Abstract
Zipper-interacting protein kinase (ZIPK) is a member of the death-associated protein kinase family associated with apoptosis in nonmuscle cells where it phosphorylates myosin regulatory light chain (RLC) to promote membrane blebbing. ZIPK mRNA and protein are abundant in heart tissue and isolated ventricular neonatal rat cardiac myocytes. An unbiased substrate search performed with purified ZIPK on heart homogenates led to the discovery of a prominent 20-kDa protein substrate identified as RLC of ventricular myosin. Biochemical analyses showed ZIPK phosphorylated cardiac RLC at Ser-15 with a Vmax value 2-fold greater than the value for smooth/nonmuscle RLC; cardiac RLC is a favorable biochemical substrate. Knockdown of ZIPK in cardiac myocytes by small interfering RNA significantly decreased the extent of RLC Ser-15 phosphorylation. Thus, ZIPK may act as a cardiac RLC kinase and thereby affect contractility.
Keywords: Enzyme Kinetics, Heart, Myosin, Protein Kinases, Protein Phosphorylation, ZIPK, Myosin Light Chain
Introduction
Myosin is the molecular motor in the thick filament of the sarcomere that binds to actin thin filaments to initiate myocyte shortening and force development when Ca2+ binds to troponin in the thin filament (1). Phosphorylation of sarcomeric proteins modulates this Ca2+-dependent contraction process, including phosphorylation of Ser-15 in cardiac myosin RLC3 (2–4). The basal phosphorylation of RLC (40–50%) in beating hearts is maintained by slow rates of phosphorylation and dephosphorylation and increases the Ca2+ sensitivity of contraction (2, 5). This modulatory role on contractile function is conferred through structural alterations that affect myosin cross bridges binding to actin thin filaments (6–10).
Studies with transgenic and knock-out mice have shown that although RLC phosphorylation is not necessary for striated muscle development, it is necessary for optimal contractile performance (10–13). Cardiac abnormalities arise from age-dependent increased load and depressed ventricular ejection in transgenic mice overexpressing a nonphosphorylatable cardiac RLC, and physiological and pathophysiological hypertrophic responses are attenuated in hearts with increased RLC phosphorylation, confirming the importance of phosphorylation in long term modulation of cardiac function (11, 14, 15). In addition, RLC phosphorylation appears to be involved in positive inotropic responses to long term β-adrenergic stimulation (15).
Although the importance of cardiac RLC phosphorylation is clear, the identity of its protein kinase(s) remains is not determined. The unique amino acid sequence adjacent to the phosphorylation site makes cardiac RLC a distinct substrate from smooth/nonmuscle RLCs (16). Smooth muscle myosin light chain kinase (MLCK) is not a good kinase for cardiac RLC, and skeletal MLCK is not significantly present in cardiac myocytes (10, 17, 18). A novel cardiac muscle-specific myosin light chain kinase (cardiac MLCK) was recently identified (17, 18). Its knockdown in NRCM did not completely ablate cardiac RLC phosphorylation, which raises the possibility of an alternate RLC kinase.
ZIPK, also known as DAPK3, is a highly conserved Ca2+-independent serine/threonine protein kinase belonging to the death-associated protein kinase (DAPK) family (19). ZIPK is comprised of an N-terminal kinase domain (amino acids 13–275) and a C-terminal domain of unknown function that contains a leucine zipper and possibly an autoinhibitory loop (19, 20). Northern blots of various tissues show that ZIPK is ubiquitously expressed, including in heart (21, 22). Previous investigations have focused on its respective roles in apoptosis in nonmuscle cells and contraction in smooth muscle, where studies on the latter were stimulated by identification of smooth muscle RLC and the regulatory subunit of smooth muscle myosin light chain phosphatase (MYPT1) as substrates for ZIPK (21, 23, 24).
Therefore, we initiated studies to identify protein substrates for ZIPK in heart. We found that cardiac RLC is phosphorylated by ZIPK at Ser-15 in vitro and in vivo, implicating a role for this protein kinase in modulating cardiac contractility.
