Abstract
Usa1p is a recently discovered member of the HRD ubiquitin ligase complex. The HRD pathway is a conserved route of ubiquitin-dependent, endoplasmic reticulum (ER)-associated degradation (ERAD) of numerous lumenal (ERAD-L) and membrane-anchored (ERAD-M) substrates. We have investigated Usa1p to understand its importance in HRD complex action. Usa1p was required for the optimal function of the Hrd1p E3 ubiquitin ligase; its loss caused deficient degradation of both membrane-associated and lumenal proteins. Furthermore, Usa1p functioned in regulation of Hrd1p by two mechanisms. First, Hrd1p self-degradation, which serves to limit the levels of uncomplexed E3, is absolutely dependent on Usa1p and the ubiquitin-like (Ubl) domain of Usa1p. We found that Usa1p allows Hrd1p degradation by promoting trans interactions between Hrd1p molecules. The Ubl domain of Usa1p was required specifically for Hrd1p self-ubiquitination but not for degradation of either ERAD-L or ERAD-M substrates. In addition, Usa1p was able to attenuate the activity-dependent toxicity of Hrd1p without compromising substrate degradation, indicating a separate role in ligase regulation that operates in parallel to stability control. Many of the described actions of Usa1p are distinct from those of Der1p, which is recruited to the HRD complex by Usa1p. Thus, this novel, conserved factor is broadly involved in the function and regulation of the HRD pathway of ERAD.
Keywords: Protein/Degradation, Protein/Turnover, Subcellular Organelles/Endoplasmic Reticulum, E3 Ubiquitin Ligase, Ubiquitin, ERAD, HRD Pathway, Hrd1, Usa1
Introduction
ER3-associated degradation (ERAD) is a conserved process by which eukaryotic cells target and degrade ER-resident proteins by the ubiquitin-proteasome pathway. ERAD pathways play a major role in the destruction of misfolded or unassembled ER proteins, including both lumenal and integral-membrane substrates. In addition, normal proteins are regulated by this pathway, the most prominent case being the sterol pathway-regulated degradation of HMG-CoA reductase in both yeast and mammals (1, 2). Eukaryotic ERAD is brought about by the action of multiple pathways of ubiquitin-mediated degradation that operate at the ER surface (3, 4).
Covalent addition of ubiquitin to proteins brings about their recognition and degradation by the cytosolic 26S proteasome. Protein ubiquitination occurs by a cascade of enzymes that add 7.6-kDa ubiquitin to the targeted protein. The E1 ubiquitin-activating enzyme first forms a high energy bond with ubiquitin in an ATP-dependent reaction, and then the ubiquitin is transferred to an E2, or ubiquitin-conjugating enzyme. E2-bound ubiquitin is next transferred from the charged E2 to the target protein by the action of a ubiquitin ligase, or E3, that ensures specificity of transfer to the proper degradation substrate. The action of the E3 is iterative, causing the construction of a substrate-bound multiubiquitin chain that is recognized by the 26S proteasome (5). Several E3 ubiquitin ligases are involved in the destruction of ER proteins. Thus, ERAD is a composite of ubiquitination pathways with distinct ligases that use both separate and common components to effect recognition, ubiquitination, and delivery of ER substrates to the cytosolic proteasome. It now appears that distinct complexes of proteins are responsible for these separate ERAD pathways (3, 4).
Yeast Hrd1p is one of several highly conserved ER-localized E3 ligases that mediate ERAD in eukaryotes (6, 7). Hrd1p has an N-terminal multispanning membrane domain that anchors it in the ER and a C-terminal domain with a RING-H2 motif found in many E3 ligases. At its natural levels Hrd1p exists in complex with several other proteins. Initial studies spawned from the genetic analysis of the HRD pathway revealed Hrd3p as a stoichiometric binding partner (7). Hrd3p promotes Hrd1p stability, appears to enhance Hrd1p-dependent recognition of some substrates, and recruits factors that mediate substrate detection and delivery to the HRD complex (8–11). More recent proteomic studies revealed that a number of proteins reside in the HRD complex, including Hrd1p, Hrd3p, Der1p, Ubx2p, the Cdc48-Npl4-Ufd1 complex, Kar2p, Yos9p, and Usa1p (3, 4). Of these, only Usa1p was novel and was, thus, the most poorly characterized of the HRD complex components.
Because the Usa1p protein is uniquely found in the HRD complex and has corresponding proteins in other eukaryotes, it might be expected to play a pivotal role in HRD function, albeit one not revealed by genetic approaches. Usa1p is predicted to be a two-transmembrane-spanning protein, with a large N-terminal cytoplasmic region containing a ubiquitin-like (Ubl) domain. Usa1p has been shown to be required for degradation of lumenal ERAD substrates and to link Der1p to the HRD complex (3). These studies posited that Usa1p was uniquely required for HRD-mediated degradation of lumenal ERAD, or ERAD-L, substrates (3). We have more fully investigated the function of Usa1p in the HRD complex and found that it is in fact broadly involved in ERAD function and HRD ligase regulation.
As reported, we observed that Usa1p was required for degradation of lumenal substrates (3). In addition, we show that Usa1p is also required for optimal degradation of membrane-bound (ERAD-M) substrates. Although many proteins are involved in the function of the HRD complex and have been relatively well characterized, much less is known about the regulation of this pathway. In these studies we have found that Usa1p plays a multifaceted role in regulation of Hrd1p. Usa1p was absolutely required for Hrd1p self-catalyzed degradation, and the Ubl domain of Usa1p uniquely functioned in this capacity. Usa1p-dependent Hrd1p self-degradation had mechanistic features distinct from all other ERAD substrates. In addition to governing autoregulation, phenotypic studies revealed that Usa1p had a separate role in modulating Hrd1p activity, limiting the toxicity of Hrd1p while preserving its ability to degrade bona fide ERAD substrates.
Taken together, our studies show that Usa1p has a vital role in the HRD complex at multiple levels, consistent with this highly conserved presence in all eukaryotes. It is important for degradation of all classes of ERAD substrates and has thus far unique roles in the regulation of HRD ligase stability and specificity.
EXPERIMENTAL PROCEDURES
Plasmids and DNA Methods
The following plasmids were previously described: pRH469 (Hmg2p-GFP) (12), pRH244 (6myc-Hmg2p) (13), pRH423 (1myc-Hmg2p) (14), pRH 2038 (TDH3-Der1p) (15), pRH 808 (TDH3-Hrd1p) (6). Plasmids expressing HA-CPY* (pRH1377) and KWW-HA (pRH1960) were obtained from Davis Ng (16). Details about plasmids constructed for this study can be found in supplemental Table S1. All plasmids were constructed with standard molecular biology techniques as previously described (17). PCR was carried out as previously described (17), and plasmids constructed using PCR were sequence-verified (Eton Bioscience, Inc.).
Yeast Strains and Media
Yeast strains were grown in minimal medium supplemented with 20% dextrose and appropriate amino acids at 30 °C as previously described (18). All strains were isogenic and are tabulated in supplemental Table S2). More information about strain construction is provided in the supplemental Materials and Methods.
Dilution Assays
Yeast strains were grown to high density (A600 > 1.5). Beginning with an A600 of 0.35, 5-fold serial dilutions were made and spotted onto the appropriate drop-out plates followed by incubation at the indicated temperatures for 2 days (30 and 35 °C) or 3 days (37 °C).
