Abstract
We previously found that pigeon IgG possesses unique N-glycan structures that contain the Galα1–4Galβ1–4Galβ1–4GlcNAc sequence at their nonreducing termini. This sequence is most likely produced by putative α1,4- and β1,4-galactosyltransferases (GalTs), which are responsible for the biosynthesis of the Galα1–4Gal and Galβ1–4Gal sequences on the N-glycans, respectively. Because no such glycan structures have been found in mammalian glycoproteins, the biosynthetic enzymes that produce these glycans are likely to have distinct substrate specificities from the known mammalian GalTs. To study these enzymes, we cloned the pigeon liver cDNAs encoding α4GalT and β4GalT by expression cloning and characterized these enzymes using the recombinant proteins. The deduced amino acid sequence of pigeon α4GalT has 58.2% identity to human α4GalT and 68.0 and 66.6% identity to putative α4GalTs from chicken and zebra finch, respectively. Unlike human and putative chicken α4GalTs, which possess globotriosylceramide synthase activity, pigeon α4GalT preferred to catalyze formation of the Galα1–4Gal sequence on glycoproteins. In contrast, the sequence of pigeon β4GalT revealed a type II transmembrane protein consisting of 438 amino acid residues, with no significant homology to the glycosyltransferases so far identified from mammals and chicken. However, hypothetical proteins from zebra finch (78.8% identity), frogs (58.9–60.4%), zebrafish (37.1–43.0%), and spotted green pufferfish (43.3%) were similar to pigeon β4GalT, suggesting that the pigeon β4GalT gene was inherited from the common ancestors of these vertebrates. The sequence analysis revealed that pigeon β4GalT and its homologs form a new family of glycosyltransferases.
Keywords: Glycoproteins, Carbohydrate Biosynthesis, Carbohydrate Structure, Enzyme Structure, Enzymes, Gb3 Synthase, P1 Antigen, Glycan Diversity, Glycosyltransferase, Molecular Evolution
Introduction
The structures of glycans attached to glycoproteins or glycolipids vary greatly with various glycosidic linkages, including sialylations, fucosylations, galactosylations, and/or other glycan modifications. Different glycan structures are often expressed in a species-specific fashion in animals and plants and are thought to be involved in species-specific interactions between hosts and foreign organisms, such as bacteria, viruses, and parasites (1, 2). The various kinds of species-specific glycans seem to have been generated, maintained, and/or lost during the evolution and diversification of animals, but the evolutional process of glycan diversity remains to be elucidated due to the lack of sufficient information (3).
Galα1–4Gal is known as a minimum glycan sequence that is essential for the cell surface attachment of certain bacterial adhesins, e.g. P-adhesin of uropathogenic Escherichia coli, and certain enterotoxins, e.g. Shiga toxins (see Ref. 4 and references therein). Mammals express Galα1–4Gal on glycolipids, such as globotriosylceramide (Gb3,2 Galα1–4Galβ1–4Glc-Cer) and P1 antigen (Galα1–4Galβ1–4GlcNAcβ1–3Galβ1–4Glc-Cer), but not on glycoproteins, except for those found in hydatid fluids caused by tapeworm (5, 6). In contrast, we found that pigeon egg white is a rich source of glycoproteins that possess the Galα1–4Galβ1–4GlcNAc sequence at the nonreducing termini of N-glycans (4). It was demonstrated that pigeon egg white glycoproteins are useful in preventing infection with uropathogenic E. coli (7). We also found that the Galα1–4Gal sequence was absent from the egg white glycoproteins of two large taxa of avians, i.e. Ratitae3 (traditionally called Palaeognathae, e.g. ostrich, rhea, emu, and tinamou) and Galloanserae (e.g. chicken and duck), but was present in the majority of Neoaves (e.g. pigeon, swiftlet, parrot, and gull) (8–11). However, it is unclear how avian species acquired the ability to produce Galα1–4Gal on glycoproteins.
Pigeon IgG possesses another unique glycan sequence, Galα1–4Galβ1–4Galβ1–4GlcNAc, at the nonreducing termini of N-glycans (12). The same sequence is found in O-glycans from the salivary gland mucin of the Chinese swiftlet (13). This fact indicates that these birds express not only Galα1–4Gal but also Galβ1–4Gal sequence. Unlike Galα1–4Gal, the distribution of the Galβ1–4Gal sequence among various avian species remains to be investigated. Among mammals, it has been reported that pigs express glycolipids containing the Galβ1–4Galβ1–4Glc-Cer sequence at an internal position (14), but Galβ1–4Gal is not found in glycoproteins in mammals. In contrast, high titers of natural antibodies against Galβ1–4Gal were detected in human sera (15), suggesting that there may be some (Galβ1–4Gal)-containing antigens, either microbes or macromolecules in human habitats.
Galα1–4Gal and Galβ1–4Gal sequences are most likely biosynthesized in pigeon by the actions of putative UDP-Gal:β-galactoside α1,4-galactosyltransferase (α4GalT(Gal)4) and UDP-Gal:β-galactoside β1,4-galactosyltransferase (β4GalT(Gal)), respectively, which have not been reported so far. Because the Galα1–4Gal sequence is rarely attached to glycoproteins in mammals, the putative α4GalT(Gal) in pigeon is likely to have distinct acceptor substrate specificities from those of mammals. Although mammalian α4GalT(Gal)s, also known as Gb3 synthases, have been cloned from human (16, 17), mouse (18), and rat (19), they utilize lactosylceramide (Galβ1–4Glc-Cer) as the preferred acceptor substrate to produce Gb3. Only when the expression level of the enzyme is sufficiently high, human Gb3 synthase can also synthesize P1 antigen using lacto-N-neotetraosylceramide (Galβ1- 4GlcNAcβ1–3Galβ1–4Glc-Cer) as a substrate (20). Another putative enzyme in pigeon, β4GalT(Gal), which transfers Gal to Gal residues by forming a β-1,4-linkage on glycans, has also not been well studied to date. Because of its different acceptor substrate specificities, β4GalT(Gal) is distinct from β4GalT(GlcNAc)s, which produce Galβ1–4GlcNAc sequences found commonly in mammals and many other animals (21).
In this study, to understand the molecular basis of species-specific glycan differentiation, we isolated cDNAs encoding pigeon α4GalT(Gal) and β4GalT(Gal), which are responsible for the production of Galα1–4Gal and Galβ1–4Gal on glycoproteins, respectively. The deduced amino acid sequence of pigeon α4GalT(Gal) is homologous to those of human and putative chicken α4GalT(Gal)s. However, by comparing these α4GalT(Gal)s, we demonstrated that pigeon α4GalT(Gal) possesses distinct substrate specificities from those of mammals and of chicken. In contrast, the deduced amino acid sequence of pigeon β4GalT(Gal) does not resemble those of known members of the β4GalT(GlcNAc) family, which produce Galβ1–4GlcNAc in vertebrates, or those of other known glycosyltransferases. However, genes encoding “hypothetical proteins” with sequence similarity to pigeon β4GalT(Gal) were found in the molecular data bases of frogs and fishes as well as zebra finch, suggesting that the expression of Galβ1–4Gal in birds is genetically related to that in these vertebrates. Moreover, the results of sequence analysis revealed that pigeon β4GalT(Gal) and its homologs form a new family of glycosyltransferases.
EXPERIMENTAL PROCEDURES
Additional Experimental Procedures are described in the supplemental material.
