Abstract
We report a technique to evaluate the same tumor microenvironment over multiple intravital imaging sessions in living mice. Individual tumor cells expressing photoswitchable proteins in an orthotopic mammary carcinoma are optically marked and followed for extended times through a Mammary Imaging Window (MIW). We demonstrate, for the first time, that two distinct microenvironments in the same orthotopic mammary tumor affect differently the invasion and intravasation of tumor cells.
Keywords: Tumor microenvironment, photoswitching, photoactivatable fluorescent proteins, migration, invasion, intravasation, intravital imaging, metastasis, mammary imaging window
The early steps of metastasis are characterized by tumor cells invading the stroma (invasion) and entering the blood (intravasation)1,2. Short term tracking of individual cells inside fluorescent tumors by intravital imaging has revealed dramatic heterogeneity in tumor cell invasion and intravasation3. However, long term tracking of individual cells is required to quantify these behaviors and to determine the fates of cells in specific tumor microenvironments. Imaging techniques that rely on surgical dissection to expose the imaging site have limitations for long-term experiments such (1) tissue dehydration, impaired thermoregulatory control and/or animal survival upon surgical dissection, (2) possible effects of prolonged anesthesia exposure, and (3) a limited field of view. These limitations can be overcome by studying tumors through a dorsal skinfold chamber4. The use of dorsal skinfold chambers, however, limits the experiments to tumor-models based on cell lines, and for many tumors a non-orthotopic environment. For example, invasion and intravasation of breast tumor cells is highly dependent on the specific local microenvironment5 which may not exist in the non-mammary environments such as the dorsal skinfold chamber site4. Here, we describe the integration of a Mammary Imaging Window (MIW) with photoswitchable fluorescent protein labeling, which enabled tracking of selected tumor cell subpopulations in different breast tumor microenvironments. Using this technique, we were able to monitor cell migration for greater than 24 hours.
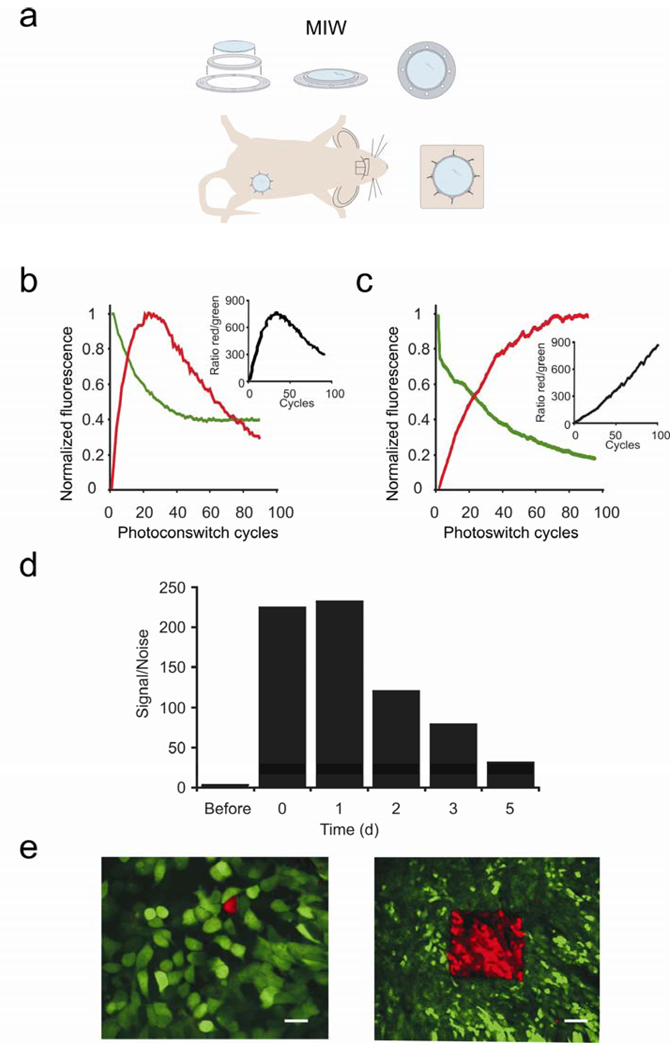
To image orthotopic (in the natural, mammary gland environment) breast tumors intravitally at high resolution for prolonged times, we developed a MIW that can be placed on top of the mammary gland of a mouse (Fig. 1a). The protocol for these animal studies was approved by the Institutional Animal Care and Use Committee for the Albert Einstein College of Medicine. The MIW consists of two plastic rings which form a mount for a glass coverslip. The mount has holes which facilitate suturing into the skin, whereas the glass coverslip assures the optimal working distance and refraction index for high resolution imaging (for more details on the equipment used for imaging, see Supplementary Fig. 1 online). While surgical dissection of the skin overlaying the imaging site allows for several hours of data to be collected and is usually a terminal procedure, imaging through the MIW extended the imaging time to multiple days (up to 21 days). Tumors with MIW implants did not show inflammation, or a change in growth and microenvironments scored at 1–9 days after the implantation procedure (Supplementary Fig. 2 online). In order to locate the same subpopulation of cells in each of the imaging sessions, reference points were required6. In the fast