Degradation Assays and Unfolded Protein Response (UPR) Measurements
Cycloheximide chase assays were performed by the addition of cycloheximide to log-phase cultures followed by lysis at the indicated times, as previously described (17, 18) and in supplemental Materials and Methods. Equal loading of gels was confirmed by India ink staining of the nitrocellulose membranes (18). GFP levels were analyzed in living cells (10,000 cells per sample) by flow microfluorimetry of log-phase cultures (12) using a FACScalibur machine (BD Biosciences) and CellQuest software.
Immunoprecipitation
Immunoprecipitations were performed as described (19, 20), with minor variations detailed in the supplemental Materials and Methods. In vivo Hrd1p ubiquitination was assayed by immunoprecipitating Hrd1p from pdr5Δ null strains after treatment of log-phase cultures (A600 < 0.25) with the proteasome inhibitor MG132 (benzyloxycarbonyl-Leu-Leu-aldehyde; Sigma) for 2.25 h before lysis.
Antibodies and Immunoblotting
Immunoblotting for proteins in cellular lysates or in ubiquitination assays was performed as described (19, 20) with variations and reagent descriptions detailed in supplemental “Materials and Methods”.
RESULTS
The ER-resident, integral membrane protein Hmg2p is the prototype substrate of the HRD pathway. Hmg2p is the isozyme of yeast HMG-CoA reductase that undergoes regulated degradation by the HRD pathway in response to changing levels of sterol pathway signals (21). Using this ERAD-M substrate, it was reported that Usa1p was not involved in this branch of ERAD (3). We first confirmed that active Hmg2p underwent normal degradation in a usa1Δ null mutant as measured by cycloheximide chase (Fig. 1A). Comparison of the degradation of 1myc-Hmg2p in wild type or usa1Δ cells revealed no discernable difference in half-life, as reported (3). We also tested the related substrate 6myc-Hmg2p, which because of an insertion of six tandem myc tags in the transmembrane region, is constitutively degraded by the HRD pathway. That is, the 6myc-Hmg2p degradation rate is constant and unresponsive to changes in the sterol pathway signals that control Hmg2p stability (13). In all other ways the unregulated 6myc substrate is identical to wild type Hmg2p in its requirements for HRD pathway degradation. In contrast to Hmg2p, 6myc-Hmg2p showed clear stabilization in the usa1Δ (Fig. 1B), implying that Usa1p did have a role in ERAD-M.
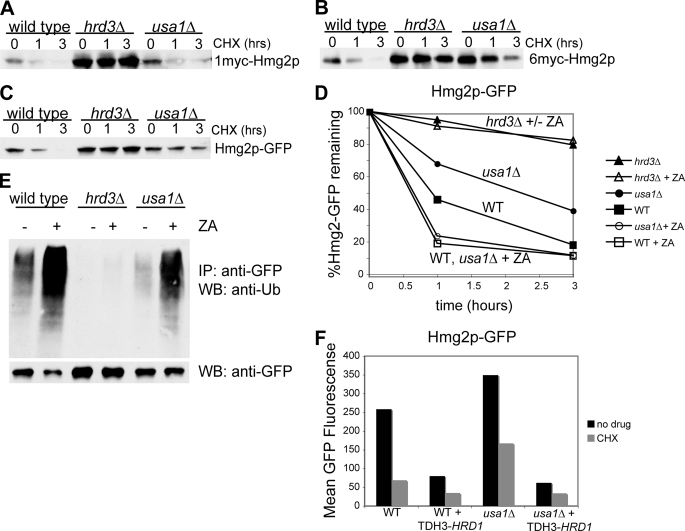
FIGURE 1.
Usa1p is required for optimal degradation of ERAD-M substrates. Log-phase cultures of either WT, hrd3Δ, or usa1Δ cells expressing normally regulated, catalytically active 1myc-Hmg2p (A), or the unregulated variant 6myc-Hmg2p (B) were subjected to cycloheximide (CHX) chase for the indicated times followed by lysis and immunoblotting to evaluate protein stability. The hrd3Δ strain served as a positive control that stabilizes both substrates. C, cycloheximide chase of Hmg2p-GFP is shown. Log-phase cultures of either WT, hrd3Δ, or usa1Δ cells expressing the normally regulated but catalytically inactive Hmg2p-GFP were subjected to cycloheximide chase. D, degradation of Hmg2p-GFP was evaluated by flow cytometry of live cells at the indicated times after cycloheximide addition using 10,000 cells per point in this and all subsequent flow cytometric experiments. For each strain used (WT (squares), hrd3Δ (triangle), or usa1Δ (circles)) the experiment was run with (open symbols) or without (solid symbols)) 10 μg/ml ZA to evaluate the effect of elevating degradation signal on Hmg2p-GFP degradation. E, shown is ubiquitination of Hmg2p-GFP in WT, hrd3Δ, or usa1Δ strains. Log-phase cultures of the indicated strains expressing Hmg2p-GFP were incubated for 5 min with or without 10 μg/ml ZA followed by lysis, immunoprecipitation (IP), and immunoblotting (WB) for Hmg2p-GFP (bottom row) or ubiquitin immunoreactivity. The − lanes indicate the ubiquitination state of Hmg2p normally present in each strain without drug treatment. F, Hrd1p overexpression suppressed the Hmg2p-GFP degradation defect of usa1Δ strains. Wild type or usa1Δ strains harboring either empty vector or a HRD1-overexpressing plasmid (TDH3-HRD1) were subject to cycloheximide chase for 2 h (gray bars) and compared with strains that were not treated with drug (black bars) by flow cytometry.
What is the cause of this difference in degradation? The normal Hmg2p in Fig. 1A was responsive to increases in sterol pathway activity provided by its own catalytic domain, whereas 6myc-Hmg2p was not. Thus, one explanation for the differing response to the usa1Δ null mutant is that the normal Hmg2p used in the original analysis had its degradation rate sufficiently enhanced by its own catalytic activity to overcome a partial loss of HRD pathway activity in the usa1Δ, whereas the unregulated 6myc-Hmg2p, although also catalytically active, could not similarly respond. To test this idea, we examined the effect of a usa1Δ on the normally regulated but catalytically inactive Hmg2p-GFP, which has an intact Hmg2p transmembrane domain, with the catalytic region replaced by GFP. The use of Hmg2p-GFP allows quantitative examination of degradation by cycloheximide chase with flow microfluorimetry (21, 22). Like 6myc-Hmg2p, Hmg2p-GFP was stabilized by the usa1Δ null mutant when compared with wild type in a cycloheximide chase (Fig. 1C). In the strain used to test Hmg2p-GFP, the HMG-CoA reductase activity was provided by the native HMG-CoA reductase genes, which produce ample activity for life but less activity than that from the full-length, myc-tagged Hmg2p used in Fig. 1A. Thus, the sterol pathway-derived degradation signals in the Hmg2p-GFP strain were lower than in the 1myc-Hmg2p strain of Fig. 1A and allowed the difference between wild type and usa1Δ cells to be observed. Consistent with this idea, treating the cells with the drug zaragozic acid (ZA), which increases the cellular signal for Hmg2p degradation (21), restored wild type degradation of Hmg2p-GFP in the usa1Δ strain (Fig. 1D). Degradation of Hmg2-GFP was about 2-fold slower in the usa1Δ as compared with the wild type strain (closed symbols). A hrd3Δ null strain was included as a control, showing the expected severe block in degradation of Hmg2p-GFP and illustrating that the usa1Δ defect in membrane substrate degradation resulted in an intermediate phenotype. Treatment of either a wild type or a usa1Δ strain with ZA (open symbols) resulted in equivalent Hmg2p-GFP degradation in both the wild type and usa1Δ strains. Normally regulated Hmg2p was stabilized by a usa1Δ, but the partial block to the Hrd1p pathway can be overcome by sufficient elevation of its sterol pathway degradation signal either by the in cis catalytic activity in the case of 1myc-Hmg2p or by pharmacological elevation of the signal with ZA in the case of Hmg2p-GFP. Thus, a usa1Δ null mutant imposes a partial defect in degradation of ERAD-M substrates.
