Abstract
Insulin resistance can occur in response to many different external insults, including chronic exposure to insulin itself as well as other agonists such as dexamethasone. It is generally thought that such defects arise due to a defect(s) at an early stage in the insulin signaling cascade. One model suggests that this involves activation of the mammalian target of rapamycin/S6 kinase pathway, which inactivates insulin receptor substrate via Ser/Thr phosphorylation. However, we have recently shown that insulin receptor substrate is not a major node for insulin resistance defects. To explore the mechanism of insulin resistance, we have developed a novel system to activate Akt independently of its upstream effectors as well as other insulin-responsive pathways such as mitogen-activated protein kinase. 3T3-L1 adipocytes were rendered insulin-resistant either with chronic insulin or dexamethasone treatment, but conditional activation of Akt2 stimulated hemagglutinin-tagged glucose transporter 4 translocation to the same extent in these insulin-resistant and control cells. However, addition of insulin to cells in which Akt was conditionally activated resulted in a reversion to the insulin-resistant state, indicating a feedforward inhibitory mechanism activated by insulin itself. This effect was overcome with wortmannin, implicating a role for phosphatidylinositol 3-kinase in this inhibitory process. We conclude that in chronic insulin- and dexamethasone-treated cells, acute activation with insulin itself is required to activate a feedforward inhibitory pathway likely emanating from phosphatidylinositol 3-kinase that converges on a target downstream of Akt to cause insulin resistance.
Keywords: Metabolism, Phosphorylation, Signal Transduction/Protein Kinases, Transport/Glucose, Insulin, AS160, GLUT4 Translocation, Insulin Resistance
Introduction
Insulin resistance is a hallmark of type 2 diabetes. It is characterized by a reduced ability of target tissues, such as the liver, skeletal muscle, and adipose tissues, to respond to insulin (1). A major compensatory consequence of insulin resistance is the development of hyperinsulinemia, which is able to prevent hyperglycemia at the potential expense of beta cell function. Thus, prolonged insulin resistance may be accompanied by prolonged hyperinsulinemia, ultimately leading to the demise of the beta cell, resulting in diabetes (2). Hence, it is widely considered that strategies that can reverse insulin resistance will have major benefit in preventing the onset of metabolic disease.
Numerous insults are known to cause insulin resistance, but defining the precise mechanism(s) by which such insults impact upon the normal insulin action pathway remain unclear. Despite this, the components of the insulin signaling pathway that control metabolic processes such as glucose transporter 4 (GLUT4)2 translocation have been identified. The phosphatidylinositol 3-kinase (PI3-kinase)/Akt pathway plays a crucial role in insulin-stimulated GLUT4 translocation (3). Insulin binding to the insulin receptor triggers tyrosine phosphorylation of insulin receptor substrate (IRS) proteins. This promotes binding of Src homology 2) domain-containing signaling proteins, which includes PI3-kinase, Grb-2, and SHP2 (4). Upon association with IRS proteins, PI3-kinase is activated, leading to the generation of phosphatidylinositol 3,4,5-triphosphate at the plasma membrane (PM) leading to activation of Akt. Full activation of Akt requires phosphorylation on Thr308 by PDK1 and Ser473 by the mTOR·Rictor complex (5, 6). Active Akt phosphorylates multiple downstream effectors that promote diverse biological responses, including stimulation of glucose transport, protein and glycogen synthesis, and the regulation of gene expression (3). Of particular interest among the Akt substrates is AS160, which plays an important role in insulin-stimulated GLUT4 translocation to the PM (7, 8).
In several models of insulin resistance, it has been shown that reduced glucose uptake is due to defects in insulin signaling. A number of studies suggest that this is due to a defect at the level of IRS (9–12) whereas others suggest that the defect lies further downstream (13–15).
To explore the mechanism(s) of insulin resistance in 3T3-L1 adipocytes chronically exposed to insulin or dexamethasone, we have utilized a previously described novel system involving a rapalog-inducible heterodimerization strategy to rapidly activate Akt2 alone without activating its upstream effectors (16). Rapalog-induced HA-GLUT4 translocation in 3T3-L1 adipocytes was not inhibited in insulin-resistant cells, but was significantly impaired when rapalog was combined with the acute readdition of insulin. This inhibitory effect was overriden by inhibition of PI3-kinase. This, combined with the observation that insulin resistance was not accompanied by a major defect in AS160 phosphorylation, leads us to propose the following model. Many of the common insults that cause insulin resistance such as hyperinsulinemia or dexamethasone activate a signaling pathway that emanates from the PI3-kinase node. Chronic stimulation of this pathway leads to the induction of a feedforward inhibitory signal that is readily activated by acute elevation of insulin itself. This negative signal causes insulin resistance by impacting upon elements in the insulin action cascade downstream of Akt to inhibit GLUT4 translocation.
EXPERIMENTAL PROCEDURES
Materials
Polyclonal rabbit antibody raised against total Akt, monoclonal rabbit antibody raised against pSer473 Akt (193H12) (for immunofluorescence), and mouse monoclonal antibody raised against pSer473 Akt (for Western blotting) were purchased from Cell Signaling Technologies (Beverly, MA). Polyclonal rabbit antibody raised against 14-3-3β was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Polyclonal AS160 antibody was from Upstate Biotechnology (Lake Placid, NY). Antibodies against mouse monoclonal GLUT4 and rabbit polyclonal syntaxin 4 were described previously (17). Horseradish peroxidase-conjugated secondary antibodies were from Amersham Biosciences, Alexa Fluor 488-conjugated secondary antibody was from Molecular Probes, and Cy2-conjugated secondary antibody was obtained from Jackson ImmunoResearch (West Grove, PA). Polyclonal sheep antibody raised against pThr642 AS160 was obtained from Peter Shepherd (Symansis, Auckland, New Zealand). Paraformaldehyde was from ProSciTech (Thuringowa, Australia). Insulin, PKCζ pseudosubstrate, and Rac inhibitor were purchased from Calbiochem. MAP kinase inhibitor (PD98059) was from Cell Signaling Technologies. Dulbecco's modified Eagle's medium (DMEM) and newborn calf serum were from Invitrogen. Fetal calf serum was from Trace Scientific (Melbourne, Australia), and antibiotics were from Invitrogen. Bovine serum albumin (BSA) was from Bovogen (Essendon, Australia). bicinchoninic acid reagent and SuperSignal West Pico chemiluminescent substrate were from Pierce. Complete protease inhibitor mixture tablets were from Roche Applied Science. All other materials were obtained from Sigma.