EXPERIMENTAL PROCEDURES
Quantitative Real-time PCR
mRNA was prepared from C57 mouse tissues or NRCM by TRIzol and phenol-chloroform (Invitrogen) extraction, and cDNA was prepared using SuperScript III for QPCR (Invitrogen). Template cDNA (50 ng) was used in each 10-μl reaction performed in triplicate with SYBR GreenER (Invitrogen) in an ABI PRISM 7200 sequence detector (Applied Biosystems). Mouse cyclophilin B was used as an internal control. Relative tissue ZIPK mRNA quantities were normalized to that measured from mouse urinary bladder. For knockdown, ZIPK mRNA amount was normalized to the amount obtained as negative control with scrambled siRNA-transfected NRCM. There were no differences in ZIPK mRNA amounts between untreated and negative control for NRCM.
Substrate Search
Wild-type C57 mouse tissues were homogenized in 30 × weight to volume ratio (100 mm MOPS, pH 7.4, 300 mm NaCl, 10 mm EGTA, 1 mm dithiothreitol, 1% Triton X-100, protease inhibitor cocktail (Sigma)) and cleared by centrifugation (10,000 × g for 5 min at 4 °C). The supernatant fraction was desalted using Sephadex G-25 mini-columns (PD-10, from GE Healthcare) to remove endogenous ATP and preincubated at 30 °C for 20 min to dephosphorylate proteins. The processed supernatant fraction (25 μl) was incubated with constitutively active ZIPK in a 50-μl total assay mixture containing 10 mm MgCl2, 1 μg of ZIPK, 0.5 μl of phosphatase inhibitor cocktail I and II (Sigma), and 0.1 mm ATP for 5 min at 30 °C. The reaction was stopped by adding 10 μl of 6× Laemmli sample buffer and boiling for 5 min. Samples were resolved by 4–20% gradient SDS-PAGE and transferred to PVDF membrane, which was then fixed in 0.4% glutaraldehyde for 15 min at room temperature. After rinsing the membrane in phosphate-buffered saline, it was exposed to film overnight. The membrane was stained with MemCode (Pierce) to obtain an image of the transferred bands and confirm equal loading of samples. Membrane was destained and blotted with RLC antibodies (anti-cardiac RLC antibody 1:10,000, anti-smooth muscle RLC antibody 1:5,000).
Mouse Cardiac Myosin Purification
Myosin was purified from C57 mouse ventricles using low salt precipitation steps at 4 °C, similar to the original protocol by Murakami et al. (25).
Recombinant RLCs and ZIPK Purification
Glutathione S-transferase-tagged constitutively active mouse ZIPK (amino acids 1–320), which lacked the autoinhibitory C terminus (20), was expressed and purified following steps specified by the pGEX vector plasmid instruction manual (GE Healthcare). C-terminal His6 tag was constructed into human ventricular cardiac RLC in pBad22 expression vector for subsequent purification using HisPur cobalt resin (Pierce). N-terminal His6-tagged human smooth muscle RLC construct was a generous gift from Christine Cremo (University of Nevada, Reno) (26).
Kinase Activity Assay
ZIPK (25 ng) was assayed for activity in 10 mm MOPS, pH 7.4, 10 mm MgCl2, 100 mm NaCl, 3 mm EGTA, 1 mm dithiothreitol, and 0.2 mm [γ-32P]ATP (100–300 cpm/pmol) with purified substrates in 50-μl total volume. Reaction mixtures were preincubated for 5 min, and the kinase activity was measured by the addition of [γ-32P]ATP as described previously (27). Km and Vmax values were calculated by nonlinear fit to the Michaelis-Menten equation using the GraphPad Prism 5.0 software.
Mass Spectrometry
Human cardiac and smooth muscle RLCs (1 μg each) were phosphorylated by 1 ng of ZIPK for 5–15 min, as described above. Mono- and diphosphorylated RLCs were separated by urea/glycerol-PAGE and stained with GelCode Blue Safe (Pierce). Monophosphorylated bands were subjected to trypsin digestion in the gel followed by LC/MS/MS using an ABI QStar XL mass spectrometer (Applied Biosystems) at the Protein Chemistry Core at University of Texas Southwestern. Sequence coverage was 80% for smooth muscle RLC and 93% for cardiac RLC.