changing tissue topology of the tumor, the use of fixed reference points is limited, and therefore we used photoswitchable fluorescent proteins7, 8 as photomarkers of the cells of interest. These proteins represent a new group of GFP-like fluorophores which allow labeling and tracking of a single cell or a group of cells9, 10, 11, 12. We stably expressed the photoswitchable protein Dendra2 in the metastatic breast cancer line MTLn3. Dendra2 resembles GFP in its spectrum prior to photoswitching, but exposure to blue light (e.g. 405 nm) can induce an irreversible red shift >150 nm in the excitation and emission spectra of the chromophore13. Following the photoswitch, the red fluorescence stably increases up to 250 fold both in vitro and (Fig. 1b) and in vivo (Fig. 1c), resulting in red/green contrast of up to 850 and allowing us to track cells marked in this way. Five days after photoswitching, the red fluorescence of the photoswitched cells is still 31 fold higher than the red fluorescence of non-switched cells, which enables us to recognize the highlighted cells in vivo for extended times after the photoswitch (Fig. 1d).
Figure 1.
The Mammary Imaging Window (MIW) allows for long-term, high resolution imaging of the orthotopic tumors. (a) Components and the assembly of the MIW: a coverslip is mounted on a plastic frame consisting of two plastic rings and surgically implanted on top of the mammary gland or mammary tumor. (b,c) Average increase in red and decrease in green signal for Dendra2, as measured in a region of interest, in cells in vitro (b) or in vivo (c) upon photoswitching. The values were normalized to the highest fluorescent level in red and the initial fluorescent level in green. The insets show the ratio of the non-normalized red and green fluorescence. (d) Cells within Dendra2-tumors were photoswitched through the MIW and the red fluorescence was quantified before, immediately after (0 days) photoswitching and for the 5 subsequent days. The values were normalized to the red fluorescence level before photoswitching. (e) Photoswitching of Dendra2-expressing MTLn3 tumor cells in vivo can be done in regions of interest ranging from one cell (left panel, scale bar 10 µm) to hundreds of cells (right panel, scale bar 75 µm) through the MIW. Shown are combined images from the green and red channels using an OR-function: Only the pixels in the red channel that are above background are shown, and for all other pixels, the green channel is shown.
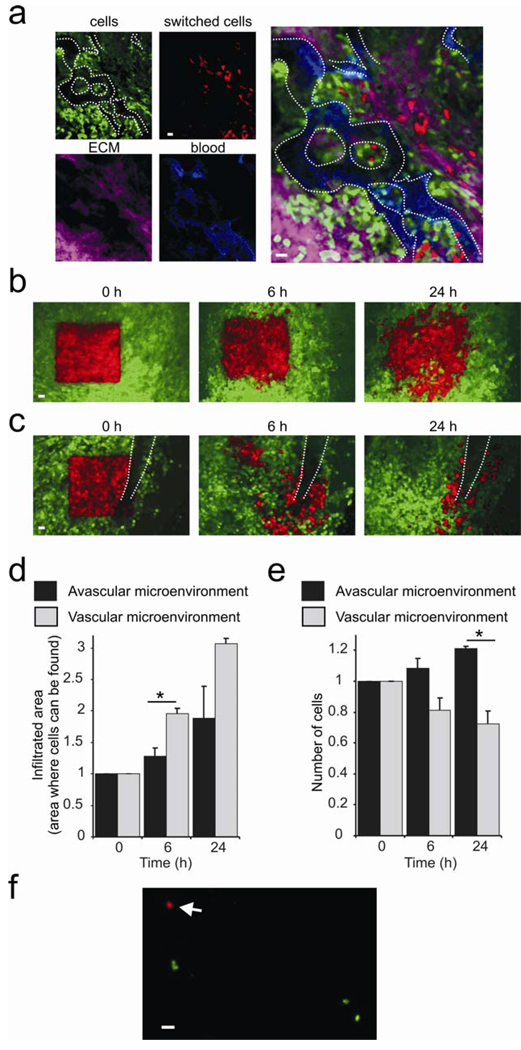
Regions containing one to hundreds of cells can be photoswitched and imaged through the MIW (Fig. 1e, Supplementary Movies 1 and 2 online, Supplementary Fig. 3 online). As cells in the tumor migrate and invade, the distribution of these cells relative to blood vessels and other tumor cells changed over time. By selectively photoswitching the fluorophore in a group of cells as demonstrated in Fig. 1e, the changes in distribution of cells in the tumor microenvironment was visualized. Twenty four hours after photo-switching, we recorded images of the non-photoswitched tumor cells (green), photoswitched cells (red), extracellular matrix (collagen was visualized with reflectance in Fig. 2a, or second-harmonic-generation in Supplementary Movie 3 online) and blood vessels (fluorescent dextrans were used for vessel labeling, see Supplementary Movie 4 online). Interestingly, some photoswitched regions showed dramatic migration and invasion of the surrounding microenvironment (Fig. 2a).
