Abstract
Small guanosine triphosphatases (GTPases) become activated when GDP is replaced by GTP at the highly conserved nucleotide binding site. This process is intrinsically very slow in most GTPases but is significantly accelerated by guanine nucleotide exchange factors (GEFs). Nucleotide exchange in small GTPases has been widely studied using spectroscopy with fluorescently tagged nucleotides. However, this method suffers from effects of the bulky fluorescent moiety covalently attached to the nucleotide. Here, we have used a newly developed real-time NMR-based assay to monitor small GTPase RhoA nucleotide exchange by probing the RhoA conformation. We compared RhoA nucleotide exchange from GDP to GTP and GTP analogues in the absence and presence of the catalytic DH-PH domain of PDZ-RhoGEF (DH-PHPRG). Using the non-hydrolyzable analogue guanosine-5′-O-(3-thiotriphosphate), which we found to be a reliable mimic of GTP, we obtained an intrinsic nucleotide exchange rate of 5.5 × 10−4 min−1. This reaction is markedly accelerated to 1179 × 10−4 min−1 in the presence of DH-PHPRG at a ratio of 1:8,000 relative to RhoA. Mutagenesis studies confirmed the importance of Arg-868 near a conserved region (CR3) of the Dbl homology (DH) domain and revealed that Glu-741 in CR1 is critical for full activity of DH-PHPRG, together suggesting that the catalytic mechanism of PDZ-RhoGEF is similar to Tiam1. Mutation of the single RhoA (E97A) residue that contacts the pleckstrin homology (PH) domain rendered the mutant 10-fold less sensitive to the activity of DH-PHPRG. Interestingly, this mutation does not affect RhoA activation by leukemia-associated RhoGEF (LARG), indicating that the PH domains of these two homologous GEFs may play different roles.
Keywords: Diseases/Cancer/Oncogene, Enzymes/Catalysis, Enzymes/Kinetics, Enzymes/Mechanisms, Enzymes/Structure, G Proteins/Low Molecular Weight, Methods/NMR, Protein/Protein-Protein Interactions
Introduction
The small GTPase3 RhoA, a member of the Ras superfamily, plays a crucial role in cellular processes including proliferation, movement, cell shape, as well as cell-cell and cell-matrix interactions (1). RhoA acts as a molecular switch, cycling between the inactive GDP- and activated GTP-bound states (Fig. 1A). Akin to other small GTPases, RhoA contains a phosphate binding loop (P-loop) and two switch regions that undergo conformational changes upon nucleotide cycling and mediate interactions with effector proteins. Together these three regions interact with the nucleotide phosphate groups and a magnesium ion that is required for high affinity nucleotide binding (low nm to sub nm Kd). GTPase-activating proteins catalyze nucleotide hydrolysis, thus inactivating GTPases, whereas guanine nucleotide exchange factors (GEFs) activate GTPases by stimulating nucleotide exchange (2). PDZ-RhoGEF, like its homologues LARG and p115-RhoGEF, is specific to RhoA and is activated by the Gα12/13 subunit of G-protein coupled receptors via its regulator of G-protein signaling domain. Interestingly, PDZ-RhoGEF single nucleotide polymorphisms have been associated with type II diabetes (3) and lung cancer (4).
FIGURE 1.
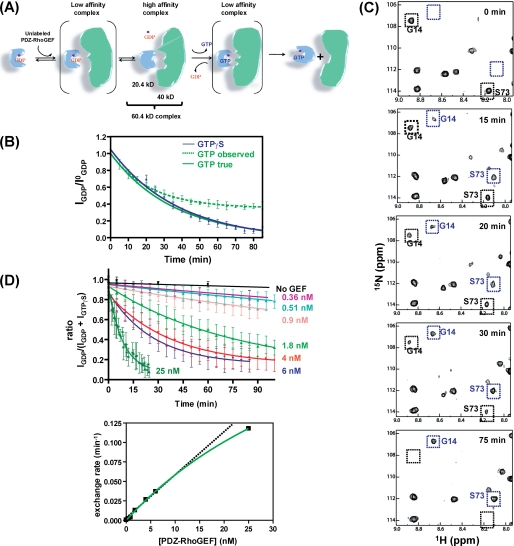
RhoA nucleotide exchange in the presence of DH-PHPRG monitored by NMR. A, schematic of RhoA nucleotide exchange mediated by DH-PHPRG. 15N-labeled RhoA (blue, 20.4 kDa) with GDP and Mg2+ (purple) bound is in an inactive state. DH-PHPRG (green, 43 kDa) interacts with residues in the RhoA switch regions, promoting the release of GDP. Subsequently, GTP binds RhoA, and the GEF is released to produce the activated form of RhoA. B, kinetics of RhoA-GDP nucleotide exchange to GTP (green dashed line) and GTPγS (blue) measured by the real-time NMR assay. A theoretical GTP exchange curve corrected for hydrolysis (see supplemental material) is also presented (green solid line) Error bars indicate S.D. for values reported by multiple peaks. C, snapshots of 1H-15N HSQC spectra during the 75-min time course of RhoA-GDP to GTPγS nucleotide exchange, in the presence of DH-PHPRG (6 nm). Black and blue boxes indicate the positions of the RhoA-GDP and RhoA-GTPγS cross peaks, respectively, for Gly-14 and Ser-73. D, RhoA-GDP to GTPγS nucleotide exchange, with increasing concentration of DH-PHPRG. The intrinsic nucleotide exchange rate is shown in black. Nucleotide exchange rates k (10−4 min−1) are 5.5, 19, 24, 39, 130, 267, 371, and 1,179 for GEF concentrations of 0, 0.36, 0.51, 0.9, 1.8, 4, 6, 25 nm, respectively. Lower panel, the hyperbolic (green curve) dependence, on GEF concentration, of the nucleotide exchange rate is indicative of a two-step binding model.