Neonatal Rat Cardiac Myocyte Preparation, Gene Silencing by RNA Interference, and Overexpression by Adenovirus
All procedures were performed in accordance with the Institutional Animal Care and Use Guidelines at University of Texas Southwestern Medical Center at Dallas. Ventricular NRCM were isolated and cultured using procedures described previously (28, 29) and transfected with 10 nm siRNA 2 days after plating using Lipofectamine RNAiMax (Invitrogen). Negative control and ZIPK-targeted siRNA (ABI siRNA sequence number s64814) were selected from Silencer Select predesigned sequences from Applied Biosystems. The amount of siRNA used in these studies was optimized by titration studies for effective knockdown of ZIPK as determined by QPCR and immunoblot analyses. At all concentrations tested (0–500 nm), there were no obvious morphological changes in the myocytes or effects on the expression amounts of related kinases as compared with vehicle and negative control samples. For adenovirus-mediated protein overexpression, cells were infected 48 h after plating for 2 h with adenovirus expressing full-length GFP-tagged ZIPK or empty vector at a multiplicity of infection of 25 plaque-forming units/cell. Cells were then cultured for an additional 48 h in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. For measurements of cardiac RLC phosphorylation, NRCM were overlaid with 10% trichloroacetic acid, quick-frozen in liquid nitrogen, and scraped after thawing. Precipitated protein was collected by centrifugation, washed in diethyl ether, and completely solubilized in urea sample buffer (8 m urea, 20 mm Tris, 23 mm glycine, 0.2 mm EDTA, 5% saturated sucrose, 10 mm dithiothreitol). For cardiac MLCK and smooth muscle MLCK Western blots, cells were scraped in cold phosphate-buffered saline and lysed in 50 mm Tris, 100 mm KCl, 5 mm β-mercaptoethanol, 1% Nonidet P-40, protease inhibitor cocktail (Sigma), pH 7.5. Solubilized samples were quantified by Bradford assay and boiled in equal volume of 2× Laemmli buffer prior to SDS-PAGE.
Western Blots and Antibodies
Tissue and myocyte protein samples were subjected to SDS-PAGE with proteins transferred to PVDF membrane and blotted by standard procedures. Antibody to mouse ZIPK was generated by Lampire Biological Laboratories with a mixture antigen of full-length mouse ZIPK and two unique N-terminal and C-terminal peptides. Antibodies to RLCs were reported previously (14, 30). Antibodies to GAPDH (Santa Cruz Biotechnology) and phospho-cardiac RLC (generous gift of N. Epstein, NHLBI, National Institutes of Health) were also used.
Statistical Analyses
All error bars shown in figures represent S.E. Significance of mRNA levels in tissues was determined by one-way analysis of variance followed by Dunnett's multiple comparison test. Changes in ZIPK protein knockdown and RLC phosphorylation were analyzed by paired t test. In all figure legends, n = number of independent experiments performed.
RESULTS
ZIPK mRNA and Protein Are Expressed in the Heart
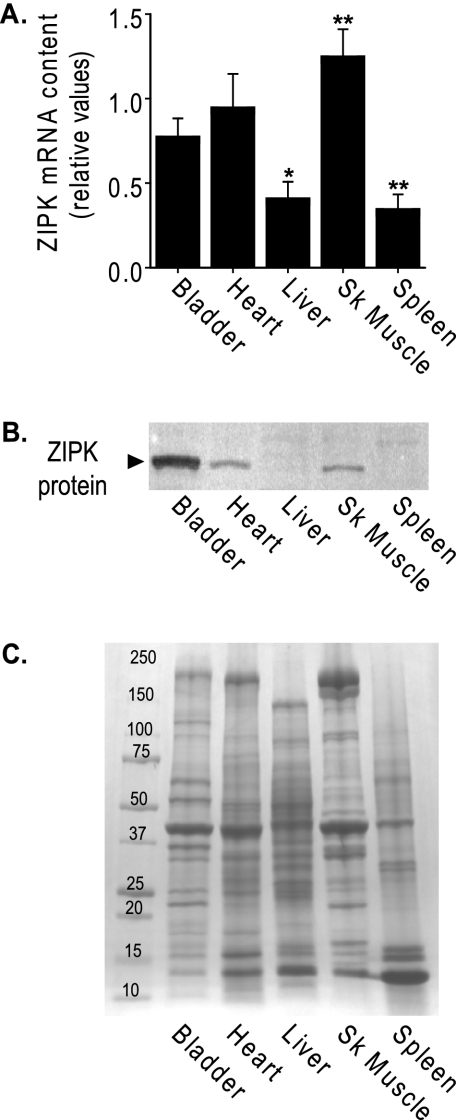
QPCR results showed ZIPK mRNA was abundant in mouse striated muscles relative to the amount in urinary bladder smooth muscle (Fig. 1A). Significantly less mRNA was expressed in liver and spleen. Western blots of the corresponding tissues show that the 52-kDa ZIPK protein was present in the muscle tissues, with the largest amount expressed in smooth muscle (Fig. 1B) when compared under conditions of equal total protein loaded (Fig. 1C).
