Summary
We show that the secreted antigen, IbpA, of the respiratory pathogen Histophilus somni induces cytotoxicity in mammalian cells via its Fic domains. Fic domains are defined by a core HPFxxGNGR motif and are conserved from bacteria to humans. We demonstrate that the Fic domains of IbpA catalyze a unique reversible adenylylation event that uses ATP to add an adenosine monophosphate (AMP) moiety to a conserved tyrosine residue in the switch I region of Rho GTPases. This modification requires the conserved histidine of the Fic core motif and renders Rho GTPases inactive. We further demonstrate that the only human protein containing a Fic domain, HYPE (Huntingtin yeast-interacting protein E), also adenylylates Rho GTPases in vitro. Thus, Fic domain containing proteins are a new class of enzymes that mediate bacterial pathogenesis as well as a previously unrecognized eukaryotic post-translational modification that may regulate key signaling events.
Introduction
Histophilus somni poses a major infectious threat to livestock (Tagawa et al., 2005). It produces a large fibrillar surface antigen called IbpA (immunoglobulin binding protein A), which is expressed as a series of proteins from 76 to 350 kDa. The IbpA protein bears sequence identity to Bordetella pertussis filamentous hemagglutinin (FhaB), an essential virulence factor that mediates adhesion to the host respiratory epithelium (Jacob-Dubuisson et al., 2001). The COOH-terminus of IbpA is homologous to the Yersinia type III secreted effector, YopT, which causes host cell cytotoxicity (Shao et al., 2002). Convalescent serum from symptomatic animals infected with H. somni recognizes IbpA, while serum from asymptomatic animals does not (Yarnall and Corbeil, 1989). As such, the presence of IbpA directly correlates with H. somni virulence. We hypothesized that IbpA’s filamentous hemagglutinin-like domain mediates attachment to host cells, while its COOH-terminus containing the YopT homology sequence could serve as a cytotoxic effector when internalized into host cells. Indeed, histological characterization of in vivo pathogenesis indicates that H. somni causes endothelial cell damage (Gogolewski et al., 1987a).
We previously demonstrated that Yersinia YopT functions as a cysteine protease that cleaves and inactivates Rho GTPases (Shao et al., 2002). In the present study, we report that the YopT-like domain of IbpA does not disrupt the actin cytoskeleton despite its conservation of the key catalytic C/H/D triad that defines members of the YopT family. Instead, we discovered an unexpected virulence determinant in IbpA encoded by its Fic (filamentation induced by c-AMP) domains. Although Fic domains are found in proteins from bacteria to humans, their activity has remained unknown until recently, where one Fic domain containing protein was shown to catalyze an adenosine monophosphate (AMP) modification on threonine residues of Rho GTPases (Yarbrough et al., 2009). We now show that the Fic domains of IbpA also induce cytotoxicity by targeting the host GTPases, RhoA, Rac and Cdc42; however, they block signaling by these GTPase by catalyzing the addition of AMP to a tyrosine (Tyr) residue in the GTPase switch I region. This reaction, and cytotoxicity, depends on the presence of a conserved histidine (His) in the Fic domain’s core motif HPFxxGNGR. We extend these findings to show that the sole human protein with a Fic motif, HYPE, which has been previously reported to interact with huntingtin (Faber et al., 1998), is also capable of adding AMP to RhoA, Rac, and Cdc42 in vitro. These findings identify a new class of enzymes that mediate bacterial pathogenesis and suggest that addition of AMP may be an underappreciated post-translational modification in higher eukaryotes.
Results
Identification of a YopT homolog in H. somni
Psi-BLAST analysis of Yersinia pestis YopT identified a highly conserved YopT-like domain in H. somni that is located near the COOH-terminus of a 4095 amino acid protein called IbpA. YopT is a CLAN 58 cysteine protease that requires a conserved C/H/D catalytic triad for function (Shao et al., 2002). Accordingly, the YopT homology domain of IbpA contains this same catalytic triad (Figure 1). In addition, IbpA contains a filamentous hemagglutinin (FHA)-like domain near the N-terminus. The FHA domain is preceded by an N-terminal signal sequence important for secretion of this protein, followed by a coiled coil (CC) domain, and two Fic (filamentation induced by c-AMP) domains immediately preceding the YopT-like sequence (Figure 1). Unlike Yersinia YopT that is secreted by a type III secretory mechanism (Cornelis and Van Gijsegem, 2000), IbpA is secreted from the bacteria using a two-partner secretion system, forms a fibrillar network on the bacterial surface, and likely serves as an adhesin to allow bacterial attachment to the host respiratory epithelium (Tagawa et al., 2005). The CC domain is important for the fibrillar structure of large bacterial surface proteins (Fischetti, 1989). Very little is known about the Fic containing domains. In E. coli, the Fic protein is suggested to comprise a regulatory circuit that controls bacterial cell division, as a deletion of the fic gene results in aberrant septation, giving the bacteria a filament-like appearance (Utsumi et al., 1982).
Figure 1. Architecture of H. somni IbpA.

IbpA contains an N-terminal secretion signal (hatched rectangle) followed by a filamentous hemagglutinin-like domain (FHA-like, blue box). Internal coiled-coils (CC, blue boxes) precede two Fic containing domains (FIC, light orange boxes). The YopT homology domain is present at the C-terminus (YopT, blue box) and the invariant catalytic C/H/D residues of IbpA and Y. pestis YopT are indicated. The invariant residues of IbpA’s Fic motif, HPFAEGNGR, are noted. The IbpA-COOH, Fic1 and Fic2 sequences are shown. All sequences are numbered from the initiator methionine for IbpA.
Expression of IbpA-COOH results in disruption of the host actin cytoskeleton
We sought to determine if the YopT-like domain of IbpA functioned similarly to YopT of Yersinia species. To this end, we expressed amino acids 3222–4087 of IbpA as an enhanced green fluorescent protein (EGFP)-tagged fusion protein (designated IbpA-COOH) in HeLa cells (Figure 1). HeLa cells transfected with the EGFP vector control did not show any signs of cytoskeletal collapse (Figure 2-A,B). However, expression of IbpA-COOH in HeLa cells resulted in plasma membrane retraction and disruption of actin stress fibers at eight hours post transfection (Figure 2-E,F). As shown previously, transient transfection of HeLa cells with Yersinia EGFP-YopT also resulted in drastic cytoskeletal collapse (Figure 2-C,D; (Shao et al., 2002)). Interestingly, the cell rounding phenotype associated with YopT was much more dramatic than that seen with IbpA-COOH, which led us to question whether the YopT-like sequence of IbpA functioned in a manner similar to the Yersinia YopT.
Figure 2. The Fic containing domains of IbpA are cytotoxic.
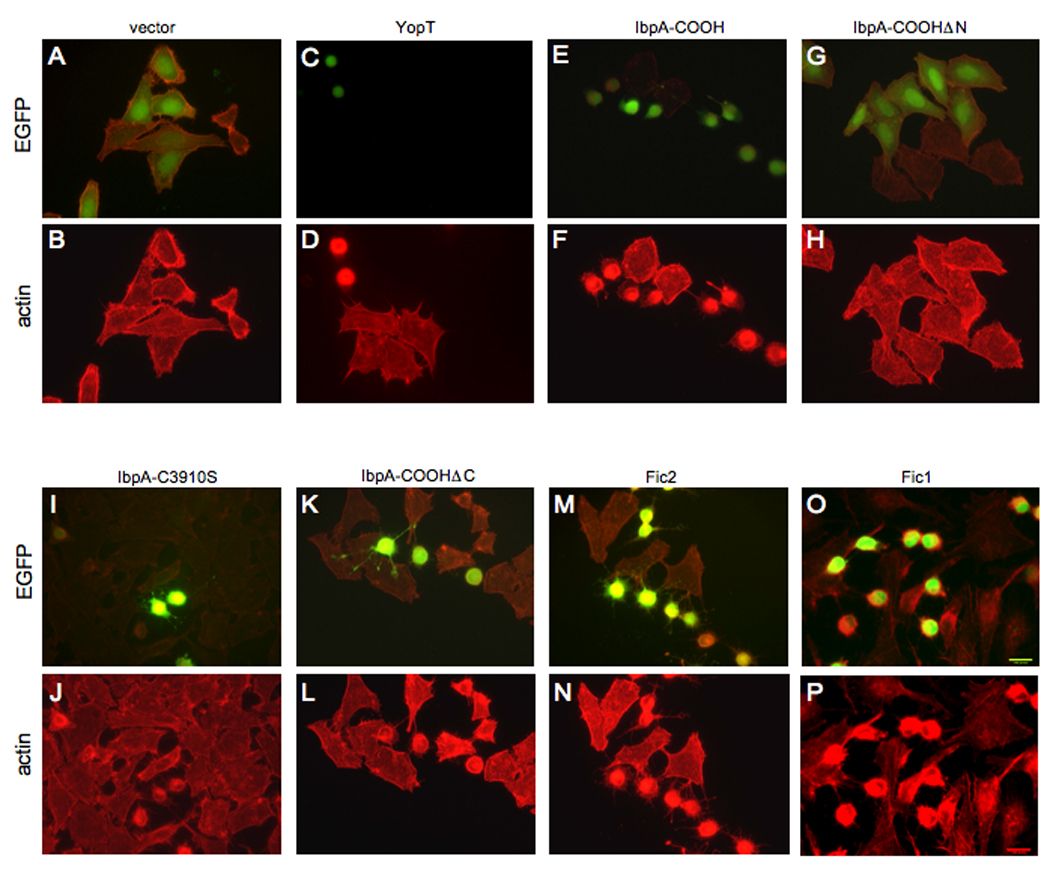
Immunofluorescence microscopy was performed on HeLa cells transfected with the EGFP vector control and EGFP-YopT, EGFP-IbpA-COOH, or the designated EGFP-IbpA deletion constructs. Cell transfection, IbpA construct expression, and cell morphology of the indicated constructs were visualized by EGFP fluorescence (A, C, E, G, I, K, M, O). Actin filaments were visualized by rhodamine phalloidin staining (B, D, F, H, J, L, N,P). Expression of EGFP alone is not cytotoxic (A, B), while expression of both EGFP-YopT (C, D) and EGFP-IbpA-COOH (E, F) induces cytotoxicity. This cytotoxic activity was further narrowed down to the Fic2 (M, N) and Fic1 (O, P) domains of IbpA.