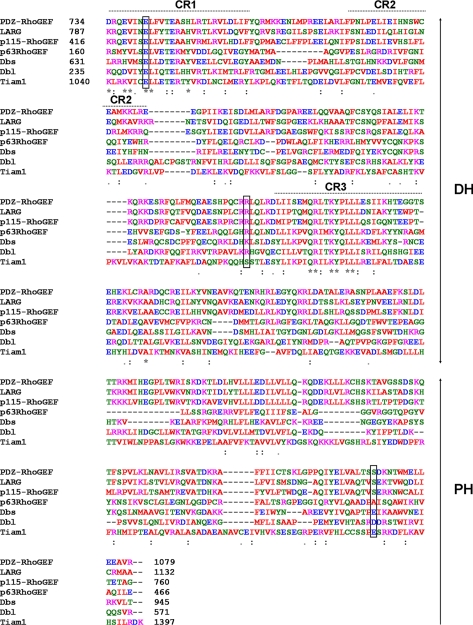
RhoGEFs are multidomain proteins (5), most of which contain a conserved catalytic Dbl homology (DH) domain with an associated pleckstrin homology (PH) domain (6, 7). Interactions between the DH domains and Rho GTPases induce structural changes of the nucleotide binding pocket and stabilize the nucleotide-free form of Rho (6). Although the DH domain is required for GEF activity, the PH domain plays multiple roles including stabilizing the DH domain, directing its subcellular localization, and regulating GEF activity (6, 7). DH domains are comprised of a helix bundle in which three highly conserved regions (CR1, CR2, and CR3) constitute the core domain. The DH domain interacts extensively, through CR1 and CR3, with the switch regions of the cognate GTPase as illustrated in the crystal structure of PDZ-RhoGEF (DH-PHPRG) in complex with RhoA (1XCG) (7). A highly conserved glutamate in CR1 and a conserved basic residue near CR3 (Fig. 2) have been implicated in the formation of the GEF-GTPase complex and in the nucleotide exchange catalysis in many RhoGEFs (6–8). Although mutations of the CR1 and CR3 region were found to affect the GEF activity of Tiam1 and Trio (6, 8, 9), the importance of the CR1 for PDZ-RhoGEF activity has not been investigated.
FIGURE 2.
Sequence alignment of DH-PH domains of human RhoGEFs. PDZ-RhoGEF (O15085, residues 734–923), LARG (Q9NZN5, residues 787–977), p115-RhoGEF (Q92888, residues 416–605), Dbl-GEF (Q92974,, residues 235–432), p63-RhoGEF (Q86VW2, residues 160–336), Dbs-GEF (O15068, residues 631–811), and Tiam1 (Q13009, residues 1,040–1,234) were aligned using Clustal_W2. Hydrophobic, acidic, basic, and polar residues are indicated in red, blue, magenta, and green, respectively. The stars mark residues conserved throughout Rho guanine nucleotide exchange factors. Colons correspond to conserved substitutions and, periods correspond to semiconserved substitutions. Conserved residues examined in this study (Glu-741, Arg-868, and Ser-1065) are boxed. CR1, CR2, and CR3 are three highly conserved regions of the DH domain.
The PH domain is comprised of seven antiparallel β-strands topped by a helix containing a partially conserved residue (Ser-1065 in PDZ-RhoGEF) that interacts with Glu-97 of RhoA (7). The DH domain alone is ∼41-fold less active than the DH-PH domain of PDZ-RhoGEF (7), but it is also less stable; thus the importance of the RhoA PH domain interaction for GEF activity is not clear.
We recently developed an NMR-based, real-time assay to measure the kinetics of GTP hydrolysis by the small GTPase Rheb by monitoring nucleotide-dependent changes in the NMR spectra of the GTPase protein (10). In this report, the NMR GTPase assay was extended to monitor intrinsic and PDZ-RhoGEF-mediated RhoA nucleotide exchange using native GDP and GTP as substrates. Our data show that PDZ-RhoGEF favors RhoA activation by catalyzing the exchange from GDP to GTP, severalfold more efficiently than the reverse reaction. Our mutagenesis studies demonstrate that Arg-868 near the CR3 and Glu-741 in the CR1 are critical for the full activity of PDZ-RhoGEF. We also found that RhoA Glu-97 is required for full activation by DH-PHPRG, suggesting that the PH domain plays an important role in catalysis. While investigating PDZ-RhoGEF-mediated RhoA nucleotide exchange reactions using different nucleotides, we discovered that GTPγS better mimics GTP than the analogues mant-GTP or GMPPNP.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification of RhoA and DH- PHPRG
A construct encoding human RhoA residues 1–181 was cloned into pET28A using NdeI and EcoRI restriction endonucleases and expressed in Escherichia coli BL21(DE3) cells as a His6-tagged protein The cells were grown in isotope-supplemented M9 media with 100 μg/ml kanamycin and induced at A600 = 0.8 with 0.25 mm isopropyl β-d-1-thiogalactopyranoside overnight at 15 °C. To prepare samples for resonance assignment, 2 g/liter [13C]glucose and 1 g/liter [15N]ammonium chloride were added to the media, whereas unlabeled glucose and [15N]ammonium chloride were used to prepare samples for the nucleotide exchange assays. The cells were harvested by centrifugation, resuspended with lysis buffer (50 mm Tris, pH 8, 150 mm NaCl, 5 mm MgCl2, 10% glycerol, 10 mm imidazole, 10 mm β-mercaptoethanol, 0.1% Nonidet P-40, 0.1 mm GDP, 1 mm phenylmethylsulfonyl fluoride), and sonicated, and the supernatant was stirred with nickel-nitrilotriacetic acid agarose (Qiagen) for 1 h at 4 °C. The resin was washed, and the protein was eluted with 50 mm Tris (pH = 8), 150 mm NaCl, 10% glycerol, 250 mm imidazole, 10 mm β-mercapto-ethanol, 5 mm MgCl2. Thrombin was used