Abstract
The inflammasome is a multi-protein complex that mediates activation of caspase-1 which promotes the secretion of the proinflammatory cytokines IL-1β and IL-18 as well as pyroptosis, a form of cell death induced by bacterial pathogens. Members of the Nod-like receptor family including NLRP1, NLRP3 and NLRC4 and the adaptor ASC are critical components of the inflammasome by linking microbial and endogenous danger signals to caspase-1 activation. Several diseases are associated with the dysregulated activation of caspase-1 and IL-1β secretion. Thus, understanding of inflammasome pathways may provide insights into disease pathogenesis that might serve as potential targets for therapeutic intervention.
The eradication of invading microorganisms is essential for the survival of multicellular organisms including humans. To ensure the removal of harmful pathogens, eukaryotic hosts have evolved an arsenal of defense mechanisms to sense and destroy invading microbes. The innate immune system is responsible for the initial task of recognizing and eradicating potentially dangerous microorganisms. Unlike the adaptive immune system that relies on a diverse and specific repertoire of clonally selected lymphocytes, innate immune cells display broad anti-microbial functions which are activated rapidly upon encountering microbes1. A critical property of the innate immune system is its ability to discriminate microbes from “self” through the recognition of conserved microbial structures called pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharides (LPS), peptidoglycan (PGN), flagellin and microbial nucleic acids2. The sensing of PAMPSs is mediated by germ-line encoded innate immune receptors which include Toll-like receptors (TLRs) and Nod-like receptors (NLRs). Unlike membrane-bound TLRs that sense PAMPs on the cell surface or within endosomes, NLRs recognize microbial molecules in the host cytosol3, 4. Upon microbial recognition, both TLRs and NLRs induce the activation of host signaling pathways which lead to innate and adaptive immune responses1, 5.
The NLR family is composed of 23 family members in humans whereas the mouse genome contains at least 34 NLR genes6. Homologs of NLRs are present in plants (R genes) and animals including phylogenetically more primitive organisms such as the zebrafish and the sea urchin, although they are not found in insects or worms7, 8. Most NLRs exhibit a tripartite structure consisting of a variable N-terminal domain, a centrally located nucleotide-binding oligomerization (NOD) domain that mediates self-oligomerization and a C-terminal leucine-rich repeat that detects PAMPs. The N-terminal domain includes several protein interactions modules such as caspase recruitment domain (CARD), pyrin domain (PYD), or baculovirus inhibitor repeat (BIR) which are critical for downstream signaling through the recruitment of adaptors or effector molecules8. NOD1 and NOD2 were the first identified NLRs9. Both NOD1 and NOD2 sense bacterial molecules produced during the synthesis, degradation, and remodeling of PGN, a major component of bacterial cell walls. NOD2 detects muramyl dipeptide (MDP), which is found in nearly all Gram-positive and Gram-negative organisms whereas NOD1 recognizes γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP)-containing PGN fragments from most Gram-negative bacteria and certain Gram-positive bacteria10. In response to the their microbial agonists, NOD1 and NOD2 induce the activation of NF-κB and MAPKs which drive the transcription of numerous genes involved in both innate and adaptive immune responses11–13. In contrast, several NLRs including NLRC4 (also called IPAF), NLRP1, NLRP3 (also called Cryopyrin or NALP3) and NAIPs are involved in the assembly of a multi-protein platform, named the inflammasome, that is responsible for the activation of caspase-114. In this review, we will discuss recent studies on the inflammasome including its regulation by NLRs and its role in host defense and inflammatory disease.
The inflammasomes
Caspases are a family of intracellular cysteine proteases that cleave a limited number of substrates after aspartic acid residues. The function of caspases has been extensively studied in apoptosis where they play an essential role in the activation and implementation of cellular demise15. However, several caspases including the human caspase-1, -4, and -5 and the murine caspase-1, -11 and -12 are involved in the processing and secretion of pro-inflammatory molecules and are often refereed as “pro-inflammatory caspases”14 15. Caspase-1, the first caspase to be identified, is present in the cytosol of phagocytic cells as an inactive zymogen16, 17. Upon stimulation by a myriad of microbial and endogenous signals, the dormant procaspase-1 zymogen is self activated by proteolytic cleavage into the enzymatically active heterodimer composed of two 10- and 20-kDa subunits18. Active caspase-1 is essential for the cleavage of pro-IL-1β and pro-IL-18 into their mature, biologically active forms. Mature IL-1β is implicated in multiple immune reactions including the recruitment of inflammatory cells to the sites of infection; whereas IL-18 is important for the production of interferon-γ and enhancement of the cytolytic activity of NK cells19. In addition, caspase-1 cleaves other cellular protein substrates including pro-caspase-7 and mediates unconventional protein secretion as well as membrane repair and cell survival in response to bacterial pore forming toxins through mechanisms that remain largely undefined20–22.
A critical step in caspase-1 activation is the assembly of large macromolecular complex through CARD-CARD and PYD-PYD protein-protein interactions to form a scaffold for procaspase-1 recruitment and activation. This molecular platform for caspase-1 activation which includes NLR family members and the adaptor ASC has been termed the “inflammasome” as an analogy to the “apoptosome” which drives caspase-9 activation via Apaf-1 during apoptosis23. Based on the mechanism of caspase-9 activation induced by Apaf-1, it is postulated that NLRs drive caspase-1 activation through their oligomerization and induced proximity of caspase-1 molecules8, 23. The inflammasome was initially described using extracts from human THP-1 monocytic cells and found to contain NLRP1, caspase-1, caspase-5, and the adaptor proteins ASC and CARDINAL23. This initial inflammasome was induced by temperature shift using low K+ buffer23, but whether NLR-specific inflammasome formation occurs in response to physiological stimuli such as bacterial infection remains to be determined.
