Abstract
FGF19 and FGF21, unique members of the fibroblast growth factor (FGF) family, are hormones that regulate glucose, lipid, and energy homeostasis. Increased hepatocyte proliferation and liver tumor formation have also been observed in FGF19 transgenic mice. Here, we report that, in contrast to FGF19, FGF21 does not induce hepatocyte proliferation in vivo. To identify the mechanism for FGF19-induced hepatocyte proliferation, we explored similarities and differences in receptor specificity between FGF19 and FGF21. We find that although both are able to activate FGF receptors (FGFRs) 1c, 2c, and 3c, only FGF19 activates FGFR4, the predominant receptor in the liver. Using a C-terminal truncation mutant of FGF19 and a series of FGF19/FGF21 chimeric molecules, we determined that amino acids residues 38–42 of FGF19 are sufficient to confer both FGFR4 activation and increased hepatocyte proliferation in vivo to FGF21. These data suggest that activation of FGFR4 is the mechanism whereby FGF19 can increase hepatocyte proliferation and induce hepatocellular carcinoma formation.
Keywords: Cancer/Liver, Cell/Hepatocyte, Diseases/Diabetes, Diseases/Metabolic, Hepatocyte, FGF19, FGF21, FGFR4, β-Klotho, Fibroblast Growth Factors
Introduction
FGF19, FGF21, and FGF23 form a unique subfamily of fibroblast growth factors (FGFs).3 Unlike other FGFs, all three have been shown to function as endocrine hormones in the regulation of various metabolic processes (1). FGF23 originates in bone and regulates phosphate homeostasis in kidney (2), FGF21 is expressed predominantly in liver and signals in adipose tissue (3), and FGF19 is secreted from ileum and functions as an enterohepatic signal for the regulation of bile acid metabolism (4).
FGF19 and FGF21 have similar effects on regulating glucose, lipid, and energy metabolism. Both FGF19 and FGF21 transgenic mice are resistant to diet-induced obesity, have decreased body fat mass and improved insulin sensitivity, glucose disposal, and plasma lipid parameters (5–8). Administration of recombinant FGF19 or FGF21 protein to diabetic mice resulted in the reduction of serum glucose and insulin levels, improved glucose tolerance, and reduced liver steatosis and body weight (7, 8). In addition, FGF21 improved glucose, insulin, and lipid profiles and reduced body weight in diabetic rhesus monkeys (9). Taken together, these observations suggest the potential utility of FGF19 and FGF21 for the treatment of diabetes and obesity (1).
Because of the sequence and structural homology between FGF19 and FGF21 with the other FGFs, most of which have well established roles in cell proliferation and mitogenesis, whether FGF19 and FGF21 could induce cell proliferation have also been investigated. In the case of FGF19, hepatocellular carcinoma (HCC) formation was observed in transgenic mice overexpressing FGF19 in skeletal muscle (10). In addition, increases in the proliferation of pericentral hepatocytes, as measured by enhanced BrdU labeling, was observed both in FGF19 transgenic animals and wild-type mice administered recombinant FGF19 (10). Because constitutive hepatocellular proliferation may be a prerequisite for transformation (11), it is interesting to note that cell lineage analysis of FGF19-induced tumors suggests that dysplastic and neoplastic hepatocytes originated adjacent to central veins, where increased proliferation was localized as determined by BrdU labeling (10). These results suggest that FGF19 induced hepatocyte proliferation may ultimately lead to HCC formation (10). FGFR4 has been proposed to play a role in the observed induction of hepatocyte proliferation and carcinogenesis by FGF19 (10); however, contradicting evidence proposing a protective role for FGFR4 in suppressing hepatoma progression has also been proposed (12). Therefore, the mechanism for FGF19 induced hepatocyte mitogenesis has not been elucidated, and the receptor responsible for this activity remains unclear.
Receptor utilization for this subfamily has been elucidated recently. Both FGF19 and FGF21 utilize β-Klotho, a single transmembrane protein, as a co-receptor in addition to FGFRs for signaling (13). Potential differences in FGFR utilization between FGF19 and FGF21 have been reported (14–16). However, whether FGF21 causes hepatocyte proliferation and whether the reported differences in receptor specificity between FGF19 and FGF21 contribute to mitogenicity are not clear.