The Fic domains are responsible for disruption of the host actin cytoskeleton
To define the region of IbpA-COOH responsible for cytoskeletal collapse, we analyzed deletion mutants and site-specific mutants of IbpA-COOH in tissue culture assays. Transient transfection of HeLa cells with an N-terminal deletion mutant, IbpA-COOHΔN (amino acids 3781–4087), which expresses only the Yop-T like domain of IbpA, surprisingly, failed to disrupt the actin cytoskeleton (Figure 2-G,H). In addition, expression of IbpA-C3910S in which the homologous cysteine residue critical for YopT's catalytic activity is mutated still caused host cell rounding, suggesting that IbpA-COOH is not a cysteine-protease harboring the same substrate specificity as YopT (Figure 2-I,J). In agreement with these data, expression of IbpA-COOHΔC (amino acids 3222–3781) in which the YopT-like sequences are deleted but the Fic sequences are expressed, still disrupted the host cytoskeleton (Figure 2-K,L). We conclude that the Fic containing domains of IbpA, and not the YopT homology region, are responsible for the observed cytoskeletal collapse.
The Fic consensus sequence is present within two highly homologous direct repeat sequences of H. somni IbpA, designated Fic1 and Fic2 (Figure1). To assess if these direct repeats can individually disrupt the actin cytoskeleton, we transiently transfected HeLa cells with constructs encoding the EGFP-tagged fusion proteins of Fic2 (Figure 2-M,N) or Fic1 (Figure 2-O,P). In each case, the Fic domain containing fusion protein caused host cell cytotoxicity in a manner similar to IbpA-COOH.
Fic domains are conserved in bacteria and eukaryotes
Iterated Psi-BLAST analysis using H. somni Fic2 as an index protein against non-redundant and EST based databases retrieved several proteins with amino acid sequences showing significant identity to Fic2. These proteins shared a consensus motif of HPFxxGNGR, with a conserved predicted secondary structure of an α–β–α–β topology around the consensus motif (Figure 3A). These Fic domains are found throughout evolution in bacterial pathogens, prophages and eukaryotes including humans, with a notable exception in yeast and plants. Figure 3A shows an alignment of selected proteins that define the diversity of these Fic domains. Interestingly, the Fic domains of IbpA have a high degree of similarity to the Fic domains found in eukaryotes. A single copy of the Fic domain is found in the human genome. This protein, known as HYPE, interacts with the glutamine-rich N-terminus of huntingtin (residues 1–425) and contains a tetratricopeptide repeat (TPR) that could serve as a cellular targeting sequence (Faber et al., 1998).
Figure 3. Identification of a Fic core motif.
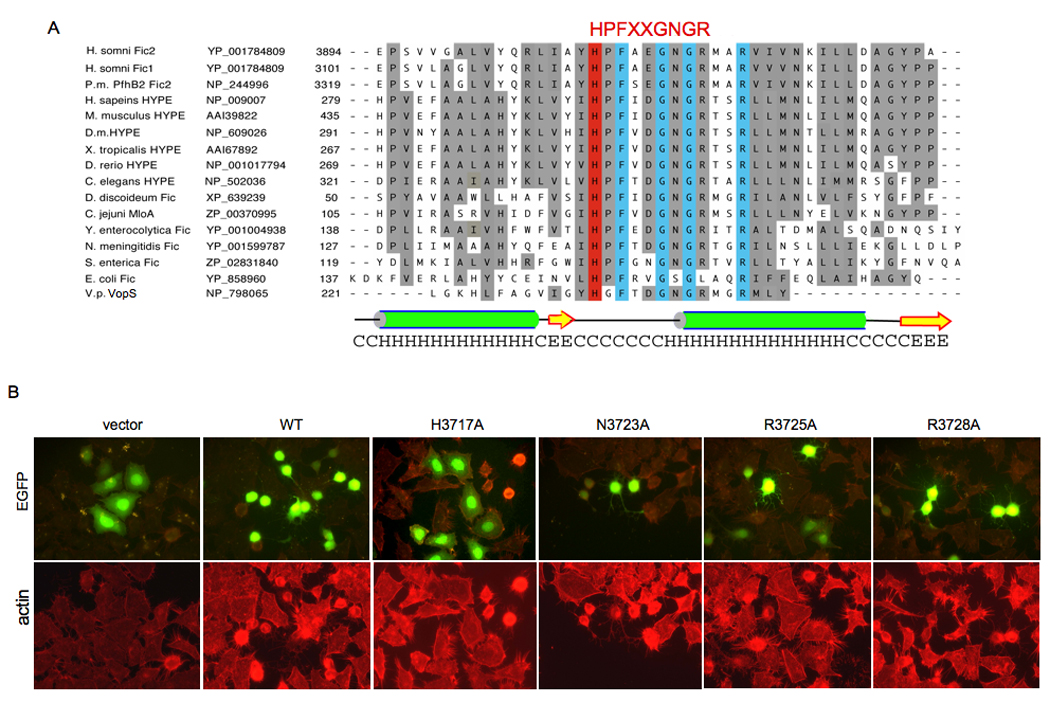
(A) Multiple sequence alignment of the Fic motifs. The Fic family was identified by BLAST searches and aligned by Clustal-W and Promals3D web programs (Pei et al., 2008), and manually adjusted using secondary structure predictions from PSIPRED (McGuffin et al., 2000). The two Fic containing domains of IbpA, Fic1 and Fic2 bear greatest sequence similarity to the two Fic containing domains of P. multocida, PfhB2, followed by mammalian HYPE. Selected Fic motifs displaying their diverse evolutionary conservation in descending order of similarity to IbpA’s Fic2 domain are shown. Listed from left to right are species, protein, accession number, and amino acids. Identities are boxed in blue. Similarities are shaded grey. The Fic HPFxxGNGR active site motif and the catalytic His residue are highlighted in red. The conserved predicted secondary structure for the Fic motif based on the prediction for Fic2 is diagramed; alpha-helices (H) are green; beta-strands (E) are yellow. The species listed are Histophilus somni, Pasteurella multocida (P.m.), Homo sapiens, Mus musculus, Drosophila melanogaster (D.m.), Xenopus tropicalis, Danio rerio, Caenorhabditis elegans, Dictyostelium discoideum, Campylobacter jejuni, Yersinia enterocolytica, Neisseria meningitidis, Salmonella enterica, Escherichia coli, and Vibrio parahemolyticus (V.p.).
(B) Histidine 3717 is critical for Fic2 cytotoxicity. The invariant putative active site residues identified in the Fic family members were mutated in the context of EGFP-Fic2 and phenotypically assayed in HeLa cells. Cell transfection, mutant Fic2 expression, and cell morphology were visualized by EGFP fluorescence (top panels). Actin filaments were visualized by rhodamine phalloidin staining (bottom panels). Wild type (WT) Fic2 and its N3723A, R3725A, and R3728A mutants remained cytotoxic, but the vector control and the Fic2 H3717A mutant were not cytotoxic.