FIGURE 1.
Expression of ZIPK mRNA and protein in tissues. A, quantitative comparison of ZIPK mRNA in mouse tissues by QPCR. Relative quantities are shown as ratios of amounts found in each tissue relative to the amount in cerebellum, after normalization to an internal control, mouse cyclophilin B. Bars ± S.E. represent the average of separate experiments using tissues from different wild-type C57 mice. Measurements were performed in triplicate (* = p < 0.05 and ** = p < 0.001 versus bladder), n = 5 for all except skeletal muscle (SK Muscle, n = 4). B, ZIPK protein expression in mouse tissues. Total tissue proteins (∼10 μg/lane) were separated by SDS-PAGE and ZIPK protein (▶) shown by Western blotting. C, Coomassie Blue-stained gradient gel shows equal loading of total protein. Mass markers (kDa) are indicated.
Previous publications report a discrepancy in the size of ZIPK found in cells and smooth muscle tissues (20, 22, 23). The predicted molecular mass of 52 kDa was observed in cells in culture, but only a smaller 32-kDa form was detected in some smooth muscle tissues. We find only a 52-kDa form in mouse smooth muscle that comigrates with full-length recombinant ZIPK. Additionally, we find a 52-kDa immunoreactive ZIPK in heart tissue. We have no explanation for the 32-kDa form reported by others but consider the possibility that the kinase may have been subject to partial proteolysis.
RLCs Are Substrates for ZIPK
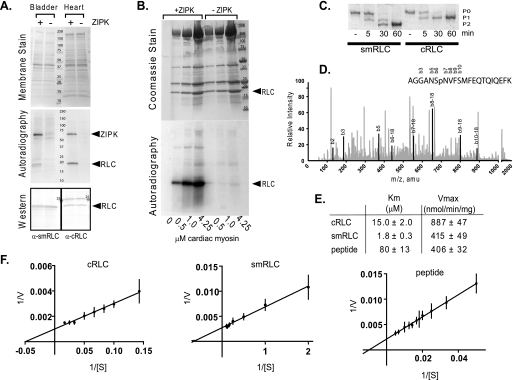
A substrate search with and without constitutively active ZIPK (1–320) added to bladder and heart homogenates identified two proteins that were robustly phosphorylated, ZIPK itself and an ∼20-kDa protein (Fig. 2A, autoradiography panel). Western blot of the same membrane, subjected to autoradiography, showed precise overlap of the respective smooth and cardiac muscle RLC bands with the 32P-labeled bands on film (Fig. 2A).
FIGURE 2.