In this report, we show that, in contrast to FGF19, FGF21 does not increase hepatocyte proliferation in vivo. In addition, using a series of novel FGF19 and FGF21 truncation and chimeric molecules, we have identified a region on FGF19 that is responsible for FGFR4 activation and propose that FGFR4 activation is the mechanism whereby FGF19 can increase hepatocyte proliferation and induce hepatocellular carcinoma formation.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant FGF19 and Chimeric Proteins
Wild-type FGF19 (encoding residues 23–216, without secretory leader peptide sequence) and chimeras were cloned into the pET30 vector (Novagen). DNA constructs were transformed into BL21(DE3) Escherichia coli (Novagen). Protein expression was induced with isopropyl-1-thio-β-d-galactopyranoside at 37 °C. The purification process was the same as previously described (17). FGF21 (29–208, without secretory leader peptide) was purified as previously described (8). The sequences of all chimeras are as follows: FGF19/21-1: M::hFGF19(23–80)::hFGF21(81–208) (FGF19/21-1 sequence is composed of methionine, followed by human FGF19 sequences 23–80, then followed by human FGF21 sequences 81–208); FGF19/21-2: M::hFGF19(23–49)::hFGF21(51-208); FGF19/21-3: M::hFGF19(23–42)::hFGF21(44–208); FGF19/21-4: M::hFGF19(23–37)::hFGF21(41–208); FGF19/21-5: M::hFGF19(23–32)::hFGF21(36–208); FGF21/1938–42: M::hFGF21(28–41)::hFGF19(38–42)::hFGF21(45–208).
Cell Culture and Transfections
L6 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. Cells were transfected with expression vectors using the Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's protocol.
Analysis of FGF Signaling
Analysis of FGF signaling in L6 cells were performed as described previously (15). Cells were collected 10 min after treatment with FGF19 or chimeric proteins, snap frozen in liquid nitrogen, homogenized in the lysis buffer, and subjected to Western blot analysis using anti-phospho-p44/42 MAP kinase (ERK1/2) antibody and anti-ERK antibody (Cell Signaling).
Glucose Uptake Assay
3T3L1 preadipocytes (ATCC CL-173) were cultured and induced to differentiate. Glucose uptake was assayed as previously described (7).
In Vivo Hepatocyte BrdU Labeling
On day 1 of the study, an osmotic minipump (ALZET®, model 1007D) containing 5-bromo-2′-deoxyuridine (BrdU, Sigma) (16 mg/ml) was implanted subcutaneously into each 8-week-old female FVB mouse (Charles River Laboratories, Charles River, MA). Each mouse was given an IP injection of either phosphate-buffered saline (PBS, vehicle), or test proteins as indicated daily at 2 mg/kg/day beginning on day 2 of the study and continuing for 6 consecutive days. Samples of liver and duodenum were collected from each mouse on the day following the last IP injection and placed in 10% neutral-buffered formalin in preparation for paraffin-embedding, sectioning, and light microscopic evaluation. Sections of all collected tissues were stained by an immunohistochemical method described below to visualize BrdU incorporation as a marker of mitotic activity. Tissue sections were examined at random by routine light microscopy without knowledge of treatment group. The number of hepatocyte nuclei stained for BrdU incorporation was assigned a score on a semiquantitative scale where 0 is defined as no increase above expected levels in vehicle-treated (control) mice and ± is equivocal, 1 is minimal, 2 is mild, 3 is moderate, and 4 is a marked increase above control levels. The localization (centrilobular or diffusely scattered through hepatic lobules) of the hepatocytes stained for BrdU incorporation was also recorded. Only hepatocyte nuclei (large, round nuclei clearly within hepatocytes) were considered for semiquantitative scoring of BrdU labeling. Nuclei of other cells types (e.g. bile duct epithelium, Kupffer cells, endothelial cells, and infiltrating leukocytes) were sometimes labeled with BrdU; however, these cells and their nuclei are morphologically distinct from hepatocytes and hepatocyte nuclei and have different anatomic localizations. These nuclei were not considered for the scoring of BrdU labeling in hepatocytes.