to cleave the His tag during dialysis against 50 mm Tris (pH 8), 150 mm NaCl, 2% glycerol, 5 mm MgCl2, 2 mm dithiothreitol, and 2 mm TCEP. Final purification of RhoA was achieved by gel filtration chromatography (Superdex S75 20/60, GE Healthcare) with 20 mm HEPES (pH 7.0), 100 mm NaCl, 5 mm MgCl2, and 2 mm TCEP. A construct encoding the DH and PH domains of human PDZ-RhoGEF (DH-PHPRG, residues 713–1,081) was subcloned into the vector PGEX 4T between the BamHI and EcoRI restriction endonuclease sites. DH-PHPRG was expressed as a glutathione S-transferase fusion protein in E. coli BL21(DE3) Codon+ cells in Luria-Bertani broth supplemented with ampicillin and chloramphenicol (100 and 50 μg/ml, respectively) by induction with 0.25 mm isopropyl β-d-1-thiogalactopyranoside at 15 °C overnight. Following cell lysis by sonication, the fusion protein was enriched by binding to glutathione-Sepharose resin (GE Healthcare), eluted with 20 mm reduced glutathione, cleaved with thrombin overnight, and dialyzed against 10 mm Tris, pH 7.5, 150 mm NaCl, 2% glycerol, 2 mm dithiothreitol. Glutathione S-transferase was removed by an ion exchange HiTrap SP column using 20 mm Tris, pH 7.5, 1 mm dithiothreitol, and a 50–500 mm NaCl gradient. PDZ-RhoGEF DH-PH (713–1,081) domain was purified to homogeneity by gel filtration chromatography (Superdex 75 20/60) and concentrated in an Amicon 30,000 nominal molecular weight limit centrifugal concentrator. The mutants RhoA-E97A, DH-PHPRG-E741A, DH-PHPRG-E741D, and DH-PHPRG-R868G were generated using QuikChange mutagenesis (Stratagene) according to the manufacturer's instructions.
Circular Dichroism
CD spectra were recorded for the wild-type and mutant forms of DH-PHPRG in 20 mm HEPES buffer, pH 7, 100 mm NaCl, 5 mm MgCl2, 2 mm TCEP using a cell with a 1-mm path length, at room temperature, on a Jasco J-815 CD spectrometer.
NMR Resonance Assignments
NMR experiments were performed at 20 °C on a 800-MHz Bruker AVANCE II spectrometer, equipped with a 5-mm TCI CryoProbe, and on a four-channel Varian Inova 600-MHz spectrometer equipped with z axis pulsed field gradient units and room temperature shielded triple resonance probes. NMR data were processed and analyzed using the NMRPipe (11) and NMRView programs (12). Chemical shift assignments were obtained using a 15N, 13C double-labeled sample of 0.3 mm RhoA-GDP or RhoA-GTPγS in 20 mm HEPES buffer, 100 mm NaCl, 5 mm MgCl2, 2 mm TCEP, 10% D2O, pH 7. Amide backbones 1H, 15N, CO, Hα, Cα, and Cβ were assigned using two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) and the conventional three-dimensional triple-resonance experiments (13, 14), HNCACB, CBCA(CO)NH, HN(CA)CO, HNCO, HNHA, and three-dimensional 15N-edited-nuclear Overhauser effect spectroscopy HSQC with a mixing time of 100 ms.
Nucleotide Exchange Assays
RhoA nucleotide exchange was monitored using NMR spectroscopy by acquiring successive 1H-15N HSQC spectra (two scans, 5-min acquisition time) at 20 °C over a period of time. DH-PHPRG was added to 15N-labeled GDP-bound RhoA (or 15N-labeled GTPγS-bound RhoA) along with a 5-fold molar excess of nucleotide (GTP or analogue) at time 0. Nine assigned RhoA amides, with distinct chemical shifts in the GDP- and GTPγS-bound form, were used to evaluate the fraction of GDP-bound RhoA remaining in each spectrum. The data were fitted to a single phase exponential decay function to obtain the exchange rate kex for non-hydrolyzable analogues. For hydrolyzable nucleotides, kex was determined by fitting the data to an equation that considers both the nucleotide exchange and hydrolysis (see supplemental material for a more detailed methodology).
Concentration dependence of the GEF activity of DH-PHPRG was assessed using 0.2 mm 15N-labeled RhoA GDP-bound, 1 mm GTPγS, and unlabeled DH-PHPRG at concentrations from 0.36 to 25 nm. DH-PHPRG mutation studies were performed with 0.2 mm [15N]-RhoA-GDP and 1.8 nm DH-PHPRG (wild-type or mutants E741A, E741D, or R868G) and 1 mm GTPγS. Each of these mutants was determined to be well folded on the basis of circular dichroism spectra (supplemental Fig. S1). The nucleotide exchange assays of wild-type RhoA (0.2 mm) and the E97A mutant (0.2 mm) were compared using 4 nm DH-PHPRG and a 5-fold molar excess of GTPγS.
A competition assay was carried out to assess the relative affinities of RhoA for different nucleotides. A series of 1H-15N HSQC spectra were recorded over 24 h following the addition of DH-PHPRG (4 nm) and a mixture of GTP, GTPγS, GMPPNP, and mant-GTP (each 1 mm) to 15N-labeled RhoA-GDP (0.2 mm). Binding of these nucleotides was assessed using the cross-peak of Gln-29, which exhibits distinct chemical shifts with each nucleotide.
