Abstract
The relationship between cognition and a functional polymorphism in the catechol-O-methlytransferase (COMT) gene, val108/158met, is one of debate in the literature. Furthermore, based on the dopaminergic differences associated with the COMT val108/158met genotype, neural differences during cognition may be present, regardless of genotypic differences in cognitive performance. To investigate these issues the current study aimed to 1) examine the effects of COMT genotype using a large sample of healthy individuals (n = 496–1218) and multiple cognitive measures, and using a subset of the sample (n = 22), 2) examine whether COMT genotype effects medial temporal lobe (MTL) and frontal activity during successful relational memory processing, and 3) investigate group differences in functional connectivity associated with successful relational memory processing. Results revealed no significant group difference in cognitive performance between COMT genotypes in any of the 19 cognitive measures. However, in the subset sample, COMT val homozygotes exhibited significantly decreased MTL and increased prefrontal activity during both successful relational encoding and retrieval, and reduced connectivity between these regions compared with met homozygotes. Taken together, the results suggest that although the COMT val108/158met genotype has no effect on cognitive behavioral measures in healthy individuals, it is associated with differences in neural process underlying cognitive output.
Keywords: COMT, fMRI, genetic neuroimaging, relational memory
Introduction
The field of genetic neuroimaging in humans aims to characterize the functional effects of common genetic polymorphisms on both cognition and brain activity by comparing individuals of different genotypes. Several studies have focused on catechol-O-methyl-transferase (COMT). COMT is densely expressed throughout both the prefrontal cortex (PFC) and limbic system (e.g., Hong et al. 1998; Matsumoto et al. 2003) where it metabolizes released dopamine, thereby playing a critical role in the regulation of dopamine (DA) levels (Li et al. 2003; Matsumoto et al. 2003). Dopamine regulates neuronal firing in both the PFC and hippocampus (Li et al. 2003), and recent positron emission tomography studies have suggested that a distributed frontal–temporal dopaminergic network is involved in higher-order cognitive functions (Aalto et al. 2005; Takahashi et al. 2007). Thus, variation in COMT-mediated dopamine metabolism has been hypothesized to impact cognitive processes mediated by these regions.
One common variant in the COMT gene is a single nucleotide polymorphism that causes a valine-to-methionine substitution, val108/158met (rs4680). This substitution leads to a three- to fourfold decrease in the activity of the enzyme (Lotta et al. 1995; Chen et al. 2004), thereby increasing dopamine availability, leading to increased signal-to-noise ratio and improvements in signal detection during cognitive tasks, both throughout the cortex (Sawaguchi et al. 1990; Servan-Schreiber et al. 1990; Winterer et al. 2006) and specifically within the PFC (Sawaguchi et al. 1990; Yang and Mogenson 1990).
Consistent with this evidence, a number of genetic association studies have suggested that the COMT val allele confers poorer performance on tasks dependent on PFC and medial temporal lobe (MTL) function such as working memory, declarative memory, and executive functioning (e.g., Goldberg et al. 2003; de Frias et al. 2004; Bruder et al. 2005; Diaz-Asper et al. 2008), although other studies have found no association between cognition and COMT (e.g., Tsai et al. 2003; Stefanis et al. 2004; Ho et al. 2005; Schott et al. 2006; Winterer et al. 2006). Unfortunately, the published effects of COMT on cognitive performance have been clouded by small sample sizes, lack of correction for population stratification and lack of correction for testing of multiple cognitive measures. One of the goals of the current study was to examine the effects of COMT val108/158met on a large battery of cognitive tasks, in a large sample size, correcting both for population stratification and for multiple testing.
However, based on the dopaminergic differences associated with the COMT val108/158met genotype, neural changes during cognition may be present, regardless of any genotypic differences in cognitive performance. A good test of this idea is the examination of a cognitive task critically dependent on a PFC–MTL network, such as relational memory. Relational memory is a ubiquitous form of memory that requires not only the memory for individual items, but memory for associations amongst the items and the context in which they were presented (for a review see Johnson et al. 1993). The formation and retrieval of such associations has been shown to rely upon both the MTL (and specifically the hippocampus) (e.g., Sperling et al. 2001; Davachi 2006; Dennis et al. 2008; Eichenbaum et al. 2007) and also the PFC (e.g., Moscovitch 1992; Johnson et al. 1993; Cabeza et al. 1997; Prince et al. 2005). In addition to the individual contribution of these regions to relational memory, enhanced connectivity between the MTL and PFC is associated with successful memory performance (e.g., Ranganath et al. 2005; Dickerson et al. 2007). Enhanced MTL–PFC connectivity can also be expected to be found in a system whose individual components are functioning efficiently compared with one hampered by processing deficits. Given the reliance of relational memory on both PFC and MTL functioning, genetic variants affecting dopamine availability in these regions could have a profound impact on the neural correlates and connectivity associated with relational memory in these regions.
Although no study to date has examined the effects of the COMT val108/158 polymorphism on neural signal-to-noise and MTL/PFC processing during a relational memory task, several neuroimaging studies have reported increased PFC and reduced MTL activity associated with the COMT val allele during item memory (Egan et al. 2001; Bertolino et al. 2006; Schott et al. 2006). This heightened PFC activity has been suggested to be due to inefficient frontal processing, and attributed to the lower signal-to-noise in frontal activity seen in COMT val carriers (i.e., Winterer et al. 2006). However, the inefficiency interpretation is currently not well-justified, because the subjects that underwent functional magnetic resonance imaging (fMRI) in the Egan et al. (2001) study did not show differences in performance accuracy (despite exhibiting differences in neural activity), and Schott et al. (2006) reported no differences in memory performance.
Analysis of the functional coupling between the MTL and PFC has met with mixed results. Bertolino et al. (2006) found activity in the hippocampus and ventrolateral PFC to be negatively associated, whereas Schott et al. (2006) found increased functional coupling between the left hippocampus and bilateral PFC regions. Despite the directional group differences, both studies reported memory-associated functional connectivity changes to be greater in met homozygotes than in COMT val carriers.
However, there are several factors to consider when interpreting the foregoing results. First, both the Bertolino et al. (2006) and Egan et al. (2001) studies utilized blocked designs (i.e., designs in which trials are not separated by jittered intervals and hence do not allow the identification of activity specific to one trial and the distinction between activations for successful vs. unsuccessful trials) to assess differences in memory activity between groups. Consequently, it is unclear whether the role of increased PFC activity in the 2 studies is related to memory success per se, or simply task-related measures. Moreover, Bertolino et al. (2006) also observed poorer cognitive performance in COMT val carriers, thereby confounding observed fMRI differences between the genotype groups. Additionally, although the Schott et al. (2006) study employed an event-related design and found no cognitive performance differences between groups, it only examined encoding of item memory, a relative simpler and less demanding task than relational memory.
Finally, regarding the connectivity findings, it should also be noted that although the Bertolino et al. (2006) result is based on sustained activations at retrieval, the Schott et al. (2006) analysis used event-related activations at encoding. Thus, a complete picture of the effects of COMT genotype on MTL–PFC functional connectivity remains unclear. The current study aims to expand on previous studies by examining the impact of the COMT val158met genotype on functional activity in the PFC and MTL during both successful encoding and retrieval of a relational memory task.
Given the outstanding questions concerning both cognition and neural functioning in relation to COMT val 108/158 met genotype, the current study had 3 main goals. The first was to examine the effect of COMT on cognition using a large sample and multiple measures of cognition, while correcting for population stratification and multiple testing. The second goal was to examine whether COMT genotype affects MTL and PFC activity during successful relational memory (a demanding task shown to require both PFC and MTL involvement) performance. Encoding success activity (ESA) was identified by directly comparing study-phase activity during subsequently remembered trials to that observed during subsequently forgotten trials (Paller and Wagner 2002). Retrieval success activity (RSA) was similarly identified by directly comparing test-phase activity on remembered trials to that of forgotten trials. Specifically, we aimed to investigate whether previously reported differences in MTL and PFC activity associated with the val allele were related to poorer learning in those subjects, or to differences in brain activity independent of learning. Finally, the third goal was to investigate group differences in functional connectivity associated with successful relational memory processing during both encoding and retrieval.
Materials and Methods
Participants
Cognitive performance was measured in participants from the greater Durham, NC, area, ranging from 18 to 84 years of age (see Supplementary Table 1 for participants’ demographics). Twenty-two of the foregoing participants took part in the imaging portion of the study—11 individuals homozygous for the COMT val allele and 11 homozygous for the COMT met allele. The participants selected for imaging were matched as closely as possible on age and gender, as well as North American Adult Reading Test (NAART) score and tests of spatial span and spatial working memory taken from the Cambridge Neuropsychological Test Automated Battery (CANTABeclipse, version 2.0; Cambridge Cognition, Ltd., Cambridge, UK). The imaging participants were also balanced by brain-derived neurotrophic factor (BDNF) val66met status, which has also been implicated in previous imaging studies (e.g., Egan et al. 2003; [COMT val homozygotes: 5 BDNF val/met and 6 BDNF val/val; COMT met homozygotes: 6 BDNF val/met and 5 val/val]). All participants gave written informed consent and received financial compensation. All experimental procedures were approved by the Duke University Institutional Review Board for the ethical treatment of human participants.