Cell Culture and Retrovirus Transduction
3T3-L1 fibroblasts (ATCC, Manassas, VA) were cultured and differentiated to adipocytes as described previously (8). To generate 3T3-L1 adipocytes stably expressing HA-GLUT4, fibroblasts were infected with pBabepuro-HA-GLUT4 retrovirus. After a 24-h recovery period, infected cells were selected in DMEM containing 10% fetal calf serum and 2 μg/ml puromycin. Polyclonal pools of puromycin-resistant 3T3-L1 fibroblasts were then grown to confluence and subsequently differentiated into adipocytes as described above. To generate 3T3-L1 adipocytes stably expressing both HA-GLUT4 with either PDGFR WT or PDGFR Y740/Y751, puromycin-resistant 3T3-L1 fibroblasts expressing HA-GLUT4 were subsequently infected with the pLXSN PDGFR retroviruses as above. Infected cells were selected in DMEM containing 10% fetal calf serum, 2 μg/ml puromycin, and 800 μg/ml geneticin. Polyclonal pools of puromycin and geneticin-resistant 3T3-L1 fibroblasts were then grown to confluence and differentiated into adipocytes as described above.
Lentivirus Preparation and Transduction
Recombinant lentivirus (FRB-Akt2-Myc, Myr-FKBP WT-V5, and Myr-FKBP A2-V5) were produced by co-transfecting HEK-293 FT cells (Invitrogen) with the lentivirus expression and packaging plasmids using the calcium phosphate transfection method as described previously (18, 19). Infectious lentiviruses were harvested at 24 h and 48 h after transfection and centrifuged at 1620 × g for 10 min. The supernatants collected were concentrated 10-fold using Amicon Ultracel concentration tubes (100-kDa pore size; Millipore). 3T3-L1 adipocytes at day 5 of differentiation were infected with 50 μl/well for 96-well plates or 200 μl/well for 24-well plates of concentrated FRB-Akt2-Myc lentivirus containing 4 μg/ml Polybrene. After 16–24 h, the concentrated lentivirus was removed and infected again with either concentrated Myr-FKBP WT-V5 or Myr-FKBP A2-V5 lentivirus. Experiments were performed at 5 days after transduction.
Insulin Resistance Treatments
Chronic insulin-induced insulin resistance was generated in 3T3-L1 adipocytes by incubation the cells with 10 nm insulin in DMEM-high glucose containing 0.2% BSA for 24 h at 37 °C. Because 3T3-L1 adipocytes exhibit substantial “insulinase” activity (20), fresh insulin-supplemented DMEM was replaced every 4 h as possible (at 1100, 1500, and 1900 h on day 1, and 0700 h the following day). At 1100 h on the second day, cells were washed three times with PBS and incubated in serum- and bicarbonate-free DMEM containing 20 mm HEPES, pH 7.4, and 0.2% BSA for 90 min in a 37 °C waterbath before acute stimulation with insulin, rapalog, or PDGF.
Glucocorticoid-induced insulin resistance was created in 3T3-L1 adipocytes with 1 μm dexamethasone in DMEM-high glucose containing 0.2% BSA for 24 h at 37 °C. Following treatment, cells were washed three times with PBS and incubated in serum- and bicarbonate-free DMEM containing 20 mm HEPES, pH 7.4, and 0.2% BSA for 90 min in a 37 °C waterbath before acute stimulation with insulin or rapalog.
Subcellular Fractionation
After stimulation, 3T3-L1 adipocytes were washed with ice-cold PBS and harvested in ice-cold HES buffer (20 mm HEPES, pH 7.4, 1 mm EDTA, 250 mm sucrose) containing Complete protease inhibitor mixture and phosphatase inhibitors (2 mm sodium orthovanadate, 1 mm sodium pyrophosphate, 10 mm sodium fluoride). The cells were lysed with 12 passes through a 22-gauge needle and 6 passes through a 27-gauge needle. Cell lysates were then centrifuged at 500 × g for 10 min at 4 °C to remove unbroken cells. The supernatant was centrifuged at 10,080 × g for 20 min at 4 °C to yield two fractions: the pellet fraction consisting of PM and mitochondria/nuclei, and the supernatant fraction consisting of cytosol, low density microsomes, and high density microsomes. The supernatant was then centrifuged at 15,750 × g for 20 min at 4 °C to obtain the pellet high density microsome fraction. The supernatant was again centrifuged at 175,000 × g for 75 min at 4 °C to obtain the cytosol fraction (supernatant) and the low density microsome fraction (pellet). To obtain PM fraction, the pellet from the first ultracentrifuge spin was resuspended in HES buffer containing phosphatase and protease inhibitors and layered over high sucrose HES buffer (20 mm HEPES, pH 7.4, 1 mm EDTA, 1.12 m sucrose) and centrifuged at 78,925 × g for 60 min at 4 °C. The PM fraction was collected above the sucrose layer, and the pellet was the mitochondria/nuclei fraction. All of the fractions were resuspended in HES buffer containing phosphatase and protease inhibitors. Protein concentration for each fraction was performed using bicinchoninic acid assay. Samples were made up in SDS sample buffer and then kept at −20 °C.