RESULTS AND DISCUSSION
RhoA Nucleotide Exchange Stimulated with DH-PHPRG
To explore the possibility of using the NMR assay for RhoA nucleotide exchange, we first sought to assign backbone resonances of RhoA. Using the standard triple-resonance approach, we were able to assign nearly 90% of the RhoA-GDP backbone amide 1H and 15N resonances (supplemental Fig. S2a), as well as the Cα, Cβ, and CO resonances. A few residues in switch I, switch II, and the P-loop could not be assigned due to severe peak broadening. The secondary structure of RhoA-GDP predicted by the chemical shift index approach (15) is consistent with the crystal structure (16) (supplemental Fig. S3a). We also assigned 111 out of 132 amide resonances present in the 1H-15N HSQC of RhoA in the GTPγS-bound state (supplemental Fig. S2b). A significant number of amide cross peaks, particularly those in the P-loop and switch regions, were missing in the spectrum. Peak intensities for some of the broadened amides increased when the temperature was elevated from 20 to 40 °C (supplemental Fig. S4, dash boxed insets), indicative of intermediate chemical exchange on the NMR time scale. Similar phenomena were previously reported for Rheb-GMPPNP (10) and c-Ha-Ras (17). These results suggest that the nucleotide binding site of RhoA is in equilibrium between two or more conformational states when bound to GTPγS. Comparison of the 1H-15N HSQC spectra of RhoA-GDP and RhoA-GTPγS shows that amides located in the P-loop, the switch regions, and amides near the binding site display the largest chemical shift changes (supplemental Fig. S3b). Although rapid hydrolysis precluded independent assignment of the GTP-bound form, assignments for most peaks in the HSQC spectrum of RhoA-GTP could be transferred from that of RhoA-GTPγS, which is markedly similar (supplemental Fig. S5).
We used 1H-15N HSQC spectra to monitor RhoA intrinsic nucleotide exchange from GDP to GTP. When a 10-fold excess of GTP was added to 15N-labeled GDP-loaded RhoA, the 1H-15N HSQC spectrum remained nearly identical to the spectrum of the GDP-loaded form for several days, indicating that intrinsic exchange is slow relative to intrinsic hydrolysis. With the addition of DH-PHPRG, RhoA-GTP peaks appeared and RhoA-GDP peak intensities decreased, indicative of nucleotide exchange. With a GEF:RhoA ratio of 1:50,000 the observed nucleotide exchange reaction plateaus at IGDP/I0GDP ∼0.4 after 1 h (Fig. 1B, dotted line) due to GTP hydrolysis. A true kex rate of 286 × 10−4 min−1 was obtained by fitting the observed data to an equation (see supplemental Equation 2) that considers both exchange and hydrolysis. This true GTP kex value was used to plot an exponential decay curve representing the exchange component (Fig. 1B, green curve) and was compared with the exchange rate constants obtained with three analogues, GTPγS, GMPPNP, and mant-GTP. The results revealed that GTPγS exchanges with kinetics similar to GTP (kex = 265.5 × 10−4 min−1, Fig. 1B, blue line), whereas GMPPNP and mant-GTP produced significantly slower rate constants (see “Effects of GTP Analogues on the Structure and Exchange Kinetics of RhoA”). Therefore GTPγS was substituted for GTP in the following nucleotide exchange assays, eliminating the GTP hydrolysis component.
To determine a GEF concentration range amenable to our nucleotide exchange assay, we performed a series of nucleotide exchange assays on [15N]-RhoA-GDP (0.2 mm) with increasing DH-PHPRG concentrations (0.36 nm to 25 nm). A 1H-15N HSQC spectrum (two scans, 5 min) of [15N]-RhoA-GDP was recorded before adding DH-PHPRG and 1 mm GTPγS. Spectra collected successively at 5-min intervals exhibit decreasing intensities of RhoA-GDP peaks and appearance of RhoA-GTPγS peaks (Fig. 1C) until only the latter are visible. The fraction of RhoA in the GDP-bound state (IGDP/(IGDP+IGTPγS)) was calculated from peak heights in each spectrum and fitted to an exponential decay curve to extract an exchange rate constant for each DH-PHPRG concentration. We found a rate of 5.5 × 10−4 min−1 for the intrinsic nucleotide exchange and observed 3.5–214-fold stimulation of exchange for these GEF concentrations, which represent ratios from 1:500,000 to 1:8,000 relative to RhoA (Fig. 1D). Akin to RhoA-p190DHPH (18), the rates follow a hyperbolic dependence on GEF concentration, indicative of a two-step model that includes the formation of a low affinity ternary complex of GEF and nucleotide-bound GTPase prior to the formation of the high affinity complex of GEF and nucleotide-free GTPase (Fig. 1A) (19). In previous fluorescence-based studies, the PDZ-RhoGEF DH-PH domain (8) was shown to stimulate exchange 124-fold at a 1:10 ratio relative to RhoA, which is 4–11 times faster than Tiam (20), Dbs (20), and LARG (21). Here, we obtained a greater enhancement (214-fold) in nucleotide exchange with a substantially lower ratio of GEF to RhoA (1:8,000). This may be due in part to the inhibitory effect of mant-labeled nucleotides on GEF activity (30) as well as the high protein concentrations used for the NMR assay.
Does DH-PHPRG Stimulate the Nucleotide Exchange of Active and Inactive Forms of RhoA Equally?
It has been assumed that GEFs promote nucleotide exchange with GDP and GTP equally and that the activation of small GTPases is determined by the higher cellular concentration of GTP (2). However, it has been shown that some GEFs, e.g. CDC25 (22) and ITSN1L (19), further favor GTPase activation by more efficiently catalyzing the exchange of GDP to GTP. Here, we examined the activity of DH-PHPRG in the reverse reaction with the replacement of GTPγS bound to RhoA with GDP.