Because Usa1p is part of the HRD ubiquitin ligase complex, we directly tested its role in Hrd1p-mediated Hmg2p-GFP ubiquitination by immunoprecipitation of the substrate followed by immunoblotting for either the attached ubiquitin or the substrate itself as indicated (Fig. 1E). In the usa1Δ strain, Hmg2p-GFP ubiquitination was significantly decreased (compare the wild type and usa1Δ − lanes, Fig. 1E). The addition of ZA to increase the degradation signal greatly increased Hmg2p-GFP ubiquitination in both the wild type and usa1Δ strains, consistent with the ability of increased degradation signal to overcome the usa1Δ-stabilizing effect on Hmg2p-GFP shown in Fig. 1D. Taken together, these experiments indicated that Usa1p was required for optimal function of Hrd1p toward ERAD-M substrates; the usa1Δ null mutant does not cause a complete loss of Hrd1p activity. We would predict that numerous ERAD-M substrates should show some degree of stabilization in a usa1Δ null mutant. Below, we show that the degradation of another ERAD-M substrate, Sec61-2p, was also impaired in a usa1Δ strain (see Fig. 4B, right).
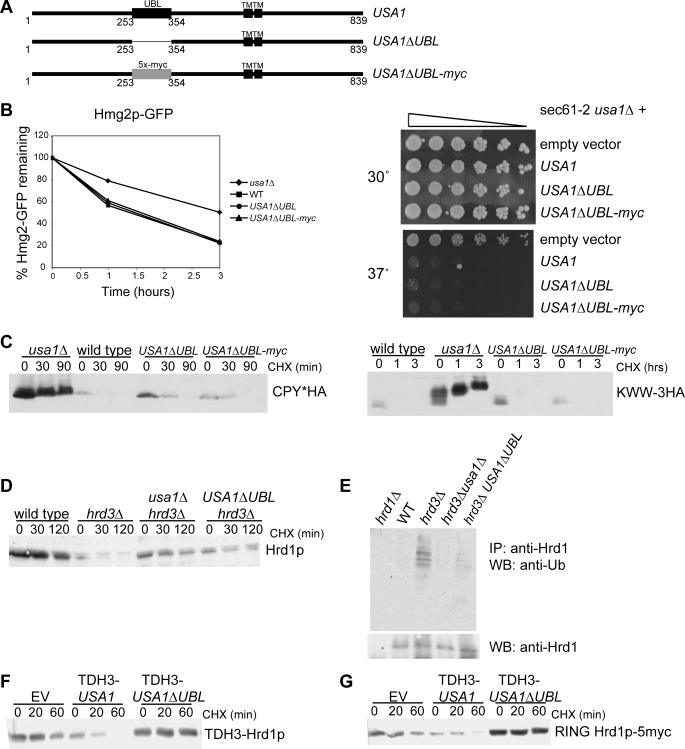
FIGURE 4.
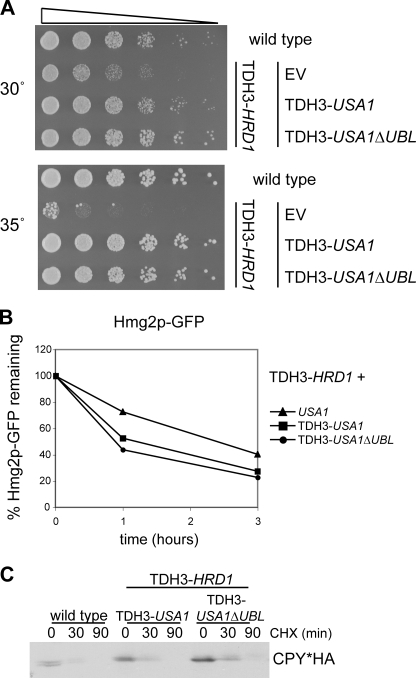
Role of the Ubl domain in ERAD and Hrd1p self-degradation. A, shown is a schematic of USA1 and USA1ΔUBL. Two versions of USA1ΔUBL were made, one with a clean deletion of 100 amino acids (USA1ΔUBL) and one with 5 copies of the myc epitope tag in place of the deleted sequence (USA1ΔUBL-myc). B, Hmg2p-GFP degradation in wild type, usa1Δ, USA1ΔUBL, and USA1ΔUBL-myc strains (left) was analyzed by flow cytometry of log-phase cultures treated with cycloheximide for the indicated times. Sec61-2p degradation (right) was analyzed by dilution assay in wild type, usa1Δ, USA1ΔUBL, and USA1ΔUBL-myc strains. 5-Fold serial dilutions were plated and grown at the indicated temperatures. C, CPY* degradation (left) and KWW degradation (right) in wild type, usa1Δ, USA1ΔUBL, and USA1ΔUBL-myc strains was analyzed by cycloheximide (CHX) chase for the indicated time points followed by lysis and immunoblotting for the HA epitope tag. D, Hrd1p degradation was evaluated by cycloheximide chase. Hrd1p degradation was compared in wild type, hrd3Δ, USA1ΔUBL hrd3Δ, and usa1Δhrd3Δ strains. E, Hrd1p self-ubiquitination was evaluated in the same strains as in D. Log-phase cultures were treated with MG132 to allow ubiquitinated Hrd1p to accumulate followed by immunoprecipitation (IP) of the Hrd1p and immunoblotting (WB) for ubiquitin or Hrd1p. Equal amounts of Hrd1p were loaded to directly compare ubiquitination levels. F, degradation of overexpressed Hrd1p (TDH3-Hrd1p) in strains with empty vector (EV), overexpressed USA1 (TDH3-USA1), and overexpressed USA1ΔUBL (TDH3-USA1ΔUBL) was analyzed by cycloheximide chase followed by immunoblotting for Hrd1p. G, degradation of overexpressed RING Hrd1p-5myc in strains with empty vector, overexpressed USA1 (TDH3-USA1), and overexpressed USA1ΔUBL (TDH3-USA1ΔUBL) was analyzed by cycloheximide chase followed by immunoblotting for the myc epitope.
Loss of Usa1p caused a diminution in Hrd1p activity. Thus, elevation of Hrd1p levels would be expected to suppress the degradation defect imposed by a usa1Δ null mutant. When Hrd1p was expressed from the strong TDH3 promoter, the degradation of Hmg2p-GFP was hastened and comparable with that seen in a wild type strain with similarly elevated levels of Hrd1p (Fig. 1F). The usa1Δ can, thus, be described as rendering Hrd1p hypomorphic toward its ERAD-M substrates, so that increasing the efficiency of recognition by altering degradation signals or by elevating Hrd1p overcomes the deficiency in substrate degradation.
The recent proteomic studies on the HRD complex indicate that Usa1p functions to couple Der1p, the prototype member of the derlin family, to the HRD complex (3). If this were the sole function of Usa1p, then a usa1Δ would be expected to phenocopy a der1Δ null. We directly examined this idea by comparing der1Δ and usa1Δ null mutants and found they are distinct by a number of criteria.