The conserved His3717 in Fic2 is essential for cytotoxicity
Alignment of the Fic domain containing proteins revealed a common consensus sequence of HPFxxGNGR. We reasoned that one or more of these highly conserved residues would be critical for activity and generated point mutations in the context of the Fic2 protein for H3717A, N3723A, R3725A, and R3728A. Transient transfection of these mutated sequences into HeLa cells revealed that only the H3717A mutation abolished the ability of Fic2 to disrupt the host actin cytoskeleton (Figure 3B). Thus, H3717 likely represents a critical catalytic residue for Fic activity.
Expression of Fic2 inhibits RhoA and Rac from binding to their downstream effectors
The cytoskeletal collapse induced by the expression of the Fic domain was reminiscent of proteins targeting the Rho family of GTPases. However, immunoprecipitation of transiently transfected Fic2 from HeLa cells failed to pull down RhoA, Rac or Cdc42, indicating the lack of a sustained direct interaction between the Fic domain and Rho GTPases (data not shown). Further, coexpression of constitutively active or dominant negative versions of RhoA (RhoA-G14V or T19N, respectively), Rac (RacG12V or T17N, respectively), or Cdc42 (Cdc42G14V or T17N, respectively) with Fic2 failed to reverse Fic2-mediated cytoskeletal collapse (data not shown). We, therefore, sought to determine if expression of Fic2 in mammalian cells blocked signaling by precluding the binding of RhoA, Rac, and Cdc42 to their downstream effectors, rhotekin and PAK, respectively (Benard et al., 1999; Ren and Schwartz, 2000; Sander et al., 1998). The rhotekin protein exhibits a selective affinity for the GTP-bound form of RhoA. The rhotekin protein binding domain (Rht-PBD) expressed as a GST fusion protein was purified from bacteria and incubated with extracts prepared from HeLa cells expressing constitutively active RhoA-G14V or the dominant negative RhoA-T19N in the presence of Fic2 or Fic2-H3717A. As expected, Rht-PBD interacted with RhoA-G14V but not with RhoA-T19N (Figure 4A). Interestingly, activated RhoA failed to bind the Rht-PBD when co-expressed with Fic2, but bound Rht-PBD when co-expressed with the inactivated Fic2-H3717A. Similarly, co-expression of Fic1 with activated RhoA prevented RhoA-G14V from interacting with the Rht-PBD (Figure 4A). These data suggest that Fic activity blocks signaling to one or more downstream targets of RhoA. Likewise, Fic2 and Fic1 reduced binding of Rac to one of its downstream effectors, PAK (Figure 4B). The PAK protein exhibits a selective affinity for the GTP-bound form of Rac or Cdc42. We expressed the PAK PBD as a GST fusion protein in bacteria and subjected it to the pulldown analyses described above. The PAK-PBD interacted only with RacG12V. This interaction was significantly reduced by co-expression of RacG12V with Fic2 or Fic1 but not with Fic2-H3717A (Figure 4B). Together, these results suggest that Fic1/2 elicits disruption of the host cytoskeleton by modifying activated RhoA and Rac, thereby preventing them from interacting with their downstream effectors.
Figure 4. Fic2 expression in HeLa cells inhibits the ability of activated RhoA and Rac to bind to downstream effectors.
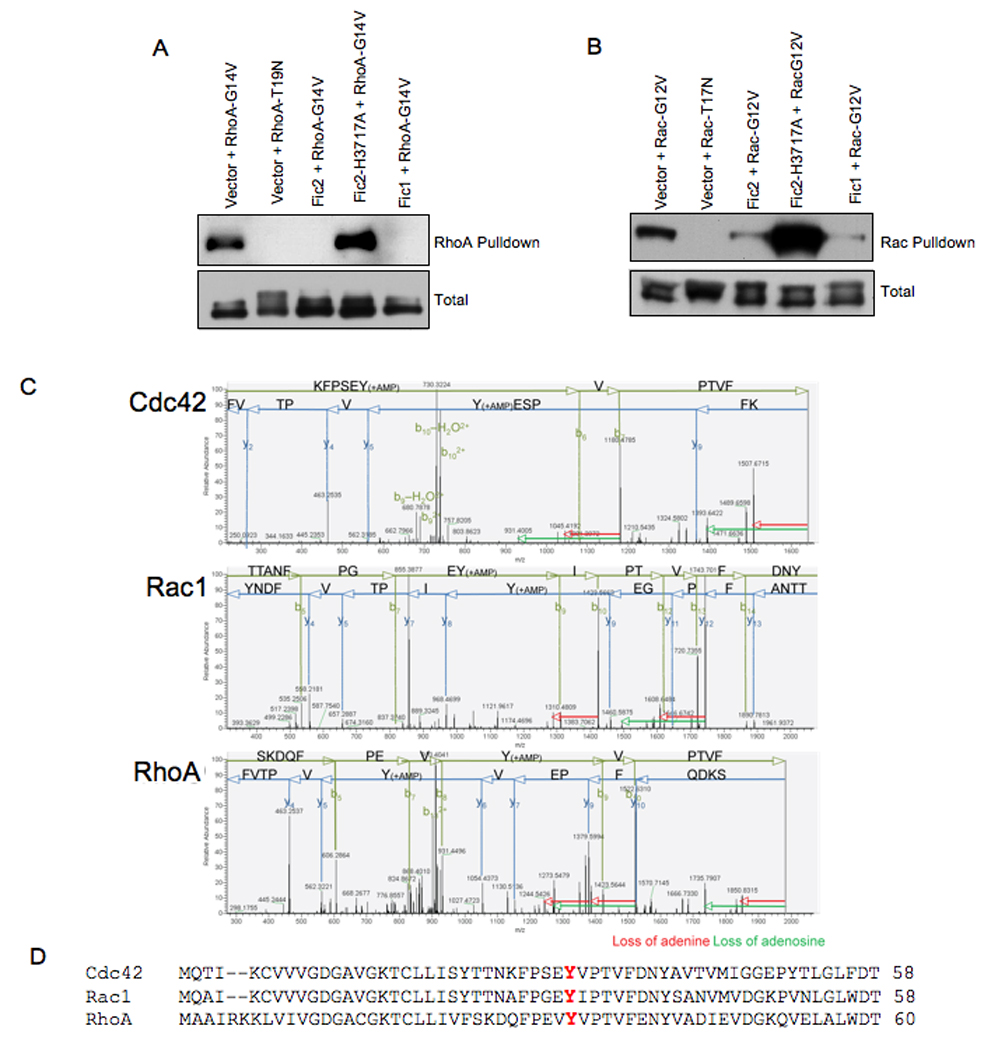
(A) Fic2 expression inhibits the binding of activated RhoA to rhotekin. HeLa cells expressing EGFP-Fic2, EGFP-Fic2-H3717A, or EGFP-Fic1 in conjunction with the constitutively GTP-bound 3xHA-tagged RhoA (G14V) or the constitutively GDP-bound 3xHA-tagged RhoA (T19N) were lysed and used for affinity precipitation with 30 µg Rht-PBD. The proteins bound to the beads as well as total extract samples were separated on SDS-PAGE, transferred to nitrocellulose membrane, and blotted with HA.11 antibody.
(B) Fic2 expression reduces the binding of activated Rac to its downstream effector, PAK. HeLa cells expressing EGFP-Fic2, EGFP-Fic2-H3717A, or EGFP-Fic1 in conjunction with the constitutively GTP-bound 3xHA-tagged Rac (G12V) or the constitutively GDP-bound 3xHA-tagged Rac (T19N) were lysed and used for affinity precipitation with 30 µg PAK-PBD. The proteins were visualized as described above.
(C) MS/MS fragmentation patterns. MS/MS fragmentation acquired on the ThermoLTQ Orbitrap mass spectrometer of AMP modified peptides of Cdc42, Rac and RhoA. Key fragments are annotated in either green (b ions) or blue (y ions). The loss of adenine (red arrow) or the loss of adenosine (green arrow) are indicated from the fragment losing the molecules.
(D) Alignment of the switch I region of Rho GTPases. The adenylylated Tyr (Y) is shown in red.