In general, to detect the activity of α4GalT(Gal) or β4GalT(Gal) and to determine their biochemical properties, 2-aminopyridine (PA)-derivatized biantennary N-glycans (PA-N-glycan A in Fig. 1) was used as an acceptor substrate and analyzed by normal phase HPLC using an Amide-80 column (Tosoh Co., Tokyo, Japan). Synthetic glycans, such as p-nitrophenyl β-d-lactopyranoside (Lac-pNP), were also used as acceptor substrate for kinetic analysis, NMR measurements, and methylation analysis. To determine the specificity of acceptor substrates, free monosaccharides or oligosaccharides were incubated with UDP-[3H]Gal in the presence of pigeon α4GalT(Gal) or β4GalT(Gal).
FIGURE 1.
Structures of PA-oligosaccharides used in this study. PA-N-glycan A was prepared from human γ-globulin and utilized as an acceptor substrate of pigeon α4GalT(Gal) and β4GalT(Gal). PA-N-glycans B and C were prepared from PA-N-glycan A by the action of pigeon α4GalT(Gal) and β4GalT(Gal), respectively. PA-N-glycan D was prepared from PA-N-glycan A by β4-galactosidase digestion.
RESULTS
Expression Cloning of Pigeon Liver α4GalT(Gal) and β4GalT(Gal) cDNAs
We selected 293T cells to be the host recipient cells and expected Galα1–4Galβ1–4GlcNAc structures to be expressed on the cell surfaces when cDNA encoding α4GalT(Gal) was transfected into these cells. Pigeon liver was utilized as a source of mRNA to construct the cDNA library for expression cloning, because we detected (Galα1–4Gal)-containing glycoproteins in this tissue (12). After the selection using anti-P1 mAb (specific for Galα1–4Galβ1–4GlcNAc) by cell sorting, the plasmid designated as pcDNA-pigeon- α4GalT(Gal) was isolated.
Pigeon β4GalT(Gal) cDNA was also isolated from the pigeon liver cDNA library by expression cloning, after selection using anti-(Galβ1–4Gal) mAb. The isolated plasmid was designated as pcDNA-pigeon-β4GalT(Gal).
Sequences of the Pigeon α4GalT(Gal) and β4GalT(Gal)
The entire nucleotide sequence of the inserted cDNA in pcDNA-pigeon-α4GalT(Gal) consisted of a single 2263-bp open reading frame encoding a protein of 360 amino acids (supplemental Fig. S1). As found in many other Golgi-resident glycosyltransferases, which possess type II transmembrane topology, the cytoplasmic tail of pigeon α4GalT(Gal) is most likely located toward the N terminus and followed by the transmembrane domain, whereas the putative stem region and the catalytic domain are toward the C terminus. Three potential N-glycosylation sites were present at the stem and/or catalytic regions, and the DXD motif was found in the catalytic domain. The gene products with the highest identity for pigeon α4GalT(Gal) were found in NCBI Protein Database for two avian species, chicken and zebra finch (Table 1 and supplemental Fig. S2). There are three sequences in zebra finch and one sequence in chicken, and these sequences were annotated “similar to α1,4-galactosyltransferase.” The BLAST analysis also revealed human (16, 17), mouse (18), and rat (19) α4GalT(Gal)s as being homologous proteins, as well as some other predicted proteins from mammals that were annotated similar to α1,4-galactosyltransferase. The pigeon α4GalT(Gal) also has some homology with the human α1,4-N-acetylglucosaminyltransferase (α1,4-N-acetylglucosaminyltransferase, 36.3% identity; accession number AAD48406) and putative α1,4-N-acetylglucosaminyltransferase from chicken (34.4%, accession number XP_426692) and zebra finch (31.5%, accession number XP_002189476). These results suggest that the newly isolated clone from pigeon liver also belongs to the α4GalT family.
TABLE 1.
Proteins or hypothetical proteins in vertebrates similar to pigeon α4GalT(Gal)/β4GalT(Gal)
| Classes, species | α4GalT(Gal)a |
β4GalT(Gal) |
||||
|---|---|---|---|---|---|---|
| Accession no.b | Enzyme activity | Identityc | Accession no.b | Enzyme activity | Identityc | |
| % | % | |||||
| Mammals | ||||||
| Human | BAA95915 | Gb3 synthase | 58.2 | Not found | ||
| Mouse | ABA25853 | Gb3 synthase | 56.2 | Not found | ||
| Rat | AAF82758 | Gb3 synthase | 55.9 | Not found | ||
| Othersd | (See below) | Unknown | 53.1–58.2 | Not found | ||
| Birds | ||||||
| Chicken | XP_416448.2 | Gb3 synthasee | 68.0 | Not found | ||
| Zebra finch | XP_002189441 | Unknown | 65.7 | XP_002189371 | Unknown | 78.8 |
| XP_002189476 | Unknown | 66.6 | ||||
| XP_002189509 | Unknown | 65.5 | ||||
| Amphibians | ||||||
| African clawed frog | Not found | NP_001089439 | Unknown | 59.6 | ||
| Western clawed frog | NP_001120129 | Unknown | 60.4 | |||
| NP_001120419 | Unknown | 58.9 | ||||
| Fishes | ||||||
| Zebrafish | Not found | NP_001116761 | Unknown | 42.4 | ||
| NP_001116796 | Unknown | 43.0 | ||||
| XP_001334398 | Unknown | 37.2 | ||||
| Pufferfish | CAG10330 | Unknown | 43.3 | |||
a Proteins or hypothetical proteins more similar to human α 1,4-N-acetylglucosaminyltransferase than to human Gb3 synthase are not listed in this table.
b Protein data were obtained from NCBI Protein Database in September 2009.
c Identity (%) was based on amino acid sequences.
d Other mammals possessing putative proteins “similar to Gb3 synthase” were as follows: platypus, XP_001519949; dog, XP_538343; horse, XP_001500792; rhesus monkey, XP_001107622; orangutan, CAH91488; and chimpanzee, BAA94504.
e Chicken putative α4GalT(Gal) was demonstrated to possess the activity of Gb3 synthase in this study (see Fig. 5).
The entire sequence of the insert of pcDNA-pigeon- β4GalT(Gal) revealed a 1690-bp fragment with a single open reading frame encoding a protein of 438 amino acids with type II transmembrane topology (supplemental Fig. S3). Five potential N-glycosylation sites were present at the stem and/or catalytic regions, and the DXD motif was found in the catalytic domain. The gene products in the NCBI Protein Database with the highest identity with pigeon β4GalT(Gal) were the hypothetical proteins from zebra finch, one of the birds belonging to Passeriformes (Table 1 and supplemental Fig. S4). It is notable that all the other gene products with high levels of identity are hypothetical proteins from frogs and fishes, as shown in Table 1. The sequence of pigeon β4GalT(Gal) also has some identity (20–26%) with hypothetical proteins from several insects, such as the honeybee, jewel wasp, red flour beetle, and fruit fly, and from nematodes. In contrast, no homologous proteins were found in the NCBI-DNA/Protein Databases (as of September 2009) of mammals and chicken. None of the proteins with known functions, including β4GalT(GlcNAc), showed significant homology with pigeon β4GalT(Gal) in a BLAST search.