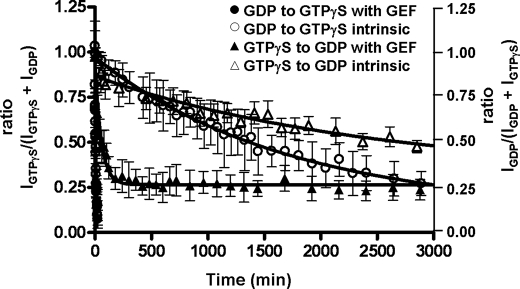
Although DH-PHPRG (25 nm) accelerates the nucleotide exchange reaction from RhoA-GDP to RhoA-GTPγS by a factor of 214 in the presence of a 5-fold molar excess of GTPγS, the reverse reaction from RhoA-GTPγS to RhoA-GDP is only stimulated by a factor of 31 (kex = 178 × 10−4 min−1, intrinsic rate kex = 5.8 × 10−4 min−1) in the presence of DH-PHPRG and a 50-fold molar excess of GDP (Fig. 3). Despite the large excess of GDP, the reaction reaches a plateau at IGTPγS ∼0.25 due to the very efficient PDZ-RhoGEF favoring the exchange reaction toward the RhoA activated form and as well as the higher affinity of RhoA for GTPγS (Fig. 3). The inefficient exchange from GTPγS to GDP in the absence of GEF indicates a higher RhoA affinity for GTPγS. The discrepancy in the -fold stimulation for the forward and reverse reactions suggests that the affinity of DH-PHPRG for RhoA is affected by nucleotide-dependent structural changes in RhoA. These results indicate that the GEF activity of DH-PHPRG depends on the nature of the bound nucleotide. Our results show that like CDC25 (22) and ITSN1L (19), PDZ-RhoGEF acts as a positive regulator by efficiently catalyzing nucleotide exchange in the forward direction (GDP to GTP), therefore driving the equilibrium toward the activation of the GTPase RhoA.
FIGURE 3.
PDZ-RhoGEF preferentially catalyzes nucleotide exchange in the direction of activation (GDP to GTPγS). Plots of the nucleotide exchange for RhoA-GDP to GTPγS (circles) and RhoA-GTPγS to GDP (triangles) (intrinsic, open; GEF-mediated, filled) are shown. A 5-fold molar excess of GTPγS was added to 15N-labeled RhoA-GDP, and a 50-fold molar excess of GDP was added to 15N-labeled RhoA-GTPγS. Error bars indicate S.D. for values reported by multiple peaks.
DH-PHPRG Mutations in the CR1 and CR3 Regions of the DH Domain Dramatically Reduce GEF Activity
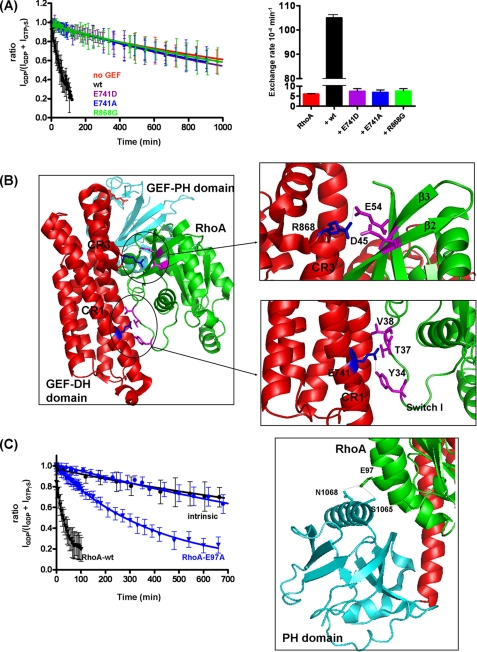
Using our NMR GEF assay combined with site-directed mutagenesis, we carried out a structure-function analysis of PDZ-RhoGEF. Based on the crystal structure (7), DH-PHPRG Arg-868 is hydrogen-bonded to RhoA Asp-45 and Glu-54, important residues for recognition. We first tested the effect of the mutation R868G in DH-PHPRG, which resulted in a dramatic reduction in GEF activity. At a DH-PHPRG (R868G) concentration of 1.8 nm (Fig. 4A, green curve), no GEF activity was detected, whereas a 19-fold increase was observed with the wild type (Fig. 4A, black curve). This is consistent with the crystal structure of PDZ-RhoGEF in complex with RhoA, in which the side chain of Arg-868 forms hydrogen bonds with Asp-45 and Glu-54 of RhoA (Fig. 4B) and was required for GEF activity (7, 8).
FIGURE 4.
Mutations in conserved regions of DH-PHPRG severely decrease GEF activity, and E97A mutation decreases the activation of RhoA. A, RhoA nucleotide exchange (GDP to GTPγS) stimulated by wild-type (wt) DH-PHPRG (1.8 nm) versus mutants E741D, E741A, and R868G. Rates derived from these curves are displayed in a histogram in the right panel. The experiments were also performed with 4 nm DH-PHPRG, in which wild-type, E741D, E741A, R868G, and intrinsic nucleotide exchange rates are 207 × 10−4 ± 3.3 × 10−5 min−1, 6.4 × 10−4 ± 1 × 10−4 min−1, 14.4 × 10−4 ± 1 × 10−4 min−1, 11.1 × 10−4 ± 1 × 10−4 min−1 and 4.4 × 10−4 ± 2 × 10−4 min−1, respectively. B, interface of PDZ-RhoGEF DH (red) and PH domains (cyan) with RhoA (green). DH-PHPRG Glu-741 (in CR1), Arg-868 (near CR3), and RhoA switch I residues Tyr-34, Thr-37, and Val-38 are highlighted (PDB code: 1XCG). PyMOL software was used to create the schematic representations. C, mutation of Glu-97 reduces the ability of DH-PHPRG to activate RhoA. RhoA nucleotide exchange to GTPγS in the absence (circles) or presence (triangles) of DH-PHPRG (4 nm). Black and blue curves correspond to wild-type RhoA and RhoA-E97A, respectively. Right panel, RhoA Glu-97 forms hydrogen bonds with Ser-1065 and Asn-1068 of the PH domain. In A and C, error bars indicate S.D. for values reported by multiple peaks.