Genotyping
Genotyping of the COMT val108/158met (rs4680) and BDNF val66met (rs6265) single-nucleotide polymorphism was carried out using the Illumina HumanHap550 or 610quad whole-genome chips (Illumina, San Diego, CA).
Cognitive Testing
The Cantab battery was administered by trained personnel, according to a script specified word for word by the Cantab producers. The timing and presentation of the tests were maintained with perfect consistency due to the computer-based administration. The Attix battery was administered only by personnel with training by a professional neuropsychologist, and again the tests were administered according to a set script. For both batteries, spot checks and continual training refreshers were performed to ensure internal consistency in the administration. Cantab scores were automatically computed and extracted from the Cantab software. Test results for the Attix battery were recorded on paper, and scores were initially determined by the test administrator, and in every case were double scored by trained personnel. To gain information about relevant nongenetic influences on the cognitive tests, each participant was asked to complete a questionnaire, which included questions of age, education, ethnicity, and current issues with substance abuse and neurological disorders. Participants completing the Attix battery were also asked to fill out a questionnaire on daily activities, familiarity with the testing battery, strategies used during testing, and depression as measured by the Beck Depression Inventory (BDI)-II (Beck and Steer 1993).
Materials
Participants taking part in the genetic association study for cognition underwent testing in either one or both of 2 cognitive batteries. For the Cantab battery we selected tests designed to mimic tests used in the animal literature to study learning and memory, for which there are large bodies of data on the effect of brain lesions, and pharmacological and genetic interventions. For the Attix battery we selected standardized tests that have published normative data and favorable psychometric properties, have been extensively validated, and are known to be sensitive to human cognitive function. The computer-based test battery used tests from CANTAB (CANTABeclipse, version 2.0; Cambridge Cognition, Ltd) including paired associates learning (PAL), spatial working memory (SWM), verbal recognition memory—immediate recall (VRM), intra-extradimensional set shifting (IED), rapid visual processing (RVP), spatial span (SSP), and spatial recognition memory (SRM). The Attix battery comprised Green's story recall (Green 2005)—immediate and delayed, the trail making test (trails A and B) (1944), WAIS-III digit span (forward and backward), digit symbol, and symbol search (Wechsler 1997), controlled oral word association test (COWAT) (Benton et al. 1978), semantic fluency: animals (Rosen 1980), and Stroop color-word interference (Golden 1975). For the CANTAB battery, those measures that showed the greatest variability in a homogenous subset of 18–21 years old Caucasian undergraduates were extracted for analysis, thereby excluding measures in which variation depended largely on nongenetic contributors such as age, education or race/ethnicity. From these, redundant measures were excluded and a final set of 8 measures was selected for genetic analysis: PAL first trial memory score; SWM total mistakes between trials, and strategy; VRM immediate free recall; IED extradimensional shift errors; RVP probability of a hit; SSP span length; SRM percent correct. For the Attix battery, standard scores (e.g., the number of story components correctly remembered) of the 11 measures were used directly as genetic phenotypes. Measures were normalized by transformation where necessary.
Analysis
Association of the cognitive measures with COMT val158met genotype was tested in STATA (StatCorp 2005) using a regression model that included only covariates that had a significant effect on the model (shown in Table 1). Standard covariates tested included age, sex, whether English was the first language learned, test battery, test type (no-ceiling or normal for Cantab), whether the full battery or the shorter battery was tested, tester, education and (non-Cantab tests only) BDI score.
Table 1.
Genetic association in healthy COMT val108/158met (rs4680) individuals for 19 measures from 2 cognitive test batteries using an additive regression model
| n | beta | P value^ | n metmet | n valmet | n valval | Mean (SD) mm | Mean (SD) vm | Mean (SD) vv | Covariatesa | |
| Cantab immediate verbal recallb | 1159 | −0.01 | 0.629 | 250 | 548 | 361 | 9.88 (2.59) | 10.09 (2.54) | 10.12 (2.62) | 1, 2, 3, 4 |
| Cantab spatial recognition memoryb | 1152 | 0.01 | 0.48 | 247 | 544 | 361 | 85.12 (9.35) | 84.69 (9.96) | 85.65 (10.09) | 2, 3 |
| Cantab paired associates learning first trial memory | 656 | 0.58 | 0.047 | 157 | 301 | 198 | 21.14 (6.09) | 22.08 (6.08) | 22.73 (5.42) | — |
| Cantab spatial working memory between trial errors | 656 | −0.12 | 0.86 | 157 | 301 | 198 | 8.04 (10.15) | 8.30 (9.42) | 7.59 (9.55) | 1, 2, 3 |
| Cantab spatial working memory strategy | 656 | −0.06 | 0.877 | 157 | 301 | 198 | 25.78 (6.25) | 26.31 (6.89) | 26.36 (6.95) | 2, 3 |
| Cantab spatial span | 652 | −0.04 | 0.541 | 157 | 297 | 198 | 7.58 (1.43) | 7.45 (1.38) | 7.65 (1.48) | 2, 3 |
| Cantab rapid visual processingb | 651 | 0 | 0.924 | 156 | 299 | 196 | 0.82 (0.13) | 0.81 (0.15) | 0.83 (0.14) | 2,3 |
| Cantab Intra–Extra dimensional set shifting | 629 | −0.25 | 0.526 | 149 | 291 | 189 | 5.30 (7.69) | 4.91 (6.79) | 5.35 (7.59) | 1, 2 |
| WAIS-III Digit Symbol | 1218 | 0.02 | 0.988 | 246 | 581 | 391 | 90.13 (14.94) | 92.43 (15.08) | 90.91 (15.39) | 4, 8, 9 12, 13, 14 |
| Stroop color-word | 1212 | 0.43 | 0.669 | 245 | 578 | 389 | 51.71 (10.89) | 51.67 (10.97) | 50.76 (11.72) | 1, 4, 8, 9, 13 |
| Green's story recall test immediate recall | 595 | 0.25 | 0.638 | 111 | 291 | 193 | 53.68 (9.63) | 53.51 (9.45) | 51.42 (10.63) | 1, 4, 6, 7, 8 |
| Semantic fluency: animals | 499 | 0.42 | 0.221 | 100 | 236 | 163 | 24.67 (6.00) | 24.57 (5.85) | 23.09 (6.25) | 1, 4, 11, 16, 18, 19, 20, 21 |
| TrailsAb | 499 | 0.01 | 0.006 | 100 | 236 | 163 | 24.74 (11.25) | 21.59 (7.17) | 22.39 (8.79) | 4, 11, 12, 16, 17 |
| WAIS-III digit span backward | 499 | −0.1 | 0.239 | 100 | 236 | 163 | 5.51 (1.34) | 5.73 (1.43) | 5.59 (1.51) | 1, 4, 5 |
| WAIS-III digit span forward | 499 | 0.27 | 0.763 | 100 | 236 | 163 | 7.14 (1.31) | 7.20 (1.36) | 7.14 (1.33) | 1, 4, 5, 12 |
| Green's story recall test delayed recall | 496 | 0.6 | 0.371 | 100 | 236 | 160 | 44.52 (12.15) | 44.42 (11.51) | 40.90 (12.96) | 1, 4, 6, 7 |
| TrailsBb | 496 | 0 | 0.337 | 100 | 235 | 161 | 50.29 (24.62) | 46.90 (35.26) | 51.19 (27.07) | 1, 4, 11, 12, 16 |
| WAIS-III symbol search | 496 | 0.45 | 0.32 | 100 | 235 | 161 | 40.53 (9.10) | 41.14 (8.93) | 39.56 (10.12) | 4, 11, 15 |
| COWAT | 499 | 0.06 | 0.927 | 100 | 236 | 163 | 43.92 (10.80) | 44.44 (10.94) | 43.41 (11.39) | 1, 4, 11, 16, 22, 10 |
Note: ^P value corrected for multiple testing would be 0.05/19 = 0.0026. ESL: English as a second language. 1 = English as a Second Language; 2 = site; 3 = test version; 4 = education; 5 = whether a grouping strategy was used on the test; 6 = hours per week reading books; 7 = whether visualized events in story; 8 = whether took full battery; 9 = whether Duke student; 10 = age not included as a covariate; 11 = BDI scores; 12 = who tested them; 13 = whether seen the test before; 14 = handedness; 15 = whether currently play number games such as Sudoku; 16 = whether currently play word games such as crosswords; 17 = whether were placed in classes for gifted students; 18 = whether listed animals in alphabetical order; 19 = whether listed subcategories of animals (dog, poodle, greyhound…); 20 = whether named animals by thinking of a habitat; 21 = whether often do an activity similar to this test or have animals as a large part of life (such as Veterinarians); 22 = whether often do an activity similar to this test.