Structural Analysis of the Glycans Produced by the Action of Pigeon α4GalT(Gal) and β4GalT(Gal)
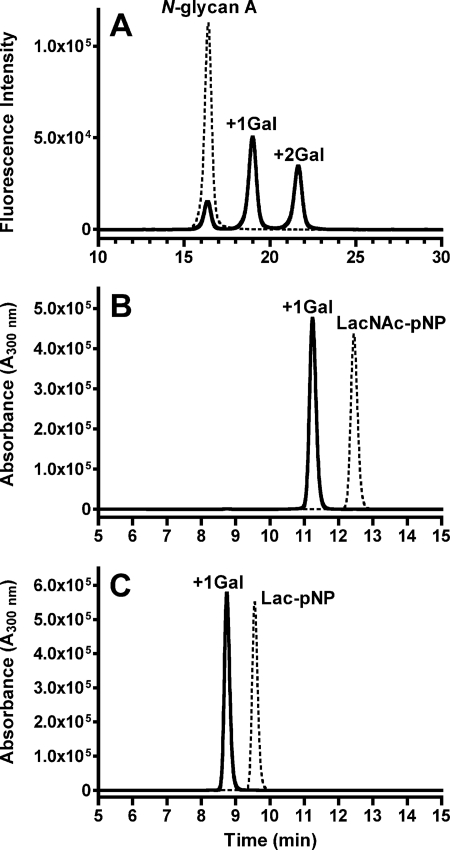
Triton X-100 extracts (0.2 and 1 mg of protein/ml) of 293T cells transfected with pcDNA-pigeon-α4GalT(Gal) transferred one or two Gal residues from UDP-Gal to PA-derivatized oligosaccharides with a β4-galactosylated biantennary structure (PA-N-glycan A in Fig. 1), as revealed by normal phase HPLC analysis (Fig. 2), whereas the extracts of wild-type cells or mock transfectants did not transfer the Gal residues under the same conditions (data not shown). No Gal transfer was observed in the absence of UDP-Gal. Affinity-isolated FLAG-tagged soluble pigeon α4GalT(Gal) also transferred one or two Gal residues to the PA-N-glycan A substrate in the presence of UDP-Gal (data not shown). Matrix-assisted laser desorption/ionization time of flight-mass spectrometry analysis revealed that the m/z values of the substrate and the final product were 1888 and 2213, respectively, confirming that the two Gal residues were added to the substrate. When the product of soluble pigeon α4GalT(Gal) was digested with α-galactosidase from green coffee beans, m/z values were changed to 1888, which was exactly the same as those of PA-N-glycan A (Fig. 1). These results suggest that two α-Gal residues were added to the substrate by the action of the recombinant enzyme, and the structure of the product is most likely PA-N-glycan B (Fig. 1). No products of α4GalT(Gal) were detected when UDP-GalNAc instead of UDP-Gal was used as a donor, whereas products with UDP-Glc (14% of those with UDP-Gal) or UDP-GlcNAc (1.8%) were detected. FLAG-tagged soluble pigeon α4GalT(Gal) also transferred one Gal residue to synthetic monosaccharide or disaccharide substrates, such as p-nitrophenyl 2-acetamide-2-deoxy-4-O-(β-d-galactopyranosyl)-β-d-glucopyranoside (Galβ1- 4GlcNAcβ1-pNP, LacNAc-pNP) (Fig. 2B) and p-nitrophenyl β-d-lactopyranoside (Galβ1–4Glcβ1-pNP, Lac-pNP) (Fig. 2C), and Galβ1-pNP (data not shown). The enzymatic products were 162 mass units larger than those of the substrates (data not shown), confirming that the products contain one additional Gal residue. The linkage of newly formed Gal-Gal sequence on Lac-pNP was confirmed to be Galα1–4Gal by NMR (supplemental Fig. S5) and methylation analysis (supplemental Fig. S6), as described in supplemental material. The results clearly indicate the activities of recombinant enzyme that acts as α4GalT(Gal).
FIGURE 2.
HPLC analysis of the glycans produced by the action of pigeon α4GalT(Gal). A, Triton X-100 extracts of 293T cells (0.2 mg of protein/ml) transfected with full-length pigeon α4GalT(Gal) cDNA (A) were incubated with the substrate (PA-N-glycan A) in the presence (solid line) or absence (broken line) of UDP-Gal. The reaction mixtures were analyzed with normal phase HPLC using an Amide-80 column. B and C, HPLC profiles of the reaction mixtures of FLAG-tagged soluble pigeon α4GalT(Gal) with LacNAc-pNP (B) or Lac-pNP (C) in the presence (solid lines) or absence (broken lines) of UDP-Gal. The reaction mixtures were analyzed with reversed-phase HPLC using an ODS column.
The enzymatic activity of pigeon β4GalT(Gal) was detected using the cell lysates of transfectants and FLAG-tagged soluble form of pigeon β4GalT(Gal) by the same in vitro assay as for pigeon α4GalT(Gal) as described above. The HPLC profiles of the products from PA-N-glycans or pNP-glycans by the action of β4GalT(Gal) (data not shown) were similar to those of α4GalT(Gal) as shown in Fig. 2. However, the products by β4GalT(Gal) were digested with β4-galactosidase from Streptococcus pneumoniae. For instance, the product from PA-N-glycan A (Fig. 1) by the β4GalT(Gal), which is most likely PA-N-glycan C (m/z = 2213), was digested with β4-galactosidase and became PA-N-glycan D (m/z = 1564). These results suggest that two β-Gal residues were added to the substrate, PA-N-glycan A, by the action of the recombinant enzyme. No products of β4GalT(Gal) were detected when it was incubated with UDP-GalNAc, UDP-Glc, or UDP-GlcNAc instead of UDP-Gal. FLAG-tagged soluble pigeon β4GalT(Gal) also transferred one Gal residue to synthetic disaccharide substrates, such as LacNAc-pNP and Lac-pNP (data not shown but similar to Fig. 2, B and C, respectively), but not to Galβ1-pNP (data not shown). The linkage of the newly formed Gal-Gal sequence on Lac-pNP was confirmed to be Galβ1–4Gal by NMR (supplemental Fig. S5) and methylation analysis (supplemental Fig. S6), as described in supplemental material. The results clearly indicate the activities of recombinant enzyme that acts as β4GalT(Gal).
Biochemical Properties of Pigeon α4GalT(Gal) and β4GalT(Gal)
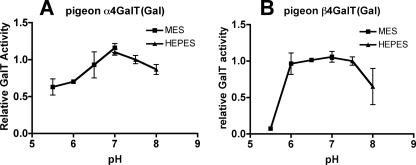
As shown in Fig. 3, the highest activities of the α4GalT(Gal) and β4GalT(Gal) were observed under neutral conditions around pH 7.0. The cation-dependent enzymatic activities of α4GalT(Gal) and β4GalT(Gal) were observed as summarized in Table 2.
FIGURE 3.
Determination of the optimum pH for pigeon α4GalT(Gal) and β4GalT(Gal) activities. FLAG-tagged soluble pigeon α4GalT(Gal) or β4GalT(Gal) was incubated with the PA-derivatized substrate (PA-N-glycan A) and UDP-Gal in MES- or HEPES-buffered solutions of various pH values. Relative α4GalT(Gal) or β4GalT(Gal) activity is indicated, with the activity at pH 7.5 being set to 1. The results indicate the means ± S.D. of duplicate experiments.
TABLE 2.
Effects of metal ions on pigeon α4GalT(Gal)/β4GalT(Gal) activities
| Additiona | Activityb |
|
|---|---|---|
| α4GalT(Gal) | β4GalT(Gal) | |
| % | ||
| MnCl2 | 100 ± 6.2 | 100 ± 0.1 |
| CoCl2 | 49 ± 1.9 | 134 ± 1.8 |
| MgCl2 | 29 ± 0.8 | 41 ± 0.6 |
| NiCl2 | 13 ± 0 | 6.6 ± 0.2 |
| CaCl2 | 6.3 ± 1.1 | 27 ± 2.2 |
| ZnCl2 | NDc | ND |
| None | 10 ± 2.4 | 11 ± 0.1 |
| EDTA | ND | ND |
a Final concentrations of divalent cations and EDTA were 20 mm.
b Relative activities of 100% were calculated as a percentage of the incorporation obtained with the addition of MnCl2 (set to 100%). The values represent the means ± S.D. of duplicate samples.
c ND means not detected.