Three inflammasomes named after the NLR involved, that is, NLRP1, NLRP3, and NLRC4 have been partially characterized. Common to these inflammasomes is the role of ASC as the adaptor protein that links these NLRs to caspase-124. In humans, Pyrin-containing NLRs such as NLRP3 and NLRP1 associate with caspase-1 through the adaptor molecule ASC23, 25–27. In contrast, NLRC4 is thought to directly associate with caspase-1 via CARD-CARD interactions. However, genetic studies show conclusively that ASC is required for NLRC4-dependent caspase-1 activation28–30. These results suggest that ASC is somehow required for the interaction between NLRC4 and caspase-1 or that ASC mediates another critical step which is important for inflammasome activation. The NLRP1 inflammasome is the only caspase-1-activating platform that has been reconstituted in vitro with purified proteins31. The existence of the NLRP3- and NLRC4-inflammasome has been largely defined by the ability of both NLRP3 and NLRC4 to activate caspase-1 in an ASC-dependent manner in response to specific stimuli28–30, 32–35. There is evidence that NLRP2 associates with procaspase-1 and promotes IL-1β production, but whether this is a bonafide inflammasome that functions to activate caspase-1 in response to specific microbial stimuli remains to be determined36. Similarly, mouse Naip5 can act in concert with Nlrc4 to promote the activation of caspase-1 in response to flagellin from Legionella pneumophila37, 38. However, the mechanism by which Naip5 acts to control the Nlrc4 inflammasome in response to L. pneumophila infection remains poorly defined.
NLRC4 inflammasome
Several Gram-negative bacteria, including Salmonella typhimurium, L. pneumophila, Pseudomonas aeruginosa, and Shigella flexneri induce caspase- 1 activation and pyroptosis via the NLRC4 inflammasome (Fig. 1). The activation of the NLRC4 inflammasome requires a functional type III secretion system (T3SS) for S. typhimurium28, 29, S. flexneri39, and P. aeruginosa40, 41 or type IV secretion system (T4SS) in the case of L. pneumophila35, 37. These bacterial secretion systems can form pores in host membranes and mediate translocation of virulence factors (effectors proteins) into the host cell cytosol42. Remarkably, S. typhimurium, L. pneumophila and P. aeruginosa appear to induce the activation of caspase-1 through the cytosolic delivery of flagellin which triggers the activation of the NLRC4 inflammasome. The activation of caspase-1 through the NLRC4 inflammasome can be recapitulated by the delivery of purified flagellin into the host cytosol with cationic liposomes or expression systems28, 29, 38. Thus, small amounts of flagellin leaked via the T3SS or T4SS into the host cytosol may trigger NLRC4 activation during bacterial infection. In agreement with this observation, the T3SS of S. typhimurium promotes the translocation of flagellin into the macrophage cytosol43. However, it is also possible that the T3SS creates an organelle that permits flagellin transport into the host cytosol, perhaps by an endogenous host peptide transport apparatus.
Figure 1.
The NLRC4 inflammasome. Infection of macrophages with several Gram-negative bacteria including Salmonella, Legionella and Pseudomonas activates caspase-1 through NLRC4 and ASC. A critical step is the cytosolic delivery of flagellin via bacterial T3SS or T4SS. Shigella activates the NLRC4 inflammasome independent of flagellin through an unknown microbial molecule. Activation of caspase-1 via NLRC4 leads to processing and secretion of IL-1β and IL-18 as well as other activities.
Caspase-1 activation via NLRC4 is TLR5 independent, in that it proceeds unabated in TLR5 deficient macrophages28, 29, 35. These results indicate that flagellin is recognized by at least two different sensors; TLR5 senses extracellular flagellin whereas NLRC4 recognizes cytosolic flagellin. Recent evidence indicates that the NLR protein Naip5 is also important for the recognition of the C-terminal portion of flagellin from L. pneumophila and contributes to the activation of the NLRC4 inflammasome38. It was proposed that Naip5 physically associates with NLRC4 to form an inflammasome complex in response to L. pneumophila37. In contrast, caspase-1 activation induced by S. typhimurium and P. aeruginosa infection which is also triggered by cytosolic flagellin is Naip5 independent38. Because Naip5 polymorphisms can influence the susceptibility to L. pneumophila independently of caspase-1 activation44, it will be important to investigate whether Naip5 has an additional role independently of NLRC4 in restricting the growth of L. pneumophila in macrophages. There is also evidence for a flagellin-independent pathway that activates the NLRC4 inflammasome after infection with certain aflagellated bacteria. For example, S. flexneri induces caspase-1 activation and IL-1β secretion in a NLRC4 and ASC-dependent manner39. Similarly, Mycobacterium tuberculosis, another aflagellated bacterium, activates caspase-1 via NLRC445. As a strategy to avoid host defenses, M. tuberculosis inhibits inflammasome activation by producing zmp1, a Zn2+ metalloprotease, by a mechanism that remains poorly understood45. As multiple PAMPs can be recognized by the same TLR46 it is likely that other microbial molecules distinct from flagellin can also induce the activation of NLRC4.
