidisciplinary management and outcome of treatment for soft tissue sarcomas. Ann Oncol. 2004;15:307–315. doi: 10.1093/annonc/mdh058. [DOI] [PubMed] [Google Scholar]
- 32.Rydholm A. Centralization of soft tissue sarcoma. The southern Sweden experience. Acta Orthop Scand Suppl. 1997;273:4–8. doi: 10.1080/17453674.1997.11744695. [DOI] [PubMed] [Google Scholar]
- 33.Clasby R, Tilling K, Smith MA, et al. Variable management of soft tissue sarcoma: regional audit with implications for specialist care. Br J Surg. 1997;84:1692–1696. doi: 10.1002/bjs.1800841213. [DOI] [PubMed] [Google Scholar]
- 34.van Dalen T, Hoekstra HJ, van Geel AN, et al. Locoregional recurrence of retroperitoneal soft tissue sarcoma: second chance of cure for selected patients. Eur J Surg Oncol. 2001;27:564–568. doi: 10.1053/ejso.2001.1166. [DOI] [PubMed] [Google Scholar]