Substrate Specificity of the Pigeon α4GalT(Gal) and β4GalT(Gal)
Several kinds of monosaccharides or oligosaccharides and UDP-[3H]Gal were incubated with FLAG-tagged soluble pigeon α4GalT(Gal) or β4GalT(Gal).
When α4GalT(Gal) was added to the reaction mixtures, most of the monosaccharides or oligosaccharides containing Gal residues transferred additional Gal residues (Table 3). In contrast, no incorporation of Gal was observed into monosaccharides or oligosaccharides that do not possess any Gal residues, such as GalNAc, GlcNAc, Man, Glc, Fuc, or maltooligosaccharides. As expected, LacNAc was one of the preferred acceptors and was used as a standard to indicate the relative activities of the other acceptor substrates. Galβ1–4Gal and Galβ1–4Galβ1–4Glc were notably better acceptors than LacNAc. Galβ1–4Glc and Galβ1–4Man sequences were, however, less preferred substrates, although they both possess the same Galβ1–4Hex sequence as Galβ1–4Gal. β-Galactosides were clearly better substrates than α-galactosides, as seen in Galβ1-methyl versus Galα1-methyl activity or Galβ1–4Gal versus Galα1–4Gal activity. A type II linkage (Galβ1–4GlcNAc) was preferred to a type I (Galβ1–3GlcNAc) linkage, and the Galβ1–6GlcNAc linkage was even better than the type II linkage, although the Galβ1–6GlcNAc linkage was not as common as the type II linkage in vertebrates. Fucosylation at either inner GlcNAc (Lea and Lex) or outer Gal (H-trisaccharide and lacto-N-fucopentaose I) significantly decreased the incorporation of Gal residues, suggesting that fucosylation sterically hinders the action of pigeon α4GalT(Gal).
TABLE 3.
Acceptor substrate specificity of pigeon α4GalT(Gal)/β4GalT(Gal)
| Acceptors | Name | Activityab |
|
|---|---|---|---|
| α4GalT(Gal) | β4GalT(Gal) | ||
| % | |||
| Galβ1–4GlcNAc | N-Acetyllactosamine | 100 ± 12 | 100 ± 6.4 |
| Galβ1–4Glc | Lactose | 59 ± 3.7 | 28 ± 3.6 |
| Gal | 5.4 ± 0.9 | NDc | |
| Galβ1-methyl | Methyl-β-galactoside | 57 ± 2.0 | ND |
| Galα1-methyl | Methyl-α-galactoside | ND | ND |
| Galβ1–3GlcNAc | 8.2 ± 0.8 | ND | |
| Galβ1–3GalNAc | 30 ± 0.2 | ND | |
| Galα1–3Gal | 10 ± 1.0 | ND | |
| Galβ1–4Gal | 321 ± 8.8 | ND | |
| Galβ1–4Man | 11 ± 0.9 | 4.0 ± 2.0 | |
| Galα1–4Gal | 20 ± 0.2 | ND | |
| Galβ1–6GlcNAc | 121 ± 4.2 | ND | |
| Galα1–6Glc | Melibiose | ND | ND |
| Galβ1–4Galβ1–4Glc | β4′-Galactosyllactose | 258 ± 38 | ND |
| Galα1–4Galβ1–4Glc | 2.4 ± 0.5 | ND | |
| Galβ1–3(Fucα1–4)GlcNAc | Lea | 1.4 ± 0.6 | ND |
| Galβ1–4(Fucα1–3)GlcNAc | Lex | 9.5 ± 0.7 | 2.8 ± 3.1 |
| Fucα1–2Galβ1–4GlcNAc | H-trisaccharide | 0.5 ± 0.1 | 2.7 ± 1.5 |
| Galβ1–3GlcNAcβ1–3Galβ1–4Glc | Lacto-N-tetraose | 13 ± 0.4 | 0.9 ± 0.7 |
| Galβ1–4GlcNAcβ1–3Galβ1–4Glc | Lacto-N-neotetraose | 71 ± 3.5 | 107 ± 1.6 |
| Fucα1–2Galβ1–3GlcNAcβ1–3Galβ1–4Glc | Lacto-N-fucopentaose I | 2.7 ± 0.5 | 2.9 ± 1.1 |
a Relative activities were calculated as a percentage of the incorporation obtained with Galβ1–4GlcNAc. The values represent the means ± S.D. of duplicate samples.
b No activities for both α4GalT(Gal) and β4GalT(Gal) were detected when GalNAc, GlcNAc, Man, Glc, Fuc, maltose, maltotriose, and maltotetraose were used as acceptor substrates.
c ND means not detected.
The kinetic parameters of pigeon α4GalT(Gal) were also compared using several pNP-saccharides as the acceptor substrates. As shown in supplemental Table S2, the apparent Km values for Galβ1–4Galβ1–4GlcNAcβ1-pNP and Galβ1–4Galβ1–4Glcβ1-pNP were 2.9- and 6.2-fold lower, respectively, than that for Galβ1–4GlcNAcβ1-pNP, although the apparent kcat values of these three substrates are almost equivalent. The results suggest that different Km values reflect the different reactivity to these acceptor substrates. However, significant substrate inhibition for pigeon α4GalT(Gal) was also observed, especially when Galβ1–4Galβ1–4GlcNAcβ1-pNP or Galβ1–4Galβ1–4Glcβ1-pNP was used as acceptor substrates as indicated with lower Ki values (supplemental Table S2).
When FLAG-tagged soluble pigeon β4GalT(Gal) was added into the reaction mixture, Galβ1–4GlcNAc (LacNAc) and Galβ1–4GlcNAcβ1–3Galβ1–4Glc (lacto-N-neotetraose) were the preferred acceptor substrates among the tested saccharides (Table 3). Galβ1–4Glc (Lac) was also recognized as an acceptor, but only about a quarter of the relative activity was detected for this compound. Compared with pigeon α4GalT(Gal), the acceptor substrate specificity of pigeon β4GalT(Gal) was restricted almost exclusively to Galβ1–4Glc(NAc) sequences. No Gal residues were transferred onto either Galβ1–3GlcNAc or Galβ1–3GalNAc, suggesting that the type I linkage and the core 1 structure on O-glycans were low preference substrates of pigeon β4GalT(Gal). Fucosylations either onto inner GlcNAc (i.e. Lex) or outer Gal (i.e. H-trisaccharide) on LacNAc significantly reduced the incorporation of Gal residues. The kinetic parameters of pigeon β4GalT(Gal) revealed that the apparent Km values for Galβ1–4GlcNAcβ1-pNP were 7.3-fold lower than that for Galβ1–4Glcβ1-pNP (supplemental Table S2), supporting the results that pigeon β4GalT(Gal) prefers Galβ1–4GlcNAc than Galβ1–4Glc as an acceptor.
Distribution of the Pigeon α4GalT(Gal) and β4GalT(Gal) Transcripts in Various Tissues
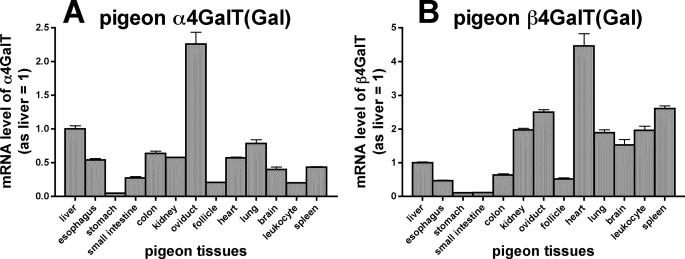
Real time PCR analysis using gene-specific primers revealed that both α4GalT(Gal) and β4GalT(Gal) mRNA/cDNAs were detected in all tissues examined (Fig. 4). However, the expression levels varied from tissue to tissue.