The numbers refer to the covariates that were significant of the up to 60 tested in the regression models, age and sex were included in all models.
Normalized by transformation.
For the Attix battery, using data from the supplemental questionnaire, a further 60 covariates were tested for association with each phenotype. The covariates were added one at a time to a multivariate regression model of all subjects that also included age, education, ethnicity, tester, BDI score, ESL, and sex, in STATA. From these analyses, all covariates that made intuitive sense and contributed to the model with a P value below 0.008 were included in the genetic analysis. Also included were Eigenstrat axes (Price et al. 2006) sufficient to account for the population structure in each data set (n = 18 for the Cantab tests, after automatic outlier removal which included all subjects with African ancestry; n = 5 for the Attix battery, including all subjects) (StatCorp 2005). In line with previous studies, COMT val108/158met genotype was tested using an additive genetic model (Diaz-Asper et al. 2008).
Imaging Task
Materials
Relational memory was examined by assessing memory for face–scene pairings. Face stimuli consisted of 172 faces gathered from the adult face database from Dr Denise Park's lab (Minear and Park 2004). Scene stimuli consisted of 172 indoor and outdoor scenes gathered from the internet. Using Adobe Photoshop CS2 version 9.0.2 and Irfanview 4.0 (http://www.irfanview.com/), face stimuli were edited to a uniform size (320 × 240 pixels) and background (black), and scene stimuli were standardized to 576 × 432 pixels. Face–scene pairs were created using a custom MatLab (version 6.1) script that overlaid faces on scenes, and images were standardized to 576 × 432 pixels.
Procedure
Blood draws and genotyping, as well as neuropsychological testing were completed on separate testing days prior to the scanning session. For the imaging session, stimuli were presented via a mirror in the head coil and a rear projection system using a PC computer with Cogent, a stimulus presentation toolbox within Matlab. Button responses and response times were recorded using a magnetically shielded 4-button box held in the participant's right hand.
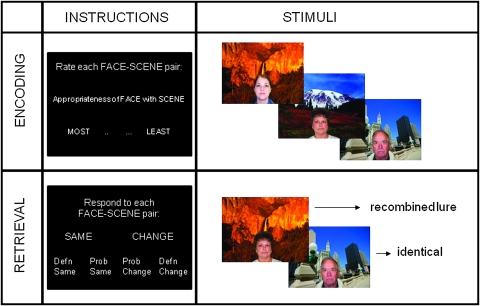
The relational memory task consisted of 8 runs, alternating between encoding and retrieval. During each encoding run participants saw 48 face–scene pairings and were asked to decide “How appropriate is the fit?” between the face and scene on a 4-point scale ranging from “most” to “least.” The goal of using the goodness of fit task during encoding was to assure that participants were attending to both the face and the scene, as well as their relationship with one another (see Fig. 1).
Figure 1.
Task instructions and example of stimuli used in the relational memory task. During encoding participants were asked to rate the appropriateness of the face–scene pair, on a 4-point scale of most-to-least appropriate. During retrieval they were asked to determine whether the face–scene pair they were viewing was the same or changed from that which was presented during encoding. They were asked to respond either “definitely same,” “probably same,” “probably change,” or “definitely change.”
During each retrieval run, 36 intact face–scene pairings were shown as well as 12 lures that were recombination of previous face–scene pairings. Participants made memory judgments to each pairing using a 4-point scale: “definitely same,” “probably same,” “probably changed,” or “definitely changed.” Because the lures consisted of recombined studied items, participants had to encode and remember relationships among the items (i.e., specific face–scene pairings)—memory for the items alone was not sufficient to support correct performance on the recognition test. Intertrial jitter ranged from 750 to 1500 ms to facilitate deconvolution of the hemodynamic response (Dale 1999).
Image Acquisition
Images were collected on a General Electric 3.0 Tesla Signa Excite HD short bore scanner (Milwaukee, WI) equipped with an 8-channel head coil. Following acquisition of the high-resolution anatomical images (450-ms repetition time (TR), a 3-ms echo time (TE), a 24-cm field of view (FOV), a 2562 matrix, and a slice thickness of 1.9 mm), whole-brain functional images were acquired parallel to the anterior–posterior commissure plane using an inverse spiral sequence (direction = interleaved, matrix = 642, FOV = 24 cm, TR = 2000 ms, TE = 30 ms, sections = 34, thickness = 3.8 mm, interscan spacing = 0). The 8 task runs each lasted 4 min 8 s, and 2-min break was given between each run.
Image Processing
Functional data were preprocessed and analyzed with SPM2 (Statistical Parametric Mapping; Wellcome Department of Cognitive Neurology, http://www.fil.ion.ucl.ac.uk/spm). Time-series data were corrected for differences in slice acquisition times and realigned. Functional images were spatially normalized to a standard stereotaxic space using the Montreal Neurological Institute (MNI) templates implemented in SPM2 and resliced to a resolution of 3.75 mm3. The coordinates were later converted to Talairach space (Talairach and Tournoux 1988). Finally, the volumes were spatially smoothed using an 8-mm isotropic Gaussian kernel. The hemodynamic response for each trial was modeled using the canonical hemodynamic response function. Head motion was assessed prior to preprocessing. No individual moved more than 3 mm in any direction, in any run. Thus, no data were eliminated in either group due to motion artifacts.
Statistical Analyses
During encoding, subsequently remembered trials were defined as encoding trials that led to a “definitely same” recognition response and subsequently forgotten trials were defined as encoding trials that led to a “changed” recognition response. These 2 trial types were modeled separately. ESA was identified by directly comparing subsequently remembered to subsequently forgotten trials. Retrieval trials were similarly defined and modeled, with RSA identified by directly comparing remembered to forgotten trials. Lures (and subsequent lures), trials leading to a “probably same” recognition response, as well as confounding factors such as head motion and scanner drift, were also included in the model and treated as regressors of no interest.
Based upon our a priori hypotheses focusing on group differences in the MTL and PFC we constructed and applied masks of each region of interest (ROI) using an anatomical library (Tzourio-Mazoyer et al. 2002) available within SPM2. Within each group, ESA was identified within the focal PFC and MTL ROIs by directly comparing subsequently remembered vs. forgotten trials and subsequently masking the resulting activations with the group effects. Thus, activations were required to pass 2 thresholds: 1) they had to show a significant ESA effect (remembered > forgotten) within one of the groups (P < 0.05, and a minimum of 10 contiguous voxels); and 2) a group difference in the ESA effect (P < 0.05, and a minimum of 10 contiguous voxels). The conjoint probability following inclusive masking approaches P = 0.0025 (Fisher 1950; Lazar et al. 2002), but this estimate should be taken with caution given that the contrasts were not completely independent. This analysis approach was used in order to ensure that group differences were associated specifically with those activations exhibiting greater activity for remembered than forgotten trials in a given genotype group. Similar analyses were conducted for retrieval (i.e., significant RSA within one group plus significant group effects).
In order to identify activations outside of our a priori regions of interest that also exhibited between group differences, whole-brain between group contrasts were conducted at P < 0.001, uncorrected, with a minimum cluster size of 10 contiguous voxels.
Connectivity Analysis
To assess functional connectivity during the relational memory task (at both encoding and retrieval), we used the MTL as a seed region and examined correlations in whole-brain activity with MTL activity. The seed MTL voxels used were those which showed the greatest group differences in the left MTL for ESA (−15, −12, −22) and right MTL for RSA (26, −5, −29) analyses. Laterality of seed voxels were based upon general memory research indicating encoding to be left lateralized and retrieval, right lateralized (i.e., Tulving et al. 1994). Each trial in the model was uniquely coded so that the time-series activity of the seed region could be correlated with the rest of the voxels in the brain. The validity of this design has been confirmed in previous studies (Rissman et al. 2004; Daselaar, Fleck, and Cabeza 2006; Daselaar, Fleck, Dobbins et al. 2006; Dennis et al. 2008). The goal of this exploratory analysis was to assess connectivity for ESA and RSA; therefore, the analysis was constrained to only high-confidence remembered trials during encoding and retrieval. As a second step, group averages were calculated by employing a 1-sample t-test on the resulting correlation maps (random effects). Group differences were again calculated using a multiple contrast approach. The between group 2-sample t-test was conducted at P < 0.05 with a minimum cluster size of 30 voxels.