Figure 2.
Photoswitching through the MIW is a tool for studying orthotopic tumor microenvironments. (a) By using Dendra2 as the label for tumor cells (red and green), Texas Red dextran for blood vessels (blue) and reflectance (purple) for extracellular matrix (ECM), one can define vascular microenvironments and monitor chosen cells inside them. (b,c) Non-photoswitched cells (green) and photoswitched cells (red) are shown at 0h, 6h and 24h after the photoswitch in avascular (b) and vascular (c) microenvironments (visible vessel indicated by white dotted lines). Shown are combined images from the green and red channels using an OR-function: Only the pixels in the red channel that are above background are shown, and for all other pixels, the green channel is shown. Scale bars 30 µm. The relative infiltration areas (d) and numbers (e) of photoswitched cells over time in vascular and avascular microenvironments. Error bars represent s.e.m., asterisk represents P < 0.05, and n > 20. (f) Detection of photoswitched cells in the lung. Lungs of an animal which had a large vascular area (20–40 mm2) of the primary tumor photoswitched were examined ex vivo 24h after photoswitching in green and red channels by epifluorescence microscopy. Arrow points to a red tumor cell photoswitched in and disseminated from the primary tumor. We determined 0.009 +/− 0.007 (s.e.m.) red cells and 1.4 +/− 0.33 green cells per mm2 lung, resulting in a green/red ratio of 152 +/−0.81. See the last section of the supplementary materials for more details. Scale bar 20 µm.
The tumor perivascular microenvironment (tumor tissue surrounding blood vessels) is enriched in tumor-associated-macrophages and extracellular matrix, which supports metastatic behavior, including inhibition of proliferation and stimulation of migration, invasion and intravasation5,14. This suggests the existence of distinct mammary tumor microenvironments within the same tumor with different rates of invasion and intravasation. However, the long term implications of these observations required the ability to revisit distinct microenvironments inside the same mammary gland over a 24- hour period. In order to quantify invasion and intravasation within distinct mammary gland microenvironments, we photoswitched square regions (~300 cells) in different tumor microenvironments of the same orthotopically grown tumor (Supplementary Fig. 3 online), focusing on regions lacking (Fig. 2b) and containing (Fig. 2c) detectable blood vessels. The location of the photoswitched red cells was determined by acquiring Z-stacks of red and green images of the same regions at 0, 6 and 24 hours after photoswitching. In regions not containing detectable vessels (Fig. 2b) there was limited migration (Fig. 2d) and the number of cells increased (Fig. 2e), suggesting that this microenvironment does not support metastatic behavior. In contrast, cells in the vascular microenvironment (Fig. 2c) have infiltrated larger areas (Fig. 2d) and even migrated to sites outside of the field of view. Moreover, cells in this vascular microenvironment lined up along the blood vessel (Fig. 2c), with a concomitant decrease in the number of red tumor cells (Fig. 2e), and the appearance of red tumor cells in the lung (Fig. 2f). From these experiments, we conclude that cell behavior is determined by the surrounding microenvironment, and that the vascular microenvironment promotes invasion and intravasation of tumor cells.