FIGURE 4.
Expression profiles of pigeon α4GalT(Gal) and β4GalT(Gal) mRNA detected by real time PCR. mRNA was isolated from various tissues of pigeon and analyzed by real time PCR using gene-specific primers. The mRNA amounts used in the experiments were first normalized against the amount of β-actin, a housekeeping gene, and then the relative mRNA levels of pigeon α4GalT(Gal) or β4GalT(Gal) were calculated, with the level in liver being set to 1. The results indicate the means ± S.D. of triplicate experiments.
Comparison of the Substrate Preferences of Pigeon, Human, and Chicken α4GalT(Gal)s in Vivo and in Vitro
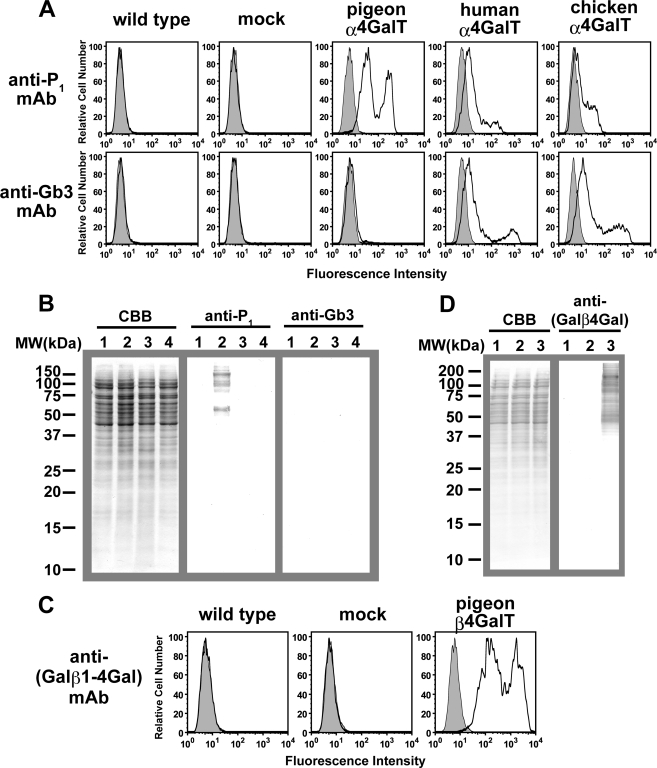
Wild-type 293T cells and cells transfected with mock or full-length cDNAs encoding pigeon, human, and putative chicken α4GalT(Gal)s were compared by immunostaining with anti-P1 mAb or anti-Gb3 mAb. The FACS analysis revealed that pigeon α4GalT(Gal) transfectants were stained with anti-P1 mAb on the cell surface, whereas no staining was observed on wild-type cells and mock transfectants (Fig. 5A). The pigeon α4GalT(Gal) transfectants did not stain with anti-Gb3 mAb. In contrast, 293T cells transfected with cDNA encoding human α4GalT(Gal) (also named Gb3 synthase) stained slightly with anti-P1 mAb but stained stronger with anti-Gb3 mAb. Similarly, the putative chicken α4GalT(Gal) transfectants stained strongly with anti-Gb3 mAb, suggesting that this chicken α4GalT(Gal) possesses Gb3 synthase activity. These results suggest that pigeon α4GalT(Gal) is unlikely to act as a Gb3 synthase, although 293T cells express the precursor of Gb3, i.e. lactosylceramide.
FIGURE 5.
Expression of α4GalT(Gal) or β4GalT(Gal) in 293T cells. A and B, detection of P1 antigen and Gb3 on cells transfected with pigeon, human, or chicken α4GalT(Gal) DNA. Wild-type and 293T cells transfected with mock, pigeon, human, or chicken α4GalT(Gal) DNA were analyzed with FACS (A) or Western blotting (B). A, cell surfaces were stained with anti-P1 mAb or anti-Gb3 mAb, and fluorescein isothiocyanate-goat anti-mouse IgM was used as the secondary antibody. Histograms in gray indicate the results in the absence of primary antibodies. B, Western blotting analysis of cell extracts from cells transfected with mock (lane 1), pigeon (lane 2), human (lane 3), or chicken (lane 4) α4GalT(Gal) DNA. Proteins blotted onto polyvinylidene difluoride membranes were stained with Coomassie Brilliant Blue R-250 (CBB), anti-P1 mAb, or anti-Gb3 mAb, as indicated. C and D, detection of Galβ1–4Gal on cells transfected with pigeon β4GalT(Gal) cDNA. Wild-type and 293T cells transfected with mock or pigeon β4GalT(Gal) cDNA were analyzed with FACS (C) or Western blotting (D). C, cell surfaces were stained with anti-(Galβ1–4Gal) mAb, and fluorescein isothiocyanate-goat anti-mouse IgG was used as the secondary antibody. Histograms in gray indicate the results in the absence of primary antibodies. D, Western blotting analysis of cell extracts from cells transfected with wild-type (lane 1), mock (lane 2), or pigeon β4GalT(Gal) (lane 3) cDNA. Proteins blotted onto polyvinylidene difluoride membranes were stained with Coomassie Brilliant Blue R-250 or anti-(Galβ1–4Gal) mAb.
Although proteins extracted from wild-type 293T cells (data not shown) and from mock, pigeon, human, and chicken α4GalT(Gal) transfectant cells on the Western blotting membrane had equal levels of staining with Coomassie Brilliant Blue R-250 (CBB, Fig. 5B), only pigeon α4GalT(Gal) transfectants stained with anti-P1 mAb. Proteins extracted from human α4GalT(Gal) transfectants did not stain with either anti-P1 or anti-Gb3 mAb, confirming that the human α4GalT(Gal) transfers Gal residues onto glycolipids rather than onto glycoproteins. These results strongly suggest that the preferred substrates, either on glycoproteins or glycolipids, differed significantly between pigeon and human α4GalT(Gal)s. Protein extracts of chicken α4GalT(Gal) transfectants also failed to stain with either anti-P1 or anti-Gb3 mAb, suggesting that the substrate preferences of chicken α4GalT(Gal) are closer to those of human α4GalT(Gal) than to those of pigeon α4GalT(Gal).
The substrate preferences of pigeon, human, and chicken α4GalT(Gal)s were also compared by in vitro GalT assays. Although cell extracts of pigeon α4GalT(Gal) transfectants transferred one or two Gal residues to the PA-derivatized N-glycan A, as shown in Fig. 2A, no products were detected on the HPLC analysis when cell extracts (0.2 and 1 mg of protein/ml for the assay) of human or chicken α4GalT(Gal) transfectants were used as enzyme sources (data not shown). Similarly, affinity-isolated FLAG-tagged soluble pigeon α4GalT(Gal)s (0.1 μg/25 μl/reaction) transferred Gal residues to PA-N-glycan A, LacNAc-pNP, or Lac-pNP after incubation at 37 °C for 4 h, although no products were detected when the same amount of FLAG-tagged soluble human or chicken α4GalT(Gal)s were incorporated in the reaction mixtures (data not shown).