Results
Genetic Association Results
Table 1 displays the genetic association results for the 19 cognitive measures tested against COMT val108/158met genotype in an additive regression model corrected for population structure as well as other significant covariates (additional descriptive statistics concerning the sample and the are provided in Supplemental Table 1). The P values shown are before correction for multiple hypothesis testing (19 tests). Two phenotypes showed nominal associations with COMT genotype before correction: PAL first trial learning P = 0.047 (borderline) and Trails A P = 0.006. After correction for multiple hypothesis testing, these P values were nonsignificant (0.94 and 0.12, respectively). It is worth noting that although Trails A measures sensorimotor speed and is likely related to dopamine-striatal functions, the possibility of a potential link between COMT genotype and sensorimotor speed is not supported by other tests in the battery, such as Cantab's Rapid Visual Processing test. Thus in this data set we show no association between cognitive outcome and COMT genotype in a wide range of cognitive tests including working memory (verbal and spatial), episodic memory, attention, and executive function.
Results for Cognitive Performance in the Imaging Subgroup
Table 2 reports demographic data, selected test results from the Cognitive Assessment, and both the hit and false alarm rate for each group of participants in the imaging study (n = 11 val homozygotes; n = 11 met homozygotes). Paired t-tests revealed that both groups made significantly more hits than false alarms (val homozygotes: t(10) = 9.16, P < 0.001; met homozygotes: t(10) = 7.46, P < 0.001), indicating that both groups were successful in correctly recognizing face–scene associations. Unpaired t-tests on each measure revealed no significant between group differences in either hit rate, false alarm rate, or response bias (d′), indicating that both groups exhibited similar levels of memory performance. Thus, differences in neural (fMRI) activity between COMT val and met homozygotes are not confounded by differences in cognitive performance.
Table 2.
Demographics and cognitive functioning of the 22 participants in the fMRI study by COMT homozygote group
| met/met | val/val | P | |
| n | 11 | 11 | |
| Male/female | 5/6 | 6/5 | |
| Agea | 24.64 (7.68) | 20.45 (3.62) | 0.12 |
| NAARTa | 17.60 (4.09) | 16.60 (6.39) | 0.69 |
| Cantab spatial spana | 8.18 (0.60) | 8.18 (1.25) | 1 |
| Cantab spatial span errorsa | 12.27 (7.42) | 7.91 (6.22) | 0.15 |
| Cantab spatial working memory total errorsa | 7.55 (11.73) | 4.55 (4.70) | 0.44 |
| Cantab Spatial Working Memory Strategya | 25.55 (6.15) | 23.45 (3.64) | 0.34 |
| Relational memory task during fMRI | |||
| Hit rate | 0.75 (0.08) | 0.72 (0.09) | 0.68 |
| False alarm rate | 0.36 (0.14) | 0.26 (0.14) | 0.26 |
| d‘ | 1.06 (0.50) | 1.30 (0.52) | 0.40 |
Subjects were preselected to match for these variables.
fMRI Results
Medial Temporal Lobe
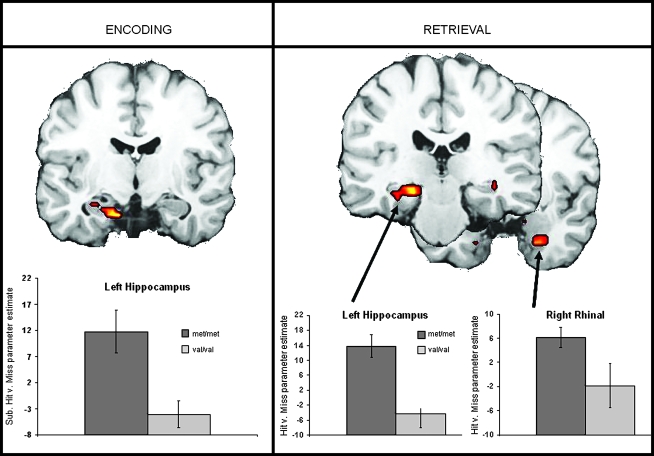
Compared with COMT met homozygotes, COMT val homozygotes exhibited significantly weaker MTL activity associated with both ESA and RSA (Fig. 2 and Table 3), specifically in the left hippocampus for ESA and in the left hippocampus and bilateral parahippocampal gyrus (PHG) for RSA.
Figure 2.
Weaker ESA and RSA for COMT val homozygotes compared with met homozygotes.
Table 3.
Significant group differences in fMRI activity associated with successful encoding and retrieval within the MTL and PFC
| Coordinates |
|||||||
| H | BA | x | y | z | T | Voxels | |
| met/met > val/vala | |||||||
| ESA | |||||||
| Hippocampus/PHG | L | −15 | −12 | −22 | 3.27 | 12 | |
| RSA | |||||||
| Hippocampus/PHG | L | −19 | −30 | −11 | 3.84 | 32 | |
| PHG | L | 28/36 | −23 | −9 | −28 | 2.57 | 10 |
| R | 28/34 | 26 | −5 | −29 | 3.64 | 26 | |
| PHG | R | 36 | 41 | −19 | −12 | 3.43 | 10 |
| Ventromedial PFC | L | 47/11 | −23 | 25 | −14 | 3.13 | 33 |
| R | 47 | 15 | 7 | −13 | 3.92 | 23 | |
| val/val > met/metb | |||||||
| ESA | |||||||
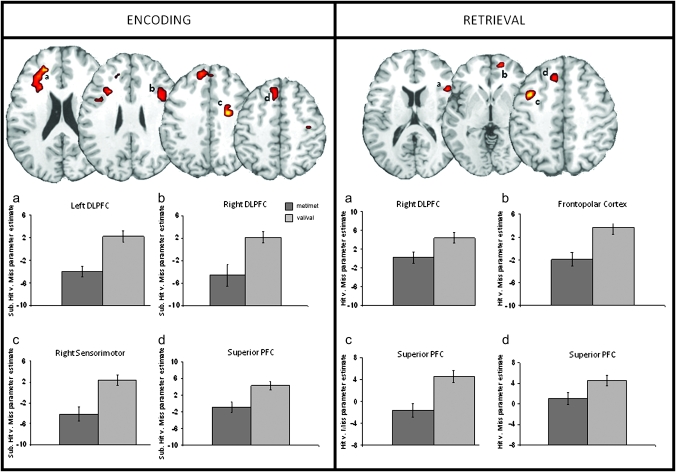
| Dorsolateral PFC | L | 46/9/10 | −19 | 41 | 15 | 4.59 | 116 |
| R | 46/9 | 38 | 16 | 24 | 3.14 | 35 | |
| Superior PFC | L | 8 | −8 | 32 | 44 | 2.43 | 23 |
| Motor/premotor | R | 6/9 | 26 | −5 | 39 | 3.82 | 30 |
| R | 6/4 | 45 | −5 | 39 | 3.2 | 29 | |
| R | 6 | 8 | 11 | 62 | 2.51 | 15 | |
| RSA | |||||||
| Dorsolateral PFC | R | 45 | 30 | 23 | 16 | 2.53 | 25 |
| R | 44/45 | 45 | 19 | 10 | 2.51 | 16 | |
| Superior PFC | L | 8/6 | −38 | 10 | 45 | 3.56 | 36 |
| L | 8 | −11 | −20 | 50 | 3.16 | 29 | |
| L | 9 | −11 | 42 | 29 | 2.29 | 13 | |
| Frontopolar cortex | R | 10 | 11 | 55 | −6 | 2.47 | 19 |
| Motor/premotor | L | 6/4 | −11 | 28 | 48 | 3.2 | 31 |
| R | 6/4 | 11 | −30 | 65 | 3.07 | 17 | |
Regions within the MTL and PFC where individuals homozygous for the met allele showed increased ESA and RSA compared with individual homogygous for the val allele.
Regions within the MTL and PFC where individuals homozygous for the val allele showed increased ESA and RSA compared with individual homogygous for the met allele. Results include all significant group differences within the MTL and PFC significant at P < 0.05, k > 10 voxels masked with the effects of interest at P < 0.05, k > 10 voxels (conjoint probability P < 0.0025). H: hemisphere; BA: Brodmann's area.
Prefrontal Cortex
Compared with COMT met homozygotes, COMT val homozygotes exhibited significantly greater PFC activity associated with both ESA and RSA (see Fig. 3 and Table 3). Specifically, these effects were seen in the bilateral dorsolateral PFC, left superior PFC, and right sensorimotor regions for ESA and in the right dorsolateral PFC, left superior PFC, right frontopolar, and bilateral motor/premotor regions for RSA.
Figure 3.
Greater ESA and RSA for COMT val homozygotes compared with met homozygotes. DLPFC: dorsolateral prefrontal cortex.
Other Brain Regions
Only one contrast exhibited a significant between group difference: met carriers exhibited greater RSA in the right fusiform gyrus (Talariach coordinates: 49, −16, −25; t(20) = 6.46; voxels = 18).