Cellular incorporation of BrdU was detected by digesting deparaffinized tissue sections with 0.1% protease (Sigma) and treating the sections with 2N hydrochloric acid. Sections were blocked with CAS BLOCK (Zymed Laboratories Inc., San Francisco, CA), incubated with rat antibody to BrdU (Accurate, Westbury, NY; catalogue no. OBT0030, lot no. H9180), and bound rat antibody was detected with biotinylated rabbit antibody to rat IgG (Vector Laboratories, Burlingame, CA; catalogue no. BA 4001, lot no. S0907). Tissue sections were quenched with Peroxidase Blocking Solution (DAKO Corp., Carpinteria, CA) and retained biotin was detected with Vectastain Elite ABC kit (Vector Laboratories). Reaction sites were visualized with DAB+Substrate-Chromagen System (DAKO Corp.) followed by DAB enhancer (Invitrogen, Carlsbad, CA). Sections were counterstained with hematoxylin.
RESULTS
FGF21 Treatment Does Not Increase Hepatocyte Proliferation in Vivo
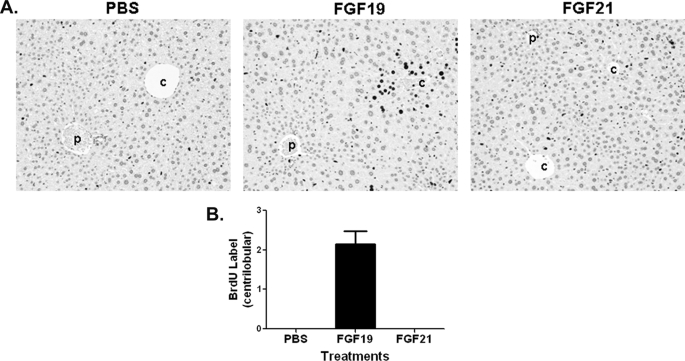
Because FGF21 and FGF19 belong to the same FGF subfamily and share significant similarities with respect to receptor/co-receptor requirements and metabolic effects, we wanted to directly compare their effects on hepatocyte proliferation. Using an in vivo BrdU labeling method similar to that described previously (10), we compared the effects of FGF21 and FGF19 on hepatocyte proliferation. As shown in Fig. 1, histopathological examination of liver sections from FGF19-treated animals showed increased BrdU-labeled hepatocytes concentrating in centrilobular regions of hepatic lobules, consistent with published observations (10). In contrast, livers from FGF21-treated animals did not show increased numbers of BrdU-labeled hepatocytes in pericentral regions, nor was increased BrdU labeling noted in any other area of the liver. These results suggest that FGF21 is distinct from FGF19 in its mitogenic activity on hepatocytes and that FGF21 does not enhance hepatocyte proliferation under the conditions tested.
FIGURE 1.
FGF21 treatment does not increase hepatocyte proliferation. A, BrdU immunostaining of livers from female FVB mice after an 8-day BrdU infusion by osmotic minipump. Mice received daily injections of PBS (left), 2 mg/kg recombinant FGF19 (middle), or 2 mg/kg FGF21 (right) beginning on day 2 of the study and continuing for 6 consecutive days. Stained nuclei of hepatocytes in the liver from the FGF19-treated mouse are oriented around central veins (c) and away from portal veins (p). Hematoxylin counterstain is shown. B, semiquantitative analysis of BrdU-positive hepatocytes from A. Scores assigned to BrdU incorporation for these animals were based on a semiquantitative scale described under “Experimental Procedures.” Solid bars represent group mean score with S.E. (n = 8 for each group).