Loss of ERAD components stimulates the UPR in yeast (23, 24), in which a buildup of misfolded ER proteins activates a transcriptional response mediated by the unfolded protein response element in target genes. We used this response as a means of comparing the effects of null mutations in USA1 or DER1. Strains with an unfolded protein response element-driven GFP reporter were prepared with either or both nulls and compared with a wild type strain by flow cytometry (Fig. 2A). Both single mutants showed a similar activation of the UPR as measured by mean cellular fluorescence, and the usa1Δder1Δ double mutant showed no additional stimulation of the UPR even though the range of the UPR reporter is much greater than the 2-fold induction of UPR seen in the usa1Δder1Δ double mutant (supplemental Fig. S1). Thus, by this measure it would appear that the two genes function to alleviate ER stress in a common pathway, consistent with Usa1p mediating the action of Der1p. However, direct degradation studies showed distinct, separable functions for these two proteins.
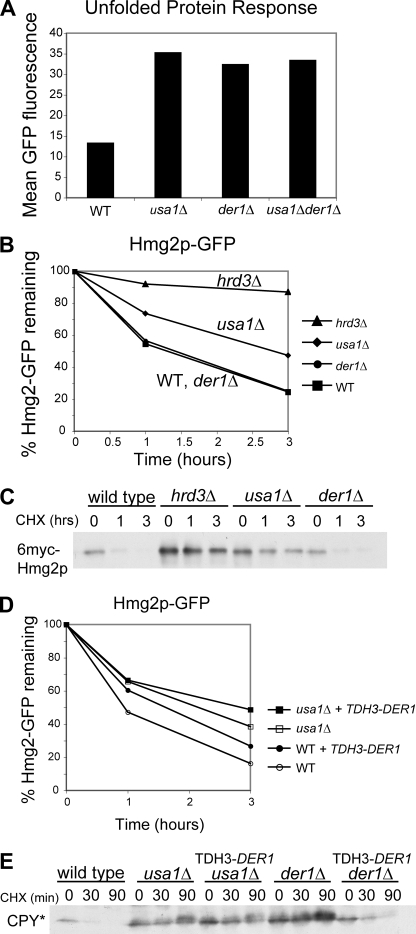
FIGURE 2.
Usa1p and Der1p have distinct roles in ERAD. A, shown is UPR caused by usa1Δ or der1Δ null mutants. Otherwise identical WT, usa1Δ, der1Δ, or usa1Δder1Δ strains harboring the 4x UPRE-GFP unfolded protein response reporter were compared by flow cytometry of log-phase cultures for mean fluorescence to evaluate the level of the UPR in each genetic circumstance. B, shown is degradation of Hmg2p-GFP in WT, hrd3Δ, usa1Δ, or der1Δ strains, as measured by flow cytometry after the addition of cycloheximide at the indicated times. C, shown is degradation of 6myc-Hmg2p in the same strains as panel B, measured by cycloheximide (CHX) chase at the indicated times followed by immunoblotting for the myc epitope tag. D, shown is the effect of Der1p overexpression on Hmg2p-GFP levels in wild type, der1Δ, or usa1Δ strains. Each strain type with empty vector or a DER1-overexpressing plasmid (TDH3-DER1) was compared in log phase for Hmg2p-GFP levels by flow cytometry. E, shown is the effect of Der1p overexpression on the lumenal substrate CPY*HA in WT, usa1Δ, or der1Δ strains. Each strain type with empty vector or a DER1-overexpressing plasmid was subjected to cycloheximide chase at the indicated times and immunoblotted for the HA tag. Note that the der1Δ phenotype is suppressed by the DER1-overexpressing plasmid.
Der1p has been described as being required only for the degradation of lumenal ERAD substrates, such as CPY*, with no effect on membrane-bound substrates such as Hmg2p (15). Direct comparison confirmed that this is the case; a der1Δ had no effect on the degradation of Hmg2p-GFP or 6myc-Hmg2p (Fig. 2, B and C) but did stabilize CPY* (Fig. 2E). In contrast, the usa1Δ null mutant stabilized both of the ERAD-M substrates (Fig. 1, B and C) and CPY* (Ref. 3 and Fig. 2E). Furthermore, overexpression of DER1 from the strong TDH3 promoter did not suppress the degradation defect of usa1Δ for either Hmg2p-GFP (Fig. 2D) or CPY* (Fig. 2E). Thus, Usa1p has a broader role in ERAD than Der1p, whose ERAD function is restricted to degradation of ERAD-L substrates.
We next examined if Usa1p was important in Hrd1p self-degradation. Hrd1p is associated with and strongly stabilized by Hrd3p. In the absence of Hrd3p, Hrd1p undergoes rapid degradation mediated by its own RING-H2 domain, with a half-life of ∼10 min (Ref. 7 and Fig. 3A). This self-degradation indicates regulatory communication between the lumenal Hrd3p and the cytosolic RING domain and has been posited to protect the cell from levels of Hrd1p that exceed the modulatory influence of Hrd3p (4, 7). We prepared a hrd3Δ null strain to test the role of Usa1p in Hrd1p self-degradation. In the hrd3Δ background, Hrd1p showed the expected low steady-state levels and rapid degradation when compared with the HRD3 wild type strain (Fig. 3a, wild type versus hrd3Δ). In contrast, a usa1Δ alone had no effect on Hrd1p stability. However, the added presence of the usa1Δ completely inhibited Hrd1p degradation in the hrd3Δ and restored the initial steady-state levels to nearly those of the control HRD3 strain in which Hrd1p is quite stable. Thus, Usa1p was absolutely required for Hrd1p self-destruction. We developed a ubiquitination assay to test if Usa1p was required for Hrd1p self-ubiquitination (Fig. 3B). In the hrd3Δ, the steady-state levels of Hrd1p are quite low, making the detection of ubiquitinated Hrd1p difficult. Accordingly, we treated strains with the proteasome inhibitor MG132 to allow accumulation of ubiquitinated Hrd1p and then carried out immunoprecipitation with anti-Hrd1p antibodies followed by immunoblotting for ubiquitin or Hrd1p itself. To allow the action of the proteasome inhibitors, a pdr5Δ null mutation was included in all strains (25). Hrd1p works primarily with the E2 Ubc7p (6), and we confirmed that the ubiquitination seen in our assay is Ubc7p dependent, shown in supplemental Fig. S2. Because of its rapid degradation, Hrd1p levels differ from strain to strain. We directly compared the ubiquitination state of Hrd1p in the strains tested by loading the same amount of immunoprecipitated Hrd1p in each lane (Fig. 3B). Usa1p was absolutely required for Hrd1p self-ubiquitination; there was no detectable Hrd1p self-ubiquitination in the hrd3Δusa1Δ double null. Thus, Usa1p mediates Hrd1p self-ubiquitination, ultimately resulting in the destruction of Hrd1p.
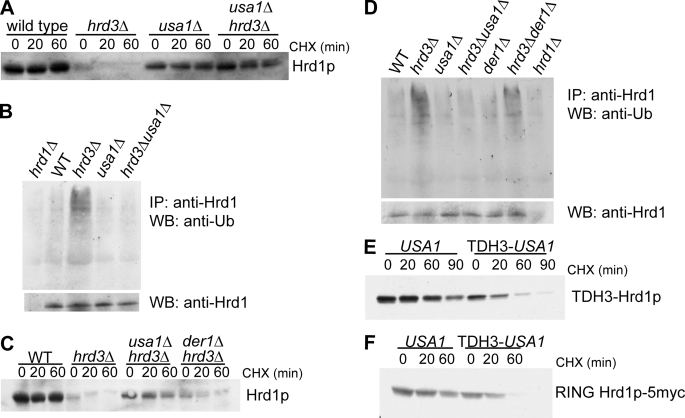
FIGURE 3.