Connectivity Results
MTL–PFC Functional Connectivity
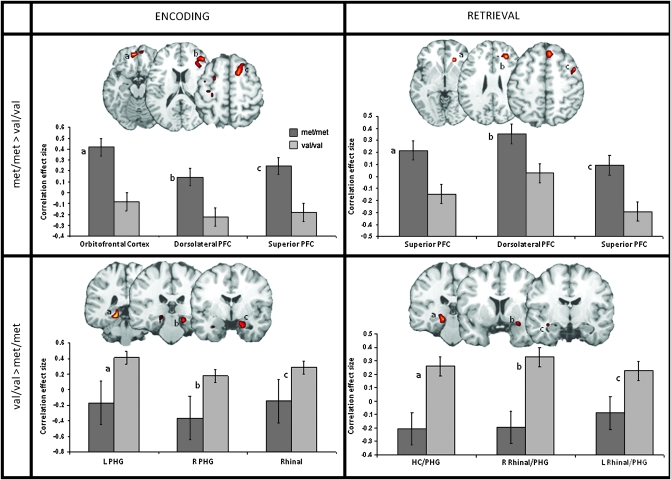
Functional connectivity analyses using ESA and RSA voxels within the MTL as seeds identified several PFC regions functionally connected to the MTL. Within all of these regions, MTL–PFC connectivity was weaker in COMT val compared with COMT met homozygotes. PFC regions showing weaker connectivity included right orbitofrontal, right dorsolateral PFC, and bilateral left ventromedial PFC for ESA, and, right dorsomedial PFC, right anterior cingulate cortex (ACC), and right superior PFC for RSA (see Fig. 4 and Table 4).
Figure 4.
Group differences in MTL connectivity for both ESA and RSA. COMT val homozygotes show weaker MTL–PFC connectivity compared with met homozygotes, and only greater connectivity limited to MTL regions. L: left; R: right; HC: hippocampus.
Table 4.
Group differences in MTL connectivity associated with both encoding and retrieval success in 22 healthy participants
| H | BA | x | y | z | T | Voxels | |
| met/met > val/vala | |||||||
| ESA | |||||||
| ^Orbitofrontal cortex | L | 11 | −11 | 47 | −18 | 4.02 | 50 |
| ^Superior PFC | R | 6 | 11 | 11 | 66 | 3.43 | 43 |
| ^ACC | R | 24/32 | 15 | 16 | 27 | 3.24 | 41 |
| ^Dorsolateral PFC | R | 10/46 | 41 | 41 | 12 | 3.28 | 30 |
| ^Sensorimotor cortex | L | 4/6 | −26 | −9 | 46 | 3.61 | 85 |
| Insula | R | 13 | 38 | −7 | 14 | 3.59 | 79 |
| Superior temporal gyrus | R | 21/22 | 56 | −44 | −1 | 5.28 | 93 |
| Temporal–parietal junction | L | 40/37/22/20 | −38 | −43 | 16 | 4.43 | 210 |
| RSA | |||||||
| ^Dorsomedial PFC | R | 9/46 | 26 | 38 | 23 | 2.9 | 103 |
| Putamen | R | 23 | 22 | 2 | 3.68 | 30 | |
| ^ACC | R | 24 | 19 | 29 | −5 | 3.42 | (10) |
| Parietofrontal cortex | R | 40/7/39/31 | 30 | −54 | 31 | 5.49 | 506 |
| ^Superior PFC | R | 8/6 | 45 | 10 | 49 | 3.28 | (71) |
| Thalamus | R | 19 | −19 | −5 | 2.97 | 50 | |
| *Anterior PHG | R | 34 | 11 | −8 | −19 | 2.27 | (8) |
| Retrosplenial cortex | M | 23 | 0 | −28 | 22 | 3.54 | 44 |
| Occipital cortex | R | 18/19 | 23 | −85 | 8 | 3.21 | 38 |
| Cerebellum | R | 23 | −31 | −30 | 2.85 | 34 | |
| val/val>met/metb | |||||||
| ESA | |||||||
| Occipitotemporal cortex | L | 17/18/27/30 | −11 | −33 | −2 | 4.68 | 262 |
| *PHG | L | 27/30 | −19 | −37 | −8 | 4.17 | (18) |
| Posterior cingulate/occipital cortex | L | 18/19/20/30/35 | −11 | −36 | 23 | 3.38 | 204 |
| *PHG | R | 35 | 19 | −30 | −11 | 2.84 | (13) |
| Anterior temporal cortex | R | 28/34/38 | 26 | 10 | −23 | 4.57 | 67 |
| *Rhinal cortex | R | 28/34 | 23 | −5 | −19 | 3.23 | (18) |
| Anterior temporal cortex | L | 38/28/20 | −41 | 5 | −39 | 3.16 | 41 |
| *Rhinal cortex | L | 38/20 | −26 | −1 | −22 | 2.26 | (6) |
| ACC | R | 32/10 | 11 | 45 | 12 | 3.19 | 47 |
| Caudate | L | −23 | 14 | −7 | 2.63 | 32 | |
| Occipitoparietal cortex | R | 40013 | 15 | −53 | 45 | 4.06 | 78 |
| Cerebellum | M | 0 | −50 | −33 | 3.98 | 84 | |
| RSA | |||||||
| *PHG/HC | L | 27/35 | −19 | −26 | −2 | 3.89 | 34 |
| Superior/middle temporal gyrus | R | 22/42 | 49 | −25 | 15 | 4.31 | 186 |
| *Rhinal cortex/PHG | R | 34 | 30 | 7 | −16 | 3.54 | (9) |
| Superior temporal gyrus | L | 29 | −41 | −32 | 16 | 3.09 | 96 |
| *Rhinal cortex/PHG | L | 34 | −30 | −4 | −13 | 1.87 | (4) |
| Orbitofrontal cortex | L | 10/9 | −8 | 63 | 7 | 3.44 | 59 |
| Posterior cingulate | L | 31 | −11 | −66 | 14 | 4.55 | 61 |
| Inferior parietal cortex | L | 3 | −56 | −20 | 40 | 3.26 | 45 |
| Cerebellum | L | −11 | −64 | −32 | 3.99 | 383 | |
Brain regions exhibiting greater MTL connectivity during encoding and retrieval for individuals homozygous for the met allele compared with individual honogygous for the val allele.
Brain regions exhibiting greater MTL connectivity during encoding and retrieval for individuals homozygous for the val allele compared with individual honogygous for the met allele. Results include all significant group differences within the MTL and PFC significant at P < 0.05, with a minimum cluster size > 30 voxels. ESA: encoding success activity; RSA: retrieval success activity; PHG: parahippocampal gyrus; PFC: prefrontal cortex; ACC: anterior cingulate cortex; H: hemisphere; BA: Brodmann's area; ^highlights PFC regions; *highlights MTL regions; brackets () indicate the voxel extent of the subpeaks, which are identified through the use of indentations in the table.
In contrast, COMT val homozygotes showed greater connectivity than COMT met homozygotes within the MTL. Specifically, greater connectivity between the MTL seed voxels and other MTL regions was seen in the right rhinal cortex and anterior PHG, left hippocampus and left PHG for ESA, and in the left hippocampus/PHG, bilateral rhinal cortex and anterior PHG for RSA.
Discussion
The current study investigated the effects of a common functional variant of the COMT gene on both cognitive and neural processes. To ascertain whether there was an association of cognition with COMT val108/158met genotype, we tested up to 1218 individuals with 19 cognitive tests, and found no effect on any single cognitive measure (including tests of working memory, attention, executive function, and episodic memory). We went on to investigate the effects of COMT val108/158met on neural activity during successful relational memory in a subset of 22 of these individuals. Focusing on the MTL and PFC, the 2 integral components of the relational memory network, the results showed that compared with COMT met allele homozygotes, COMT val homozygotes exhibited significantly greater PFC recruitment during both successful encoding and retrieval, while simultaneously exhibiting weaker MTL recruitment. In addition to these group differences in ESA and RSA, functional connectivity analysis showed that COMT val homozygotes exhibit weaker connectivity between MTL and PFC but greater connectivity within MTL regions during both stages of the relational memory task. These results indicate that, despite being similar in cognitive performance, the COMT genotype groups utilize different neural processing or cognitive strategies to achieve this performance.