Recently, it was also reported that P. aeruginosa may activate caspase-1 through a NLR4-dependent, but flagellin-independent pathway41. Consistently, infection of macrophages with high number of P. aeruginosa or S. typhimurium mutants deficient in flagellin can induce weak, but reproducible activation of caspase-1 through NLRC4 (28 and L.F. unpublished observations). These results suggest the existence of a minor alternative pathway that leads to caspase-1 activation in macrophages infected with flagellated bacteria which can be elicited under certain experimental conditions. However, the importance of the different inflammasome pathways elicited by P. aeruginosa and S. typhimurium in vivo remains to be determined.
There is evidence that activation of the NLRC4 inflammasome functions as a host defense strategy against pathogenic infections. In the case of S. typhymurium, P. aeruginosa, and S. flexneri, NLRC4-dependent activation of caspase-1 is accompanied by the secretion of IL-1β and the induction of a specific form of cell death called pyroptosis. Surprisingly, while both NLRC4 and the adaptor molecule ASC are required for caspase-1 activation and IL-1β secretion, NLRC4, but not ASC, is critical for the induction of pyroptosis30, 40. Thus, the cellular activities of NLRC4 and ASC can be dissociated. In agreement with this, NLRC4 can inhibit autophagy independently of ASC39. A possible explanation for these results is that ASC mediates cell survival thorough NF-κB activation independently of NLRC447.
The NLRC4 inflammasome is also involved in restricting the replication of L. pneumophila, an intracellular pathogen that causes Legionnaire's disease, a pneumonia that can be deadly especially in older individuals. A key event in disease pathogenesis is the replication of L. pneumophila inside host macrophages which requires the formation of a specialized vacuole that blocks the fusion of the phagosome containing the bacterium to the lysosomes48. L. pneumophila induces NLRC4-dependent caspase-1 activation via a T4SS and cytosolic flagellin35, 37. The recognition of flagellin through the Naip5 and NLRC4 inflammasome is critical for phagosome maturation and restriction of L. pneumophila replication inside macrophages35. Thus, NLRC4-dependent caspase-1 activation promotes the fusion of the L. pneumophila-containing phagosome to the lysosome for bacterial degradation35. Importantly, mice deficient in Nlrc4 exhibit an increased bacterial burden after pulmonary infection with L. pneumophila35, indicating that the inflammasome is important for disease pathogenesis. However, the mechanism by which caspase-1 activation controls phagosome maturation in response to L. pneumophila infection is presently unknown. A possibility is that caspase-1 targets substrates involved in phagosome maturation or L. pneumophila virulence factors required for lysosomal evasion.
NLRP1 inflammasome
The human NLRP1 inflammasome was the first caspase-1-activating platform to be identified23. Studies with purified NLRP1, ASC and caspase-1 revealed that NLRP1 oligomerizes with caspase-1 in the presence of MDP31. These studies suggested that caspase-1 is activated via a two-step mechanism, whereby microbial MDP induces a conformational change in NLRP1, which in turn allows it to bind nucleotide and to oligomerize, thus creating a platform for caspase-1 activation31. Because there is no evidence that MDP binds NLRP1, the mechanism that triggers NLRP1 oligomerization remains unclear. Remarkably, the adaptor molecule ASC was not essential, although its addition augmented NLRP1-mediated caspase-1 activation in vitro31. Consistent with the human studies, ASC is not required for the activation of caspase-1 mediated by Nlrp1b in mouse macrophages49, suggesting that ASC acts as an adaptor only for a subset of inflammasomes. Unlike humans that posses a single NLRP1 gene, three Nlrp1 paralogs, named Nlrp1a, -b and –c, are present in the mouse genome. Genetic studies identified Nlrp1b as the gene responsible for the pathology induced by lethal toxin (LT), a toxin secreted by Bacillus anthracis50. LT is a dimeric protein complex consisting of Protective Antigen (PA), a pore forming toxin, and Lethal factor, a protease that is delivered by the PA into the cytosol of infected cells51. Subsequent studies revealed that susceptibility to LT-induced macrophage cell death, a critical event in disease pathogenesis, is mediated by Nlrp1b-dependent activation of caspase-150. An important role for the Nalp1b inflammasome in host defense is suggested by the observation that mice harboring the Nlrp1b “susceptible” allele are more susceptible to infection with Bacillus anthracis52. The mechanism by which the Lethal factor, a metalloprotease, triggers Nlrp1 inflammasome activation remains to be determined.