FGF19, but Not FGF21, Selectively Activates FGFR4 in Liver to Induce Pericentral Hepatocyte Proliferation
We hypothesized that the observed differential effects of FGF19 and FGF21 on hepatocyte proliferation might be the result of selective activation of a liver expressed receptor by FGF19 but not by FGF21. Therefore, we next evaluated the receptor and co-receptor requirements for FGF19 and FGF21. The rat myoblast cell line L6, which expresses very low levels of endogenous FGF receptors, was co-transfected with FGFR1c, 2c, 3c, or 4 together with β-Klotho. Receptor activation was determined by Western blot analysis of phospho-ERK levels. As shown in Fig. 2, and consistent with previous findings (14, 18), both FGF19 and FGF21 were able to activate FGFR1c, 2c, and 3c, but only FGF19 induced significant ERK phosphorylation via FGFR4. Given that FGFR4 is predominantly expressed in hepatocytes, this raised the possibility that the mitogenic effects of FGF19 are mediated through FGFR4. To determine the relative expression levels of each receptor, we also used β-Klotho and FGFRs with the V5 tag on their C-terminal end. The tagged and untagged receptors showed similar responses to treatments (data not shown), using the V5-tagged receptors allowed determination of relative expression levels of each receptor by Western analysis (shown in supplemental Fig. S1).
FIGURE 2.
Receptor specificities for FGF19 and FGF21. L6 cells were co-transfected with expression vectors for FGFR1c, 2c, 3c, or 4 and β-Klotho. Following overnight serum starvation, cells were stimulated with vehicle, 50 nm recombinant FGF19 or FGF21 for 10 min and snap frozen in liquid nitrogen. Cell lysates were prepared for Western blot using antibodies against phosphorylated ERK1/2 (p-ERK) or total ERK1/2 (T-ERK). ctl, PBS treatment.
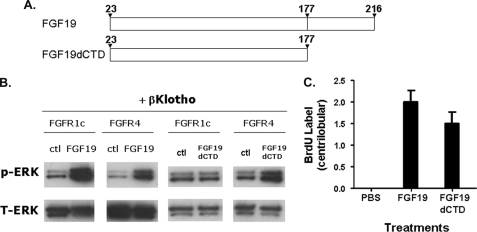
We previously showed that an interaction with β-Klotho is absolutely required for FGF19-induced activation of FGFRs 1c, 2c, and 3c, but not FGFR4 (15). Therefore, we utilized a variant of FGF19, FGF19dCTD, which is selective for FGFR4 due to deletion of the β-Klotho interaction C-terminal domain, to explore the effects of selective FGFR4 activation on hepatocyte proliferation. The structure of FGF19dCTD and its selectivity toward FGFR4 is shown in Fig. 3. Similar to our previous report (15), FGF19dCTD only activated FGFR4 but not FGFR1c (Fig. 3B). We administered FGF19dCTD to mice and evaluated BrdU staining to measure hepatocyte proliferation. Analysis of BrdU-immunostained liver sections from FGF19dCTD-treated animals also showed enhanced BrdU labeling similar to wild-type FGF19 treatment, indicating increased mitotic activity by FGF19dCTD (Fig. 3C and supplemental Fig. S2). This is the first direct evidence suggesting that selective activation of FGFR4 is sufficient to cause increased hepatocyte proliferation.
FIGURE 3.
Activities of FGF19dCTD. A, schematic diagram showing FGF19 and FGF19dCTD. B, L6 cells were transfected with FGFR1c or FGFR4 and with β-Klotho. Following overnight serum starvation, cells were stimulated with vehicle, 50 nm recombinant FGF19 or FGF19dCTD for 10 min, and snap frozen in liquid nitrogen. Cell lysates were prepared for Western blot using antibodies against phosphorylated ERK1/2 (p-ERK) or total ERK1/2 (T-ERK). C, semiquantitative analysis of BrdU immunostaining of livers from female FVB mice treated for 6 days with PBS, 2 mg/kg/day recombinant FGF19, or 2 mg/kg/day FGF19dCTD. The scores assigned to BrdU incorporation for these animals were based on a semiquantitative scale described under “Experimental Procedures.” The BrdU immunostaining of livers were showed in supplemental Fig. S2. Solid bars represent the group mean score with S.E. (n = 8 for each group).