Expression of Galβ1–4Gal on Pigeon β4GalT(Gal) Transfectant Cells
The FACS analysis revealed that full-length pigeon β4GalT(Gal) transfectants stained with anti-(Galβ1–4Gal) mAb on the cell surface, although no staining was observed on wild-type 293T cells and mock (pcDNA3.1(+)) transfectants (Fig. 5C). Similarly, only the proteins extracted from pigeon β4GalT(Gal) transfectants on the Western blotting membrane stained with anti-(Galβ1–4Gal) mAb, although those from wild-type, mock, and pigeon β4GalT(Gal) transfectant cells stained with Coomassie Brilliant Blue R-250 equally (Fig. 5D). This result suggests that glycoproteins possessing Galβ1–4Gal sequences were produced by transfection with pigeon β4GalT(Gal).
Molecular Phylogeny of α4GalT(Gal) and β4GalT(Gal)
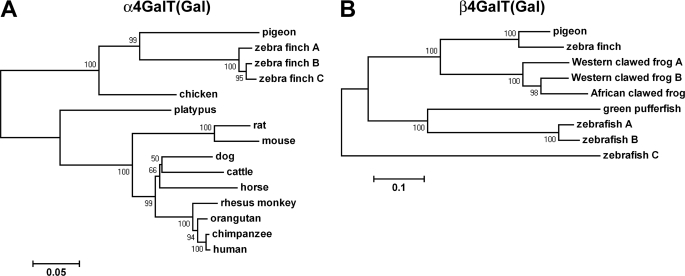
A phylogenetic analysis was performed using aligned DNA sequences encoding α4GalT(Gal)s from human, mouse, rat, chicken, and pigeon and the predicted α4GalT(Gal)s from several mammals (platypus, dog, cattle, horse, rhesus monkey, orangutan, and chimpanzee) and a bird (zebra finch), as shown in Fig. 6A. As expected, among mammals, the phylogenetic distances between α4GalT(Gal)s correlate with the taxonomy of the animals. Among birds, three putative α4GalT(Gal)s from zebra finch were the closest to pigeon α4GalT(Gal), and chicken α4GalT(Gal) was closer to these avian α4GalT(Gal) or its homolog than to those from mammals. It should be noted that chicken α4GalT(Gal), i.e. Gb3 synthase, is a much closer relative of pigeon α4GalT(Gal) than of mammals, although the substrate preferences of chicken α4GalT(Gal) are more similar to those from mammals than to those from pigeon.
FIGURE 6.
Neighbor-joining phylogenetic tree of α4GalT(Gal) or β4GalT(Gal) and their related proteins. The bootstrap value (%) based on 1000 replications is shown for each interior branch. The scale bar represents the number of substitutions per site for a unit branch length. A, phylogenetic analysis for α4GalT(Gal). Coding sequences of DNA were extracted from NCBI GenBankTM, and their accession numbers are as follows: zebra finch (Taeniopygia guttata), XM_002189405 (A), XM_002189440 (B), and XM002189473 (C); chicken (Gallus gallus), XM_416448; platypus (Ornithorhynchus anatinus), XM_001519899; rat (Rattus norvegicus), AF248544; mouse (Mus musculus), DQ143885; dog (Canis lupus familiaris), XM_538343; cattle (Bos taurus), XR_028271; horse (Equus caballus), XM_001500742; rhesus monkey (Macaca mulatta), XM_001107622; orangutan (Pongo abelii), CR859310; chimpanzee (Pan troglodytes), AB041419; human (Homo sapiens), AB037883. B, phylogenetic analysis for β4GalT(Gal) and the related proteins. Coding sequences of DNA were obtained from NCBI GenBankTM, and their accession numbers are as follows: zebra finch (Taeniopygia guttata), XM_002189335; African clawed frog (Xenopus laevis), NM_001095970; Western clawed frog (Xenopus tropicalis), NM_001126657 (A) and NM_001126947 (B); zebrafish (Dario rerio), NM_001123289 (A), NM_001123324 (B), and XM_001334362 (C); spotted green pufferfish (Tetraodon nigroviridis), CAAE01015019.
A phylogenetic analysis was also performed using aligned sequences of DNA encoding pigeon β4GalT(Gal) and its homologs from zebra finch, African clawed frog, Western clawed frog, zebrafish, and spotted green pufferfish, as shown in Fig. 6B. The β4GalT(Gal) homolog from zebra finch was the closest to pigeon β4GalT(Gal). The hypothetical proteins from frogs were closer to each other on the phylogenetic tree and relatively closer to pigeon β4GalT(Gal) than those from fishes.
DISCUSSION
Pigeon α4GalT(Gal) Possesses Distinct Substrate Preferences from Those of Human and Chicken Gb3 Synthases
The distribution of (Galα1–4Gal)-containing glycoproteins in nature was initially believed to be limited, because mammalian glycoproteins so far studied do not possess Galα1–4Gal on glycoproteins. However, we found that the expression of Galα1–4Gal on glycoproteins is correlated with the avian phylogeny (8). To investigate the enzyme catalyzing the formation of Galα1–4Gal on glycoproteins, we successfully isolated the cDNA encoding pigeon α4GalT(Gal), which had not been identified previously. This pigeon α4GalT(Gal) revealed distinct substrate specificity from that of human α4GalT(Gal), i.e. Gb3 synthase, because pigeon α4GalT(Gal) is capable of transferring Gal residues to β-galactosides on glycoproteins to form Galα1–4Gal linkages. The deduced amino acid sequence was, however, homologous to that of human, mouse, and rat Gb3 synthases (Table 1), which belong to the GT32 family in Carbohydrate-Active Enzyme Database (22–24). We also confirmed that the putative chicken α4GalT(Gal), which was identified from chicken genome/protein data bases in NCBI, acts as a Gb3 synthase, like human α4GalT(Gal). This observation is consistent with the absence of Galα1–4Gal on glycoproteins in chicken (4, 25). The high similarity between pigeon α4GalT(Gal) and chicken Gb3 synthase implies that they are most likely derived from a common ancestral gene relatively recently. To estimate the period of divergence of these genes, the sequences of α4GalT(Gal) genes from various avian species will be compared by extensive investigations.
The isolated pigeon α4GalT(Gal) will also be useful in generating the building blocks of Galα1–4Gal by chemoenzymatic synthesis. Because bindings of the pathogenic microbes (e.g. uropathogenic E. coli) or enterotoxins (e.g. Shiga toxins) to glycolipids containing the Galα1–4Gal sequence in mammalian cells are the first step of their invasion of the host cell, high affinity glycoconjugate inhibitors containing Galα1–4Gal may serve as therapeutic and preventive agents. As indicated in this study, this recombinant enzyme has the ability to transfer Gal residues to β-galactosides at the nonreducing termini of Galβ1–4GlcNAc, Galβ1–4Glc, and Galβ1–4Gal or even of monosaccharides, such as Galβ1-methyl and Galβ1-pNP. Unlike human Gb3 synthase, which has a preference for transferring Gal residues onto Galβ1–4Glc-Cer over Galβ1–4GlcNAcβ1–3Galβ1–4Glc-Cer (20), the pigeon α4GalT(Gal) has broad substrate specificity. Therefore, this enzyme has a large potential to be utilized in the production of various kinds of glycoconjugates containing Galα1–4Gal.