NLRP3 inflammasome
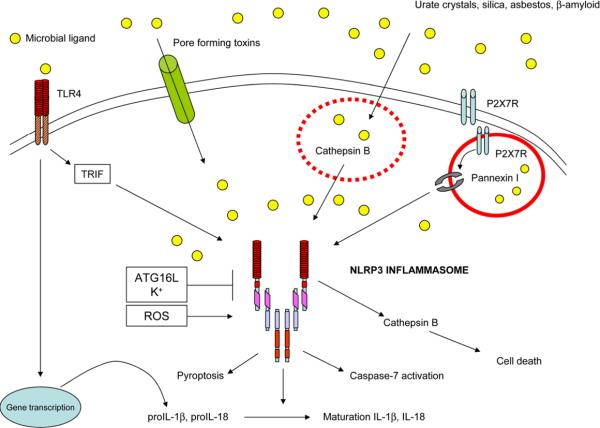
Initial studies identified NLRP3 as a critical component of the inflammasome that is spontaneously formed in cell extracts of human monocytes after hypotonic lysis23. Follow-up studies revealed that the NLRP3 inflammasome is activated by a plethora of microbial stimuli including LPS, MDP, bacterial RNA, the double-stranded RNA analog poly I:C as well as the imidazoquinoline antiviral compounds R837 and R84832, 53–57. In contrast, activation of caspase-1 by cytosolic double-stranded DNA is not mediated via NLRP3, but it involves the Pyrin domain-containing AIM2, a member of the hematopoietic interferon-inducible nuclear protein HIN-200 family, and ASC58. An important characteristic of the NLRP3 inflammasome is its activation by endogenous molecules such as urate crystals and ATP, bacterial pore-forming toxins and particulate matter including asbestos and silica14, 33, 34, 59. Significant insight into the activation of the NLRP3 inflammasome comes from the observation that microbial ligands such as LPS induce robust caspase-1 activation and IL-1β secretion after brief stimulation with high concentrations of ATP60. Extracellular ATP activates the purinergic ATP-gated P2X7 receptor (P2X7R), which acts as a cation channel to rapidly induce a complete collapse of normal ionic gradients, including the release of intracellular K+61. Recent evidence indicates that extracellular ATP also results in the opening of a pore mediated by pannexin-162–64 that induces the translocation of MDP from an intracellular vesicular compartment into the cytosol, where it induces the activation of caspase-1 via NLRP355 (Fig. 2). In contrast, the activation of the NLRP3 inflammasome triggered by cytosolic delivery of microbial molecules with pore-forming bacterial toxins or through the lipophilic DOTAP delivery system is pannexin-1 independent64. Thus, the pannexin-1 pore activated by ATP via the P2X7R may serve as a conduit to deliver microbial molecules to the host cytosol. Although MDP can mediate caspase-1 activation via NLRP355, 65, there is also evidence supporting a role for NOD2 and NLRP1 in caspase-1 activation induced by MDP and the LT of B. anthracis49. It was shown that NOD2 interacts with NLRP1 and suggested that the NOD2-NLRP1 complex mediates the activation of caspase-149. Further studies are needed to understand the discrepancies between these studies and the physiological relevance of the NOD2-NLRP1 complex in inflammasome activation.
Figure 2.
The NLRP3 inflammasome. Activation of caspase-1 through NLRP3 is induced by co-stimulation with microbial molecules such as LPS and the P2X7R, pore-forming molecules or particulate matter (e. g. silica, asbestos, urate crystals, and fibrilar β-amyloid). Stimulation of the P2X7R by extracellular ATP induces the activation of a cation channel that mediates K+ efflux, an event that has been linked to inflammasome activation. In addition, P2X7R activation promotes the opening of the pannexin-1 pore which may mediate cytosolic delivery of microbial molecules such as MDP. The mechanism of NLRP3 activation in the cytosol remains poor understood. There is evidence that it may involve, at least in part, lysosomal membrane destabilization and cathepsin B activation (in the case of silica, urate crystals and fibrilar β-amyloid). TLR stimulation also induces the activation of the NLRP3 inflammasome through TRIF when the autophagy machinery is compromised (e.g. Atg16L or Atg7 deficiency). ROS have been also implicated in the activation of the NLRP3 inflammasome.
Our knowledge about the activation of the NLRP3 inflammasome in response to microbial infection is limited. NLRP3 has been implicated in the activation of caspase-1 induced by viruses such as Sendai virus54, Influenza virus54, and certain Adenovirus strains used as vectors for gene therapy56 as well as bacteria such as L. monocytogenes and S. aureus34. Initial reports indicated that L. monocytogenes induces caspase-1 activation through NLRP3 inflammasome in a TLR2-independent manner66, suggesting a critical role of cytosolic recognition for caspase-1 activation. Accordingly L. monocytogenes-induced caspase-1 activation requires the bacterial pore-forming lysteriolysin O (LLO) that mediates the escape of the bacterium from the vacuole into the cytosol66, 67. Subsequent studies, however, could not confirm the essential role of NLRP3 in the activation of caspase-1 induced by Listeria infection68. The difference in results may be explained, at least in part, by differential expression of bacterial factors that contribute to caspase-1 activation under different experimental conditions. Consistent with this possibility, recent studies reported that L. monocytogenes induces the activation of caspase-1 via both the NLRC4 and NLRP3 inflammasomes69. The physiological relevance of these findings awaits studies that compare mice deficient in one or more inflammasomes with mice deficient in ASC or caspase-1.
Danger Signals, Crystals and the Inflammasome
Another stimulus that activates the NLRP3 inflammasome is monosodium urate crystals. Uric acid was identified by Rock and coworkers as an endogenous danger signal released by necrotic cells capable of activating adaptive immune responses70. Subsequently, it was shown that monosodium urate crystals and calcium pyrophosphate dehydrate crystals are potent activators of caspase-1 via the NLRP3 inflammasome71. A role for the inflammasome in inducing innate immune response was suggested by the finding that mice deficient in ASC exhibit impaired neutrophil recruitment after intraperitoneal injection of urate crystals71. Using a similar model of uric acid-induced peritonitis, it was shown that mice lacking the IL-1R in non-myeloid cells have decreased recruitment of neutrophils72. The urate crystal-induced pathway of caspase-1 activation appears to be important for triggering gouty inflammation. Consistently, patients with gouty attacks improved clinically when treated with IL-1Ra, a molecule that inhibits IL-1R signaling73. However, necrotic cells do not stimulate the recruitment of neutrophils via the NLRP3 inflammasome because caspase-1 deficient mice display unimpaired inflammatory responses to necrotic cells74. Further studies revealed that necrotic cells passively release IL-1α which mediates neutrophil recruitment in the peritoneal cavity via IL-1R and induction of CXCL174. These results suggest IL-1R signaling, but not the NLRP3 inflammasome, is involved in mediating acute inflammation in response to necrotic cells.