Identification of FGF19 Sequences That Are Critical for FGFR4 Activation
To provide further evidence for the involvement of FGFR4 activation in hepatocyte proliferation, we next sought to identify the region(s) of FGF19 responsible for FGFR4 signaling. FGF19 and FGF21 share significant sequence homology but differ in the ability to activate FGFR4 signaling. Results from our experiments using FGF19dCTD suggested that the C-terminal region of FGF19 is not essential for FGFR4 activation. Thus we generated a series of FGF19/FGF21 chimeric molecules sequentially replacing the N-terminal region of FGF21 with the corresponding region of FGF19 (as illustrated in Fig. 4A) to identify the region responsible for FGFR4 activation. Receptor specificity was determined in vitro by signaling and adipocyte glucose uptake assays, and mitogenic potential in hepatocyte was determined in vivo by BrdU immunostaining. To establish that all the chimeras were functional, they were tested in L6 cells transfected with FGFR1c and β-Klotho and were shown to activate ERK phosphorylation (Fig. 4B, left panel). In addition, each was found to induce glucose uptake in 3T3L1 adipocytes, where FGFR1c is the predominant receptor (Fig. 4C). Therefore expression of the chimeras yielded properly folded and active proteins.
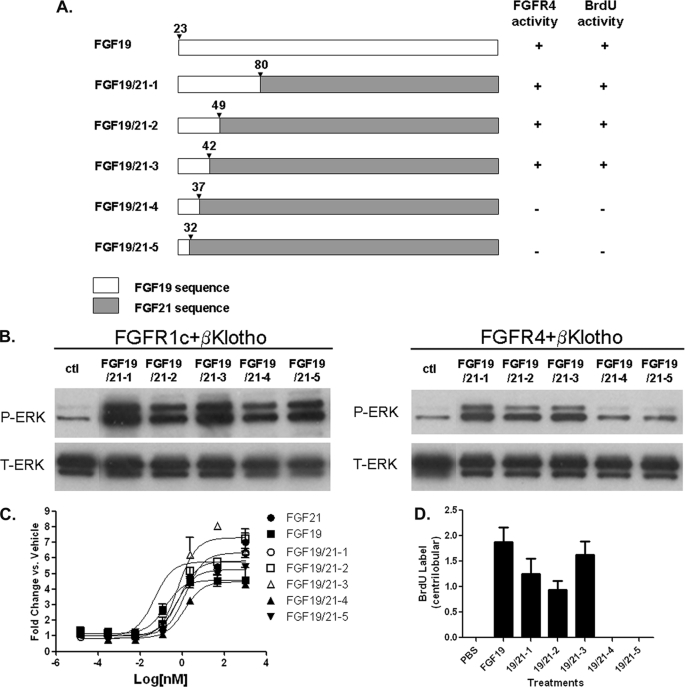
FIGURE 4.
FGF19/21 chimeric proteins. A, schematic diagram showing chimeric proteins between FGF19 and FGF21. The numbers for the last residue from N-terminal FGF19 sequences in each chimeric construct are shown. B, L6 cells were transfected with FGFR1c or FGFR4 and with β-Klotho. Following overnight serum starvation, cells were stimulated with vehicle or with 50 nm of recombinant FGF19, FGF21, or chimeric proteins for 10 min and snap frozen in liquid nitrogen. Cell lysates were prepared for Western blot using antibodies against phosphorylated ERK1/2 (p-ERK) or total ERK1/2 (T-ERK). C, differentiated 3T3-L1 adipocytes were incubated for 72 h with recombinant FGF19, FGF21 or chimeric proteins and assayed for glucose uptake. D, semiquantitative analysis of BrdU immunostaining of livers from female FVB mice treated for 6 days with PBS, 2 mg/kg/day recombinant FGF19 or 2 mg/kg/day chimeric proteins. The scores assigned to BrdU incorporation for these animals were based on a semiquantitative scale described under “Experimental Procedures”. The BrdU immunostaining of livers were showed in supplemental Fig. S3. Solid bars represent group mean score with S.E. (n = 8 for each group).