Usa1p is required for Hrd1p self-degradation. A, the effect of a usa1Δ on Hrd1p stability was evaluated in both wild type and hrd3Δ strains. The indicated strains were evaluated for Hrd1p stability by cycloheximide (CHX) chase followed by Hrd1p immunoblotting. B, shown is the effect of usa1Δ on Hrd1p self-ubiquitination observed in the absence of Hrd3p. The indicated strains were treated with the proteasome inhibitor MG132 for 2.25 h and subjected to lysis, immunoprecipitation (IP) of Hrd1p, and immunoblotting (IB) for Hrd1p (bottom panel) or ubiquitin (top panel) to evaluate Hrd1p self-ubiquitination. A hrd1Δ null strain (left lane) was included as a specificity control. Equal amounts of Hrd1p were loaded in all other lanes to allow direct comparison of ubiquitination state. All strains harbored the pdr5Δ null mutation to allow the effective use of MG132. C, comparison of the effect of usa1Δ or der1Δ on the hrd3Δ-dependent degradation of Hrd1p is shown. The indicated strains were compared for Hrd1p stability by cycloheximide chase for the indicated times. Note that the der1Δ still allowed significant degradation of Hrd1p. D, Hrd1p self-ubiquitination in the indicated strains (with added pdr5Δ) is shown. The identical Hrd1p self-ubiquitination assay as described in panel B was employed. E, degradation of TDH3-driven Hrd1p was evaluated in strains containing either empty vector or TDH3-USA1 by cycloheximide chase followed by immunoblotting for Hrd1p. F, degradation of TDH3-driven RING Hrd1p-5myc was evaluated in strains containing either empty vector or TDH3-USA1 by cycloheximide chase followed by immunoblotting for the myc epitope to detect RING-Hrd1p.
It has been reported that Der1p is required for Hrd1p degradation (26). We wondered if the powerful stabilizing effect of usa1Δ on Hrd1p self-degradation was caused by a loss of Usa1p-mediated interaction between Hrd1p and Der1p. We directly compared the effect of either a usa1Δ or der1Δ on Hrd1p degradation in a hrd3Δ strain (Fig. 3C). In the simple case of Usa1p mediating the interaction of Der1p and Hrd1p, the phenotypes would be expected to be the same or perhaps more severe in the der1Δ. In fact, the der1Δ had a modest stabilizing effect on Hrd1p; its phenotype was much weaker than that caused by usa1Δ (Fig. 3C). Similarly, der1Δ had only a small effect on Hrd1p ubiquitination in the hrd3Δ, whereas usa1Δ showed complete inhibition (Fig. 3D).
In the course these studies, we discovered an epitope-specific effect that further distinguished Der1p from Usa1p. Hrd1p with a C-terminal triple-HA tag is a reliable, functional version of Hrd1p that we have used in many studies. However, in the usa1Δ background, Hrd1p-3HA shows a reproducible and dramatic mobility shift of ∼12 kDa by gel mobility (supplemental Fig. S3, arrow). This shift was highly specific for the usa1Δ null mutant and for the HA-tagged Hrd1p; it was not observed with native Hrd1p in the previous experiments or with GFP-tagged Hrd1p. The HA-dependent shift was not affected by ubiquitin proteases or N-glycanases.4 In light of this ambiguous mobility shift, unless otherwise specified, native untagged Hrd1p was used throughout our work. Whereas the nature of the shift remains unclear, it is nevertheless another way to assess the degree of similarity between usa1Δ and der1Δ mutants. Direct comparison showed that the shift of Hrd1p-3HA was not observed in the der1Δ but was seen in the usa1Δ mutant either with or without Hrd3p present (supplemental Fig. S3, arrow). Thus, by this criterion as well, the losses of Usa1p and Der1p have distinct phenotypes.
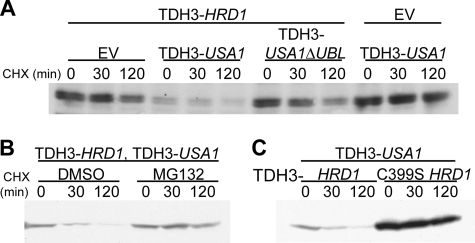
Hrd1p self-degradation in the absence of Hrd3p is extremely rapid at native levels (Fig. 2A and Refs. 7 and 26). However, when overexpressed, Hrd1p is degraded sluggishly despite being in great excess of Hrd3p. If Hrd1p self-degradation were autonomous, we would expect all Hrd1p not in complex with Hrd3p to be degraded rapidly at any level of expression. Because we had shown Usa1p to be required for Hrd1p self-ubiquitination and degradation, we reasoned that Usa1p was the “missing” component preventing rapid degradation of overexpressed Hrd1p. When we overexpressed both HRD1 and USA1 from the same strong promoter (TDH3), we observed much faster degradation of Hrd1p (Fig. 3E), indicating that Usa1p is directly required for Hrd1p self-degradation. We also investigated degradation of a TDH3-driven version of Hrd1p that lacks the transmembrane domain and includes only the C-terminal half of Hrd1p (RING Hrd1p-5myc), which is the half of Hrd1p that contains the catalytically active RING-H2 domain (7). Like the full-length Hrd1p, degradation of RING Hrd1p-5myc was increased by the addition of TDH3-driven USA1 (Fig. 3F), indicating that Usa1p interacts with the C-terminal half of Hrd1p.
Usa1p contains a cytoplasmic Ubl domain (3). We tested the Ubl domain for involvement in the Usa1p ERAD functions. We made two versions of USA1 lacking this domain. One was a clean deletion of 100 amino acids (USA1ΔUBL), whereas the other had 5 copies of the myc epitope tag in place of the deleted sequence in order to preserve the register and spacing of the protein (USA1ΔUBL-myc) (Fig. 4A). We first asked whether the Ubl domain was required for Hmg2p degradation. The different versions of USA1ΔUBL were expressed in a usa1Δ strain so that the only Usa1p present would be the mutant Usa1p. We evaluated degradation of Hmg2p-GFP in wild type, usa1Δ, USA1ΔUBL, and USA1ΔUBL-myc strains (Fig. 4B, left). Hmg2p-GFP degradation was impaired in the usa1Δ strain as shown previously (Fig. 1C) but was equivalent to wild type in the two USA1ΔUBL strains, demonstrating that the Ubl domain of Usa1p is not required for Hmg2p-GFP degradation. The unregulated 6myc-Hmg2p substrate was also unaffected by loss of the Ubl domain (data not shown). We evaluated degradation of another ERAD-M substrate, Sec61-2p, to determine whether the Ubl domain is dispensable for ERAD-M substrates in general. Sec61-2p is a mutant version of the Sec61p translocon protein. Sec61-2p is unstable at 37 °C and is degraded by the ERAD machinery, causing cell death, because Sec61p is essential (27). If ERAD is impaired, Sec61-2p is not degraded, and the cells can live at the high temperature. We took a sec61-2 usa1Δ strain and added back either empty vector, USA1, USA1ΔUBL, or USA1ΔUBL-myc and made 5-fold serial dilutions and compared growth at the permissive temperature (30 °C) and the non-permissive temperature (37 °C) (Fig. 4B, right). As expected, the strain with full-length Usa1p (and, therefore, intact ERAD) was temperature-sensitive due to Sec61-2p degradation. The empty vector strain, which has the usa1Δ null mutation (and, therefore, impaired ERAD), was able to grow normally at 37 °C, indicating that Usa1p is required for Sec61-2p degradation in addition to being required for Hmg2p degradation. The two versions of Usa1pΔUBL were also temperature-sensitive and were undistinguishable from wild type Usa1p, demonstrating that the Ubl domain is not required for the Usa1p role in ERAD-M. Usa1pΔUBL and Usa1pΔUBL-myc expressed in an otherwise wild type strain did not confer temperature sensitivity on their own (supplemental Fig. S4). Thus, the role of Usa1p in ERAD-M does not depend on the Ubl domain.