The molecular mechanism whereby urate crystals induces the activation of the NLRP3 inflammasome is not fully understood, but it may involve phagocytosis of the crystalline particles and the generation of reactive oxygen species (ROS)59. Another set of stimuli that induce the activation of the NLRP3 inflammasome is represented by silica and asbestos59, 75, 76, aluminiun hydroxide77–80 and fibrilar amyloid-β 81, a molecule implicated in the pathogenesis of Alzheimer disease. These observations are potentially important because inflammation is thought to contribute to the disease pathways elicited by these stimuli. The activation of the NALP3 inflammasome induced by some of these particulate stimuli appears to hinge, in part, on the destabilization of the lysosomal membrane and the activation of lysosomal proteases76 (Fig. 2). Intriguingly, it was shown that cathepsin B is important for the activation of caspase-1 induced by silica, while that induced by ATP was cathepsin B independent76. Conversely, NLRP3 has been implicated in the activation of a caspase-1-independent cell death pathway that involves cathepsin B82. Clearly, additional work is needed to understand the interplay between NLRP3 and cathepsin B in the regulation of innate immune responses.
Adjuvants, adaptive immunity and the inflammasome
Several groups have shown that aluminiun hydroxide (Alum), the most widely used adjuvant, activates caspase-1 and IL-1β via the NLRP383. Importantly, initial studies revealed a critical role for the NLRP3 inflammasome in mediating antigen-specific IgG1 adjuvant activity elicited by Alum77, 80. However, other authors did not observe alteration in antigen-specific IgG titers including IgG1 following intraperitoneal immunization in Nlrp3-null mice78, 79. The reason for this discrepancy is not clear, but it maybe explained, at least in part, by the different immunization protocols used. Collectively, these experiments suggest that NLRP3 is not essential, but it can influence adaptive immune responses. As the production of antibodies induced by Alum is independent of TLRs, IL-1R, and IL-18R signaling84, a major challenge for future research will be the elucidation of the pathway(s) by which Alum induces adjuvant activity.
Mechanisms of inflammasome activation
The mechanism responsible for the induction of IL-1β secretion is complex, but for simplicity it can be divided into two separate steps, namely induction of pro-IL-1β and activation of caspase-1. A first layer of complexity is represented by the fact that the proIL-1β amounts are usually low, and up-regulation of proIL-1β is required for IL-1β secretion. Induction of pro-IL-1β by microbial stimuli is mediated via TLR or NOD2 stimulation and involves NF-κB activation independently of the inflammasome30. A second layer of complexity is represented by the intricate mechanism of activation of the inflammasome itself. This is clearly demonstrated in the case of the NLRP3 inflammasome. Unlike the NLRC4 inflammasome that is triggered by cytosolic flagellin, stimulation with a microbial ligand alone is not sufficient to induce the activation of the NLRP3 inflammasome. A notable exception is the activation of the inflammasome by LPS under conditions in which the autophagic pathway is compromised85 (Fig. 2). This TLR4-induced pathway of inflammasome activation, revealed in macrophages deficient in the autophagy regulators Atg16L or Atg7, is mediated via TRIF but it remains poorly characterized85. In most cases, however, robust activation of caspase-1 requires an additional signal which can be provided by several molecules including extracellular ATP, certain bacterial toxins, and particulate agents such as silica or asbestos fibers. Activation of the P2X7R by ATP induces marked K+ efflux which may be critical for NLRP3, but not NLRC4, inflammasome activation68. Consistently, the bacterial toxins nigericin and maitotoxin, two K+ ionophores, trigger the activation of the NLRP3 inflammasome34. Furthermore, incubation of cells in buffers containing high concentrations of K+ abolishes NLRP3-mediated caspase-1 activation68, 86. However, stimulation with ATP alone, which induces K+ efflux, does not activate of caspase-1, arguing that reduction of cytosolic K+ is not sufficient to drive the activation of the NLRP3 inflammasome. Similarly, stimulation with silica, asbestos, and aluminiun hydroxide does not activate the NLRP3 inflammasome unless macrophages are pre-stimulated with microbial ligands such as LPS59, 78, 81. Collectively, these results indicate that microbial molecules provide a necessary signal that act in concert with the P2X7R, bacterial toxins or crystalline structures to induce the activation of the NLRP3 inflammasome. A possible unifying model is that microbial ligands are delivered via pores or membrane damaging molecules to the cytosol where they trigger inflammasome activation64 (Fig. 2). Consistent with this model, P2X7R stimulation induces the opening of an endogenous pore mediated by pannexin-1 that promotes the translocation of microbial molecules MDP to the cytosol55. In addition, particulate matter such as silica, aluminium hydroxide and fibrillar amyloid-β induces the rupture of lysosomal membranes and this could mediate the passage of microbial molecules into the cytosol76, 81. Alternatively, microbial stimulation and the signals provided through the P2X7R or membrane-damaging molecules may induce different signaling pathways, both of which are required for inflammasome activation. Because there is no evidence that microbial ligands bind to NLR proteins, it has been suggested that inflammasome activation is indirect. Such a mechanism is particularly attractive for the NLRP3 inflammasome given that multiple molecules without obvious homology can induce caspase-1 activation via NLRP3. A possible model is that the different molecules induce a common activity in the host cytosol which mediates NLRP3 activation. Indeed, a role for ROS, calcium-independent phospholipase A2, and cathepsin B is proposed for the activation of the NLRP3 inflammasome59, 81, 87, 88. The contribution of cathepsin B was found to be stimulus-specific in that it was observed in caspase-1 activation induced by silica and β-amyloid but not LPS81. Additional work is needed to understand the mechanism by which different stimuli trigger the activation of the NLRP3 inflammasome.