Identification of cardiac RLC as a substrate for ZIPK. A, ZIPK substrate search in mouse bladder and heart tissue homogenates incubated with (+) or without (−) purified ZIPK. Upper panel, assay samples were subjected to electrophoresis on a 4–20% gradient gel and transferred to PVDF membrane. Total proteins on membrane are shown by MemCode PVDF stain (Pierce), and mass of markers (kDa) is indicated. Middle panel, proteins phosphorylated by ZIPK were visualized by autoradiography of membrane shown in the upper panel. Lower panel, RLC band presence in lanes with (+) or without (−) added kinase was confirmed subsequently by Western blot analysis of the same membrane used in the upper and middle panels. Specific antibodies used to detect cardiac (α-cRLC) and smooth muscle RLC (α-smRLC) are indicated. B, RLC in cardiac myosin is a substrate for ZIPK. Purified ZIPK (300 ng) was incubated with 0.5–4.25 μm purified myosin (molecular mass 520 kDa) for 3 min in 50 μl of assay buffer. Top panel, Gelcode Blue Safe (Pierce)-stained 4–20% gradient gel of assay samples resolved by SDS-PAGE. Mass markers (kDa) are indicated. Dominant proteins include myosin heavy chain (200 kDa), actin (45 kDa), myosin light chain 1 (24 kDa), and RLC (20 kDa). Bottom panel, autoradiograph of gel with + and − symbols depicting the presence or absence of kinase, respectively. C, purified recombinant human smooth muscle RLC and human cardiac RLC (1 μg each) were phosphorylated with ZIPK for the indicated minutes and resolved by urea/glycerol-PAGE. Non- (P0), mono- (P1), and di- (P2) phosphorylated forms are indicated. D, phosphorylated cardiac RLC was subjected to phosphorylation site identification by LC/MS/MS where greater than 90% of protein was analyzed, and the peptide shown contained the only phosphorylated residue. Peaks that correspond to specific b residues are darkened and labeled. The b6 residue that corresponds to Ser-15 is the only site phosphorylated. amu, atomic mass units. E, the rates of phosphorylation of cardiac RLC (n = 4), smooth muscle RLC (n = 5), and RLC peptide (n = 3) by ZIPK were measured at different substrate concentrations, and average Km and Vmax values with S.E. were determined from independent nonlinear regression analysis using GraphPad Prism software. F, Lineweaver-Burk plots of the average values are shown.
Purified Mouse Myosin Is Phosphorylated by ZIPK
Mouse ventricular myosin was partially purified and phosphorylated in vitro by ZIPK. Autoradiography of the Coomassie Blue-stained gel confirmed autophosphorylation of ZIPK and phosphorylation of cardiac RLC (Fig. 2B). The smaller amount of ZIPK added to myosin resulted in less 32P incorporation with autophosphorylated ZIPK as compared with results shown in Fig. 2A.
Cardiac RLC Ser-15 Is Phosphorylated by ZIPK
Mouse cardiac RLC sequence has an additional serine residue adjacent to Ser-15 in contrast to other animal species. To eliminate the possibility that the phosphorylation of cardiac RLC is a mouse species-dependent artifact, human cardiac RLC was purified and phosphorylated with ZIPK. Urea/glycerol-PAGE showed that cardiac RLC was monophosphorylated, as compared with diphosphorylation seen with smooth muscle RLC (Fig. 2C). As doubly phosphorylated smooth muscle RLC is rarely seen in smooth muscle tissues, smooth muscle RLC phosphorylation reaction was optimized for monophosphorylation by ZIPK, and the phosphorylation sites were confirmed to be Thr-18 and Ser-19 (data not shown), as predicted by previous enzymatic studies (31). Ser-15 in human cardiac RLC was identified as the unique phospho-amino acid by LC/MS/MS (Fig. 2D).
Cardiac RLC Is a Favorable Substrate for ZIPK
The phosphorylation properties of ZIPK with human cardiac RLC, smooth muscle RLC, and peptide (smooth muscle RLC residues 11–22) from smooth muscle RLC were measured with [γ-32P]ATP to determine Km and Vmax values. Lineweaver-Burk plots of average points used for nonlinear regression analysis and corresponding calculated Km and Vmax values (Fig. 2, E and F) are shown. The Vmax value for cardiac RLC was 2-fold greater than for smooth muscle RLC, whereas the Km value of smooth muscle RLC was lower. Vmax for the peptide was similar to smooth muscle RLC, whereas the Km value was significantly greater.