No significant Difference in Cognitive Performance
Using large sample sizes, and correcting for population stratification, multiple covariates, and multiple hypothesis testing, we found no association of COMT val108/158met genotype with any of the cognitive test measures. This is in keeping with the results of a recent meta-analysis that analyzed COMT 108/158met genetic association results from 31 genetic association studies on healthy volunteers and found no association between COMT genotype and any cognitive measure except IQ (which was not measured in this study) (Barnett et al. 2008). However, these results contradict several studies which have published positive association findings (for reviews see Savitz et al. 2006; Tunbridge et al. 2006; Lewandowski 2007). Several reasons for this discrepancy may exist. Firstly, there is an effect of publication bias, in that it is easier to publish positive findings than it is to publish negative findings (see Barnett et al. 2008). Second, even when negative results are reported, the focus tends to be centered on the positive findings from any particular study (e.g., Diaz-Asper et al. 2008). This can be particularly misleading if multiple tests are used as phenotypes and genotypes are not corrected for multiple hypothesis testing. Indeed, we found 2 associations that would have appeared to be significant (P = 0.006 with trails A and P = 0.047 with PAL first trial learning) if we had not reported, and corrected for, 19 different phenotypic tests. Finally, a particular problem in previous reports may be lack of correction for population stratification. Population stratification will occur if both the average test score and the allele frequency of the polymorphism under investigation differ between population groups. It is known that the COMT val108/158met polymorphism varies significantly in frequency between populations (Palmatier et al. 1999). Previous meta-analyses have reported heterogeneity in apparent effects of COMT on cognitive measures across different samples (Barnett et al. 2008), which could reflect population stratification effects in opposite directions in different samples. This seems a more likely hypothesis than dopamine having different neural functions in different racial or ethnic groups.
Significant Differences in Memory-Related Brain Activity
Despite the lack of significant effects on an extensive battery of cognitive measures, we found significant effects of the COMT val108/158met genotype on brain activity associated with successful encoding (ESA) and retrieval (RSA) processes. In particular, in the subset of young adults investigated, we found reliable differences in 1) MTL activity, 2) PFC activity, and 3) functional connectivity between the MTL and PFC and within MTL regions.
Medial Temporal Lobe
COMT val homozygotes exhibited weaker MTL activity during both successful encoding and retrieval of the relational memory task. Along with the PFC, the MTL, including the hippocampus, is considered to be one of the key components of a network supporting relational memory processes (Johnson et al. 1993; Eichenbaum et al. 2007), mediating the binding and retrieval of different elements of an encoding episode into a coherent and complex memory trace. Reduced MTL activity during relational memory tasks is most often interpreted as reflecting a deficit in this binding process and indicative of weaker representations of the bound items (i.e., face and scene) (e.g., Mitchell et al. 2000; Dennis et al. 2008).
Given evidence that COMT affects both frontal and midbrain dopamine levels (e.g., Hong et al. 1998; Matsumoto et al. 2003), it may be the case that decreased midbrain dopamine in COMT val carriers reduces MTL functioning (i.e., weaker binding processes) associated with successful relational memory formation and retrieval. Animal research has shown that dopaminergic inputs from the ventral tegmental area (VTA) to the MTL both 1) enhance memory formation (Gasbarri et al. 1996) and 2) modulate MTL activity (Weiss et al. 2003). Based on this empirical work, Lisman and Grace (2005) proposed a model by which the VTA and hippocampus form a functional loop, regulating dopaminergic input in the MTL and thus regulating long term potentiation and learning (see also Adcock et al. 2006). Hippocampal long term potentiation has also been shown to be associated with enhanced D1 receptor activity and an increased capacity for memory storage (Frey et al. 1990) and the COMT val allele to be associated with decreased signaling of D1 dopamine receptors (Bilder et al. 2004; Seamans and Yang 2004). Although Bertolino et al. (2006) concluded that the COMT val allele was indeed associated with weaker MTL activity and this was in turn associated with poorer retrieval performance, the current findings suggest a somewhat different interpretation. MTL deficits in the current study were found to be associated not simply with memory execution (i.e., a memory task), but rather successful memory performance (remembered > forgotten) at both the encoding and retrieval phase. Moreover, given the lack of cognitive performance differences between the val108/158met genotype groups, the current findings further conclude that this reduction in binding processes does not affect cognitive behavioral outcome. Taken together, these findings suggest that COMT val homozygotes may be engaging in different but equally successful neural processing (see PFC results below) to perform the relational memory task, compared with that utilized by met homozygotes.
Prefrontal Cortex
In support of this conclusion, and contrary to the MTL findings, COMT val homozygotes exhibited greater PFC recruitment during both successful relational memory encoding and retrieval. Although previous studies have suggested that increased frontal activations in COMT val carriers is a result of inefficient processing, resulting in poorer cognitive performance (Egan et al. 2001; Bertolino et al. 2006), the current findings again suggest a different interpretation. Increased PFC activity in COMT val homozygotes was associated with successful cognitive performance at both stages of the task, arguing against previous findings that it represents inefficient processing or is detrimental to task performance. Although previous studies have shown PFC recruitment during relational memory to be critical to successful performance (e.g., Prince et al. 2005; Dennis et al. 2008), the precise level of PFC (or any region-specific) activation that is necessary for a given task cannot be specified. Thus it is difficult to determine whether more or less activity in the PFC is “normal” or “typical.” What can be determined from the current results is that while the 2 genotyped groups performed equally as well during the imaging (relational memory) task, they recruited different levels of PFC and MTL processing to do so. Moreover, this difference in overall recruitment levels between groups was also reflected in connectivity.
MTL Connectivity
Due to the essential role of the MTL in relational memory and binding, this region was chosen as a seed region for the functional connectivity analysis. Results indicated that, in addition to exhibiting group differences in overall MTL and PFC activations, COMT met and val homozygotes also showed significant differences in MTL connectivity. Interestingly, val homozygotes, despite exhibiting greater PFC activity during both phases of the memory task, show reduced functional connectivity between the PFC and MTL during both successful encoding and retrieval. Greater connectivity in COMT val compared with met homozygotes was limited to the MTL bilaterally. The fact that COMT val homozygotes exhibit weaker functional connectivity between MTL and PFC regions suggests that they are either less able to integrate processing mediated by the 2 regions, or have less of a need to integrate processing between these regions. As noted, the influence of dopamine on cognition is not limited to the PFC but acts in the midbrain as well. Recent work in rats has shown that hippocampal functioning can influence prefrontal dopamine transmission as well as hippocampal–prefrontal interactions supporting cognitive performance (Peleg-Raibstein et al. 2005). The current results suggest that these interactions are indeed enhanced during relational memory performance in COMT met carriers who have heightened levels of prefrontal synaptic dopamine and enhanced signal-to-noise processing in the PFC compared with COMT val carriers.
Interaction of Genes, Neural Recruitment, and Cognitive Performance
The current results suggest that, in healthy young adults, COMT val108/158 genotype can affect neural processing in the absence of effects on cognitive performance. The results further suggest that more than one pattern of neural recruitment can support a given cognitive process. This idea is not a new one, but has longstanding support in the neuroimaging literature. For example, in the domain of aging, alternations in neural processing regions, but not cognitive performance, between healthy young and healthy older adults is commonplace (for reviews see Cabeza 2002; Dennis and Cabeza 2008; Grady 2008), with several studies finding age-related shifts from more posterior processing (including MTL recruitment) to more anterior processing (including PFC recruitment) (for a review see Dennis and Cabeza 2008). Such age-related differences in the pattern of neural recruitment are often termed as compensatory, with the notion being that age-related changes in neural architecture or neural efficiency has necessitated older adults to recruit additional or different neural mechanisms to accomplish cognitive processes at the level of young adults. However, this compensation interpretation needs further investigation before being applied to COMT val allele carriers.
Perhaps a more parsimonious (yet not incompatible) explanation for the current set of results is that the 2 groups are engaging in different strategies, thereby using different components of the relational memory network to varying degrees in order to perform the relational memory tasks. Results suggest that the while met homozygotes rely on more automatic, MTL-based binding mechanisms, COMT val homozygotes are employing a more frontally based, top down strategy to encode and retrieve associations amongst stimuli. Although this difference in neural recruitment may not result in differences in cognitive outcome, it is likely to be based on alteration of dopamine levels throughout the cortex. Furthermore, we cannot rule out the possibility that one of these strategies is advantageous in combination with neural recruitment differences brought about by environmental stressors, disease, aging, or neurotropic drugs, and further work will be required to elucidate such effects. Additionally, the current neuroimaging results are based on findings from healthy young adults in a relatively low sample size compared with the cognitive performance results. Thus more work is necessary to provide a complete understanding of the neural differences associated with variation in the COMT genotype.
As noted above, often a confound in neuroimaging studies arises when 2 groups exhibit significant differences in both cognitive performance and functional activations, such that the former potentially influences the latter. Such a pattern is also suggestive of group differences in difficulty, again confounding regional differences in neural recruitment. Here, not only does cognitive performance on the relational memory task negate this argument, but the fact that we found no cognitive performance differences associated with COMT val108/158met genotype on a wide range of neurocognitive measures further supports the conclusion that the 2 groups maintain similar cognitive abilities, whereas exhibiting significant differences in neural recruitment accompanying memory success.