In the crystal structure (7), PDZ-RhoGEF Glu-741 directly interacts with Tyr-34, Thr-37, and Val-38 of RhoA switch I, as do the homologous residues of Tiam1 and Rac1 (6, 23). We examined the role of Glu-741 in PDZ-RhoGEF activity by mutating this residue to Ala and Asp. Not surprisingly, E741A virtually eliminated GEF activity (kex = 5.7 × 10−4 min−1) when compared with the wild type (kex = 118 × 10−4 min−1). The loss of activity associated with this mutant clearly demonstrates the importance of this carboxyl group for the catalytic activity of PDZ-RhoGEF. We then addressed how a conservative glutamate to aspartate mutation at this position (E741D) would affect the activity. To our surprise, this substitution, which retains the carboxyl group but reduces the length of the side chain by one CH2, was also sufficient to nearly abolish the enzymatic activity of PDZ-RhoGEF (kex = 6 × 10−4 min−1). These results suggest that the shorter aspartate side chain cannot preserve the hydrogen bonds formed between Glu-741 and RhoA switch I residues (Fig. 4B). We believe that the interactions between the CR1 helix α1a in PDZ-RhoGEF and the RhoA switch I loop are crucial for the catalysis.
RhoA Glu-97 Is Important for Efficient DH-PHPRG-catalyzed Nucleotide Exchange
The PH domain is critical for the function of Dbl family proteins (24–27) and has been suggested to serve multiple roles. The Trio PH domain promotes proper cellular membrane localization through interactions with phospholipids (24), whereas the Vav PH domain regulates the GEF activity of the DH domain through binding of phosphatidylinositides (25), and the Dbs PH domain assists the DH-associated domain in binding Rho GTPases (26, 27). Based on the crystal structure of GEF-GTPase complexes, the PH domains of some GEFs interact with the GTPase (e.g. PDZ-RhoGEF-RhoA (1XCG), LARG-RhoA (1X86), and Dbs-RhoA (1LB1)), whereas no interaction is seen between other GEF PH domains and their cognate GTPases (e.g. Vav-Rac1 (2VRW) and Tiam1-Rac1 (1FOE)). Although the PH domains of LARG and PDZ-RhoGEF interact with RhoA, the PDZ-RhoGEF DH domain alone is 41-fold less active than DH-PH domain (7), whereas the equivalent truncation of the PH domain in LARG causes only a 1.5-fold reduction in activity (21). Because it has been reported that the DH domain alone of PDZ-RhoGEF is unstable (7, 8), it is not clear whether the PH domain contributes directly to the activity. The crystal structure of PDZ-RhoGEF bound to nucleotide-free RhoA shows that RhoA Glu-97 interacts with a conserved serine (Ser-1065) as well as an asparagine of the PH domain (Fig. 4C). To evaluate the role of this RhoA PH domain interaction, we mutated RhoA Glu-97 to alanine and monitored the nucleotide exchange with and without DH-PHPRG, using our NMR assay. Although the intrinsic nucleotide exchange rates remain identical for both RhoA-WT (kex = 5.5 × 10−4 min−1) and RhoA-E97A (kex = 5.3 × 10−4 min−1), a 10-fold decrease in the RhoA activation by DH-PHPRG was observed with RhoA-E97A (RhoA-WT: kex = 267 × 10−4 min−1, RhoA-E97A: kex = 25.5 × 10−4 min−1) (Fig. 4C). Although this protein-docking site was also found in the PDZ-RhoGEF homologue LARG, with the conserved serine Ser-1118 (Fig. 2) hydrogen-bonded to Glu-97 of RhoA (21), the Glu-97 mutation did not affect the ability of LARG to activate RhoA (21). These results together indicate that PDZ-RhoGEF PH domain contributes to RhoA activation by a mechanism involving the H-bonds between Glu-97 and Ser-1065 and Asn-1068, presumably as seen in the crystal structure of PDZ-RhoGEF-RhoA (7).
Effects of GTP Analogues on the Structure and Exchange Kinetics of RhoA
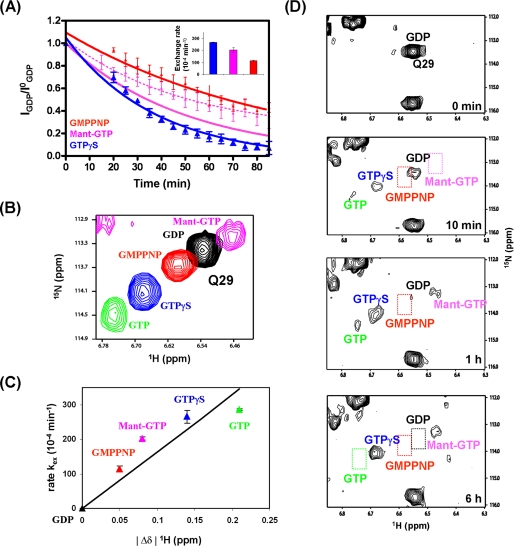
Many previous studies of GEFs have relied on assays using fluorescent nucleotide analogues (e.g. mant-GTP) (7, 8, 20, 21, 23, 27, 28). Our NMR approach has a substantial advantage over this method as it does not require a chemical modification of the nucleotide. We recently demonstrated that mant-GTP and mant-GDP alter the kinetics of the PDZ-RhoGEF-catalyzed nucleotide exchange as well as the intrinsic GTPase activity of RhoA (30). To further assess the effects of nucleotide modifications, we compared DH-PHPRG-mediated exchange kinetics of GTPγS, GMPPNP, and mant-GTP. The RhoA-GDP nucleotide exchange rates for mant-GTP (203 × 10−4min−1, corrected for hydrolysis, see supplemental material) and GMPPNP (114 × 10−4 min−1) are 30 and 55% slower than GTPγS (265.5 × 10−4 min−1), respectively (Fig. 5A). We then sought to identify aspects of RhoA structure that may account for these different exchange rates. Spectra of RhoA bound to GTPγS, GMPPNP, or mant-GTP were compared with those of RhoA-GDP and RhoA-GTP. The chemical shifts for RhoA residues located in the P-loop, switch I, switch II, β2, and β3 in the presence of GMPPNP and mant-GTP are more similar to those with GDP than GTP, whereas the fingerprint of RhoA-GTPγS is more like that of RhoA-GTP, as illustrated by Gln-29 of RhoA in Fig. 5B. These results further support that among these analogues, GTPγS provides the best mimic of GTP and that the conformation of RhoA-GTPγS most resembles the activated conformation of RhoA-GTP (Fig. 5C).