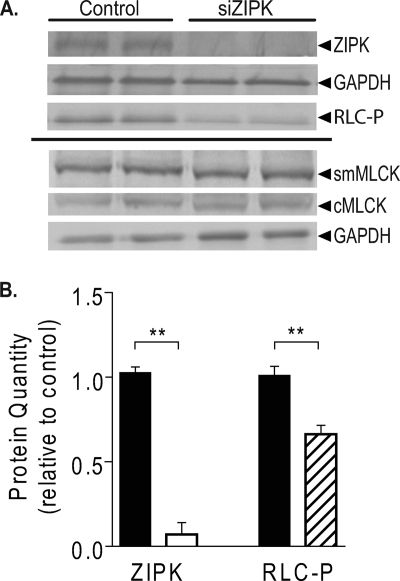
ZIPK Protein Knockdown in NRCM Decreased Cardiac RLC Phosphorylation
Transfection of NRCM with ZIPK siRNA resulted in significant reduction in ZIPK protein and phosphorylated cardiac RLC as compared with the negative control, with no significant changes in cardiac MLCK and smooth muscle MLCK, two other kinases that can phosphorylate cardiac RLC (Fig. 3A). Quantitation showed that ZIPK protein was knocked down to 7% of the negative control siRNA-transfected myocytes. ZIPK knockdown was accompanied by a 34% decrease in the extent of cardiac RLC phosphorylation.
FIGURE 3.
Effect of ZIPK knockdown on cardiac RLC phosphorylation in intact myocytes. A, NRCM were transfected with 10 nm of negative control or ZIPK-targeted siRNA (siZIPK, ABI siRNA sequence number s64814) and harvested 72 h after transfection for Western blot analysis with antibodies against ZIPK, GAPDH (load control), phospho-cardiac RLC (RLC-P), smooth muscle myosin light chain kinase (smMLCK), and cardiac myosin light chain kinase (cMLCK). B, densitometric quantitation of samples treated with siRNA for amounts of ZIPK (open) and RLC-P (hatched) normalized to negative control-siRNA transfected sample median values (filled). **, p < 0.001, n = 3.
ZIPK Protein Overexpression in NRCM Increased Cardiac RLC Phosphorylation
Overexpression of full-length ZIPK using adenoviral infection of NRCM resulted in a significant increase in phosphorylated cardiac RLC as compared with the empty virus-infected control. Quantitation showed that ZIPK overexpression was accompanied by a 16 ± 4% increase in cardiac RLC phosphorylation (p < 0.02, n = four independent experiments performed in triplicate).
DISCUSSION
ZIPK is a Ca2+-independent kinase with various substrates participating in diverse processes, ranging from promotion of apoptosis, to attenuation of inflammatory signaling, to regulation of contraction in smooth muscle (19, 21, 32, 33). The studies reported here assign ZIPK yet another role as a kinase for cardiac RLC in the heart. ZIPK message is abundant in cardiac muscle, in agreement with previous studies (21, 22). We show that ZIPK protein is present in both adult heart and NRCM. Cardiac RLC was identified as a primary substrate for ZIPK in an unbiased substrate search, and purified RLC had favorable biochemical substrate properties for the kinase. Importantly, ZIPK phosphorylates RLC in intact cardiac myosin. Finally, the biological relevance of these observations is demonstrated in intact myocytes where cardiac RLC phosphorylation is increased with ZIPK overexpression and decreased in response to diminished ZIPK amounts, whereas the amounts of two members of the MLCK family, cardiac MLCK and smooth muscle MLCK, remain unchanged.
The inability to extinguish cardiac RLC phosphorylation with a single kinase knockdown, whether cardiac MLCK (18) or ZIPK, is consistent with the idea that more than one kinase may phosphorylate cardiac RLC. Similarly, it is proposed that multiple kinases may phosphorylate smooth muscle RLC to regulate contraction, but in contrast to cardiac RLC, smooth muscle RLC is a more promiscuous substrate (16, 24). Ca2+/calmodulin-dependent smooth muscle MLCK mediates Ca2+ signaling to phosphorylate RLC to initiate contraction in smooth muscle (34). However, the addition of constitutively active ZIPK to permeabilized smooth muscles results in a contraction through the phosphorylation of smooth muscle RLC (24).