Conclusions
Contrary to previous published positive findings, but in accordance with a recent meta-analysis (Barnett et al. 2008), here we find no association between the COMT val108/158met polymorphism and cognitive performance in a large population sample. Despite the lack of cognitive performance differences, the functional imaging results indicated an effect on neural recruitment for a relational memory task. Specifically, there was greater PFC activation and reduced MTL activation in COMT val compared with met homozygotes during both ESA and RSA. Moreover, the MTL-based functional connectivity associated with ESA and RSA also differed between genotypes. Taken together, these results suggest that the genotype groups utilize different neural processing or cognitive strategies to achieve the same levels of cognitive performance. Thus, we conclude that although the COMT val108/158met polymorphism can affect neural activation patterns, it is now becoming clear that it does not have any strong and replicable influence on cognition as measured by standard neurocognitive tests used in our and other similar test batteries.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
Duke University Provost Common Fund; and National Institute on Aging grants (grant no. R01 AG019731, R01 AG23770) to R.C. and (T32 AG000029) to N.A.D.
Supplementary Material
Acknowledgments
The authors are grateful to Deborah Attix who designed the non-Cantab test battery and helped in analyzing all measures. The authors also wish to thank Phil Kragel and Lauren Warren for help in preparation of this manuscript. Conflict of Interest: None declared.
References
- Aalto S, Bruck A, Laine M, Nagren K, Rinne JO. Frontal and temporal dopamine release during working memory and attention tasks in healthy humans: a positron emission tomography study using the high-affinity dopamine D2 receptor ligand [11C]FLB 457. J Neurosci. 2005;25:2471–2477. doi: 10.1523/JNEUROSCI.2097-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adcock RA, Thangavel A, Whitfield-Gabrieli S, Knutson B, Gabrieli JD. Reward-motivated learning: mesolimbic activation precedes memory formation. Neuron. 2006;50:507–517. doi: 10.1016/j.neuron.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Barnett JH, Scoriels L, Munafo MR. Meta-analysis of the cognitive effects of the catechol-O-methyltransferase gene Val158/108Met polymorphism. Biol Psychiatry. 2008;64:137–144. doi: 10.1016/j.biopsych.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Beck AT, Steer RA. Manual for the Beck Depression Inventory. San Antonio, TX: Psychological Corporation; 1993. [Google Scholar]
- Benton A, Hamsher K, Sivan A. Multilingual Aphasia Examination. Iowa City, IA: AJA Associates, Inc; 1978. [Google Scholar]
- Bertolino A, Rubino V, Sambataro F, Blasi G, Latorre V, Fazio L, Caforio G, Petruzzella V, Kolachana B, Hariri A, et al. Prefrontal-hippocampal coupling during memory processing is modulated by COMT val158met genotype. Biol Psychiatry. 2006;60:1250–1258. doi: 10.1016/j.biopsych.2006.03.078. [DOI] [PubMed] [Google Scholar]
- Bilder RM, Volavka J, Lachman HM, Grace AA. The catechol-O-methyltransferase polymorphism: relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology. 2004;29:1943–1961. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- Bruder GE, Keilp JG, Xu H, Shikhman M, Schori E, Gorman JM, Gilliam TC. Catechol-O-methyltransferase (COMT) genotypes and working memory: associations with differing cognitive operations. Biol Psychiatry. 2005;58:901–907. doi: 10.1016/j.biopsych.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Cabeza R. Hemispheric asymmetry reduction in older adults: the HAROLD model. Psychol Aging. 2002;17:85–100. doi: 10.1037//0882-7974.17.1.85. [DOI] [PubMed] [Google Scholar]
- Cabeza R, Mangels J, Nyberg L, Habib R, Houle S, McIntosh AR, Tulving E. Brain regions differentially involved in remembering what and when: a PET study. Neuron. 1997;19:863–870. doi: 10.1016/s0896-6273(00)80967-8. [DOI] [PubMed] [Google Scholar]
- Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S, Kolachana BS, Hyde TM, Herman MM, Apud J, et al. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75:807–821. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM. Optimal experimental design for event-related fMRI. Human Brain Mapping. 1999;8:109–114. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<109::AID-HBM7>3.0.CO;2-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daselaar SM, Fleck MS, Cabeza RE. Triple dissociation in the medial temporal lobes: recollection, familiarity, and novelty. J Neurophysiol. 2006;96:1902–1911. doi: 10.1152/jn.01029.2005. [DOI] [PubMed] [Google Scholar]
- Daselaar SM, Fleck MS, Dobbins IG, Madden DJ, Cabeza R. Effects of healthy aging on hippocampal and rhinal memory functions: an event-related fMRI study. Cereb Cortex. 2006;16:1771–1782. doi: 10.1093/cercor/bhj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davachi L. Item, context and relational episodic encoding in humans. Curr Opin Neurobiol. 2006;16:693–700. doi: 10.1016/j.conb.2006.10.012. [DOI] [PubMed] [Google Scholar]
- de Frias CM, Annerbrink K, Westberg L, Eriksson E, Adolfsson R, Nilsson LG. COMT gene polymorphism is associated with declarative memory in adulthood and old age. Behav Genet. 2004;34:533–539. doi: 10.1023/B:BEGE.0000038491.06972.8c. [DOI] [PubMed] [Google Scholar]
- Dennis NA, Cabeza R. Neuroimaging of healthy cognitive aging. In: Salthouse TA, Craik FEM, editors. Handbook of aging and cognition. 3rd edn. New York: Psychological Press; 2008. pp. 1–56. [Google Scholar]
- Dennis NA, Hayes SM, Prince SE, Huettel SA, Madden DJ, Cabeza R. Effects of aging on the neural correlates of successful item and source memory encoding. J Exp Psychol Learn Mem Cogn. 2008;34:791–808. doi: 10.1037/0278-7393.34.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Asper CM, Goldberg TE, Kolachana BS, Straub RE, Egan MF, Weinberger DR. Genetic variation in catechol-O-methyltransferase: effects on working memory in schizophrenic patients, their siblings, and healthy controls. Biol Psychiatry. 2008;63:72–79. doi: 10.1016/j.biopsych.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Miller SL, Greve DN, Dale AM, Albert MS, Schacter DL, Sperling RA. Prefrontal-hippocampal-fusiform activity during encoding predicts intraindividual differences in free recall ability: an event-related functional-anatomic MRI study. Hippocampus. 2007;17:1060–1070. doi: 10.1002/hipo.20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H, Yonelinas AP, Ranganath C. The medial temporal lobe and recognition memory. Annu Rev Neurosci. 2007;30:123–152. doi: 10.1146/annurev.neuro.30.051606.094328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. Statistical methods for research workers. London: Oliver and Boyd; 1950. [Google Scholar]
- Frey U, Schroeder H, Matthies H. Dopaminergic antagonists prevent long-term maintenance of posttetanic LTP in the CA1 region of rat hippocampal slices. Brain Res. 1990;522:69–75. doi: 10.1016/0006-8993(90)91578-5. [DOI] [PubMed] [Google Scholar]
- Gasbarri A, Sulli A, Innocenzi R, Pacitti C, Brioni JD. Spatial memory impairment induced by lesion of the mesohippocampal dopaminergic system in the rat. Neuroscience. 1996;74:1037–1044. doi: 10.1016/0306-4522(96)00202-3. [DOI] [PubMed] [Google Scholar]
- Goldberg TE, Egan MF, Gscheidle T, Coppola R, Weickert T, Kolachana BS, Goldman D, Weinberger DR. Executive subprocesses in working memory: relationship to catechol-O-methyltransferase Val158Met genotype and schizophrenia. Arch Gen Psychiatry. 2003;60:889–896. doi: 10.1001/archpsyc.60.9.889. [DOI] [PubMed] [Google Scholar]
- Golden C. The measurement of creativity by the Stroop Color and Word Test. J Personal Assess. 1975;39:502–506. doi: 10.1207/s15327752jpa3905_9. [DOI] [PubMed] [Google Scholar]
- Grady CL. Cognitive neuroscience of aging. Ann N Y Acad Sci. 2008;1124:127–144. doi: 10.1196/annals.1440.009. [DOI] [PubMed] [Google Scholar]
- Green P. Story Recall Test. Edmonton (CA): Green's Publishing; 2005. [Google Scholar]