Fic2 adds an AMP moiety to a Tyr residue in RhoA, Rac, and Cdc42
We speculated that Fic2 precluded binding of RhoA and Rac to their downstream effectors by enzymatically modifying these GTPases. To test this hypothesis, we isolated GST-tagged Fic2 and incubated it with GST-tagged RhoA, Rac, or Cdc42 in an in vitro reaction containing ATP and Mg2+ (Experimental Procedures). The reaction products were analyzed by tandem mass spectrometry (MS/MS) to determine if any modified residues could be detected on RhoA, Rac or Cdc42. Interestingly, one peptide within RhoA, Rac and Cdc42 had a mass shift of 329 Da. This mass is consistent with the addition of AMP to the GTPase. Our mass spectrometry data also identified the modified residue as Y34 in RhoA, Y32 in Rac, and Y32 in Cdc42 (Figure 4C). This Tyr is conserved and constitutes a critical residue in the switch I region of the GTPases, which plays an essential role in GTPase activation (Figure 4D). These observations suggest that Fic2 represents a unique bacterial adenylylation enzyme that targets the GTPases rendering them incapable of interacting with their down stream effectors.
Fic2 uses ATP to adenylylate RhoA, Rac, and Cdc42
To confirm our mass spectrometric observations, we performed in vitro reactions in which recombinant GST-Fic2 was incubated with purified GST-RhoA, -Rac or -Cdc42 fusion protein in the presence of α32P-ATP. The 32P label was robustly transferred to RhoA, Rac, and Cdc42 by wild type Fic2 but much less efficiently when the Fic H3717A mutant was used (Figure 5A). No label was transferred when γ32P-ATP was substituted for α32P-ATP, indicating that the transfer of 32P represents addition of the AMP and not transfer of a phosphate moiety to the GTPases.
Figure 5. In vitro activity of the Fic domains.
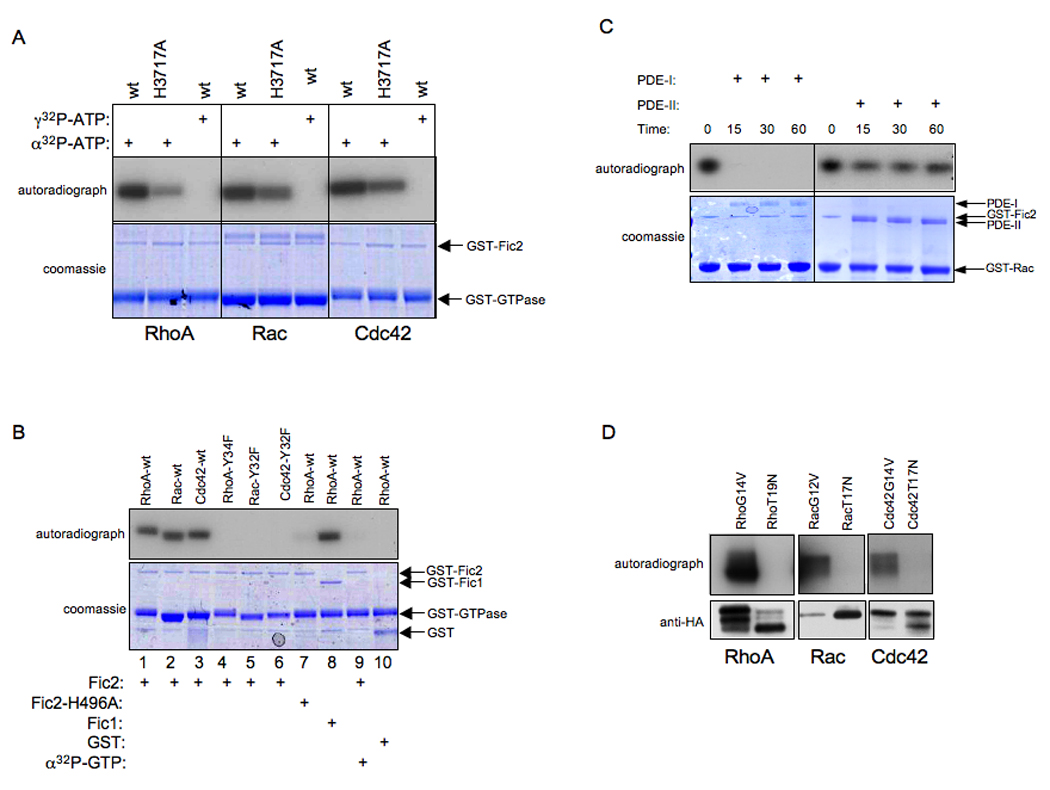
(A) Fic 2 adenylylates RhoA, Rac, and Cdc42. Bacterially expressed GST-Fic2 or GST-Fic2 H3717A was incubated with wild type RhoA, Rac, and Cdc42 expressed as GST fusion proteins in bacteria in the in vitro adenylylation assay. γ32P-ATP was substituted for α32P-ATP where indicated. Samples were separated on SDS-PAGE and visualized by autoradiography (top panel) and Coomassie staining (bottom panel). The positions of GST-Fic2 and the GST-GTPases on the gel are indicated by arrows. Wild type Fic2 adenylylates the Rho GTPases while Fic2-H3717A displays reduced activity. In addition, Fic2 is not able to transfer the γ phosphate of ATP to the substrates as indicated by the lack of transfer of radioactivity in the γ32P-ATP containing samples.
(B) Fic2 does not modify the indicated tyrosine mutants of RhoA, Rac, and Cdc42. Bacterially expressed GST, GST-Fic2, GST-Fic1 or GST-Fic2-H3717A was incubated with RhoA Y34F, Rac Y32F, and Cdc42 Y32F expressed as GST fusion proteins in bacteria in the in vitro adenylylation assay. Samples were separated on SDS-PAGE and visualized by autoradiography (top panel) and Coomassie staining (bottom panel). The positions that the GST, GST-Fic2, GST-Fic1, and the GTPases run on the gel are indicated by arrows. α32P-GTP was substituted for α32P-ATP where indicated. Fic2 is not capable of transferring a GMP residue to its substrates. In agreement with our mass spectrometry data, Fic2 adenylylates specific Tyr residues of the Rho family of GTPases.
(C) Adenylylation is reversed by the action of phosphodiesterase (PDE). Bacterially expressed GST-Fic2 was incubated with GST-Rac in an in vitro adenylylation reaction containing α32P-ATP, and subsequently treated with snake venom type I PDE (PDE-I) or bovine spleen type II PDE (PDE-II). Snake venom PDE is capable of removing the α32P-AMP moiety.
(D) Fic2 is not catalytically active against dominant negative RhoA, Rac, and Cdc42. Bacterially expressed GST-Fic2 was incubated with 3xHA RhoA (G14V), RhoA (T19N), Rac (G12V), Rac (T19N), Cdc42 (G14V), Cdc42 (T19N) isolated from 293 cells in an in vitro adenylylation reaction. Samples were separated by SDS-PAGE and visualized by autoradiography. GST-Fic2 cannot adenylylate the constitutively GDP-bound forms of the GTPases.
To further check whether Fic2 transferred an AMP moiety to the conserved Tyr residues of RhoA, Rac and Cdc42, we incubated recombinant GST-Fic2 with GST-tagged and purified RhoA-Y34F, Rac-Y32F or Cdc42-Y32F mutants in an in vitro reaction as described above. GST-RhoA-Y34F, GST-RacY32F, or GST-Cdc42Y32F were not modified by Fic2, as evidenced by the lack of 32P transfer (Figure 5B). Experiments carried out using α32P-GTP instead of α P32ATP failed to transfer 32P to the Rho GTPases. As expected, GST alone could not transfer label to the GTPases. Finally, Fic1 is also proficient in transferring the α32P-AMP moiety to RhoA, Rac and Cdc42 in a manner analogous to Fic2 (Figure 5B and data not shown). These experiments demonstrate that Fic2 and Fic1 act as adenylylation enzymes that add AMP to critical Tyr residues in the switch I region of RhoA, Rac, and Cdc42.
Phosphodiesterase treatment reverses adenylylation
Since addition of AMP to a Tyr residue results in the formation of a phosphodiester bond, we postulated that this bond would be susceptible to cleavage by phosphodiesterases (PDEs) resulting in the removal of the AMP moiety. GST-Fic2 was incubated with GST-Rac in an in vitro adenylylation reaction containing α32P-ATP. Snake venom PDE (Type I) that targets phosphodiester bonds on proteins or bovine spleen PDE (Type II) that specifically cleaves phosphodiester bonds on DNA or RNA was added to the adenylylation reaction samples for the indicated times, and analyzed for presence of radiolabeled GST-Rac. As anticipated, snake venom PDE removed the 32P moiety but bovine spleen PDE did not (Figure 5C).