It remains unclear what the upstream regulators are for the cardiac MLCK and ZIPK to produce RLC phosphorylation in heart. Although cardiac MLCK contains a Ca2+/calmodulin binding sequence, its activation by Ca2+/calmodulin is not well established as compared with the activation of skeletal and smooth muscle MLCKs (17, 18, 34). We note that overexpression of full-length autoinhibited ZIPK in NRCM consistently led to significant but small increases in cardiac RLC phosphorylation. This might be accounted for by minimal activation under standard culture conditions. Biochemically, ZIPK is phosphorylated and activated in part by Rho kinase (35). The signaling mechanism leading to activation of ZIPK in the heart may involve a RhoA/ROCK pathway, but other signaling pathways may be involved. A recent publication suggests a role for ZIPK in suppression of inflammatory signaling pathways through activation by DAPK (33, 36). ZIPK may phosphorylate cardiac RLC as a compensatory mechanism to maintain cardiac function in response to inflammatory, autophagic, or apoptotic stimuli. In light of the importance of cardiac RLC phosphorylation to cardiac performance, the case for additional studies on the role of ZIPK in the heart is compelling.
Acknowledgments
We thank Mitsuo Ikebe (University of Massachusetts Medical School) for the ZIPK clone, Danuta Szczesna-Cordary (University of Miami) for the human cardiac RLC clone, Christine Cremo (University of Nevada, Reno) for the human smooth muscle RLC clone, Jessica Mullens (University of Texas Southwestern) for adenovirus preparation, Neil Epstein for the phospho-cardiac RLC antibody (NHLBI, National Institutes of Health), and Peiguo Ding (University of Texas Southwestern) for the cardiac MLCK antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants HL080536 and HL026043 (to J. T. S.). This work was also supported by grants from the Moss Heart Fund and the Fouad A. and Val Imm Bashour Distinguished Chair in Physiology.
- RLC
- phosphorylatable regulatory light chain or myosin light chain 2 for ventricular myosin and myosin light chain 1 of smooth muscle myosin
- ZIPK
- zipper-interacting protein kinase
- DAPK
- death-associated protein kinase
- MLCK
- myosin light chain kinase
- NRCM
- neonatal rat cardiac myocytes
- LC/MS/MS
- liquid chromatography-tandem mass spectrometry
- siRNA
- small interfering RNA
- QPCR
- quantitative PCR
- PVDF
- polyvinylidene difluoride
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Kobayashi T., Solaro R. J. (2005) Annu. Rev. Physiol. 67, 39–67 [DOI] [PubMed] [Google Scholar]
- 2.Olsson M. C., Patel J. R., Fitzsimons D. P., Walker J. W., Moss R. L. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H2712–2718 [DOI] [PubMed] [Google Scholar]
- 3.Solaro R. J. (2008) J. Biol. Chem. 283, 26829–26833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweeney H. L., Stull J. T. (1986) Am. J. Physiol. 250, C657–660 [DOI] [PubMed] [Google Scholar]
- 5.Silver P. J., Buja L. M., Stull J. T. (1986) J. Mol. Cell. Cardiol. 18, 31–37 [DOI] [PubMed] [Google Scholar]
- 6.Levine R. J., Kensler R. W., Yang Z., Stull J. T., Sweeney H. L. (1996) Biophys. J. 71, 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stelzer J. E., Patel J. R., Moss R. L. (2006) J. Gen. Physiol. 128, 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweeney H. L., Bowman B. F., Stull J. T. (1993) Am. J. Physiol. 264, C1085–1095 [DOI] [PubMed] [Google Scholar]
- 9.Yang Z., Stull J. T., Levine R. J., Sweeney H. L. (1998) J. Struct. Biol. 122, 139–148 [DOI] [PubMed] [Google Scholar]
- 10.Zhi G., Ryder J. W., Huang J., Ding P., Chen Y., Zhao Y., Kamm K. E., Stull J. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17519–17524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanbe A., Fewell J. G., Gulick J., Osinska H., Lorenz J., Hall D. G., Murray L. A., Kimball T. R., Witt S. A., Robbins J. (1999) J. Biol. Chem. 274, 21085–21094 [DOI] [PubMed] [Google Scholar]