Western Blotting Analysis
After treatments, cells were washed twice with ice-cold PBS and solubilized in 2% SDS in PBS with phosphatase inhibitors (1 mm sodium pyrophosphate, 2 mm sodium vanadate, 10 mm sodium fluoride) and Complete protease inhibitor mixture. Insoluble material was removed by centrifugation at 18,000 × g for 10 min. Protein concentration was measured using the bicinchoninic acid method. Proteins were separated by SDS-PAGE for immunoblot analysis. After transferring proteins to polyvinylidene difluoride membranes, membranes were incubated in blocking buffer containing 5% skim milk in Tris-buffered saline and immunoblotted with relevant antibodies overnight at 4 °C in blocking buffer containing 5% BSA, 0.1% Tween in Tris-buffered saline. After incubation, membranes were washed and incubated with horseradish peroxidase-labeled secondary antibodies and then detected by SuperSignal West Pico chemiluminescent substrate.
Quantitative GLUT4 Translocation Assay
HA-GLUT4 translocation to the PM was measured as described previously (21). Briefly, 3T3-L1 adipocytes stably expressing HA-GLUT4 in 96-well plates were chronically treated with insulin or dexamethasone for 24 h before being serum-starved with serum- and bicarbonate-free DMEM containing 20 mm HEPES, pH 7.4, and 0.2% BSA for 90 min. Cells were then acutely stimulated with insulin, rapalog, or PDGF. After stimulation, cells were fixed and immunolabeled with monoclonal anti-HA antibody followed by Alexa Fluor 488-labeled secondary antibody in the absence or presence of saponin to analyze the amount of HA-GLUT4 at the PM or the total HA-GLUT4 content, respectively.
Confocal Laser Scanning Microscopy
3T3-L1 fibroblasts were seeded and differentiated into adipocytes on glass coverslips. Cells were exposed to 10 nm insulin for 24 h and then serum-starved for 90 min before stimulation with 10 nm insulin for 20 min. After stimulation, cells were washed in ice-cold PBS, fixed with 3% paraformaldehyde, and quenched with 50 mm glycine. Cells were then blocked and permeabilized in 2% BSA and 0.1% saponin in PBS and labeled for activated Akt using rabbit monoclonal pSer473 Akt antibody. Primary antibody was detected with anti-rabbit Cy2-conjugated secondary antibody. Optical sections were obtained through scans for Cy2 using the Leica TCS SP confocal laser scanning microscope using a 63 × 1.32 oil lens. For quantification, random images of pSer473 Akt at the base of the cells were collected with the same confocal settings. Images were analyzed using the Region detector software, and the mean fluorescence was measured.
Statistical Analysis
Data are expressed as means ± S.D. p values were calculated by two tailed Student's t test using Microsoft Office Excel 2003.
RESULTS
Use of a Conditional Akt Mutant to Study Insulin Resistance
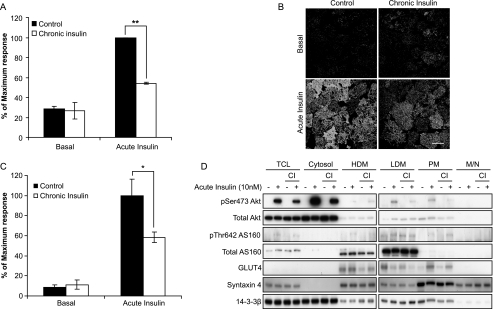
Chronic exposure of 3T3-L1 adipocytes to 10 nm insulin results in a significant reduction in insulin-stimulated GLUT4 translocation to the plasma membrane (Fig. 1A) consistent with other studies (13, 15). This impairment in insulin action occurred in parallel with a reduction in insulin-stimulated Akt Ser473 phosphorylation at both the PM (Fig. 1, B and C) and in other cellular locations (Fig. 1D). These data are consistent with other studies that have reported a defect in Akt phosphorylation in insulin-resistant conditions (9, 13). In agreement with our previous studies (13), this defect in insulin-stimulated Akt phosphorylation was not accompanied by any significant defect in insulin-dependent phosphorylation of AS160, an Akt substrate that plays a crucial role in GLUT4 trafficking (7, 8) (Fig. 1D).
FIGURE 1.
Chronic insulin exposure inhibits insulin-stimulated HA-GLUT4 translocation and Akt phosphorylation, but not AS160 phosphorylation. A, HA-GLUT4-expressing 3T3-L1 adipocytes grown in 96-well plates were chronically stimulated with 10 nm insulin for 24 h. Cells were then washed and serum-starved for 90 min prior to acute stimulation with 10 nm insulin for 20 min. The amount of HA-GLUT4 on the cell surface (nonpermeabilized cells) was expressed as a percentage of total HA-GLUT4 determined under permeabilizing conditions. Results are expressed as a maximum response of acute insulin stimulation in control cells. Results are displayed as means ± S.D. **, p < 0.01, Student's t test (n = 3). B, 3T3-L1 adipocytes were chronically stimulated with insulin and serum-starved as above. Following acute stimulation with 10 nm insulin for 20 min, cells were fixed and immunolabeled with pSer473 Akt antibody followed by Cy2-conjugated secondary antibody. Confocal slice images were taken at the base of the cell. Images are from a representative experiment (n = 3; 30–50 images/condition in each experiment). Scale bar, 40 μm. C, quantification of B. The amount of pSer473 Akt at the cell surface was determined using the Region detector program. Results are displayed as means ± S.D. *, p < 0.05, Student's t test (n = 3). D, 3T3-L1 adipocytes were chronically treated with insulin and stimulated as above. Cells were then fractionated to obtain different subcellular fractions, total cell lysates (TCL), cytosol, high density microsomes (HDM), low density microsomes (LDM), PM, and mitochondria/nuclei (M/N). Fractions were then subjected to Western blot analysis with pSer473 Akt, total Akt, pThr642 AS160, total AS160, GLUT4, syntaxin 4, and 14-3-3β antibodies. CI, chronic insulin.