While the existence of different tumor microenvironments has been reported previously 5, the quantitative analysis of such microenvironments in the same tumor with spatial and temporal resolution, is not possible with previous techniques. The combination of photoswitchable proteins with the MIW allowed for such analysis, since a distinct group of cells can be photomarked in any location in the primary tumor, and tracked over time without long term anesthesia. Furthermore, the high stability of Dendra2 enabled us to freeze fix the tissues and analyze them by microscopy without additional labeling. A limitation of Dendra2 is that a limited number of excitation wavelengths can be used. For example, since violet light causes a switch, DAPI stains should only be used after imaging other wavelengths as DAPI imaging would cause all green cells to then become red. Nevertheless, through the MIW, the formation of tumors from injection of cells in the mammary gland can be followed for days, which is not possible in surgically dissected areas. This also opens the possibility to study fluorescently-tagged proteins that have lethal effects if stably expressed, but can be studied in transiently transfected cells. For example, transiently transfected cells expressing membrane targeted GFP and injected into the mammary gland can be imaged with high resolution through the MIW (Supplementary Fig. 4a online). Although this would also be possible with the dorsal skinfold chamber4, imaging through the MIW allows studies of cell behavior in their physiological breast microenvironments and moreover, the MIW technology can be extended to tumors of transgenic origin, such as the MMTV-PyMT tumor model (Supplementary Fig. 4b online). Such tumor models allow the investigation of different stages of tumor progression15 in contrast to cell-line derived xenografts. The combined use of MIW and photomarking cells to revisit chosen subpopulations of cells is an important capability not only in tumor studies but any studies related to cell motility and morphogenesis. Visualization of infectious agents and immune responses, or the progression of chronic inflammation, are other examples of the potential applications of the technique described in this manuscript. This technique will also be helpful in monitoring of identification and proliferation of mammary stem cells, mammary gland growth and morphogenesis or testing artificial tissue heterotransplants.
Supplementary Material
Acknowledgements
This work was supported by DOD BC061403 (DK), NIH U54GM064346 (JvR), NIH CA100324 (JC, JS, JW), NIH U54CA126511 (JC, BG), NIH GM070358 and NIH GM073913 (VV). The authors thank the staff of AIF and D. Entenberg for help with microscopy, IHC facility for help with histology, M. Rottenkolber for help in fabrication of imaging box, J. Pollard (Albert Einstein College of Medicine) for the F4/80 antibody, S. Garofalo for technical assistance, and members of the Condeelis, Segall, Cox and Verkhusha labs for discussions.
Footnotes
Author contributions:
DK, BG, and JvR, JES, JC contributed equally to this manuscript, and they conceived of and designed the study, performed the imaging, and analysis and drafted the manuscript. JW advised and trained in intravital imaging, VVV provided materials and advice.
Contributor Information
John Condeelis, Email: condeeli@aecom.yu.edu.
Jeffrey E. Segall, Email: segall@aecom.yu.edu.
Jacco van Rheenen, Email: j.vanrheenen@niob.knaw.nl.
References
- 1.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer. 2003;3(12):921. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 2.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Sidani M, et al. Probing the microenvironment of mammary tumors using multiphoton microscopy. J. Mammary Gland Biol. Neoplasia. 2006;11(2):151. doi: 10.1007/s10911-006-9021-5. [DOI] [PubMed] [Google Scholar]
- 4.Lehr HA, et al. Dorsal skinfold chamber technique for intravital microscopy in nude mice. The American journal of pathology. 1993;143(4):1055. [PMC free article] [PubMed] [Google Scholar]
- 5.Wyckoff JB, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer research. 2007;67(6):2649. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 6.Bins AD, et al. Intravital imaging of fluorescent markers and FRET probes by DNA tattooing. BMC biotechnology. 2007;7:2. doi: 10.1186/1472-6750-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24(4):461. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 8.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: Photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol. 2005;6(11):885. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 9.Gray NW, Weimer RM, Bureau I, Svoboda K. Rapid Redistribution of Synaptic PSD-95 in the Neocortex In Vivo. PloS Biol. 2006;4(11):e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44(3):136. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- 11.Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc. 2006;1(2):960. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- 12.Post JN, Lidke KA, Rieger B, Arndt-Jovin DJ. One- and two-photon photoactivation of a paGFP-fusion protein in live Drosophila embryos. FEBS Lett. 2005;579(2):325. doi: 10.1016/j.febslet.2004.11.092. [DOI] [PubMed] [Google Scholar]
- 13.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nature protocols. 2007;2(8):2024. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 14.Condeelis John, Pollard Jeffrey W. Macrophages: Obligate Partners for Tumor Cell Migration, Invasion, and Metastasis. Cell. 2006;124(2):263. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. The American journal of pathology. 2003;163(5):2113. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.