FIGURE 5.
Kinetics of RhoA nucleotide exchange with GTP analogues and competition assay. A, kinetics of RhoA-GDP nucleotide exchange to GTPγS (blue), GMPPNP (red), and mant-GTP (magenta) measured individually using our NMR-based assay. The rates (10−4 min−1) are displayed with a histogram (inset) for each curve. B, overlay of five 1H-15N HSQC spectra illustrating the distinct chemical shifts of Gln-29 exhibited by RhoA when bound to GDP (black), GMPPNP (red), GTP (green), GTPγS (blue), and mant-GTP (magenta). C, Gln-29 amide proton chemical shift changes (Δδ1H, relative to RhoA-GDP) versus exchange rates for each nucleotide (colored as above).In A and C, error bars indicate S.D. D, competition assay for binding of nucleotide analogues to RhoA. Snapshots of 1H-15N HSQC spectra collected before and after the addition of premixed GTP, GTPγS, GMPPNP, and mant-GTP, each at a 5-fold molar excess over RhoA.
To more rigorously compare the individual nucleotide exchange rates using GTP and the three commonly used analogues, GTPγS, GMPPNP, and mant-GTP, we performed a real-time NMR-based competition assay. Importantly, NMR spectroscopy can distinguish RhoA bound to each of the four nucleotide variants. We added 6 nm DH-PHPRG to RhoA-GDP and initiated the exchange assay with the addition of a mixture containing GTP, GTPγS, GMPPNP, and mant-GTP (each 1 mm) (Fig. 5D). GTP and GTPγS peaks appeared first in the 1H-15N HSQC spectra, indicating their preferential recruitment by RhoA. After 1 h (Fig. 5D), a stronger GTPγS peak was observed along with weaker GTP and mant-GTP peaks, but no peak was detected for GMPPNP. It should be noted that GTP and mant-GTP peak intensities eventually decreased due to intrinsic GTPase activity of RhoA (supplemental Fig. S6). The nucleotide exchange and competition assays show that association of mant-GTP with RhoA is slower than that of GTP and GTPγS. Based on the structure of Ras in complex with mant-GTP (29), we hypothesize that similar positioning of the mant group to RhoA may interfere with the fast rotation of its Tyr-34 side chain from the position where it binds Glu-741 to the orientation that contacts the nucleotide (supplemental Fig. S7).
Our data firmly demonstrate the differences in the binding affinity of RhoA for GTP and its three commonly used analogues. Although the exact binding constants for each analogue cannot be derived from the present NMR data, NMR can distinguish small differences in the binding affinity between these ligands as translated in the exchange rates: kexGTP ≅ kexGTPγS > kexmant-GTP > kexGMPPNP. It is worth noting that these thermodynamic differences in binding correlate well with conformational changes in RhoA observed in NMR spectra (Fig. 5, B and C).
In summary, we have demonstrated that NMR spectroscopy offers an accurate and efficient measurement of GEF-mediated nucleotide exchange of a small GTPase. This methodology was applied to RhoA intrinsic and PDZ-RhoGEF-catalyzed nucleotide exchange reactions. We have shown that GTPγS mimics GTP in the RhoA exchange reaction, whereas GMPPNP and mant-GTP suffer from slower exchange rates. The intrinsic nucleotide exchange rate from GDP to GTPγS is dramatically accelerated (214-fold) by the presence of a small amount of DH-PHPRG (25 nm). This effective catalysis is achieved by highly specific interactions between RhoA and PDZ-RhoGEF DH and PH domains. Our results show that Arg-868 near CR3 and Glu-741 in CR1 play critical roles in the catalysis and that Glu-97 of RhoA is required for full GEF activity. Furthermore, PDZ-RhoGEF acts as a positive regulator that selectively catalyzes nucleotide exchange toward the activation of the GTPase RhoA. The present NMR studies provided kinetic and mechanistic insights into the enzymatic reaction of the PDZ-RhoGEF-mediated nucleotide exchange of RhoA and also posed new questions about the structural basis for the catalysis, which can be addressed by an atomic resolution structure elucidation of the binary complex.
Acknowledgment
A grant from the Canada Foundation for Innovation funded the Bruker 800- and 600-MHz NMR spectrometers.
This work was supported by a grant from the Cancer Research Society (Canada).
The backbone assignments reported in this paper have been submitted to the Biological Magnetic Resonance Data Bank under entries 16668 and 16669.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S7.