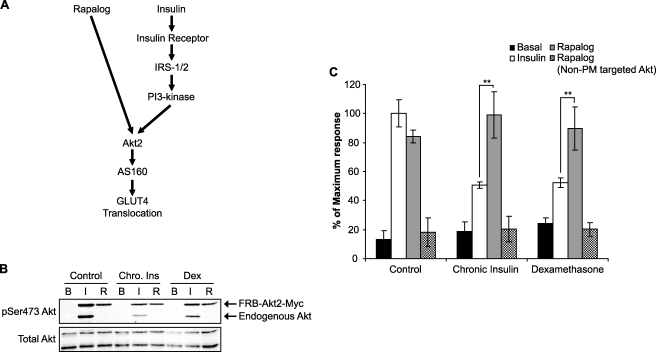
The goal of this study was to investigate the role of intermediates between IRS and Akt in contributing to insulin resistance. To explore this, we have employed a conditionally active Akt mutant that can be targeted to the PM using a regulatable heterodimerization strategy. These constructs were expressed in 3T3-L1 adipocytes using a lentivirus-mediated gene transfer technique. This system enables us to activate Akt independently of upstream elements in the insulin signaling cascade (16) (Fig. 2A). A maximum insulin response on HA-GLUT4 translocation was achieved at an insulin dose of 10 nm (data not shown), and this was used as the stimulus dose in these experiments. Insulin stimulation of 3T3-L1 adipocytes expressing the fusion proteins resulted in Ser473 phosphorylation of both endogenous Akt and FRB-Akt2-Myc, and this was reduced in chronic insulin- and dexamethasone-treated cells (Fig. 2B). Rapalog stimulated Ser473 phosphorylation on FRB-Akt2-Myc but had no effect on endogenous Akt (Fig. 2B). Rapalog-stimulated Akt phosphorylation was slightly inhibited in chronic insulin- and dexamethasone-treated 3T3-L1 adipocytes (Fig. 2B).
FIGURE 2.
Rapalog-stimulated Akt2 activation overcomes the inhibitory effect on HA-GLUT4 translocation in chronic insulin- or dexamethasone-treated cells. A, schematic diagram depicting the signaling pathways of rapalog- and insulin-stimulated GLUT4 translocation. B, 3T3-L1 adipocytes infected with the FRB-Akt2-Myc and Myr-FKBP WT-V5 lentivirus were chronically treated with 10 nm insulin (Chro. Ins) or 1 μm dexamethasone (Dex) for 24 h. Cells were then serum-starved for 90 min and stimulated with either 10 nm insulin (I) or 500 nm rapalog (R) for 30 min. Whole cell lysates were generated and immunoblotted with antibodies directed against pSer473 Akt and total Akt. C, HA-GLUT4-expressing 3T3-L1 adipocytes grown in 96-well plates were transduced with respective lentivirus (FRB-Akt2-Myc and Myr-FKBP WT-V5, or FRB-Akt2-Myc and Myr-FKBP A2-V5). At day 5 after transduction, cells were chronically stimulated with either 10 nm insulin or 1 μm dexamethasone for 24 h. Cells were then washed and serum-starved for 90 min prior to acute stimulation with 10 nm insulin or 500 nm rapalog for 30 min. The amount of HA-GLUT4 on the cell surface (nonpermeabilized cells) was expressed as a percentage of total HA-GLUT4 determined under permeabilizing conditions. Results are expressed as a maximum response of acute insulin stimulation in control cells. Results are displayed as means ± S.D. **, p < 0.01, Student's t test (n = 3). B, basal.
Insulin increased cell surface levels of HA-GLUT4 by 5–7-fold in 3T3-L1 adipocytes (Fig. 2C). Rapalog resulted in a similar degree of HA-GLUT4 translocation to that observed with maximum insulin (Fig. 2C), consistent with our previous studies (16). Chronic insulin and dexamethasone were used as two independent models of insulin resistance. In both cases, we observed an ∼50% reduction in insulin-stimulated HA-GLUT4 translocation (Fig. 2C), whereas rapalog-stimulated HA-GLUT4 translocation was not disrupted in either chronic insulin- or dexamethasone-treated cells. Because Akt is a pivotal regulator of insulin-dependent GLUT4 translocation in 3T3-L1 adipocytes (16, 22), these data are consistent with a model where the defect leading to impaired HA-GLUT4 trafficking and insulin resistance occurs upstream of Akt.
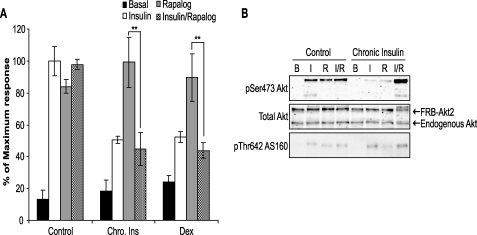
We next examined the consequences of simultaneous addition of rapalog plus insulin on GLUT4 translocation in both control and insulin-resistant cells. Addition of insulin plus rapalog to control cells resulted in a level of HA-GLUT4 translocation that was similar to that found with each agonist alone (Fig. 3A). The increase in AS160 phosphorylation on Thr642 observed with either insulin or rapalog alone was also not different from that observed with insulin plus rapalog (Fig. 3B). This supports the conclusion that insulin regulates HA-GLUT4 translocation via activation of Akt and that the rapalog system achieves a level of activation of Akt that is similar to that found with a maximum insulin dose.
FIGURE 3.