Pigeon β4GalT(Gal) Defines a New Sequence Family of Glycosyltransferases Not Found in Mammals
The protein sequence of newly isolated pigeon β4GalT(Gal) showed no significant homology with known glycosyltransferases in the Carbohydrate-Active Enzyme Database (22–24). An NCBI-BLAST search using Conserved Domain Database, which serves to identify conserved domains in protein sequences from over 12,000 model domain collections in the data base (26, 27), indicated that the sequence contains a conserved domain designated as DUF23 (accession number, pfam01697), located between amino acid residues 176 and 430. Members of the DUF23 family are found in some hypothetical proteins from various species of eukaryota, including some vertebrates (fishes, frogs, and a bird), nematodes, insects, plants, and bacteria. The DUF23 region consists of ∼300 amino acid residues containing several conserved cysteine residues and several charged residues, which are predicted to serve as catalytic residues. Therefore, proteins containing DUF23 had been expected to possess some enzymatic activities, although there was no direct evidence of this until we identified β4GalT(Gal) activity in one of the member proteins, as shown in this study. Moreover, the Conserved Domain Database indicated that DUF23 belong to the GT-A superfamily, which possesses one of the two known structural folds of nucleotide-sugar-dependent glycosyltransferases (24). Because most GT-A enzymes possess a DXD motif in which the carboxylates coordinate a divalent cation and/or ribose, this observation is consistent with the fact that the isolated pigeon β4GalT(Gal) possesses cation-dependent activity (Table 2).
Among vertebrates whose data are currently (September 2009) available in NCBI DNA/Protein Databases, some fishes, frogs, and a bird, zebra finch, but no mammals or chicken, were indicated as possessing hypothetical proteins containing DUF23. Since the presence of Galβ1–4Gal on N-glycans from zebrafish was previously reported (28, 29), the proteins containing DUF23 are candidates for β4GalT(Gal) in zebrafish. Galβ1–4Gal on N-glycans was also reported to exist in medaka (30–32) and dace (33, 34). The presence of Galβ1–4Gal on O-glycans from eggs of salmoid fishes was also reported (35–37). Several species of amphibians were reported to possess O-glycans with Galβ1–4Gal in the mucin of their egg jelly coats (38–42). Therefore, we assume that the hypothetical proteins of clawed frogs similar to pigeon β4GalT(Gal) possess the cognate enzyme activity.
Although limited numbers of mammalian species are selected for complete genomic sequence projects, DNA/protein sequences from mammalian species spanning monotreme (duck-billed platypus), marsupialia (gray short-tailed opossum), rodents (mouse and rat), dog, bovine, and primates (rhesus macaque, chimpanzee, and human) are now available in the NCBI DNA/Protein Databases (43). Despite this wide spectrum, no homologous sequences with pigeon β4GalT(Gal) or DUF23 were found in the mammalian data bases. Therefore, the genes of β4GalT(Gal) or DUF23 may have been lost in several lineages of mammals or in the common ancestors of various species of mammals after the ancestors of mammals and birds separated (∼310 million years ago) (44).
Conclusion
Species-specific glycans are often involved in interactions between hosts and foreign organisms, and even a subtle difference in glycan structure can largely influence their associations (2). For instance, Galα1–4Gal on glycolipids is expressed on cell surfaces and mediates the cellular internalization of Shiga toxins, which are activated by cellular proteases for intoxication of cells (45–47). In contrast, the presence of glycoproteins containing Galα1–4Gal in tissues and body fluids may prevent such glycolipid-mediated endocytosis and/or intracellular processing of toxins. The biological features of Galβ1–4Gal are less well understood currently, but the high titers of natural antibodies against Galβ1–4Gal in human sera (15) imply that this glycan sequence is also involved in host-foreign body interactions. It should be emphasized that the presence of Galα1–4Gal and Galβ1–4Gal is not an evolutionary relic and that there seems to be a driving force for the generation and conservation of these glycan sequences in some animals. As we have indicated, glycoprotein-specific α4GalT(Gal) was generated relatively recently in a lineage of avian species. This fact suggests that birds were able to produce novel glycan forms, and they might have entered into selective associations with some microorganisms to evade pathogenic relationships and/or to coevolve in symbiotic or commensal relationships (48). The wide distribution of Galβ1–4Gal in vertebrates and its conservation in some avian species may indicate a benefit for possessing oligosaccharides with Galβ1–4Gal. The availability of genes encoding pigeon α4GalT(Gal) and β4GalT(Gal) should help accelerate investigations into the biological necessity of producing species-specific glycans, as well as studies on the molecular evolution of these enzymes.
Addendum
While this manuscript was being reviewed, Titz et al. (49) reported the cloning of Caenorhabditis elegans N-glycan core α1,6-fucoside β1,4-galactosyltransferase, which defined a new GT family in carbohydrate-active enzymes (GT92). The amino acid sequence of this enzyme is similar to the pigeon β4GalT(Gal) (20.9% identity), suggesting that these two enzymes belong to the same GT family.
This work was supported by Grant-in-aid for Scientific Research 18770081 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Tables S1 and S2, Figs. S1–S6, and additional references.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) FJ971844 and FJ971845.
Nomenclatures for avian classification are based on Sibley and Ahlquist (50). In this paper, Neoaves does not include Galloanserae.
To distinguish glycosyltransferases based on their acceptor substrate specificities, acceptors were indicated in parentheses after the abbreviation of the enzyme, e.g., β4GalT(Gal) versus β4GalT(GlcNAc), throughout this paper.
- Gb3
- globotriosylceramide
- α4GalT
- α1,4-galactosyltransferase
- β4GalT
- β1,4-galactosyltransferase
- Cer
- ceramide
- PA
- 2-aminopyridine
- pNP
- p-nitrophenyl
- HPLC
- high performance liquid chromatography
- mAb
- monoclonal antibody
- MES
- 4-morpholineethanesulfonic acid
- FACS
- fluorescence-activated cell sorter
- Lac
- lactose.