Role of the inflammasome in human disease
Given the importance of IL-1β in mediating inflammation, it is not surprising that dysregulated inflammasome activation is implicated in the pathogenesis of a variety of inflammatory diseases. In fact, mutations in the inflammasome or inflammasome-associated pathways is linked to periodic fever syndromes, vitiligo, or Crohn's disease. In addition, inflammasome-dependent IL-1β production by specific disease-related stimuli appears to play a role in the pathogenesis of gout, pseudogout, asbestosis, silicosis and Alzheimer's disease.
Autosomal dominant mutations in NLRP3 are associated with a group of rare autoinflammatory disorders including familial cold autoinflammatory syndrome, Muckle-Wells syndrome, and neonatal-onset multisystemic inflammatory disease89. The common features of these inherited syndromes include recurring episodes of fever, skin rash and arthropathy. The disease-associated NLRP3 mutations result in enhanced caspase-1 activation and IL-1β secretion by causing constitutive activation of the NLRP3 inflammasome90, 91. Monocytes from some patients harboring NLRP3 mutations spontaneously produce IL-1β which is enhanced by LPS, but not by ATP stimulation92. These findings indicate that the requirement of ATP for inflammasome activation can be bypassed by expression of the disease-associated NLRP3 mutations. Remarkably, treatment of these patients with IL-1 receptor antagonist reverses clinical symptoms suggesting a cause-effect relationship between IL-1β production and the development of disease93, 94. As we discussed above, NLRP3 detects monosodium urate crystals and therefore, may play a critical role in the development of gout, which is triggered by the deposition of uric acid crystals in the joints. In a mouse model of monosodium urate crystal-induced inflammation with induction of peritonitis, IL-1β blockade resulted in impaired neutrophil influx in ASC deficient mice71. Indeed, treatment of gouty patients who were refractory to conventional anti-inflammatory drugs with IL-1 receptor antagonist induced an effective response95.
Vitiligo is an autoimmune disorder that involves the destruction of melanocytes resulting in patches of depigmented skin. Patients with vitiligo have an increased frequency of other autoimmune disorders, such as autoimmune thyroid disease, rheumatoid arthritis, diabetes, and lupus. NLRP1 variants are associated with vitiligo susceptibility96, although the mechanism by which NLRP1 promotes skin depigmentation is unclear.
The deregulated activation of the inflammasome has been associated to Crohn's disease via genetic variation of Atg16L1, a gene encoding a critical component of the autophagic machinery85. Upon stimulation with LPS, macrophages deficient in Atg16L1 show enhanced TRIF-dependent activation of caspase-1 and secretion of mature IL-1β and IL-18. The enhanced production of IL-1β observed in Atg16L1-deficient macrophages required both K+ efflux and the generation of ROS, suggesting that autophagy negatively regulates the NLRP3 inflammasome85. However, the mechanism by which the autophagy machinery regulates the inflammasome remains unclear. Furthermore, Atg16L1 also regulates the function of Paneth cells97, a specialized type of epithelial cells located in the crypts of the small intestine that produce anti-microbial peptides. Additional studies are needed to determine which function of Atg16L1 is critically involved in the pathogenesis of Crohn's disease.
Conclusions and Perspectives
The concept of the inflammasome was introduced less than 10 years ago. Since then there has been tremendous advances in our understanding of the activation, regulation and function of the inflammasome. A critical role for NLRs in inflammasome activation is now well established. Furthermore, microbial molecules triggering the activation of specific NLR inflammasomes have been identified. A major highlight has been the discovery of inherited diseases caused by constitutive activation of the NLRP3 inflammasome which have rapidly led to specific and effective therapy for these disorders. However, major hurdles in our understanding of the inflammasome remain including the molecular mechanisms controlling the assembly and activation of the endogenous inflammasomes, identification and validation of novel protein substrates of caspase-1, and better knowledge about the function of the inflammasomes in vivo. In addition, the role of the inflammasome in several diseases in which caspase-1 activation has been implicated including gout and Alzheimer's disease remains to be further clarified. Ultimately, successful development of therapeutics will be benefited from greater understanding of the mechanisms that govern inflammasome-associated signaling pathways.
Acknowledgments
We apologize to our colleagues whose work was not cited here due to space limitations. Work on NLR proteins in our laboratory is supported by grants from the National Institutes of Health. L. F. is recipient of a postdoctoral fellowship from the Arthritis Foundation. T. E. was supported by a Fellowship from the Jung-Stiftung für Wissenschaft und Forschung, Germany.