Acute insulin stimulation triggers an inhibitory pathway upstream of Akt2 following chronic exposure to insulin or dexamethasone. A, HA-GLUT4-expressing 3T3-L1 adipocytes grown in 96-well plates were transduced with FRB-Akt2-Myc and Myr-FKBP WT-V5 lentivirus and chronically stimulated with 10 nm insulin (Chro. Ins) or 1 μm dexamethasone (Dex) for 24 h at day 5 after transduction. Cells were then washed and serum-starved for 90 min prior to acute stimulation with either 10 nm insulin or 500 nm rapalog alone, or with both 10 nm insulin and 500 nm rapalog together for 30 min. The amount of HA-GLUT4 on the cell surface (nonpermeabilized cells) was expressed as a percentage of total HA-GLUT4 determined under permeabilizing conditions. Results are expressed as a maximum response of acute insulin stimulation in control cells. Results are displayed as means ± S.D. **, p < 0.01, Student's t test (n = 3). B, 3T3-L1 adipocytes infected with FRB-Akt2-Myc and Myr-FKBP WT-V5 lentivirus were chronically treated with 10 nm insulin for 24 h. Cells were then serum-starved for 90 min and stimulated with either 10 nm insulin (I) or 500 nm rapalog (R), or 10 nm insulin with 500 nm rapalog (I/R) for 30 min. Whole cell lysates were generated and immunoblotted with antibodies directed against pSer473 Akt, total Akt, and pThr642 AS160. B, basal.
Strikingly, co-incubation of insulin-resistant cells with insulin plus rapalog resulted in a impairment in HA-GLUT4 translocation similar to that seen when insulin was added alone (Fig. 3A). This defect could not be explained by reduced Akt signaling because phosphorylation of Akt on Ser473 Akt and AS160 phosphorylation on Thr642 were not impaired in chronic insulin-treated cells restimulated with insulin plus rapalog (Fig. 3B).
Insulin Acutely Stimulates an Insulin Resistance Pathway
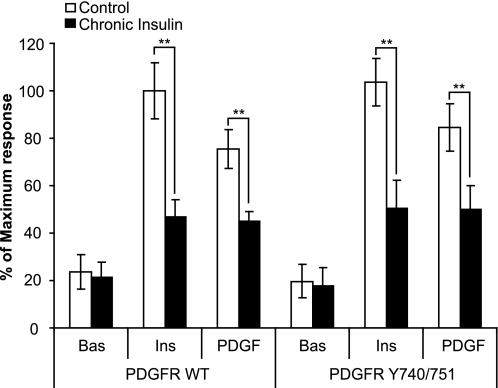
These data are not consistent with a feedback inhibitory model where insults that cause insulin resistance do so by generating an intracellular pathway that feeds back onto one of the upstream elements in the insulin-signaling cascade to inhibit flux through the entire pathway. An alternative model that better describes our data is the presence of a feedforward inhibitory pathway that may be activated by insulin itself. In this model, readdition of insulin alone or insulin plus rapalog, but not rapalog alone, would trigger this pathway, resulting in impaired HA-GLUT4 trafficking. Based upon our data with rapalog alone, whose ability to stimulate GLUT4 translocation was not impaired in insulin-resistant cells, it is unlikely that this feedforward pathway emanates from Akt itself but rather upstream of Akt. It is unlikely that this upstream insulin-regulatable inhibitory pathway emerges from IRS per se because we have shown previously that PDGF, a growth factor that acts independently of IRS, also leads to diminished HA-GLUT4 translocation when readded to cells that had been treated with either chronic insulin or dexamethasone (13). We next wanted to address the possibility that this pathway emanates from PI3-kinase. To accomplish this, we first used the heterologous PDGFR system as reported previously (13). Consistent with our previous findings, in chronic insulin-treated cells that were overexpressing the WT PDGFR, PDGF-stimulated HA-GLUT4 translocation was inhibited to the same extent as was observed with insulin (Fig. 4). Several key tyrosines in the cytoplasmic tail of the PDGFR that regulate different signaling pathways such as Src, PLCγ, RasGAP/MAP kinase, and PI3-kinase/Akt have been mapped (23). We reasoned that this system could be used to explore the role of the PI3-kinase pathway in the induction of this inhibitory insulin resistance pathway. To achieve this, we used a mutant form of the human PDGF receptor (PDGFR Y740/751). The PDGFR Y740/751 mutant retains an intact PI3-kinase-binding site but the binding sites for other associated molecules, including RasGAP, SHP2, and PLCγ, have been deleted (24). Consistent with the conclusion that the PI3-kinase/Akt pathway is necessary and sufficient for HA-GLUT4 translocation, addition of PDGF to cells expressing the PDGFR Y740/751 mutant resulted in a degree of HA-GLUT4 translocation similar to that seen with insulin (Fig. 4). Intriguingly, when cells expressing the PDGFR Y740/751 mutant were incubated with chronic insulin, HA-GLUT4 translocation in response to either acute insulin or PDGF was reduced by ∼50% (Fig. 4). Hence, this supports a model whereby the insulin-responsive inhibitory pathway arises at the level of PI3-kinase.
FIGURE 4.
Chronic insulin-induced inhibition in HA-GLUT4 translocation is independent of IRS1. 3T3-L1 adipocytes expressing HA-GLUT4 and PDGFR WT or PDGFR Y740/751 were grown in 96-well plates and chronically stimulated with 10 nm insulin for 24 h. Cells were then washed and serum-starved for 90 min prior to acute stimulation with either 10 nm insulin or 20 ng/ml PDGF for 20 min. The amount of HA-GLUT4 on the cell surface (nonpermeabilized cells) was expressed as a percentage of total HA-GLUT4 determined under permeabilizing conditions. Results are expressed as a maximum response of acute insulin stimulation in control cells. Results are displayed as means ± S.D. **, p < 0.01, Student's t test (n = 3).