REFERENCES
- 1.Varki A. (1993) Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gagneux P., Varki A. (1999) Glycobiology 9, 747–755 [DOI] [PubMed] [Google Scholar]
- 3.Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2008) Essentials of Glycobiology, 2nd Ed., pp. 281–292, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 4.Suzuki N., Khoo K. H., Chen H. C., Johnson J. R., Lee Y. C. (2001) J. Biol. Chem. 276, 23221–23229 [DOI] [PubMed] [Google Scholar]
- 5.Khoo K. H., Nieto A., Morris H. R., Dell A. (1997) Mol. Biochem. Parasitol. 86, 237–248 [DOI] [PubMed] [Google Scholar]
- 6.Cossey A., Dimichiel A., Dunstone J. (1979) Eur. J. Biochem. 98, 53–60 [DOI] [PubMed] [Google Scholar]
- 7.Johnson J. R., Berggren T. (1994) Am. J. Med. Sci. 307, 335–339 [DOI] [PubMed] [Google Scholar]
- 8.Suzuki N., Laskowski M., Jr., Lee Y. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9023–9028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki N., Laskowski M., Jr., Lee Y. C. (2006) Biochim. Biophys. Acta 1760, 538–546 [DOI] [PubMed] [Google Scholar]
- 10.Suzuki N., Lee Y. C. (2007) in Comprehensive Glycoscience: From Chemistry to Systems Biology (Kamerling J. P., Boons G. J., Lee Y. C., Suzuki A., Taniguchi N., Voragen A. G. eds) vol. 3, pp. 237–251, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 11.Suzuki N., Su T. H., Wu S. W., Yamamoto K., Khoo K. H., Lee Y. C. (2009) Glycobiology 19, 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki N., Khoo K. H., Chen C. M., Chen H. C., Lee Y. C. (2003) J. Biol. Chem. 278, 46293–46306 [DOI] [PubMed] [Google Scholar]
- 13.Wieruszeski J. M., Michalski J. C., Montreuil J., Strecker G., Peter-Katalinic J., Egge H., van Halbeek H., Mutsaers J. H., Vliegenthart J. F. (1987) J. Biol. Chem. 262, 6650–6657 [PubMed] [Google Scholar]
- 14.Slomiany A., Slomiany B. L., Horowitz M. I. (1974) J. Biol. Chem. 249, 1225–1230 [PubMed] [Google Scholar]
- 15.Bouhours D., Liaigre J., Naulet J., Bovin N. V., Bouhours J. F. (2000) Glycobiology 10, 857–864 [DOI] [PubMed] [Google Scholar]
- 16.Kojima Y., Fukumoto S., Furukawa K., Okajima T., Wiels J., Yokoyama K., Suzuki Y., Urano T., Ohta M., Furukawa K. (2000) J. Biol. Chem. 275, 15152–15156 [DOI] [PubMed] [Google Scholar]
- 17.Steffensen R., Carlier K., Wiels J., Levery S. B., Stroud M., Cedergren B., Nilsson Sojka B., Bennett E. P., Jersild C., Clausen H. (2000) J. Biol. Chem. 275, 16723–16729 [DOI] [PubMed] [Google Scholar]
- 18.Fujii Y., Numata S., Nakamura Y., Honda T., Furukawa K., Urano T., Wiels J., Uchikawa M., Ozaki N., Matsuo S., Sugiura Y., Furukawa K. (2005) Glycobiology 15, 1257–1267 [DOI] [PubMed] [Google Scholar]
- 19.Keusch J. J., Manzella S. M., Nyame K. A., Cummings R. D., Baenziger J. U. (2000) J. Biol. Chem. 275, 25315–25321 [DOI] [PubMed] [Google Scholar]
- 20.Iwamura K., Furukawa K., Uchikawa M., Sojka B. N., Kojima Y., Wiels J., Shiku H., Urano T., Furukawa K. (2003) J. Biol. Chem. 278, 44429–44438 [DOI] [PubMed] [Google Scholar]
- 21.Amado M., Almeida R., Schwientek T., Clausen H. (1999) Biochim. Biophys. Acta 1473, 35–53 [DOI] [PubMed] [Google Scholar]
- 22.Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 23.Campbell J. A., Davies G. J., Bulone V., Henrissat B. (1997) Biochem. J 326, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 25.Suzuki N., Lee Y. C. (2004) Glycobiology 14, 275–292 [DOI] [PubMed] [Google Scholar]
- 26.Marchler-Bauer A., Anderson J. B., Derbyshire M. K., DeWeese-Scott C., Gonzales N. R., Gwadz M., Hao L., He S., Hurwitz D. I., Jackson J. D., Ke Z., Krylov D., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Lu S., Marchler G. H., Mullokandov M., Song J. S., Thanki N., Yamashita R. A., Yin J. J., Zhang D., Bryant S. H. (2007) Nucleic Acids Res. 35, D237–D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A., Anderson J. B., Cherukuri P. F., DeWeese-Scott C., Geer L. Y., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Marchler G. H., Mullokandov M., Shoemaker B. A., Simonyan V., Song J. S., Thiessen P. A., Yamashita R. A., Yin J. J., Zhang D., Bryant S. H. (2005) Nucleic Acids Res. 33, D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guérardel Y., Chang L. Y., Maes E., Huang C. J., Khoo K. H. (2006) Glycobiology 16, 244–257 [DOI] [PubMed] [Google Scholar]
- 29.Moriguchi K., Takemoto T., Aoki T., Nakakita S., Natsuka S., Hase S. (2007) J. Biochem. 142, 213–227 [DOI] [PubMed] [Google Scholar]
- 30.Iwasaki M., Seko A., Kitajima K., Inoue Y., Inoue S. (1992) J. Biol. Chem. 267, 24287–24296 [PubMed] [Google Scholar]
- 31.Taguchi T., Seko A., Kitajima K., Muto Y., Inoue S., Khoo K. H., Morris H. R., Dell A., Inoue Y. (1994) J. Biol. Chem. 269, 8762–8771 [PubMed] [Google Scholar]
- 32.Taguchi T., Seko A., Kitajima K., Inoue S., Iwamatsu T., Khoo K. H., Morris H. R., Dell A., Inoue Y. (1993) J. Biol. Chem. 268, 2353–2362 [PubMed] [Google Scholar]
- 33.Taguchi T., Iwasaki M., Muto Y., Kitajima K., Inoue S., Khoo K. H., Morris H. R., Dell A., Inoue Y. (1996) Eur. J. Biochem. 238, 357–367 [DOI] [PubMed] [Google Scholar]
- 34.Inoue S., Iwasaki M., Ishii K., Kitajima K., Inoue Y. (1989) J. Biol. Chem. 264, 18520–18526 [PubMed] [Google Scholar]
- 35.Iwasaki M., Inoue S., Kitajima K., Nomoto H., Inoue Y. (1984) Biochemistry 23, 305–310 [DOI] [PubMed] [Google Scholar]
- 36.Shimamura M., Endo T., Inoue Y., Inoue S. (1983) Biochemistry 22, 959–963 [DOI] [PubMed] [Google Scholar]
- 37.Iwasaki M., Inoue S. (1985) Glycoconj. J. 2, 209–228 [Google Scholar]
- 38.Maes E., Florea D., Delplace F., Lemoine J., Plancke Y., Strecker G. (1997) Glycoconj. J. 14, 127–146 [DOI] [PubMed] [Google Scholar]
- 39.Coppin A., Maes E., Morelle W., Strecker G. (1999) Eur. J. Biochem. 266, 94–104 [DOI] [PubMed] [Google Scholar]
- 40.Mourad R., Morelle W., Neveu A., Strecker G. (2001) Eur. J. Biochem. 268, 1990–2003 [DOI] [PubMed] [Google Scholar]
- 41.Florea D., Maes E., Haddad M., Strecker G. (2002) Biochimie 84, 611–624 [DOI] [PubMed] [Google Scholar]
- 42.Coppin A., Florea D., Maes E., Cogálniceanu D., Strecker G. (2003) Biochimie 85, 53–64 [DOI] [PubMed] [Google Scholar]
- 43.Brown S. (2008) Nature 453, 138–139 [DOI] [PubMed] [Google Scholar]
- 44.Hedges S. B., Parker P. H., Sibley C. G., Kumar S. (1996) Nature 381, 226–229 [DOI] [PubMed] [Google Scholar]
- 45.Lindberg A. A., Brown J. E., Strömberg N., Westling-Ryd M., Schultz J. E., Karlsson K. A. (1987) J. Biol. Chem. 262, 1779–1785 [PubMed] [Google Scholar]
- 46.Sandvig K., Garred O., Prydz K., Kozlov J. V., Hansen S. H., van Deurs B. (1992) Nature 358, 510–512 [DOI] [PubMed] [Google Scholar]
- 47.Garred O., van Deurs B., Sandvig K. (1995) J. Biol. Chem. 270, 10817–10821 [DOI] [PubMed] [Google Scholar]
- 48.Hooper L. V., Gordon J. I. (2001) Glycobiology 11, 1R–10R [DOI] [PubMed] [Google Scholar]
- 49.Titz A., Butschi A., Henrissat B., Fan Y. Y., Hennet T., Razzazi-Fazeli E., Hengartner M. O., Wilson I. B., Kuenzler M., Aebi M. (2009) J. Biol. Chem. 284, 36223–36233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sibley C. G., Ahlquist J. E. (1990) Phylogeny and Classification of Birds: A Study in Molecular Evolution, Yale University Press, New Haven, CT [Google Scholar]