- 12.Dias F. A., Walker L. A., Arteaga G. M., Walker J. S., Vijayan K., Peña J. R., Ke Y., Fogaca R. T., Sanbe A., Robbins J., Wolska B. M. (2006) J. Mol. Cell. Cardiol. 41, 330–339 [DOI] [PubMed] [Google Scholar]
- 13.Tohtong R., Yamashita H., Graham M., Haeberle J., Simcox A., Maughan D. (1995) Nature 374, 650–653 [DOI] [PubMed] [Google Scholar]
- 14.Huang J., Shelton J. M., Richardson J. A., Kamm K. E., Stull J. T. (2008) J. Biol. Chem. 283, 19748–19756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scruggs S. B., Hinken A. C., Thawornkaiwong A., Robbins J., Walker L. A., de Tombe P. P., Geenen D. L., Buttrick P. M., Solaro R. J. (2009) J. Biol. Chem. 284, 5097–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stull J. T., Nunnally M. H., Michnoff C. H. (1986) in The Enzymes (Krebs E. G., Boyer P. D. eds) Vol. XVII, pp. 113–166, Academic Press, Orlando, FL [Google Scholar]
- 17.Chan J. Y., Takeda M., Briggs L. E., Graham M. L., Lu J. T., Horikoshi N., Weinberg E. O., Aoki H., Sato N., Chien K. R., Kasahara H. (2008) Circ. Res. 102, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seguchi O., Takashima S., Yamazaki S., Asakura M., Asano Y., Shintani Y., Wakeno M., Minamino T., Kondo H., Furukawa H., Nakamaru K., Naito A., Takahashi T., Ohtsuka T., Kawakami K., Isomura T., Kitamura S., Tomoike H., Mochizuki N., Kitakaze M. (2007) J. Clin. Invest. 117, 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haystead T. A. (2005) Cell. Signal. 17, 1313–1322 [DOI] [PubMed] [Google Scholar]
- 20.Ihara E., Edwards E., Borman M. A., Wilson D. P., Walsh M. P., MacDonald J. A. (2007) Am. J. Physiol. Cell. Physiol. 292, C1951–1959 [DOI] [PubMed] [Google Scholar]
- 21.Endo A., Surks H. K., Mochizuki S., Mochizuki N., Mendelsohn M. E. (2004) J. Biol. Chem. 279, 42055–42061 [DOI] [PubMed] [Google Scholar]
- 22.Kawai T., Matsumoto M., Takeda K., Sanjo H., Akira S. (1998) Mol. Cell. Biol. 18, 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDonald J. A., Borman M. A., Murányi A., Somlyo A. V., Hartshorne D. J., Haystead T. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihara E., MacDonald J. A. (2007) Can. J. Physiol. Pharmacol. 85, 79–87 [DOI] [PubMed] [Google Scholar]
- 25.Murakami U., Uchida K., Hiratsuka T. (1976) J. Biochem. 80, 611–619 [DOI] [PubMed] [Google Scholar]
- 26.Wu X., Clack B. A., Zhi G., Stull J. T., Cremo C. R. (1999) J. Biol. Chem. 274, 20328–20335 [DOI] [PubMed] [Google Scholar]
- 27.Ryder J. W., Lau K. S., Kamm K. E., Stull J. T. (2007) J. Biol. Chem. 282, 20447–20454 [DOI] [PubMed] [Google Scholar]
- 28.Simpson P., McGrath A., Savion S. (1982) Circ. Res. 51, 787–801 [DOI] [PubMed] [Google Scholar]
- 29.Simpson P., Savion S. (1982) Circ. Res. 50, 101–116 [DOI] [PubMed] [Google Scholar]
- 30.Ding H. L., Ryder J. W., Stull J. T., Kamm K. E. (2009) J. Biol. Chem. 284, 15541–15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niiro N., Ikebe M. (2001) J. Biol. Chem. 276, 29567–29574 [DOI] [PubMed] [Google Scholar]
- 32.Kawai T., Akira S., Reed J. C. (2003) Mol. Cell. Biol. 23, 6174–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhopadhyay R., Ray P. S., Arif A., Brady A. K., Kinter M., Fox P. L. (2008) Mol. Cell 32, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamm K. E., Stull J. T. (2001) J. Biol. Chem. 276, 4527–4530 [DOI] [PubMed] [Google Scholar]
- 35.Hagerty L., Weitzel D. H., Chambers J., Fortner C. N., Brush M. H., Loiselle D., Hosoya H., Haystead T. A. (2007) J. Biol. Chem. 282, 4884–4893 [DOI] [PubMed] [Google Scholar]
- 36.Morley S. J., Willett M. (2008) Dev. Cell 15, 639–640 [DOI] [PubMed] [Google Scholar]