We next investigated if the Ubl domain was required for ERAD-L by looking at two different substrates. Degradation of both CPY* (left) (28) and KWW (right) (29) was assayed in wild type, usa1Δ, USA1ΔUBL, and USA1ΔUBL-myc strains (Fig. 4C). As expected, these substrates were strongly stabilized in the usa1Δ strain. Degradation of both CPY* and KWW in the USA1ΔUBL strains was very similar to that in the wild type strains. Thus, the Ubl domain is not needed for ERAD of either membrane or lumenal substrates.
We evaluated the role of the Ubl domain in Hrd1p self-ubiquitination and degradation. Usa1pΔUBL was introduced as the only version of Usa1p expressed in a hrd3Δ strain and subjected to cycloheximide chase to evaluate Hrd1p stability in this case (Fig. 4D). Hrd1p was strongly stabilized in the USA1ΔUBL hrd3Δ strain as compared with the hrd3Δ alone (Fig. 4D). As expected, this stabilization was due to a loss of Hrd1p self-ubiquitination (Fig. 4E). Thus, the Ubl domain of Usa1p is necessary for Hrd1p to undergo self-ubiquitination and degradation, allowing for control of Hrd1p levels by Usa1p.
We also looked at the role of the Ubl domain in Usa1p-mediated degradation of overexpressed Hrd1p. Whereas overexpression of Usa1p restored rapid degradation of Hrd1p (Fig. 3E), the overexpression of Usa1pΔUBL had no effect on stability of overexpressed full-length Hrd1p (Fig. 4F) or the C-terminal RING domain (Fig. 4G), indicating that the Ubl domain is required for degradation of Hrd1p at all levels. In fact, it appears that overexpressed Usa1pΔUBL was partially dominant negative, as it improved the stability of overexpressed Hrd1p in these experiments.
One model for the critical role of Usa1p in Hrd1p self-degradation is that it brings two different Hrd1p molecules together so that each Hrd1p can catalyze ubiquitination of the other in trans. To test this idea, we first determined if Hrd1p is ubiquitinated in trans. We devised an assay to test this model using the C399S mutant of Hrd1p that cannot function as an E3 ligase (6). We made a C-terminal myc-tagged version of C399S Hrd1p, so that we could distinguish between the C399S Hrd1p and the untagged, wild type Hrd1p. We then performed a cycloheximide chase to evaluate degradation of the C399S Hrd1p. The only way this mutant Hrd1p could undergo HRD-dependent degradation is if another, wild type Hrd1p ubiquitinated it in trans. Overexpression of wild type Hrd1p (from the TDH3 promoter) resulted in very slow, if any, degradation of the C399S Hrd1p (Fig. 5A). However, the addition of TDH3-driven Usa1p to this same strain resulted in pronounced degradation of the C399S Hrd1p. This strongly suggested that Hrd1p does in fact undergo trans-ubiquitination and supports our model that Usa1p is required for Hrd1p to undergo trans-ubiquitination. As expected, overexpression of Usa1p alone in the absence of overexpressed Hrd1p did not cause C399S Hrd1p degradation. Importantly, the observed C399S Hrd1p degradation was proteasome-dependent as it was inhibited by MG132 (Fig. 5B). Moreover, this stimulated “trans-degradation” of C399S Hrd1p-5myc required the addition of catalytically active Hrd1p; the addition of TDH3-C399S Hrd1p did not support trans-degradation of C399S Hrd1p-5myc (Fig. 5C). Overexpression of Usa1pΔUBL was far less effective in promoting the Hrd1p-dependent degradation of C399S Hrd1p (Fig. 5A), consistent with our findings that the Ubl domain was required for Hrd1p ubiquitination and degradation. Thus, Hrd1p self-destruction appears to be mediated by trans-ubiquitination of Hrd1p brought about through the action of Usa1p.
FIGURE 5.
Hrd1p undergoes Usa1p-dependent self-ubiquitination in trans. A, degradation of C399S Hrd1p-5myc was analyzed by cycloheximide chase. Strains containing overexpressed (TDH3) HRD1 and either empty vector (EV), TDH3-USA1, TDH3-USA1ΔUBL, or a strain containing only TDH3-USA1 were subjected to cycloheximide (CHX) chase and immunoblotting for the myc epitope tag on C399S Hrd1p. B, trans-degradation of C399S Hrd1p is proteasome-dependent. A C399S Hrd1p-5myc strain containing overexpressed HRD1 and USA1 was subjected to cycloheximide chase with the addition of the proteasome inhibitor MG132. Log-phase cells were pretreated for 30 min with 25 μg/ml MG132 (or DMSO vehicle) for 15 min before addition of cycloheximide. C, trans-degradation of C399S requires catalytically active Hrd1p. A C399S Hrd1p-5myc strain containing either TDH3-HRD1 or TDH3-C399S HRD1 was subjected to cycloheximide chase for the indicated time points followed by lysis and immunoblotting for the myc epitope on C399S Hrd1p-5myc.
Overexpression of Hrd1p has been shown to cause a slow-growth phenotype in yeast in an activity-dependent manner (4). We confirmed the observation that overexpression of Hrd1p results in slow growth (Fig. 6A); this phenotype was dependent on Hrd1p activity, as overexpression of C399S Hrd1p did not cause slow growth (data not shown and Ref. 4). We reasoned that the slow growth due to having too much Hrd1p should be alleviated in the strains also overexpressing Usa1p. Indeed, the strain with both HRD1 and USA1 expressed from the TDH3 promoter exhibited normal growth (Fig. 6A), which was not surprising as Usa1p-stimulated degradation lowered the Hrd1p steady-state level. We next examined if lowering Hrd1p levels was the sole mechanism by which Usa1p protected cells from Hrd1p-mediated toxicity by testing Usa1pΔUBL, which does not promote Hrd1p degradation. Surprisingly, overexpression of Usa1pΔUBL also restored wild type growth in the strains with overexpressed Hrd1p. However, as shown in Fig. 4F, the Hrd1p in this case is stable, so the absence of Hrd1p-mediated toxicity cannot be attributed to simply lowering the levels of Hrd1p. This suggested that Usa1p has a separate regulatory action on Hrd1p. Another interpretation could be that overexpressing Usa1pΔUBL renders Hrd1p unable to catalyze any ubiquitination reactions, which would explain both the stability of Hrd1p and the lack of toxicity. However, degradation of both Hmg2p-GFP (Fig. 6B) and CPY* (Fig. 6C) proceeded normally in strains with overexpressed Usa1pΔUBL as compared with strains with overexpressed Usa1p. This indicates that Usa1p limited the toxicity of Hrd1p in a highly selective manner. Thus, Usa1p appears to function as a regulator of Hrd1p both through promotion of self-degradation and by direct regulation of its activity.
FIGURE 6.