- GTPase
- guanosine triphosphatase
- GTPγS
- guanosine-5′-O-(3-thiotriphosphate)
- GMPPNP
- guanylyl imidodiphosphate
- mant-GTP
- N-methylanthraniloyl guanosine triphosphate
- GEF
- guanine nucleotide exchange factor
- DH
- Dbl homology
- PH
- pleckstrin homology
- DH-PHPRG
- catalytic DH-PH domain of PDZ-RhoGEF
- LARG
- leukemia-associated RhoGEF
- 1H-15N HSQC
- 1H-15N heteronuclear single quantum coherence
- TCEP
- tris(2-carboxyethyl)phosphine.
REFERENCES
- 1.Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 2.Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 3.Fu M., Sabra M. M., Damcott C., Pollin T. I., Ma L., Ott S., Shelton J. C., Shi X., Reinhart L., O'Connell J., Mitchell B. D., Baier L. J., Shuldiner A. R. (2007) Diabetes 56, 1363–1368 [DOI] [PubMed] [Google Scholar]
- 4.Gu J., Wu X., Dong Q., Romeo M. J., Lin X., Gutkind J. S., Berman D. M. (2006) Cancer 106, 2716–2724 [DOI] [PubMed] [Google Scholar]
- 5.Schmidt A., Hall A. (2002) Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 6.Hoffman G. R., Cerione R. A. (2002) FEBS Lett. 513, 85–91 [DOI] [PubMed] [Google Scholar]
- 7.Derewenda U., Oleksy A., Stevenson A. S., Korczynska J., Dauter Z., Somlyo A. P., Otlewski J., Somlyo A. V., Derewenda Z. S. (2004) Structure 12, 1955–1965 [DOI] [PubMed] [Google Scholar]
- 8.Oleksy A., Opaliński Ł., Derewenda U., Derewenda Z. S., Otlewski J. (2006) J. Biol. Chem. 281, 32891–32897 [DOI] [PubMed] [Google Scholar]
- 9.Liu X., Wang H., Eberstadt M., Schnuchel A., Olejniczak E. T., Meadows R. P., Schkeryantz J. M., Janowick D. A., Harlan J. E., Harris E. A., Staunton D. E., Fesik S. W. (1998) Cell 95, 269–277 [DOI] [PubMed] [Google Scholar]
- 10.Marshall C. B., Ho J., Buerger C., Plevin M. J., Li G. Y., Li Z., Ikura M., Stambolic V. (2009) Sci. Signal. 2, ra3. [DOI] [PubMed] [Google Scholar]
- 11.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 12.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 13.Ikura M., Kay L. E., Bax A. (1990) Biochemistry 29, 4659–4667 [DOI] [PubMed] [Google Scholar]
- 14.Clore G. M., Gronenborn A. M. (1998) Curr. Opin. Chem. Biol. 2, 564–570 [DOI] [PubMed] [Google Scholar]
- 15.Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 16.Wei Y., Zhang Y., Derewenda U., Liu X., Minor W., Nakamoto R. K., Somlyo A. V., Somlyo A. P., Derewenda Z. S. (1997) Nat. Struct. Biol. 4, 699–703 [DOI] [PubMed] [Google Scholar]
- 17.Ito Y., Yamasaki K., Iwahara J., Terada T., Kamiya A., Shirouzu M., Muto Y., Kawai G., Yokoyama S., Laue E. D., Wälchli M., Shibata T., Nishimura S., Miyazawa T. (1997) Biochemistry 36, 9109–9119 [DOI] [PubMed] [Google Scholar]
- 18.Hemsath L., Ahmadian M. R. (2005) Methods 37, 173–182 [DOI] [PubMed] [Google Scholar]
- 19.Kintscher C., Groemping Y. (2009) J. Mol. Biol. 387, 270–283 [DOI] [PubMed] [Google Scholar]
- 20.Snyder J. T., Worthylake D. K., Rossman K. L., Betts L., Pruitt W. M., Siderovski D. P., Der C. J., Sondek J. (2002) Nat. Struct. Biol. 9, 468–475 [DOI] [PubMed] [Google Scholar]
- 21.Kristelly R., Gao G., Tesmer J. J. (2004) J. Biol. Chem. 279, 47352–47362 [DOI] [PubMed] [Google Scholar]
- 22.Lai C. C., Boguski M., Broek D., Powers S. (1993) Mol. Cell Biol. 13, 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worthylake D. K., Rossman K. L., Sondek J. (2000) Nature 408, 682–688 [DOI] [PubMed] [Google Scholar]
- 24.Skowronek K. R., Guo F., Zheng Y., Nassar N. (2004) J. Biol. Chem. 279, 37895–37907 [DOI] [PubMed] [Google Scholar]
- 25.Han J., Luby-Phelps K., Das B., Shu X., Xia Y., Mosteller R. D., Krishna U. M., Falck J. R., White M. A., Broek D. (1998) Science 279, 558–560 [DOI] [PubMed] [Google Scholar]
- 26.Rossman K. L., Cheng L., Mahon G. M., Rojas R. J., Snyder J. T., Whitehead I. P., Sondek J. (2003) J. Biol. Chem. 278, 18393–18400 [DOI] [PubMed] [Google Scholar]
- 27.Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. (2002) EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oleksy A., Barton H., Devedjiev Y., Purdy M., Derewenda U., Otlewski J., Derewenda Z. S. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 740–742 [DOI] [PubMed] [Google Scholar]
- 29.Scheidig A. J., Franken S. M., Corrie J. E., Reid G. P., Wittinghofer A., Pai E. F., Goody R. S. (1995) J. Mol. Biol. 253, 132–150 [DOI] [PubMed] [Google Scholar]
- 30.Mazhab-Jafari M. T., Marshall C. B., Smith M., Gasmi-Seabrook G. M., Stambolic V., Rottapel R., Neel B. G., Ikura M. (2009) J. Biol. Chem. 285, 5132–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]