Substrate specificity of the Fic domain
Our results thus far show that H. somni Fic2 (and Fic1) represents a novel bacterial virulence factor that transfers AMP to the mammalian GTPases. We hypothesized that Fic2 might show a substrate preference, catalyzing the adenylylation of the active GTPases but not the inactive GDP-bound forms. To test this hypothesis, we transiently transfected HeLa cells with HA-tagged RhoA-G14V, RhoA-T19N, Rac-G12V, Rac-T17N, Cdc42-G14V or Cdc42-T17N, immunoprecipitated these constitutively active or dominant negative versions of the GTPases, and then incubated the immunoprecipitated GTPases with wild type GST-Fic2 recombinant protein and α32P-ATP in the reaction described above. Our analysis confirmed that Fic2 modified only the constitutively active forms of RhoA, Rac and Cdc42 but did not act on their dominant negative isoforms (Figure 5D).
Relevance of the Fic domain to H. somni pathogenesis
We utilized two naturally occurring strains of H. somni to demonstrate the ability of IbpA to adenylylate Rho GTPases. H. somni strain 2336 is the etiological agent of bovine pneumonia (Tagawa et al., 2005). It expresses and secretes IbpA, which is critical for its virulence. In contrast, H. somni 129Pt is an asymptomatic strain harboring a deletion of the ibpA locus (Cole et al., 1992). Concentrated culture supernatant (CCS) prepared from H. somni 2336 is highly immunoreactive against anti-Fic2, confirming the expression of IbpA, while CCS from H. somni 129Pt lacks IbpA (Figure 6A). Importantly, as described with purified Fic1 and Fic 2, H. somni 2336 CCS AMP-modifies the purified Rho GTPases (Figure 6B). H. somni 129Pt CCS fails to adenylylate Rho GTPases, indicating that IbpA (and no other H. somni secreted protein) is likely responsible for the observed adenylylation. An easily manipulatable cell culture infection model that fully mimics H. somni pathogenesis is not available. Therefore, we used membrane free mammalian cell extract isolated from HEK293T cells as a substrate to assess the physiological target for IbpA’s Fic domains. CCS from H. somni 2336 but not 129Pt preferentially adenylylated endogenous Rho GTPases when incubated with HEK293T cell extracts, as confirmed by autoradiography and Western blot analysis for Cdc42 (Figure 6C). Therefore, the adenylylation activity of purified Fic1 and Fic2 is mimicked by CCS produced from H. somni 2336 and is undeniably attributed to the Fic domains of IbpA.
Figure 6. Importance of the Fic domain in H. somni pathogenesis.
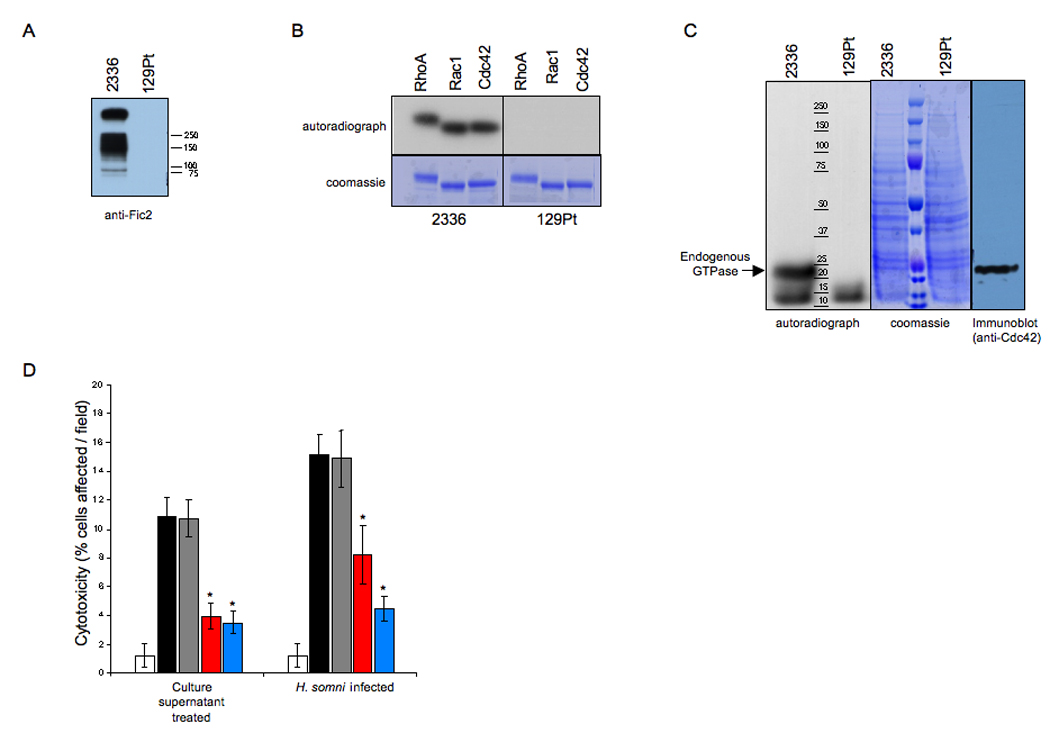
(A) CCS from H. somni 2336 but not H. somni 129Pt is immunoreactive against anti-Fic2. CCS from 2336 and 129Pt were separated on SDS-PAGE, transferred to a nitrocellulose membrane, and blotted with antibody raised against Fic2. Only CCS from 2336 was immunoreactive with anti-Fic2.
(B) CCS from 2336 adenylylates Rho GTPases. CCS from 2336 and 129Pt were used in an in vitro adenylylation reaction containing GST-RhoA, GST-Rac or GST-Cdc42. Samples were separated by SDS-PAGE and visualized by autoradiography. 2336 CCS adenylylates purified Rho GTPases.
(C) CCS from 2336 adenylylates the endogenous Rho GTPases present in HEK293T cell extract. CCS from 2336 or 129Pt was incubated with HEK293T cell extract in an in vitro adenylylation reaction. Samples were separated by SDS-PAGE, Coomassie stained and visualized by autoradiography. The migration of endogenous GTPases was determined by Western analysis using antibody to Cdc42. 2336 CCS adenylylates Rho GTPases in cellular extract.
(D) Inhibition of HeLa cell cytotoxicity with antiserum to Fic2 or convalescent serum from H. somni infected animals. Treatment of HeLa cells with CCS or live H. somni (black bars) induces cytotoxicity as compared to DMEM treated cells (white bars). Pretreatment of H. somni or CCS with preimmune rabbit serum did not reduce toxicity (grey bars). Treatment with rabbit anti-Fic2 serum (red bars) or convalescent bovine serum (blue bars) caused a significant decrease in the cytotoxic HeLa cells. Asterisk (*) denotes significant (p value ≤ 0.05) mean difference from cells without serum or those treated with preimmune serum. All groups were significantly different from untreated (medium) control group.
We extended our findings by analyzing the affect of IbpA-expressing H. somni 2336 on HeLa cell cytotoxicity. To determine whether extracellular IbpA is cytotoxic, HeLa cells were infected with live H. somni at an MOI of 100 or treated with CCS. Both treatments resulted in a rounding and cytoskeletal retraction phenotype of approximately 15% of the HeLa cells (Figure 6D). To determine whether Fic2 caused this cytotoxic phenotype, we treated HeLa cells with recombinant Fic2, but were unable to elicit a cytotoxic effect (data not shown). We reasoned that this may be due to the absence of the FHA adhesion domain mediating tight contact between the Fic2 domain and the host cell. Therefore, we assessed whether rabbit antiserum raised against recombinant GST-Fic2 could inhibit this cytotoxic phenotype. Indeed, pre-treatment of live H. somni or CCS with anti-Fic2 antiserum from H. somni infected animals that recognized IbpA showed a significant decrease in the number of rounded/retracted HeLa cells (Figure 6D). Pre-treatment of live H. somni or CCS with convalescent serum from H. somni infected calves that recognized IbpA also significantly decreased the number of rounded/retracted HeLa cells (Figure 6D). In contrast, treatment with pre-immune serum from the same rabbit failed to reduce H. somni induced cytotoxicity. These data confirm the role of Fic2 as a new bacterial virulence factor.