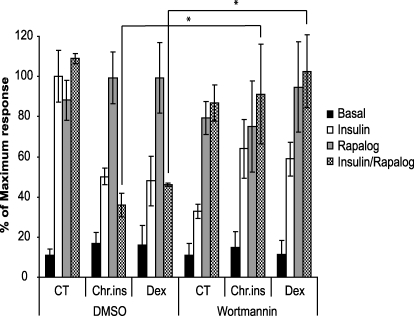
To confirm further the role of PI3-kinase in the inhibitory pathway, we performed experiments using the PI3-kinase inhibitor wortmannin. The rapalog system provides an ideal vehicle to test this because whereas insulin-dependent PI3-kinase activity is inhibited by wortmannin, rapalog-stimulated Akt activity is unaffected by this drug (16). We reasoned that because co-incubation with insulin and rapalog in chronic insulin-treated cells leads to reduced HA-GLUT4 translocation compared with that seen with rapalog alone, addition of wortmannin should overcome this inhibitory effect if the PI3-kinase pathway is the major mediator of this inhibitory pathway. Wortmannin inhibited insulin-stimulated HA-GLUT4 translocation in control cells by ∼70%, but it had no significant effect on HA-GLUT4 translocation in response to either rapalog alone or rapalog plus insulin (Fig. 5). Addition of wortmannin to cells that had been preincubated with either chronic insulin or dexamethasone, in the presence of rapalog plus insulin, caused reversal of the inhibitory effect observed with insulin plus rapalog alone (Fig. 5). Notably, a range of other inhibitors that block MAP kinase, calmodulin-dependent kinase II, Rac1, or atypical PKCζ were unable to reverse the inhibitory pathway (supplemental Fig. 1) The inhibitory effect of wortmannin on insulin-stimulated HA-GLUT4 translocation in cells preincubated with either chronic insulin or dexamethasone was somewhat blunted compared with control cells. Collectively, these data suggest that in chronic insulin- and dexamethasone-treated cells, the acute addition of insulin itself leads to the activation of a feedforward inhibitory pathway that is triggered by PI3-kinase to inhibit HA-GLUT4 translocation to the PM.
FIGURE 5.
Insulin-responsive inhibitory pathway is triggered by PI3-kinase. HA-GLUT4 expressing 3T3-L1 adipocytes grown in 96-well plates were transduced with FRB-Akt2-Myc and Myr-FKBP WT-V5 lentivirus and chronically stimulated with 10 nm insulin (Chr.ins) or 1 μm dexamethasone (Dex) for 24 h at day 5 after transduction. Cells were then washed and serum-starved for 90 min. DMSO, dimethyl sulfoxide; CT, control. 100 nm wortmannin was added 15 min prior to acute stimulation with either 10 nm insulin or 500 nm rapalog alone, or with both 10 nm insulin and 500 nm rapalog together for 30 min. The amount of HA-GLUT4 on the cell surface (nonpermeabilized cells) was expressed as a percentage of total HA-GLUT4 determined under permeabilizing conditions. Results are expressed as a maximum response of acute insulin stimulation in control cells. Results are displayed as means ± S.D. *, p < 0.05, Student's t test (n = 3).
DISCUSSION
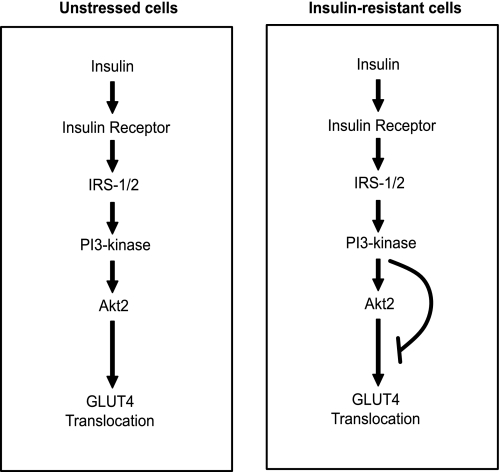
Our studies point to the existence of a feedforward inhibitory pathway in 3T3-L1 adipocytes that is activated by insulin itself. This pathway appears to be turned on by chronic exposure to either insulin or dexamethasone. Rather than acting at a site upstream of Akt, we present evidence that it impinges on a node downstream of Akt that perhaps selectively controls GLUT4 trafficking (Fig. 6). These studies indicate that this inhibitory pathway maybe a unifying feature of many insulin resistance insults, and it likely emanates from either PI3-kinase or the upstream machinery that regulates PI3-kinase.
FIGURE 6.
Schematic diagram of the mechanism of insulin resistance. In unstressed cells, insulin triggers the activation of PI3-kinase, which in turn activates Akt2 and mediates GLUT4 translocation to the PM. In cells that are made resistant with chronic exposure to insulin or dexamethasone, the readdition of acute insulin induces PI3-kinase activation. We propose that PI3-kinase activates an inhibitory pathway that is normally silenced in unstressed conditions to inhibit signaling downstream of Akt and GLUT4 translocation to the PM.
A unique feature of the present studies is that we have used a heterologous expression system to selectively turn on Akt in insulin-resistant adipocytes to determine whether there is a defect upstream or downstream of Akt. Strikingly, selective activation of Akt activity in insulin-resistant cells increased HA-GLUT4 translocation to the same extent as in control cells. One interpretation of these data is that the putative negative feedback inhibitory pathway that has been proposed (9, 10, 12) feeds back at a site upstream of Akt. Further investigation revealed that simultaneous addition of insulin and rapalog to insulin-resistant cells resulted in a level of GLUT4 translocation that was ∼50% of that observed in either control cells or insulin-resistant cells treated with rapalog alone (Fig. 3B). Thus, these data do not fit with a simple feedback inhibitory pathway but are more consistent with a feedforward inhibitory pathway.
This inhibitory pathway could be overcome with wortmannin indicating that it emanates from the PI3-kinase limb of the insulin-signaling cascade tending to exclude other auxiliary pathways, such as MAP kinase, calmodulin-dependent kinase II, or Rac1. Consistent with this, we have been unable to reverse the feedforward inhibitory effect using compounds that target each of these pathways (see supplemental Fig. 1). PI3-kinase is known to regulate a number of pathways including Akt and activation of the atypical PKC pathway (25). We have performed preliminary studies using a compound inhibitor of the atypical PKC pathway and have found no effect on the inhibitory pathway (see supplemental Fig. 1). Our studies cannot exclude a role for the two PI3-kinase upstream kinases, PDK1 and PDK2. PDK1, for example, activates many members of the AGC family, including Akt, serum and glucocorticoid-regulated kinase, protein kinase A, atypical PKCs, and S6 kinase (26). Future studies will focus on the role of these pathways in this feedforward inhibitory loop.