Usa1p abolishes toxicity of overexpressed Hrd1p. A, growth of strains was compared by making 5-fold serial dilutions and incubating at the indicated temperatures. Wild type was compared against strains containing overexpressed Hrd1p with the addition of either empty vector (EV), overexpressed USA1, or overexpressed USA1ΔUBL. B, Hmg2p-GFP degradation was evaluated in strains containing overexpressed Hrd1p with the addition of empty vector, overexpressed USA1, or overexpressed USA1ΔUBL. Log-phase cultures were treated with cycloheximide for the indicated times and analyzed by flow cytometry. C, CPY* degradation was evaluated in strains containing overexpressed Hrd1p and either overexpressed USA1 or overexpressed USA1ΔUBL and compared with a wild type strain. Log-phase cultures were treated with cycloheximide for the indicated times followed by lysis and immunoblotting for the HA epitope on CPY*HA.
DISCUSSION
Usa1p is the newest member of the consortium of proteins comprising the HRD complex in yeast. Its discovery in elegant proteomic studies identified Usa1p as an ERAD factor involved in degradation of ERAD-L substrates (3), perhaps through a structural role linking other components to Hrd1p. We have thoroughly investigated the role of Usa1p in ERAD and within the HRD complex and found that Usa1p has a significant role in ERAD and is involved in both substrate degradation and in regulation of Hrd1p.
The action of Hrd1p on membrane-bound substrates such as Hmg2p was less efficient in the absence of Usa1p. However, Usa1p was not absolutely required for degradation of ERAD-M substrates such as Hmg2p but increased the efficiency of substrate degradation. Consistent with this, a usa1Δ null mutant stabilized both regulated and unregulated versions of Hmg2p as well as Sec61-2 (Fig. 4B) and CD4,5 but not Pdr5p (Ref. 3 and data not shown) or Hmg2p in the presence of high degradation signals. Thus, the usa1Δ creates a Hrd1p hypomorph that has a decreased but not absent ability to recognize membrane substrates. There are several potential mechanisms by which Usa1p may increase the efficiency of substrate degradation by the HRD complex. One possibility is that Usa1p might assist in substrate recognition and delivery to the Hrd1p ligase. Because we have found that Usa1p abolishes Hrd1p-mediated toxicity, an intriguing idea is that Usa1p might help channel substrates to the ligase or prevent Hrd1p from accessing non-substrates. In the absence of Usa1p, Hrd1p activity would be less efficient but also less specific.
Usa1p may promote the formation of Hrd1p multimers, which could be more efficient for substrate ubiquitination than a single Hrd1p molecule. Consistent with this idea, we found that Usa1p promotes ubiquitination of one Hrd1p molecule by another, suggesting the formation of a Hrd1p dimer at least in the case of self-ubiquitination. Hrd1p complexed to Usa1p may exist primarily in dimer or multimer form, maximally efficient for substrate degradation and poised for self-ubiquitination if Hrd3p becomes limiting. Clearly, there are multiple ways that Usa1p may influence the activity of Hrd1p, and clarifying these models will be an important future activity.
Usa1p has been previously shown to mediate the association of Der1p to Hrd1p (3). However, we now show that Usa1p has a direct role in the HRD pathway distinct from Der1p recruitment. Direct comparison showed that a der1Δ null did not affect the degradation of the ERAD-M substrates stabilized by loss of Usa1p, indicating an additional role for Usa1p. If the sole function of Usa1p were to recruit Der1p, overexpression of Der1p might be expected to suppress a usa1Δ. High levels of Der1p were unable to suppress the stabilization of CPY* in a usa1Δ null mutant, again suggesting separable roles for these two proteins. Although it has been reported that Der1p is required for Hrd1p self-degradation (30), we found that the der1Δ only partially stabilized Hrd1p and still allowed self-ubiquitination, in striking contrast to the usa1Δ. This disparity in Hrd1p regulation is another readout of a direct action of Usa1p that is distinct from that of Der1p. However, the non-additivity of each null UPR is consistent with these proteins functioning in the same lumenal ERAD pathway. Thus, the full picture of Usa1p function is most likely a combination of mediating Der1p association with the HRD complex and direct actions of Usa1p on the Hrd1p ligase.
We found that Usa1p played a crucial role in Hrd1p self-degradation. Hrd1p undergoes rapid self-ubiquitination and degradation in the absence of Hrd3p that was completely abolished in a usa1Δ null mutant, indicating that Usa1p is required for Hrd1p self-destruction. Previously, it was thought that a lack of Hrd3p was both necessary and sufficient to effect degradation of Hrd1p. However, our findings now indicate that Hrd1p free of Hrd3p is unable to autonomously ubiquitinate itself and is subject to additional modes of regulation.
The effects observed for overexpressed Hrd1p further support this idea; in situations where Hrd1p is overexpressed in the absence of Hrd3p, it is degraded slowly, with a half-life of ∼1–2 h instead of the 10-min half-life observed for native Hrd1p in a hrd3Δ strain. The reason for the slow degradation of this un-partnered Hrd1p had eluded us until now, when we realized that Usa1p might become limiting in strains with overexpressed Hrd1p. Indeed, we observed that rapid degradation of Hrd1p was restored by sufficient expression of Usa1p. Moreover, Usa1p promoted degradation of Hrd1p lacking its transmembrane domain, indicating that Usa1p productively interacts with the C-terminal half of Hrd1p. Taken together, the data from both native and overexpressed Hrd1p indicate that a key function of Usa1p is to promote degradation of Hrd1p in the absence of Hrd3p, allowing for stringent control of Hrd1p levels.
We have suggested a simple model for the specific role of Usa1p in Hrd1p self-ubiquitination, in which Usa1p mediates an association that brokers efficient transfer of ubiquitin from one Hrd1p molecule to another, resulting in the remarkably fast self-degradation rate of Hrd1p when not associated with Hrd3p. This model was arrived at by co-expression of active and inactive Hrd1p. The inactive C399S mutant of Hrd1p was, not surprisingly, stable when expressed alone or in the presence of overexpressed Hrd1p. However, when Usa1p was also present at high levels, the C399S version of Hrd1p underwent rapid degradation but only when co-expressed with active Hrd1p. Thus, in the absence of Hrd3p, Usa1p works to eliminate un-partnered Hrd1p by mediating its self-ubiquitination, allowing for its ultimate degradation.
Usa1p contains a ubiquitin-like domain in its N-terminal cytoplasmic region (3), and we were interested in determining its importance in Usa1p function. Versions of Usa1p lacking the Ubl domain supported wild type degradation of both membrane and lumenal substrates, but ubiquitination and degradation of Hrd1p was severely impaired. The domain itself was required for Hrd1p degradation, because replacing the Ubl domain with five copies of the myc epitope tag (roughly equal in size to the deleted region) to preserve the register of the protein also impaired Hrd1p degradation while supporting substrate ERAD. Thus, the Ubl domain was specifically required for Hrd1p self-degradation but was not required for degradation of any other ERAD substrates besides Hrd1p, allowing us to consider the degradation of Hrd1p to be distinct from that of other ERAD substrates. As we predicted from our observation that the Ubl domain inhibits degradation of both native and overexpressed Hrd1p, we found that trans-ubiquitination of Hrd1p was impaired in a strain with Usa1pΔUBL in place of wild type Usa1p. This suggests that one function of the Ubl domain may be to mediate an association between two Hrd1p molecules to permit efficient trans-ubiquitination. However, it has recently been shown that the Ubl domain of Usa1p does not interact directly with Hrd1p and that the Ubl domain does not interact with the proteasome subunit Rpt5p (31). These findings suggest that the Usa1p Ubl domain likely does not have a role in additional downstream steps of degradation. Whereas the mechanism of Ubl domain function remains unclear, it is clear that the function of the Ubl domain of Usa1p is principally for promoting Hrd1p self-ubiquitination in a manner that appears to be distinct from that of ERAD substrates.