The human HYPE protein also functions as an adenylylation catalyst
The Fic motifs of IbpA bear high similarity to eukaryotic Fic motifs, such as the one in HYPE (Figure 3A). Therefore, we sought to determine if HYPE could function as an adenylyl transferase. We cloned and purified GST-HYPE, as well as a GST-HYPE-H295A mutant, which contains a point mutation in the putative critical His of HYPE’s Fic motif. We tested the ability of each of these proteins to modify recombinant GST-RhoA, GST-Rac and GST-Cdc42 using α32P-ATP. Amazingly, HYPE also transferred the 32P to RhoA, Rac and Cdc42, while the HYPE-H295A mutant was catalytically inactive (Figure 7A). Since it was difficult to visualize Coomassie stained GST-HYPE and GST-HYPE-H295A in these assays, we confirmed the presence of equal amounts of wild type and mutant HYPE by Western blot analysis (Figure 7B). Additionally, wild type HYPE failed to transfer an AMP moiety to RhoA-Y34F, Rac-Y32F and Cdc42-Y32F, indicating that HYPE also likely targets the conserved tyrosine of the switch I region of Rho GTPases (Figure 7C).
Figure 7. HYPE adenylylates RhoA, Rac, and Cdc42 on a Tyr residue.
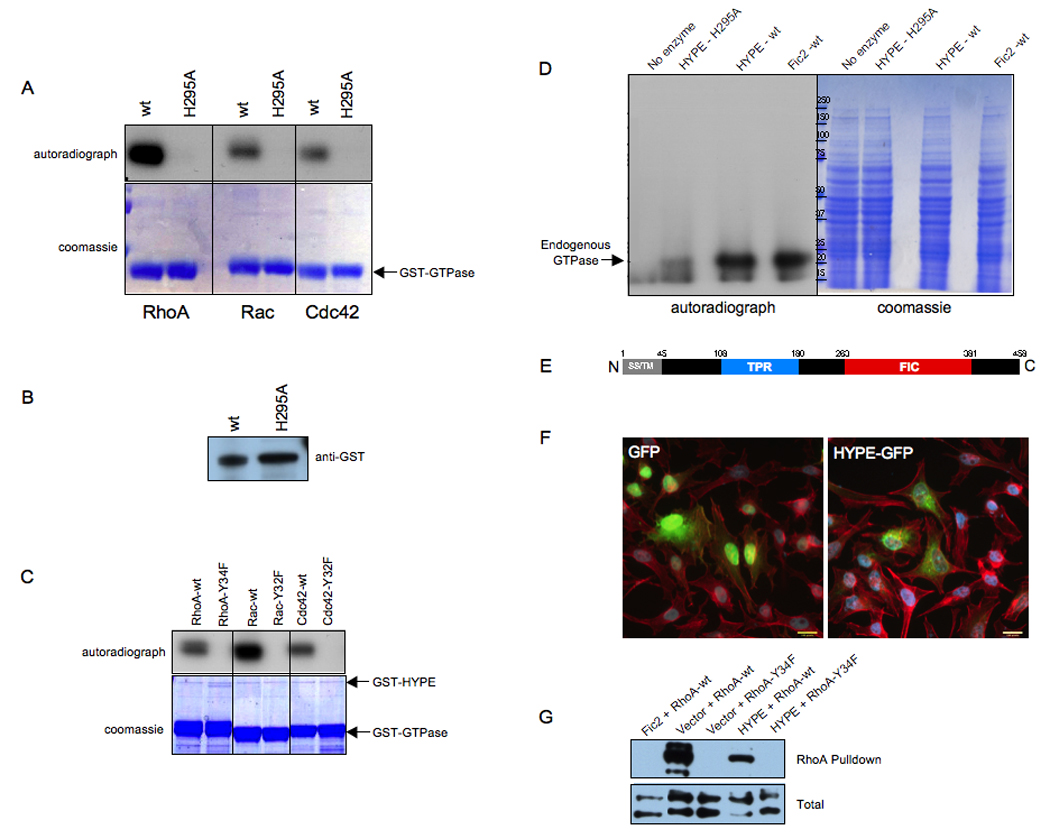
(A) HYPE adenylylates RhoA, Rac, and Cdc42. Bacterially expressed GSTHYPE or GST-HYPE H295A was incubated with wild type RhoA, Rac, and Cdc42 expressed as GST fusion proteins in bacteria in the in vitro adenylylation assay. Samples were separated on SDS-PAGE and visualized by autoradiography (top panel) and Coomassie staining (bottom panel). The GTPases are indicated by an arrow. Wild type HYPE adenylylates RhoA, Rac, and Cdc42 while the H295A mutant is catalytically inactive.
(B) GST-HYPE as visualized by western analyses. The reaction products were separated on SDS-PAGE, transferred into nitrocellulose membrane, and blotted with GST antibody.
(C) HYPE does not modify the switch I Tyr mutants of RhoA, Rac, and Cdc42. Bacterially expressed GST-HYPE was incubated with RhoA-Y34F, Rac-Y32F, and Cdc42-Y32F expressed as GST fusion proteins in bacteria in the in vitro adenylylation assay. Samples were separated on SDS-PAGE and visualized by autoradiography (top panel) and Coomassie staining (bottom panel). The positions that GST-HYPE and the GTPases run on the gel are indicated by arrows.
(D) HYPE modifies endogenous GTPases in HEK293T cell extracts. Membrane free HEK293T cell extracts were incubated with bacterially expressed GST-HYPE, GST-HYPE H295A, GST-Fic2 or without any enzyme. Samples were separated on SDS-PAGE and visualized by autoradiography (left panel) and Coomassie staining (middle panel). Like Fic2, HYPE activity is also directed against Rho GTPases in cellular extracts.
(E) Schematic representation of HYPE. The 458 amino acid protein contains a hydrophobic N-terminus (grey) consisting of a putative secretion signal (SS) and transmembrane (TM) domain, followed by a tetratricopeptide (TPR) repeat (blue) possibly involved in protein binding, and a Fic (FIC) domain (red).
(F) HYPE is not cytotoxic. Immunofluorescence microscopy was performed on HeLa cells transfected with the EGFP vector control and HYPE-GFP. Cell transfection and HYPE expression of the indicated constructs were visualized by EGFP fluorescence. Cell morphology was determined by staining actin filaments with rhodamine phalloidin. HYPE expression is not cytotoxic and does not induce a cell rounding phenotype.
(G) HYPE expression reduces binding of RhoA to its downstream effector, rhotekin. HeLa cells expressing HYPE-GFP or EGFP-Fic2 in conjunction with wild type FLAG-tagged RhoA or the FLAG-RhoAY34F mutant were lysed and used for affinity precipitation with 30 µg Rht-PBD (Experimental Procedures). The proteins bound to the beads as well as total extract samples were separated on SDS-PAGE, transferred to nitrocellulose membrane, and blotted with FLAG antibody.
We next incubated GST-HYPE with HEK293T cell extracts and α32P-ATP in our adenylylation reaction to determine HYPE’s preferred substrate. Surprisingly, a single radiolabeled band corresponding to Rho GTPases, as determined by Western blot analysis using antibody to Cdc42, was detected in this reaction (Figure 7D and Figure 6C). This band coincided with the radiolabeled Rho GTPases detected when purified Fic2 was incubated with HEK293T cell extracts, indicating that HYPE preferentially modifies Rho GTPases (Figure 7D). We, therefore, sought to determine if HYPE, like Fic1 and Fic2, caused cell rounding when ectopically expressed in mammalian cells. Unlike bacterial toxins/effectors, HYPE is an endogenous mammalian protein that is expressed in all tissues (www.symatlas.gnf.org). Additionally, HYPE contains a hydrophobic N-terminus (Figure 7E). We, therefore, reasoned that HYPE expression would be tightly controlled within the mammalian cell and/or its activity would be compartmentalized, and did not expect HYPE to induce cytotoxicity even when over-expressed within mammalian cells. As anticipated, expression of HYPE-EGFP in HeLa cells did not induce a cell rounding phenotype (Figure 7F). Accordingly, unlike Fic2, HYPE did not fully inhibit RhoA signaling, as determined by its ability to block wild type RhoA from interacting with the Rht-PBD (Figure 7G). We also tested the ability of the RhoA-Y34F mutant, which cannot be modified by Fic proteins, to signal to Rhotekin in this assay. The Tyr at position 34 of RhoA is a critical residue in the switch I region of Rho GTPases that plays an essential role in interaction with Rho GAPs, GEFs and GDIs and is therefore important for signaling. The importance of this residue is highlighted by the fact that even the most chemically conserved substitution of Tyr 34 to Phe completely abolished the ability of RhoA to bind to the Rht-PBD (Figure 7G). Thus, the Fic domains of HYPE and IbpA target a Tyr residue that is essential for Rho GTPase signaling.
Taken together our results suggest we have discovered an evolutionarily conserved enzymatic activity for the Fic domains of H. somni and human HYPE. This activity is likely present in many of the Fic-like domains found in phage, bacteria, and eukaryotic organisms.