Another possibility is that insulin may more potently activate Akt or it may lead to the activation of Akt in a different subcellular location compared with that observed with rapalog. This is intriguing because activation of Akt at discrete intracellular locations results in phosphorylation of a unique repertoire of substrates (27, 28). Thus, it will be informative to compare signaling in response to rapalog alone and rapalog plus insulin to determine where the differences may be that contribute to this inhibitory pathway. We also cannot exclude a role for different Akt isoforms in this effect. 3T3-L1 adipocytes express both Akt1 and Akt2 isoforms (29). However, evidence has been presented to indicate that Akt2 predominantly regulates metabolism whereas Akt1 may be more involved with proliferation (30, 31). We have used Akt2 as the fusion partner in our rapalog studies, and so it will be important to test an Akt1 fusion in the same system.
Insulin resistance is complex because it can be triggered by many insults, and many intracellular mechanisms have been implicated in its origin, including IRS degradation, inhibition of Akt by binding partners, and changes in the stoichiometry of the p85 to p110 subunits of PI3-kinase (9, 32, 33). We cannot exclude the possibility that the feedforward inhibitory pathway described here is relatively unique to chronic insulin- and dexamethasone-induced insulin resistance, although these two models would not be predicted to share many common features. Among the feedback inhibitory pathways, that involving activation of mTOR has received considerable attention. In this model, mTOR and/or S6 kinase inhibit insulin signaling via Ser/Thr phosphorylation of IRS1/2 (12, 34, 35). Rapalog alone stimulates mTOR activity (16), but rapalog-stimulated HA-GLUT4 translocation was not impaired in insulin-resistant cells. Hence, this is inconsistent with a major role for this feedback pathway in insulin resistance particularly given that mTOR is rapidly activated in response to acute insulin stimulation, yet insulin resistance does not ensue for many hours in response to chronic insulin treatment.
An important aspect of these studies is that the inhibitory pathway that is activated by insulin only becomes evident after prolonged incubation with insulin or dexamethasone. This implies some type of induction mechanism possibly involving changes in gene expression or protein expression. When we washed cells after chronic insulin or dexamethasone treatment, this inhibitory pathway was clearly attenuated because rapalog-stimulated HA-GLUT4 translocation was normal under these conditions. However, the inhibitory pathway was rapidly reactivated because 30-min incubation with insulin was sufficient to reinstate the inhibitory effect. Hence, this indicates that the system has been primed by prior chronic treatment, but reengagement of the entire insulin-signaling pathway is required to reactivate this primed state.
It is well recognized that intracellular signal transduction involves a complex cross-talk between multiple pathways that can engage either in a negative or positive regulatory manner. The Akt pathway comprises a relatively linear upstream relay that diverges into a multitude of biological actions downstream of Akt because of the plethora of Akt substrates. Thus, an inhibitory pathway that acts upstream of Akt will likely impact upon a multitude of biological actions that, under certain circumstances, may not be optimal. A greater degree of specificity would be achieved by selectively negatively regulating individual limbs of the Akt pathway, allowing other possibly more essential limbs to remain unaffected. The present study is consistent with the latter type of regulation whereby certain insults may induce an inhibitory pathway that can fine tune the flux through certain control points of the Akt pathway, such as metabolism, leaving other aspects of Akt function intact. It is intriguing that the GLUT4 translocation pathway is one of the processes targeted by this inhibitory pathway because previous studies have suggested that insulin resistance in humans is caused by a selective defect in GLUT4 translocation in muscle (36). It will be interesting to explore the role of this inhibitory pathway on lipid and protein metabolism as well as its role in vivo. To achieve the latter, it will be necessary to overcome the complex interplay between various parameters such as the constant supply of insulin from the beta cells as well as changes in body weight.
A major question that stems from these studies is what is the downstream target of this pathway? This intermediate is obviously able to regulate GLUT4 translocation, but we predict that it is not AS160 because its phosphorylation appeared relatively normal in insulin-resistant cells. However, there are many aspects to the control of GLUT4 trafficking that remain unknown, and so these studies should reinvigorate the search for these components.
Acknowledgments
We thank Dr. Kyle Hoehn and Dr. Cordula Hohnen-Behrens for invaluable discussions and technical help and Assoc. Prof. Greg Cooney and Dr. Lowenna Holt for critically reading the manuscript (Garvan Institute of Medical Research, Sydney, Australia). We also thank Dr. Jonathan Cooper (Fred Hutchinson Cancer Research Center, Seattle, WA) and Dr. Morris Birnbaum (University of Pennsylvania, Philadelphia, PA) for providing PDGFR plasmids, Dr. Peter Shepherd (Symansis, Auckland, New Zealand) for providing pThr642 AS160 antibody, and the ARIAD Pharmaceuticals for generously providing AP21967 and the FRB and FKBP template DNA.
This work was supported by grants from the National Health and Medical Research Council of Australia.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- GLUT4
- glucose transporter 4
- PI3-kinase
- phosphatidylinositol 3-kinase
- IRS
- insulin receptor substrate
- PM
- plasma membrane
- mTOR
- mammalian target of rapamycin
- HA
- hemagglutinin antigen
- PKC
- protein kinase C
- MAP
- mitogen-activated protein
- DMEM
- Dulbecco's modified Eagle's medium
- BSA
- bovine serum albumin
- PDGF
- platelet-derived growth factor
- PDGFR
- PDGF receptor
- WT
- wild type
- PBS
- phosphate-buffered saline
- PDK
- phosphoinositide-dependent protein kinase.