- Ho BC, Wassink TH, O'Leary DS, Sheffield VC, Andreasen NC. Catechol-O-methyl transferase Val158Met gene polymorphism in schizophrenia: working memory, frontal lobe MRI morphology and frontal cerebral blood flow. Mol Psychiatry. 2005;10(229):287–298. doi: 10.1038/sj.mp.4001616. [DOI] [PubMed] [Google Scholar]
- Hong J, Shu-Leong H, Tao X, Lap-Ping Y. Distribution of catechol-O-methyltransferase expression in human central nervous system. Neuroreport. 1998;9:2861–2864. doi: 10.1097/00001756-199808240-00033. [DOI] [PubMed] [Google Scholar]
- Johnson MK, Hashtroudi S, Lindsay DS. Source monitoring. Psychol Bull. 1993;114:3–28. doi: 10.1037/0033-2909.114.1.3. [DOI] [PubMed] [Google Scholar]
- Lazar NA, Luna B, Sweeney JA, Eddy WF. Combining brains: a survey of methods for statistical pooling of information. Neuroimage. 2002;16:538–550. doi: 10.1006/nimg.2002.1107. [DOI] [PubMed] [Google Scholar]
- Lewandowski KE. Relationship of catechol-O-methyltransferase to schizophrenia and its correlates: evidence for associations and complex interactions. Harvard Rev Psychiatry. 2007;15:233–244. doi: 10.1080/10673220701650409. [DOI] [PubMed] [Google Scholar]
- Li S, Cullen WK, Anwyl R, Rowan MJ. Dopamine-dependent facilitation of LTP induction in hippocampal CA1 by exposure to spatial novelty. Nat Neurosci. 2003;6:526–531. doi: 10.1038/nn1049. [DOI] [PubMed] [Google Scholar]
- Lisman JE, Grace AA. The hippocampal-VTA loop: controlling the entry of information into long-term memory. Neuron. 2005;46:703–713. doi: 10.1016/j.neuron.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melen K, Julkunen I, Taskinen J. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry. 1995;34:4202–4210. doi: 10.1021/bi00013a008. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Weickert CS, Akil M, Lipska BK, Hyde TM, Herman MM, Kleinman JE, Weinberger DR. Catechol O-methyltransferase mRNA expression in human and rat brain: evidence for a role in cortical neuronal function. Neuroscience. 2003;116:127–137. doi: 10.1016/s0306-4522(02)00556-0. [DOI] [PubMed] [Google Scholar]
- Minear M, Park DC. A lifespan database of adult facial stimuli. Behav Res Methods Instrum Comput. 2004;36:630–633. doi: 10.3758/bf03206543. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Johnson MK, Raye CL, D'Esposito M. fMRI evidence of age-related hippocampal dysfunction in feature binding in working memory. Brain Res Cogn Brain Res. 2000;10:197–206. doi: 10.1016/s0926-6410(00)00029-x. [DOI] [PubMed] [Google Scholar]
- Moscovitch M. Memory and working-with-memory: a component process model based on modules and central systems. J Cogn Neurosci. 1992;4:257–267. doi: 10.1162/jocn.1992.4.3.257. [DOI] [PubMed] [Google Scholar]
- Paller KA, Wagner AD. Observing the transformation of experience into memory. Trends Cogn Sci. 2002;6:93–102. doi: 10.1016/s1364-6613(00)01845-3. [DOI] [PubMed] [Google Scholar]
- Palmatier MA, Kang AM, Kidd KK. Global variation in the frequencies of functionally different catechol-O-methyltransferase alleles. Biol Psychiatry. 1999;46:557–567. doi: 10.1016/s0006-3223(99)00098-0. [DOI] [PubMed] [Google Scholar]
- Peleg-Raibstein D, Pezze MA, Ferger B, Zhang WN, Murphy CA, Feldon J, Bast T. Activation of dopaminergic neurotransmission in the medial prefrontal cortex by N-methyl-d-aspartate stimulation of the ventral hippocampus in rats. Neuroscience. 2005;132:219–232. doi: 10.1016/j.neuroscience.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Prince SE, Daselaar SM, Cabeza R. Neural correlates of relational memory: successful encoding and retrieval of semantic and perceptual associations. J Neurosci. 2005;25:1203–1210. doi: 10.1523/JNEUROSCI.2540-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath C, Heller A, Cohen MX, Brozinsky CJ, Rissman J. Functional connectivity with the hippocampus during successful memory formation. Hippocampus. 2005;15:997–1005. doi: 10.1002/hipo.20141. [DOI] [PubMed] [Google Scholar]
- Rissman J, Gazzaley A, D'Esposito M. Measuring functional connectivity during distinct stages of a cognitive task. Neuroimage. 2004;23:752–763. doi: 10.1016/j.neuroimage.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Rosen WG. Verbal fluency in aging and dementia. J Clin Neuropsychol. 1980;2:135–146. [Google Scholar]
- Savitz J, Solms M, Ramesar R. The molecular genetics of cognition: dopamine, COMT and BDNF. Genes Brain Behav. 2006;5:311–328. doi: 10.1111/j.1601-183X.2005.00163.x. [DOI] [PubMed] [Google Scholar]
- Sawaguchi T, Matsumura M, Kubota K. Catecholaminergic effects on neuronal activity related to a delayed response task in monkey prefrontal cortex. J Neurophysiol. 1990;63:1385–1400. doi: 10.1152/jn.1990.63.6.1385. [DOI] [PubMed] [Google Scholar]
- Schott BH, Seidenbecher CI, Fenker DB, Lauer CJ, Bunzeck N, Bernstein HG, Tischmeyer W, Gundelfinger ED, Heinze HJ, Duzel E. The dopaminergic midbrain participates in human episodic memory formation: evidence from genetic imaging. J Neurosci. 2006;26:1407–1417. doi: 10.1523/JNEUROSCI.3463-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamans JK, Yang CR. The principal features and mechanisms of dopamine modulation in the prefrontal cortex. Prog Neurobiol. 2004;74:1–58. doi: 10.1016/j.pneurobio.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Servan-Schreiber D, Printz H, Cohen JD. A network model of catecholamine effects: gain, signal-to-noise ratio, and behavior. Science. 1990;249:892–895. doi: 10.1126/science.2392679. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Bates JF, Cocchiarella AJ, Schacter DL, Rosen BR, Albert MS. Encoding novel face-name associations: a functional MRI study. Hum Brain Mapp. 2001;14:129–139. doi: 10.1002/hbm.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StatCorp. 2005. Stata Statistical Software: Release 9. College Station, TX: StataCorp LP. [Google Scholar]
- Stefanis NC, Van Os J, Avramopoulos D, Smyrnis N, Evdokimidis I, Hantoumi I, Stefanis CN. Variation in catechol-o-methyltransferase val158 met genotype associated with schizotypy but not cognition: a population study in 543 young men. Biol Psychiatry. 2004;56:510–515. doi: 10.1016/j.biopsych.2004.06.038. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kato M, Hayashi M, Okubo Y, Takano A, Ito H, Suhara T. Memory and frontal lobe functions; possible relations with dopamine D2 receptors in the hippocampus. Neuroimage. 2007;34:1643–1649. doi: 10.1016/j.neuroimage.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. Stuttgart, Germany: Georg Thieme; 1988. [Google Scholar]
- Tsai SJ, Yu YW, Chen TJ, Chen JY, Liou YJ, Chen MC, Hong CJ. Association study of a functional catechol-O-methyltransferase-gene polymorphism and cognitive function in healthy females. Neurosci Lett. 2003;338:123–126. doi: 10.1016/s0304-3940(02)01396-4. [DOI] [PubMed] [Google Scholar]
- Tulving E, Kapur S, Craik FI, Moscovitch M, Houle S. Hemispheric encoding/retrieval asymmetry in episodic memory: positron emission tomography findings. Proc Natl Acad Sci U S A. 1994;91:2016–2020. doi: 10.1073/pnas.91.6.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunbridge EM, Harrison PJ, Weinberger DR. Catechol-o-methyltransferase, cognition, and psychosis: Val158Met and beyond. Biol Psychiatry. 2006;60:141–151. doi: 10.1016/j.biopsych.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Adult Intelligence Scales. 3rd edn. San Antonio (TX): The Psychological Corporation; 1997. [Google Scholar]
- Weiss T, Veh RW, Heinemann U. Dopamine depresses cholinergic oscillatory network activity in rat hippocampus. Eur J Neurosci. 2003;18:2573–2580. doi: 10.1046/j.1460-9568.2003.02970.x. [DOI] [PubMed] [Google Scholar]
- Winterer G, Musso F, Vucurevic G, Stoeter P, Konrad A, Seker B, Gallinat J, Dahmen N, Weinberger DR. COMT genotype predicts BOLD signal and noise characteristics in prefrontal circuits. Neuroimage. 2006;32:1722–1732. doi: 10.1016/j.neuroimage.2006.05.058. [DOI] [PubMed] [Google Scholar]
- Yang CR, Mogenson GJ. Dopaminergic modulation of cholinergic responses in rat medial prefrontal cortex: an electrophysiological study. Brain Res. 1990;524:271–281. doi: 10.1016/0006-8993(90)90701-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.