Bibliography
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Ye Z, Ting JP. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr Opin Immunol. 2008;20:3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Franchi L, McDonald C, Kanneganti TD, Amer A, Nunez G. Nucleotide-binding oligomerization domain-like receptors: intracellular pattern recognition molecules for pathogen detection and host defense. J Immunol. 2006;177:3507–3513. doi: 10.4049/jimmunol.177.6.3507. [DOI] [PubMed] [Google Scholar]
- 5.Franchi L, et al. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 6.Ting JP, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hibino T, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 8.Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 9.Inohara N, Nunez G. The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene. 2001;20:6473–6481. doi: 10.1038/sj.onc.1204787. [DOI] [PubMed] [Google Scholar]
- 10.McDonald C, Inohara N, Nunez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem. 2005;280:20177–20180. doi: 10.1074/jbc.R500001200. [DOI] [PubMed] [Google Scholar]
- 11.Kim YG, et al. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity. 2008;28:246–257. doi: 10.1016/j.immuni.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Masumoto J, et al. Nod1 acts as an intracellular receptor to stimulate chemokine production and neutrophil recruitment in vivo. J Exp Med. 2006;203:203–213. doi: 10.1084/jem.20051229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw MH, Reimer T, Kim YG, Nunez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007;14:10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 16.Cerretti DP, et al. Molecular cloning of the interleukin-1 beta converting enzyme. Science. 1992;256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 17.Thornberry NA, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 18.Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 20.Lamkanfi M, et al. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol Cell Proteomics. 2008;7:2350–2363. doi: 10.1074/mcp.M800132-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller M, Ruegg A, Werner S, Beer HD. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008;132:818–831. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 22.Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 23.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 24.Lamkanfi M, Kanneganti TD, Franchi L, Nunez G. Caspase-1 inflammasomes in infection and inflammation. J Leukoc Biol. 2007;82:220–225. doi: 10.1189/jlb.1206756. [DOI] [PubMed] [Google Scholar]
- 25.Dowds TA, et al. Regulation of cryopyrin/Pypaf1 signaling by pyrin, the familial Mediterranean fever gene product. Biochem Biophys Res Commun. 2003;302:575–580. doi: 10.1016/s0006-291x(03)00221-3. [DOI] [PubMed] [Google Scholar]
- 26.Grenier JM, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002;530:73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874–29880. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 28.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006 doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 29.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 30.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 31.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 32.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 33.Sutterwala FS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 35.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 36.Bruey JM, et al. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-kappaB and caspase-1 activation in macrophages. J Biol Chem. 2004;279:51897–51907. doi: 10.1074/jbc.M406741200. [DOI] [PubMed] [Google Scholar]
- 37.Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 38.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franchi L, et al. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. 2007;37:3030–3039. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 41.Sutterwala FS, et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 43.Sun YH, Rolan HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–33901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 44.Lamkanfi M, et al. The Nod-like receptor family member Naip5/Birc1e restricts Legionella pneumophila growth independently of caspase-1 activation. J Immunol. 2007;178:8022–8027. doi: 10.4049/jimmunol.178.12.8022. [DOI] [PubMed] [Google Scholar]
- 45.Master SS, et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe. 2008;3:224–232. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishii KJ, Akira S. Toll or toll-free adjuvant path toward the optimal vaccine development. J Clin Immunol. 2007;27:363–371. doi: 10.1007/s10875-007-9087-x. [DOI] [PubMed] [Google Scholar]
- 47.Masumoto J, et al. ASC is an activating adaptor for NF-kappa B and caspase-8-dependent apoptosis. Biochem Biophys Res Commun. 2003;303:69–73. doi: 10.1016/s0006-291x(03)00309-7. [DOI] [PubMed] [Google Scholar]
- 48.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 49.Hsu LC, et al. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci U S A. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 51.Leppla SH, Arora N, Varughese M. Anthrax toxin fusion proteins for intracellular delivery of macromolecules. J Appl Microbiol. 1999;87:284. doi: 10.1046/j.1365-2672.1999.00890.x. [DOI] [PubMed] [Google Scholar]
- 52.Kang TJ, et al. Bacillus anthracis spores and lethal toxin induce IL-1beta via functionally distinct signaling pathways. Eur J Immunol. 2008;38:1574–1584. doi: 10.1002/eji.200838141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 55.Marina-Garcia N, et al. Pannexin-1-mediated intracellular delivery of muramyl dipeptide induces caspase-1 activation via Cryopyrin/NLRP3 independently of Nod2. J Immunol. 2008;180:4050–4057. doi: 10.4049/jimmunol.180.6.4050. [DOI] [PubMed] [Google Scholar]
- 56.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 57.Martinon F, Agostini L, Meylan E, Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr Biol. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 58.Roberts TL, et al. HIN-200 Proteins Regulate Caspase Activation in Response to Foreign Cytoplasmic DNA. Science. 2009 doi: 10.1126/science.1169841. doi10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 59.Dostert C, et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kahlenberg JM, Lundberg KC, Kertesy SB, Qu Y, Dubyak GR. Potentiation of caspase-1 activation by the P2X7 receptor is dependent on TLR signals and requires NF-kappaB-driven protein synthesis. J Immunol. 2005;175:7611–7622. doi: 10.4049/jimmunol.175.11.7611. [DOI] [PubMed] [Google Scholar]