Discussion
H. somni infection is characterized by host cell death. We hypothesized that IbpA elicits this cell death phenotype by mediating attachment via its FHA-like adhesion domain and host cell cytotoxicity via its YopT domain. Surprisingly, molecular dissection of IbpA’s cytotoxic activity revealed that its Fic domains, and not the YopT domain, were responsible for its cytotoxic phenotype. Accordingly, the cytoskeletal collapse of HeLa cells in response to transient transfection with Fic2 is less drastic than in cells transfected with YopT; Yersinia YopT mediates cell death as early as 3 hours post-transfection while Fic2-mediated cytotoxicity is observed at 8 hours post-transfection. Unlike Yersinia YopT, IbpA is not injected into host cells by a type III secretion system but is secreted into the environment by a two-partner secretion pathway, similar to FHA-like proteins of Gram negative bacteria (Jacob-Dubuisson et al., 2001). These large surface-tethered antigens create a fibrillar network around the bacterium and are also shed into the surrounding medium. We speculate that following H. somni attachment to the host cell, portions of IbpA containing the Fic domains are pinocytosed by the host cell, thereby exposing the cell cytoplasm to Fic activity. In agreement with this hypothesis, CCS from IbpA-expressing H. somni is capable of adenylylating Rho GTPases and anti-Fic2 antibodies are capable of neutralizing Fic cytotoxicity in HeLa cells treated with H. somni CCS. We are currently addressing the mechanism(s) by which IbpA’s Fic domains enter the mammalian cells.
Fic domain proteins in non-pathogenic bacteria have been implicated in the regulation of bacterial cell division and programmed death, though the mechanisms of action are not known. A temperature sensitive mutation of the fic gene in E. coli leads to filamentation, characterized by the absence of septum formation during cell division (Utsumi et al., 1982). The phenotype depends on CRP (cAMP receptor protein), a cAMP binding transcription factor (Utsumi et al., 1982). A role for fic in folate biosynthesis has also been proposed, since addition of folate or its precursor PABA to fic mutant cultures blocks filamentation in E. coli (Komano et al., 1991). All Fic domain-containing proteins share the consensus motif HPFxxGNGR. We show that Fic proteins add an AMP moiety to RhoA, Rac, and Cdc42, for which the conserved His of the HPFxxGNGR consensus motif serves as a catalytic residue. Further, this function of Fic domains appears to be evolutionarily conserved, since H. somni Fic 1 and Fic2 as well as mammalian HYPE display this ability to adenylylate GTPases. It remains to be determined if Fic domains from non-pathogenic bacteria also function in a similar enzymatic manner on a bacterial target.
The covalent attachment of AMP to proteins as a means to regulate their function is rare. The activity of E. coli glutamine synthetase was shown to be regulated by the covalent attachment and subsequent detachment of adenylyl groups to specific Tyr residues in each subunit (Shapiro and Stadtman, 1968). This reaction is catalyzed by an adenylyl transferase (ATase,EC 2.7.7.749) that shares no sequence identity with the Fic domain containing proteins. It is intriguing that the Fic domains of H. somni also utilize the adenylylation of Tyr residues to control the activity of the GTPases and that this plays an important role in pathogenesis. Interestingly, a type III secreted effector, VopS, from Vibrio parahemolyticus contains a putative Fic motif that does not have a conserved HPFxxGNGR sequence, but an HGFxxGNGR sequence. Yarbrough and colleagues recently demonstrated that, despite its non-conserved core Fic motif, VopS adds an AMP moiety to the conserved threonine residue of the switch I region of Rho GTPases, thereby blocking their ability to bind to downstream effectors (Yarbrough et al., 2009).
It is also notable that all of the interacting partners of the GTPases, irrespective of their function, bind to a common set of conserved amino acids that are clustered on the surface of the switch regions (Dvorsky and Ahmadian, 2004). We have demonstrated that Fic adenylylation of Y34 in RhoA and Y32 in Rac located in the switch I regions blocks RhoA’a ability to bind the rhotekin-PBD and significantly reduces Rac’s ability to bind PAK-PBD, respectively. We have also observed that Fic2 is not able to adenylylate the dominant negative forms of RhoA, Rac, and Cdc42. This is most likely due to the conformational change that occurs in these proteins when they bind GDP as opposed to GTP. Additional studies are underway to assess how Tyr adenylylation affects the exchange of GTP for GDP, the GTPase activities, and the binding of GDIs to the Rho GTPases.
Our report is also the first to assign an enzymatic function to human HYPE. In our in vitro reaction, HYPE adenylylated the GTPases thereby potentially inactivating them. HYPE is ubiquitously expressed in mammalian cells with highest transcript levels detected in the pituitary, pancreatic islets, liver and prefrontal cortex (www.symatlas.gnf.org). It is perplexing that a mammalian protein would function to inactivate GTPases, which regulate key signaling pathways within the mammalian cell. It is possible that RhoA, Rac and Cdc42 do not represent the only physiological substrates for HYPE. As mentioned earlier, HYPE interacts with huntingtin (Faber et al., 1998), and could function to adenylylate huntingtin. However, incubation of purified HYPE with HEK293T cell extracts did not result in a radiolabeled band corresponding to huntingtin. We speculate that this could be because 1) endogenous huntingtin levels are significantly lower than that of all Rho GTPases combined, 2) HYPE works preferentially on only the abnormal forms of huntingtin, or 3) HYPE is indirectly involved in huntingtin regulation. Experiments are underway to test these possibilities.
Our characterization of the adenylylation activity for Fic domains represents a new paradigm to an old reaction and underscores the importance of adenylylation as a novel post-translational modification in signaling. It has identified several exciting avenues for study, including a deeper understanding of bacterial pathogenesis as well as exploring this modification in higher eukaryotes.
Experimental Procedures
Cell culture, Transfection, and Cell Microscopy
HeLa cells, grown in 6-well plates in DMEM containing 10% (v/v) fetal bovine serum with 100µg/ml penicillin/streptomycin (GIBCO) at 37°C in a 5% CO2 incubator, were transfected with 0.2µg EGFP-N1 and 1µg of IbpA-COOH or its derivatives and 3µl FuGENE6 (Roche Molecular Biochemicals) as recommended by the manufacturer. To assess cell morphology, actin stress fibers were stained with rhodamine phalloidin and visualized by microscopy 8–24 h after transfection.
Protein Expression and Purification
GST-fusion proteins were expressed in E. coli BL21 RILP (Stratagene) in 2xYT medium containing 100µg/ml of ampicillin (pET-GSTx, pGEX-2T, pGEX-4T3) or kanamycin (pET41a) to a density of 0.6 A600. Protein expression was induced for 3–5h at 30°C with 0.4mM isopropyl-β-D-thiogalactopyranoside (IPTG). Cells were lysed in GST-lysis buffer (Profinia, BIO-RAD) and purified according to the manufacturer's instructions for GST-fusion proteins. GST-tagged Rht-PBD (amino acids 7–89 rhotekin) and PAK-PBD (amino acids 67–150 PAK) were isolated on GST-bind resin (Novagen), quantitated and stored as described (Pellegrin and Mellor, 2008).
Rho GTPase Activation Assays and Western Analyses
HeLa cells were transfected with 6µg of vector, Fic2-EGFP, Fic2-H3717A-EGFP, or Fic1-EGFP along with 4µg of 3XHA-tagged activated RhoA-G14V or Rac-G12V or dominant negative RhoA-T19N or Rac-T17N for 15 h before extract preparation. The assay was carried out as described (Pellegrin and Mellor, 2008) using the Rht-PBD and PAK-PBD beads. Proteins were separated by 12% SDS-PAGE, transferred to nitrocellulose membrane, and blotted with anti-HA.11 (Covance) followed by anti-rabbit HRP secondary (Amersham Bioscience). Luminescence was detected with the SuperSignal chemiluminescence kit from Pierce.
Immunoprecipitations
After transfection for 16–20 h, HeLa cells were lysed with modified RIPA buffer (50mM Tris-HCl, pH 8.0, 150mM NaCl, 1% Nonidet P40, containing 1.0mM pefabloc (Roche), 1.0mM benzimidine hydrochloride, 1µM leupeptin, 1µM E64). The homogenate was centrifuged at 14,000 × g, 4°C for 20 min. and the supernatants were incubated with HA-agarose beads (Roche) for 2–4 h at 4°C with gentle rotation. Beads were washed 4 times with ice-cold modified RIPA buffer, twice with modified RIPA buffer containing 500mM NaCl, and twice with 1X adenylylation reaction buffer (described below). Extracts were prepared from 3–4 100mm dishes of HeLa cells for each Rho GTPase and the beads were resuspended in 50µl of 1X adenylylation buffer for use in the in vitro assays.