We next tested the chimeras for their ability to activate FGFR4. In contrast to FGFR1c, these novel chimeric molecules displayed differences in FGFR4 selectivity. In L6 cells transfected with FGFR4 and β-Klotho, ERK-phosphorylation was only observed with FGF19/21-1, -2, and -3, which share FGF19 amino acid sequences 23–42. ERK phosphorylation was not observed with FGF19/21-4 and -5, which lack FGF19 sequences from residue 38 (Fig. 4B, right panel). These results suggest that critical FGFR4-activating residues are likely contained within FGF19 residues 38–42.
The effects of chimeric molecules on hepatocyte proliferation were then tested using the in vivo BrdU incorporation assay. Examination of BrdU-immunostained liver sections from treated animals showed that, like FGF19, chimeras FGF19/21-1, -2, and -3 all exhibited increased BrdU labeling in the pericentral hepatocytes. However, such increases were not observed with animals treated with FGF19/21-4 and -5 (Fig. 4D and supplemental Fig. S3). The lack of enhanced BrdU labeling in FGF19/21-4 and -5 animals is unlikely due to differential stability of these proteins in vivo, because all FGF19/21 chimeric proteins showed similar pharmacokinetic properties (supplemental Fig. S4). Therefore, enhanced BrdU labeling directly correlated with FGFR4 activation (summarized in Fig. 4A), and once again revealed the potential importance of FGF19 residues 38–42 in FGFR4 activation and induction of hepatocyte proliferation.
Residues 38–42 from FGF19 Are Sufficient to Confer FGFR4 Activation and Increased Hepatocyte Proliferation
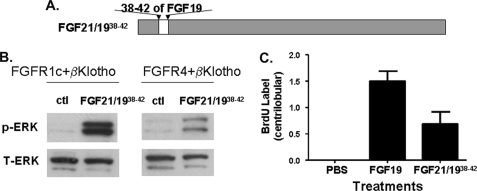
To further test whether these five residues from FGF19 are sufficient to confer FGFR4 activation, a new chimera, FGF21/1938–42, was generated, which contained residues 38–42 from FGF19 in place of the corresponding residues in FGF21 (Fig. 5A). Similar to FGF19 and FGF21, FGF21/1938–42 induced ERK-phosphorylation in L6 cells transfected with FGFR1c and β-Klotho (Fig. 5B) and was active in adipocyte glucose uptake assays (data not shown). However, in contrast to FGF21 but similar to FGF19, FGF21/1938–42 induced ERK-phosphorylation in L6 cells transfected with FGFR4 and β-Klotho (Fig. 5B). Histopathological examination of liver sections from FGF21/1938–42-treated animals showed enhanced BrdU labeling in pericentral hepatocytes similar to FGF19 treatment (Fig. 5C and supplemental Fig. S5). These results suggest that introduction of these 5 residues from FGF19 conferred a gain-of-function phenotype on FGF21 with respect to FGFR4 activation in vitro and induction of hepatocyte proliferation in vivo. Based on these results, we would speculate that these 5 amino acid residues may form direct interactions with FGFR4; however, this interaction is too weak to be detected under the experimental conditions that we have used for ELISA and Biacore binding studies (data not shown). The availability of a more sensitive detection system may provide further insights into FGF19/FGFR4 interaction and specificity determination.
FIGURE 5.
Activities of FGF21/1938–42. A, schematic diagram showing FGF21/1938–42. B, L6 cells were transfected with FGFR1c or FGFR4 together with β-Klotho. Following overnight serum starvation, cells were stimulated with vehicle, 50 nm recombinant FGF19, FGF21, or FGF21/1938–42 for 10 min, and snap frozen in liquid nitrogen. Cell lysates were prepared for Western blot using antibodies against phosphorylated ERK1/2 (p-ERK) or total ERK1/2 (T-ERK). C, semiquantitative analysis of BrdU immunostaining of livers from female FVB mice treated for 6 days with PBS, 2 mg/kg/day recombinant FGF19, or 2 mg/kg/day FGF21/1938–42. The scores assigned to BrdU incorporation for these animals were based on a semiquantitative scale described under “Experimental Procedures.” The BrdU immunostaining of livers are shown in supplemental Fig. S5. Solid bars represent the group mean score with S.E. (n = 8 for each group).