- 61.Ferrari D, et al. The P2X7 receptor: a key player in IL-1 processing and release. J Immunol. 2006;176:3877–3883. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 62.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. Embo J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pelegrin P, Barroso-Gutierrez C, Surprenant A. P2X7 Receptor Differentially Couples to Distinct Release Pathways for IL-1{beta} in Mouse Macrophage. J Immunol. 2008;180:7147–7157. doi: 10.4049/jimmunol.180.11.7147. [DOI] [PubMed] [Google Scholar]
- 64.Kanneganti TD, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity. 2007;26:433–443. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Pan Q, et al. MDP-induced interleukin-1beta processing requires Nod2 and CIAS1/NALP3. J Leukoc Biol. 2007;82:177–183. doi: 10.1189/jlb.1006627. [DOI] [PubMed] [Google Scholar]
- 66.Ozoren N, et al. Distinct roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J Immunol. 2006;176:4337–4342. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- 67.Cervantes J, Nagata T, Uchijima M, Shibata K, Koide Y. Intracytosolic Listeria monocytogenes induces cell death through caspase-1 activation in murine macrophages. Cell Microbiol. 2008;10:41–52. doi: 10.1111/j.1462-5822.2007.01012.x. [DOI] [PubMed] [Google Scholar]
- 68.Franchi L, Kanneganti TD, Dubyak GR, Nunez G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. J Biol Chem. 2007;282:18810–18818. doi: 10.1074/jbc.M610762200. [DOI] [PubMed] [Google Scholar]
- 69.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–7564. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Y, Evans JE, Rock KL. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature. 2003;425:516–521. doi: 10.1038/nature01991. [DOI] [PubMed] [Google Scholar]
- 71.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 72.Chen CJ, et al. MyD88-dependent IL-1 receptor signaling is essential for gouty inflammation stimulated by monosodium urate crystals. J Clin Invest. 2006;116:2262–2271. doi: 10.1172/JCI28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.So A, De Smedt T, Revaz S, Tschopp J. A pilot study of IL-1 inhibition by anakinra in acute gout. Arthritis Res Ther. 2007;9:R28. doi: 10.1186/ar2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eigenbrod T, Park JH, Harder J, Iwakura Y, Nunez G. Cutting Edge: Critical Role for Mesothelial Cells in Necrosis-Induced Inflammation through the Recognition of IL-1{alpha} Released from Dying Cells. J Immunol. 2008;181:8194–8198. doi: 10.4049/jimmunol.181.12.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cassel SL, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci U S A. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008 doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franchi L, Nunez G. The Nlrp3 inflammasome is critical for aluminium hydroxide-mediated IL-1beta secretion but dispensable for adjuvant activity. Eur J Immunol. 2008;38:2085–2089. doi: 10.1002/eji.200838549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kool M, et al. Cutting Edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181:3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- 80.Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum's adjuvant effect are mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willingham SB, et al. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe. 2007;2:147–159. doi: 10.1016/j.chom.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lindblad EB. Aluminium compounds for use in vaccines. Immunol Cell Biol. 2004;82:497–505. doi: 10.1111/j.0818-9641.2004.01286.x. [DOI] [PubMed] [Google Scholar]
- 84.Gavin AL, et al. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 2006;314:1936–1938. doi: 10.1126/science.1135299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 86.Petrilli V, et al. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007 doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 87.Cruz CM, et al. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282:2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walev I, et al. Potassium regulates IL-1 beta processing via calcium-independent phospholipase A2. J Immunol. 2000;164:5120–5124. doi: 10.4049/jimmunol.164.10.5120. [DOI] [PubMed] [Google Scholar]
- 89.Ting JP, Kastner DL, Hoffman HM. CATERPILLERs, pyrin and hereditary immunological disorders. Nat Rev Immunol. 2006;6:183–195. doi: 10.1038/nri1788. [DOI] [PubMed] [Google Scholar]
- 90.Agostini L, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 91.Dowds TA, Masumoto J, Zhu L, Inohara N, Nunez G. Cryopyrin-induced interleukin 1beta secretion in monocytic cells: enhanced activity of disease-associated mutants and requirement for ASC. J Biol Chem. 2004;279:21924–21928. doi: 10.1074/jbc.M401178200. [DOI] [PubMed] [Google Scholar]
- 92.Gattorno M, et al. Pattern of interleukin-1beta secretion in response to lipopolysaccharide and ATP before and after interleukin-1 blockade in patients with CIAS1 mutations. Arthritis Rheum. 2007;56:3138–3148. doi: 10.1002/art.22842. [DOI] [PubMed] [Google Scholar]
- 93.Hawkins PN, Lachmann HJ, McDermott MF. Interleukin-1-receptor antagonist in the Muckle-Wells syndrome. N Engl J Med. 2003;348:2583–2584. doi: 10.1056/NEJM200306193482523. [DOI] [PubMed] [Google Scholar]
- 94.Hawkins PN, Lachmann HJ, Aganna E, McDermott MF. Spectrum of clinical features in Muckle-Wells syndrome and response to anakinra. Arthritis Rheum. 2004;50:607–612. doi: 10.1002/art.20033. [DOI] [PubMed] [Google Scholar]
- 95.Sokolovska A, Hem SL, HogenEsch H. Activation of dendritic cells and induction of CD4(+) T cell differentiation by aluminum-containing adjuvants. Vaccine. 2007;25:4575–4585. doi: 10.1016/j.vaccine.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 96.Jin Y, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 97.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]