Overexpression of Hrd1p causes a slow growth phenotype in yeast (Ref. 4 and Fig. 6A) that is dependent upon Hrd1p activity (Ref. 4 and data not shown). In these strains, as previously discussed, Hrd1p undergoes slow degradation. The addition of overexpressed Usa1p restored normal kinetics of Hrd1p degradation, thus lowering its levels. It is, therefore, not surprising that in these strains the Hrd1p-mediated toxicity was alleviated. Surprisingly, overexpression of Usa1pΔUBL, which does not promote Hrd1p degradation, also abolished the toxicity of overexpressed Hrd1p. Even though levels of Hrd1p are high in this case, there was no overall detriment to these strains, immediately suggesting two possibilities. One is that Hrd1p is rendered non-functional by the overexpressed Usa1pΔUBL, explaining why there is neither Hrd1p degradation nor a growth deficit. Importantly, substrate degradation proceeded normally in the presence of Usa1pΔUBL, indicating that Hrd1p functions normally and ruling out this possibility. The second and more intriguing interpretation suggested by the preservation of substrate degradation is that Usa1p somehow restrains Hrd1p, preventing promiscuous ubiquitination of inappropriate ER-localized substrates and thereby preventing Hrd1p toxic effects. This could occur either by Usa1p directly regulating or activating Hrd1p or by Usa1p playing a role in governing which substrates are presented to Hrd1p for ubiquitination. One way in which Usa1p could control access to Hrd1p would be to alter its localization. However, indirect immunofluorescence of overexpressed Hrd1p and RING Hrd1p-5myc in the presence and absence of overexpressed Usa1p showed no obvious changes in the ER-localization of Hrd1p,4 although we cannot rule out the possibility of more subtle changes brought about by Usa1p.
Taken together these studies indicate that Usa1p has an important and primary role in the action and regulation of Hrd1p. This relationship is likely to be conserved as the Herp protein appears to be the mammalian counterpart to Usa1p and will even complement some of the function of Usa1p in the yeast null mutant (3). Usa1p is needed for optimal function of Hrd1p, and it is absolutely required for Hrd1p self-degradation. Thus, Usa1p effects are dichotomous; loss of Usa1p decreases Hrd1p efficiency but could also increase its steady-state levels in circumstances where self-degradation modulates Hrd1p levels. Moreover, Usa1p appears to play a crucial role in regulation of Hrd1p, preventing cellular toxicity by curbing Hrd1p activity when appropriate. The importance of these Usa1-type regulators of the HRD complex will be of interest both in normal circumstances and in cases where the Hrd1p ligase is elevated or overabundant, such as in rheumatoid arthritis (32), or in cases where ER stress elevates the levels of the HRD complex (33).
Acknowledgments
We thank Davis Ng for plasmids and the Michael David laboratory (University of California San Diego Division of Biology) for use of the FACScalibur flow cytometer. We thank Thomas Sommer, Ernst Jarosch, and their colleagues (Max Delbrück Center, Berlin) for generously sharing information before publication as well as numerous reagents. S. M. Carroll thanks Anthony Carroll for timely supplements of glucose and other nutrients. In addition, R. Y. Hampton thanks David Holway for running commentary.
This work was supported, in whole or in part, by National Institutes of Health Grant GM51996-06 (NIDDK, to R. Y. H.). This work was also supported by the American Heart Association (an Established Investigator award (to R. Y. H.)).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2, “Materials and Methods,” additional references, and Figs. S1–S4.
S. M. Carroll and R. Y. Hampton, unpublished observation.
T. Sommer, personal communication.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- ERAD-L
- ERAD-lumenal
- ERAD-M
- ERAD membrane-anchored
- Ubl
- ubiquitin-like
- HMG
- 3-hydroxy-3-methylglutaryl
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin ligase
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- ZA
- zaragozic acid
- WT
- wild type
- UPR
- unfolded protein response.
REFERENCES
- 1.Hampton R. Y., Garza R. M. (2009) Chem. Rev. 109, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song B. L., Javitt N. B., DeBose-Boyd R. A. (2005) Cell Metab. 1, 179–189 [DOI] [PubMed] [Google Scholar]
- 3.Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 4.Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 5.Hochstrasser M. (2009) Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y. (2001) Nat. Cell Biol. 3, 24–29 [DOI] [PubMed] [Google Scholar]
- 7.Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C., Hampton R. Y. (2000) J. Cell Biol. 151, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhamidipati A., Denic V., Quan E. M., Weissman J. S. (2005) Mol. Cell 19, 741–751 [DOI] [PubMed] [Google Scholar]
- 9.Kim W., Spear E. D., Ng D. T. (2005) Mol. Cell 19, 753–764 [DOI] [PubMed] [Google Scholar]
- 10.Gauss R., Jarosch E., Sommer T., Hirsch C. (2006) Nat. Cell Biol. 8, 849–854 [DOI] [PubMed] [Google Scholar]
- 11.Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008) Nat. Cell Biol. 10, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner R. G., Hampton R. Y. (1999) EMBO J. 18, 5994–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampton R. Y., Gardner R. G., Rine J. (1996) Mol. Biol. Cell 7, 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hampton R. Y., Bhakta H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12944–12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato B. K., Hampton R. Y. (2006) Yeast 23, 1053–1064 [DOI] [PubMed] [Google Scholar]
- 16.Ng D. T., Spear E. D., Walter P. (2000) J. Cell Biol. 150, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner R., Cronin S., Leader B., Rine J., Hampton R., Leder B. (1998) Mol. Biol. Cell 9, 2611–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampton R. Y., Rine J. (1994) J. Cell Biol. 125, 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. (2001) Mol. Biol. Cell 12, 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner R. G., Shearer A. G., Hampton R. Y. (2001) Mol. Cell. Biol. 21, 4276–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner R. G., Hampton R. Y. (1999) J. Biol. Chem. 274, 31671–31678 [DOI] [PubMed] [Google Scholar]
- 22.Cronin S. R., Hampton R. Y. (1999) Methods in Enzymology 302, 58–73 [DOI] [PubMed] [Google Scholar]
- 23.Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 24.Bays N. W., Hampton R. Y. (2002) Curr. Biol. 12, R366–R371 [DOI] [PubMed] [Google Scholar]
- 25.Emter R., Heese-Peck A., Kralli A. (2002) FEBS Lett. 521, 57–61 [DOI] [PubMed] [Google Scholar]
- 26.Plemper R. K., Deak P. M., Otto R. T., Wolf D. H. (1999) FEBS Lett. 443, 241–245 [DOI] [PubMed] [Google Scholar]
- 27.Biederer T., Volkwein C., Sommer T. (1996) EMBO J. 15, 2069–2076 [PMC free article] [PubMed] [Google Scholar]
- 28.Finger A., Knop M., Wolf D. H. (1993) Eur. J. Biochem. 218, 565–574 [DOI] [PubMed] [Google Scholar]
- 29.Vashist S., Ng D. T. (2004) J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plemper R. K., Bordallo J., Deak P. M., Taxis C., Hitt R., Wolf D. H. (1999) J. Cell Sci. 112, 4123–4134 [DOI] [PubMed] [Google Scholar]
- 31.Kim I., Li Y., Muniz P., Rao H. (2009) PLoS One 4, e7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K., Aratani S., Fujita H., Zhang L., Ikeda R., Fujii R., Miura N., Komiya S., Nishioka K., Maruyama I., Fukamizu A., Nakajima T. (2003) Genes Dev. 17, 2436–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007) Dev. Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]