In vitro adenylylation assays
Approximately 100ng of GST-Fic1, GST-Fic2, GST-HYPE, or their His mutants or 25X CCS from H. somni strains 2336 and 129Pt were incubated with 500ng of GST-RhoA, Rac, or Cdc42 or their Tyr mutants or 60µg of HEK293T cells extracts in 40µl adenylylation reactions containing 25mM Tris-HCl, pH 7.5, 3.0mM MgCl2, 1mM DTT, 0.5mM EDTA and 5µCi α32P-ATP for 30 min - 1 h at 30°C. γ32P-ATP and α32P-GTP replaced the α32P-ATP in specified reactions. Reactions were stopped by adding Nupage loading buffer (Invitrogen). Reaction products were separated on 12% SDS gels and visualized by autoradiography and western blot analysis with antibody to Cdc42 (Santa Cruz Biotechnology, Inc., CA) where indicated. In indicated reactions, HA-tagged activated and dominant negative GTPases immunoprecipitated from HeLa or 293T cells were substituted for the bacterially expressed GTPases. Mammalian cell extracts were prepared by dounce homogenizing HEK293T cells harvested at 90–100% confluency in a hypotonic buffer (10mM HEPES, pH 7.9; 1.5mM MgCl2; 10mM KCl; 0.5mM DTT; 1mM pefabloc; 1mM benzimidine hydrochloride; 1µM leupeptide; 1µM E64) and collecting the supernatant from a 13000 rpm centrifugation.
Phosphodiesterase assays
Approximately 50ng of GST-Fic2 was incubated with 250ng of GST-Rac in a 40µl adenylylation reaction containing 5µCi α P32-ATP (as described above). A 10µl aliquot was removed to represent the zero time point. The remainder of the reaction was treated with 1 unit of snake venom type I PDE or bovine spleen type II PDE (Worthington Biochemical Corp., Lakewood, NJ) at room temperature. 10ul aliquots were removed at 15, 30 and 60 minutes following PDE treatment. The reactions were stopped by adding Nupage loading buffer (Invitrogen). Reaction products were separated on 12% SDS gels and visualized by autoradiography.
Mass Spectrometry
50pmol of modified Cdc42, Rac and RhoA protein prepared in an in vitro adenylylation reaction using non-radioactive ATP were diluted in 50µl of 50mM ammonium bicarbonate, reduced with 2mM DTT, and alkylated with 4mM iodoacetamide for 30min. Following tryptic digests, 2µl of the peptide mixture was analyzed by automated microcapillary liquid chromatography-tandem mass spectrometry (Supplemental procedures). The MS/MS data was searched with Inspect (Tanner et al., 2005) against an E.coli database with Cdc42, Rac and RhoA added with optional modifications: +16 on methionine, +57 on cysteine, +329Da on either tyrosine, lysine, threonine or histidine. Only peptides with at least a pvalue of 0.01 were analyzed further. For verification purpose, the peptides were also introduced through a HPLC gradient into a Thermo LTQ Orbitrap mass spectrometer. Full masses (MS) spectra were recorded on the peptides over a 400–2000 m/z range at 30,000 resolution, followed by targeted MS/MS at 7,500 resolution on AMP-modified peptides at m/z 821.86 for Cdc42, m/z 1139.47 for Rac and m/z 992.24 for RhoA.
Cytotoxicity assays
Histophilus somni strain 2336 was cultured on Brain/Heart Infusion (BHI) agar with 5% calf blood in Alsevers’ solution (Colorado Serum Company, Denver, CO) at 37°C in a candled jar. Concentrated culture supernatant (CCS) was prepared as described (Supplemental procedures). CCS was added to cells at a final concentration of 20X in DMEM. HeLa cells were incubated with bacteria or CCS for 4 hours, washed with PBS, fixed with 4% paraformaldehyde, and stained with rhodamine phalloidin (actin) and DAPI (Molecular Probes, Eugene, OR). The number of rounded or retracted cells in ten microscope fields were manually counted and reported as a percentage of the total number of cells in the field as determined by DAPI stained nuclei. Cells retracted or rounded due to mitosis as assessed by the DAPI signal were excluded from the calculations.
Inhibition of cytotoxicity with DR2 specific antibody
Antibody against GST-Fic2 was raised in rabbits (Cocalico Biologicals, PA). Inhibition of cytotoxicity was also determined with a serum pool obtained from two convalescent calves (identification numbers E5 and E7) 5 weeks after experimental H. somni pneumonia (Gogolewski et al., 1987b). The rabbit or the bovine convalescent serum was incubated at a 1:100 dilution in DMEM for 45 minutes (at room temperature for H. somni or 4°C for CCS) prior to addition to HeLa cells, and cytotoxicity determined as described above.
Statistical analysis
Data were analyzed with one-way ANOVA using GraphPad Prism program. Treatment groups were compared using Bonferroni’s multiple comparison tests.
Supplementary Material
Acknowledgments
We are grateful to Dr. Kim Orth for sharing unpublished findings on AMPylation. We thank Jason Lehmann and Sandra Wiley for technical assistance. This work was funded by grants from the National Institutes of Health (AI060662) and USDA (2005-35204-16257).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- Cole SP, Guiney DG, Corbeil LB. Two linked genes for outer membrane proteins are absent in four non-disease strains of Haemophilus somnus. Mol Microbiol. 1992;6:1895–1902. doi: 10.1111/j.1365-2958.1992.tb01362.x. [DOI] [PubMed] [Google Scholar]
- Cornelis GR, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- Dvorsky R, Ahmadian MR. Always look on the bright site of Rho: structural implications for a conserved intermolecular interface. EMBO Rep. 2004;5:1130–1136. doi: 10.1038/sj.embor.7400293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- Fischetti VA. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolewski RP, Leathers CW, Liggitt HD, Corbeil LB. Experimental Haemophilus somnus pneumonia in calves and immunoperoxidase localization of bacteria. Vet Pathol. 1987a;24:250–256. doi: 10.1177/030098588702400309. [DOI] [PubMed] [Google Scholar]
- Gogolewski RP, Kania SA, Inzana TJ, Widders PR, Liggitt HD, Corbeil LB. Protective ability and specificity of convalescent serum from calves with Haemophilus somnus pneumonia. Infect Immun. 1987b;55:1403–1411. doi: 10.1128/iai.55.6.1403-1411.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Locht C, Antoine R. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol Microbiol. 2001;40:306–313. doi: 10.1046/j.1365-2958.2001.02278.x. [DOI] [PubMed] [Google Scholar]
- Komano T, Utsumi R, Kawamukai M. Functional analysis of the fic gene involved in regulation of cell division. Res Microbiol. 1991;142:269–277. doi: 10.1016/0923-2508(91)90040-h. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. Rho GTPase activation assays. Curr Protoc Cell Biol. 2008;Chapter 14 doi: 10.1002/0471143030.cb1408s38. Unit 14 18. [DOI] [PubMed] [Google Scholar]
- Ren XD, Schwartz MA. Determination of GTP loading on Rho. Methods Enzymol. 2000;325:264–272. doi: 10.1016/s0076-6879(00)25448-7. [DOI] [PubMed] [Google Scholar]
- Sander EE, van Delft S, ten Klooster JP, Reid T, van der Kammen RA, Michiels F, Collard JG. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J Cell Biol. 1998;143:1385–1398. doi: 10.1083/jcb.143.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- Shapiro BM, Stadtman ER. 5'-adenylyl-O-tyrosine. The novel phosphodiester residue of adenylylated glutamine synthetase from Escherichia coli. J Biol Chem. 1968;243:3769–3771. [PubMed] [Google Scholar]
- Tagawa Y, Sanders JD, Uchida I, Bastida-Corcuera FD, Kawashima K, Corbeil LB. Genetic and functional analysis of Haemophilus somnus high molecular weight-immunoglobulin binding proteins. Microb Pathog. 2005;39:159–170. doi: 10.1016/j.micpath.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- Utsumi R, Nakamoto Y, Kawamukai M, Himeno M, Komano T. Involvement of cyclic AMP and its receptor protein in filamentation of an Escherichia coli fic mutant. J Bacteriol. 1982;151:807–812. doi: 10.1128/jb.151.2.807-812.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough ML, Yan L, Kinch LN, Grishin NV, Ball HL, Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- Yarnall M, Corbeil LB. Antibody response to Haemophilus somnus Fc receptor. J Clin Microbiol. 1989;27:111–117. doi: 10.1128/jcm.27.1.111-117.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