DISCUSSION
Both FGF19 and FGF21 are novel hormones that regulate glucose, lipid, and energy homeostasis (5, 6, 7, 8). FGF19 is also able to induce hepatocyte proliferation and cause formation of liver tumors in mice; however, the mechanism has not been determined previously (10). In this report, using BrdU labeling as a measure for in vivo mitotic activity, we show that, in contrast to FGF19, FGF21 does not induce hepatocyte proliferation when compared directly with FGF19. These experiments suggest functional differentiation between FGF19 and FGF21. Given that increased hepatocyte proliferation is believed to be a prerequisite for neoplastic transformation (11), these results may suggest that FGF21 would not cause HCC in rodents.
To better understand the mechanism leading to the proliferative activity observed with FGF19, we initially focused our attention on the differences in receptor specificity observed between FGF19 and FGF21. Both FGF19 and FGF21 activate FGFR1c, 2c and 3c, however, only FGF19 activates FGFR4 in vitro as measured by ERK phosphorylation (Fig. 2). Because FGFR4 is the predominant FGF receptor expressed in the liver, we hypothesized that the unique specificity of FGF19 toward FGFR4 might be responsible for its involvement in liver tumorigenesis. The first direct evidence supporting this hypothesis was derived from experiments with FGF19dCTD, a C-terminally truncated variant of FGF19 protein. The inability of FGF19dCTD to bind β-Klotho explains its lack of activity at FGFRs 1c, 2c, and 3c (Fig. 3) (15). However, FGF19dCTD retained its ability to activate FGFR4 in a β-Klotho-independent manner. Therefore, this variant is an FGFR4 specific activator. When tested in vivo for its effect on hepatocyte proliferation, FGF19dCTD increased BrdU incorporation in pericentral hepatocytes similar to FGF19, suggesting activation of FGFR4 alone is sufficient to promote hepatocyte proliferation.
To provide further evidence supporting this hypothesis and to identify region(s) in FGF19 responsible for FGFR4 activation, we generated chimeric proteins between FGF19 and FGF21 and tested their activity in signaling and proliferation assays (Fig. 4). These analyses showed an absolute correlation between the ability to activate FGFR4, measured by ERK phosphorylation in vitro, and increased pericentral hepatocyte proliferation measured by BrdU labeling in liver in vivo. This provides further evidence for the hypothesis that liver FGFR4 activation and hepatocyte proliferation are linked. In addition, this study identified a 5 amino acid region (residues 38–42) on FGF19 that is important and potentially sufficient for FGFR4 activation. This was further supported by experiments with FGF21/1938–42 in which only these 5 amino acids from FGF19 were substituted into FGF21. The substitution of these 5 amino acids was sufficient to confer a gain-of-function phenotype on FGF21 with respect to both FGFR4 activation and increased hepatocyte proliferation (Fig. 5).
Taken together: 1) the lack of increased hepatocyte proliferation by FGF21 treatment in direct comparison with FGF19; 2) the ability of FGF19dCTD, an FGFR4 specific activator, to increase pericentral hepatocyte BrdU incorporation; 3) the absolute correlation among FGF19/FGF21 chimeric molecules in their ability to induce FGFR4 activation and to increase pericentral hepatocyte BrdU staining; and finally 4) the ability of just 5 residues from FGF19 to confer FGFR4 activation and increased BrdU labeling in pericentral hepatocytes, these observations provide compelling evidence that FGFR4 activation in hepatocytes leads to increased proliferation which may cause HCC.