REFERENCES
- 1.Reaven G. M. (1995) Physiol. Rev. 75, 473–486 [DOI] [PubMed] [Google Scholar]
- 2.Saltiel A. R. (2001) Cell 104, 517–529 [DOI] [PubMed] [Google Scholar]
- 3.Whiteman E. L., Cho H., Birnbaum M. J. (2002) Trends Endocrinol. Metab. 13, 444–451 [DOI] [PubMed] [Google Scholar]
- 4.White M. F. (1998) Mol. Cell. Biochem. 182, 3–11 [PubMed] [Google Scholar]
- 5.Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 6.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 7.Sano H., Kane S., Sano E., Mîinea C. P., Asara J. M., Lane W. S., Garner C. W., Lienhard G. E. (2003) J. Biol. Chem. 278, 14599–14602 [DOI] [PubMed] [Google Scholar]
- 8.Larance M., Ramm G., Stöckli J., van Dam E. M., Winata S., Wasinger V., Simpson F., Graham M., Junutula J. R., Guilhaus M., James D. E. (2005) J. Biol. Chem. 280, 37803–37813 [DOI] [PubMed] [Google Scholar]
- 9.Haruta T., Uno T., Kawahara J., Takano A., Egawa K., Sharma P. M., Olefsky J. M., Kobayashi M. (2000) Mol. Endocrinol. 14, 783–794 [DOI] [PubMed] [Google Scholar]
- 10.Carlson C. J., White M. F., Rondinone C. M. (2004) Biochem. Biophys. Res. Commun. 316, 533–539 [DOI] [PubMed] [Google Scholar]
- 11.Ozes O. N., Akca H., Mayo L. D., Gustin J. A., Maehama T., Dixon J. E., Donner D. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4640–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirola L., Bonnafous S., Johnston A. M., Chaussade C., Portis F., Van Obberghen E. (2003) J. Biol. Chem. 278, 15641–15651 [DOI] [PubMed] [Google Scholar]
- 13.Hoehn K. L., Hohnen-Behrens C., Cederberg A., Wu L. E., Turner N., Yuasa T., Ebina Y., James D. E. (2008) Cell Metab. 7, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho P. C., Lin Y. W., Tsui Y. C., Gupta P., Wei L. N. (2009) Cell Metab. 10, 516–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G., Raman P., Bhonagiri P., Strawbridge A. B., Pattar G. R., Elmendorf J. S. (2004) J. Biol. Chem. 279, 39705–39709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng Y., Ramm G., Lopez J. A., James D. E. (2008) Cell Metab. 7, 348–356 [DOI] [PubMed] [Google Scholar]
- 17.Shewan A. M., van Dam E. M., Martin S., Luen T. B., Hong W., Bryant N. J., James D. E. (2003) Mol. Biol. Cell 14, 973–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M., Rossi J. J. (2005) Methods Mol. Biol. 309, 261–272 [DOI] [PubMed] [Google Scholar]
- 19.Carlotti F., Bazuine M., Kekarainen T., Seppen J., Pognonec P., Maassen J. A., Hoeben R. C. (2004) Mol. Ther. 9, 209–217 [DOI] [PubMed] [Google Scholar]
- 20.Thomson M. J., Williams M. G., Frost S. C. (1997) J. Biol. Chem. 272, 7759–7764 [DOI] [PubMed] [Google Scholar]
- 21.Govers R., Coster A. C., James D. E. (2004) Mol. Cell. Biol. 24, 6456–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Z. Y., Zhou Q. L., Coleman K. A., Chouinard M., Boese Q., Czech M. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7569–7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claesson-Welsh L. (1994) J. Biol. Chem. 269, 32023–32026 [PubMed] [Google Scholar]
- 24.Whiteman E. L., Chen J. J., Birnbaum M. J. (2003) Endocrinology 144, 3811–3820 [DOI] [PubMed] [Google Scholar]
- 25.Cantley L. C. (2002) Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 26.Storz P., Toker A. (2002) Front. Biosci. 7, d886–d902 [DOI] [PubMed] [Google Scholar]
- 27.Schenck A., Goto-Silva L., Collinet C., Rhinn M., Giner A., Habermann B., Brand M., Zerial M. (2008) Cell 133, 486–497 [DOI] [PubMed] [Google Scholar]
- 28.Adam R. M., Mukhopadhyay N. K., Kim J., Di Vizio D., Cinar B., Boucher K., Solomon K. R., Freeman M. R. (2007) Cancer Res. 67, 6238–6246 [DOI] [PubMed] [Google Scholar]
- 29.Hill M. M., Clark S. F., Tucker D. F., Birnbaum M. J., James D. E., Macaulay S. L. (1999) Mol. Cell. Biol. 19, 7771–7781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho H., Mu J., Kim J. K., Thorvaldsen J. L., Chu Q., Crenshaw E. B., 3rd, Kaestner K. H., Bartolomei M. S., Shulman G. I., Birnbaum M. J. (2001) Science 292, 1728–1731 [DOI] [PubMed] [Google Scholar]
- 31.Cho H., Thorvaldsen J. L., Chu Q., Feng F., Birnbaum M. J. (2001) J. Biol. Chem. 276, 38349–38352 [DOI] [PubMed] [Google Scholar]
- 32.Cheng K. K., Iglesias M. A., Lam K. S., Wang Y., Sweeney G., Zhu W., Vanhoutte P. M., Kraegen E. W., Xu A. (2009) Cell Metab. 9, 417–427 [DOI] [PubMed] [Google Scholar]
- 33.Bandyopadhyay G. K., Yu J. G., Ofrecio J., Olefsky J. M. (2005) Diabetes 54, 2351–2359 [DOI] [PubMed] [Google Scholar]
- 34.Li J., DeFea K., Roth R. A. (1999) J. Biol. Chem. 274, 9351–9356 [DOI] [PubMed] [Google Scholar]
- 35.Tremblay F., Marette A. (2001) J. Biol. Chem. 276, 38052–38060 [DOI] [PubMed] [Google Scholar]
- 36.Petersen K. F., Shulman G. I. (2006) Am. J. Med. 119, S10–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]