Acknowledgments
We thank David Penny, Yi Liu, and Barbara Felder for technical support. We thank Suzanne Coberly, Jie Tang, Helene Baribault, Grant Shimamoto, Hossein Salimi-Moosavi, Ming Wang, Bei Shan, Wen-Chen Yeh, Nigel Walker, Luke Li, Jackie Sheng, Kevin Moore, and Tom Boone for helpful discussions, and we thank Scott Silbiger, Margrit Schwarz, and Peter Coward for editing this manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- FGF
- fibroblast growth factor
- PBS
- phosphate-buffered saline
- BrdU
- bromodeoxyuridine
- FGFR
- FGF receptor
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Fukumoto S. (2008) Endocr. J. 55, 23–31 [DOI] [PubMed] [Google Scholar]
- 2.Fukumoto S., Yamashita T. (2007) Bone 40, 1190–1195 [DOI] [PubMed] [Google Scholar]
- 3.Ogawa Y., Kurosu H., Yamamoto M., Nandi A., Rosenblatt K. P., Goetz R., Eliseenkova A. V., Mohammadi M., Kuro-o M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2005) Cell Metab. 2, 217–225 [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson E., Fu L., John L., Hultgren B., Huang X., Renz M., Stephan J. P., Tsai S. P., Powell-Braxton L., French D., Stewart T. A. (2002) Endocrinology 143, 1741–1747 [DOI] [PubMed] [Google Scholar]
- 6.Fu L., John L. M., Adams S. H., Yu X. X., Tomlinson E., Renz M., Williams P. M., Soriano R., Corpuz R., Moffat B., Vandlen R., Simmons L., Foster J., Stephan J. P., Tsai S. P., Stewart T. A. (2004) Endocrinology 145, 2594–2603 [DOI] [PubMed] [Google Scholar]
- 7.Kharitonenkov A., Shiyanova T. L., Koester A., Ford A. M., Micanovic R., Galbreath E. J., Sandusky G. E., Hammond L. J., Moyers J. S., Owens R. A., Gromada J., Brozinick J. T., Hawkins E. D., Wroblewski V. J., Li D. S., Mehrbod F., Jaskunas S. R., Shanafelt A. B. (2005) J. Clin. Invest. 115, 1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J., Lloyd D. J., Hale C., Stanislaus S., Chen M., Sivits G., Vonderfecht S., Hecht R., Li Y. S., Lindberg R. A., Chen J. L., Jung D. Y., Zhang Z., Ko H. J., Kim J. K., Véniant M. M. (2009) Diabetes 58, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kharitonenkov A., Wroblewski V. J., Koester A., Chen Y. F., Clutinger C. K., Tigno X. T., Hansen B. C., Shanafelt A. B., Etgen G. J. (2007) Endocrinology 148, 774–781 [DOI] [PubMed] [Google Scholar]
- 10.Nicholes K., Guillet S., Tomlinson E., Hillan K., Wright B., Frantz G. D., Pham T. A., Dillard-Telm L., Tsai S. P., Stephan J. P., Stinson J., Stewart T., French D. M. (2002) Am. J. Pathol. 160, 2295–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fausto N. (1999) Sem. Liver Dis. 19, 243–252 [DOI] [PubMed] [Google Scholar]
- 12.Huang X., Yang C., Jin C., Luo Y., Wang F., McKeehan W. L. (2009) Mol. Carcinogenesis 48, 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurosu H., Kuro-O M. (2009) Mol. Cell. Endocrinol. 299, 72–78 [DOI] [PubMed] [Google Scholar]
- 14.Kurosu H., Choi M., Ogawa Y., Dickson A. S., Goetz R., Eliseenkova A. V., Mohammadi M., Rosenblatt K. P., Kliewer S. A., Kuro-o M. (2007) J. Biol. Chem. 282, 26687–26695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X., Ge H., Lemon B., Weiszmann J., Gupte J., Hawkins N., Li X., Tang J., Lindberg R., Li Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14379–14384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X., Li Y. (2009) Aging 1, 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X., Lemon B., Li X., Gupte J., Weiszmann J., Stevens J., Hawkins N., Shen W., Lindberg R., Chen J. L., Tian H., Li Y. (2008) J. Biol. Chem. 283, 33304–33309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin B. C., Wang M., Blackmore C., Desnoyers L. R. (2007) J. Biol. Chem. 282, 27277–27284 [DOI] [PubMed] [Google Scholar]