Abstract
p21CIP1/WAF1 belongs to the CIP/KIP family of Cdk inhibitors, and its expression is tightly controlled during the cell cycle, mainly by transcriptional and post-translational mechanisms. Fine regulation of p21CIP1/WAF1 levels is critical for cell cycle control and for cellular response to stress. In the present work, we describe a novel mechanism to modulate p21CIP1/WAF1 levels mediated by the human GTSE-1 (G2 and S phase-expressed-1) protein. Our results provide evidence that hGTSE-1 protects p21CIP1/WAF1 from proteasome-dependent degradation as part of a functional complex containing the Hsp90-binding TPR protein WISp39. We further show that the hGTSE-1 N-terminal portion is sufficient for p21CIP1/WAF1 binding and stabilization. Finally, we demonstrate that hGTSE-1 mediated-p21CIP1/WAF1 stabilization is clearly involved in the ability of cells to counteract cytotoxicity induced by the microtubule poison paclitaxel.
Keywords: Apoptosis, Cell/Cycle, Drug Resistance/Cancer Therapy/Chemotherapy, Protein/Protein-Protein Interactions, Protein/Stability
Introduction
p21CIP1/WAF1 (hereafter referred to as p21) was originally identified as a p53-responsive gene and by its capacity to halt the cell cycle progression by binding Cdk-cyclin6 complexes and proliferating cell nuclear antigen. Interaction with such partners confers to p21 a role in mediating biological outcomes such as cell cycle arrest in response to stress (1), differentiation (2), and senescence (3). It is now evident that p21 expression can be induced by other transcription factors and that it can favor proliferation by promoting the assembly and consequent activity of certain Cdk-cyclin complexes (4, 5).
Because p21 is a short lived and highly unstructured protein (6), modulation of its degradation rate significantly contributes to the regulation of its intracellular levels. Regulation of p21 stability has been demonstrated to occur through proteasome-mediated and ubiquitin-dependent or -independent mechanisms (7, 8) and to be affected by phosphorylation, binding partners (9), and the Hsp90-WISp39 chaperone complex (10).
p21 also exerts cell cycle-unrelated functions, such as blocking the apoptotic pathway through inhibition of c-Jun N-terminal kinase (JNK), apoptosis signal-regulating kinase 1, and procaspase-3 and affecting adhesion and migration through Rho-associated kinase inhibition in the cytoplasm (11). Because p21 levels may influence its activities, elucidation of the mechanisms underlying this process becomes critical.
We have previously shown that human GTSE-1 (G2 and S phase-expressed-1) protein, as well as its mouse homologue, are microtubule-localized proteins whose expression in nontransformed cells is almost undetectable in G1, increases during S phase, and peaks in the G2 phase of the cell cycle (12–14). However, in transformed cells, although cell cycle regulation is still observed, low levels of hGTSE-1 are also detected in G1 when compared with S/G2 phases (see Ref. 15 and “Results”). During mitosis, it is hyperphosphorylated (13), probably leading to its subsequent degradation by the anaphase-promoting complex-Cdh1 complex (16). Although hGTSE-1 shuttles between the cytoplasm and the nucleus, it is stabilized in the nucleus following DNA damage (17). We also demonstrated that hGTSE-1 is able to bind and relocalize p53 to the cytoplasm, to down-regulate its protein levels, and to repress its transactivation function. The consequence of hGTSE-1 action on p53 results in inhibition of DNA damage-induced p53-dependent apoptosis (15, 17). The present study reveals a new p53-independent role of hGTSE-1 in inducing cell resistance to paclitaxel-induced apoptosis through the regulation of p21 protein levels as a component of the p21-stabilizing machinery (Hsp90-WISp39).
EXPERIMENTAL PROCEDURES
Cell Lines and Treatments
All cell lines were cultured at 37 °C in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml) with the exception of the H1299 cell line that was cultured in RPMI 1640 medium. JPIC/U (full-length hGTSE-1 inducible U2OS monoclonal cell line) and JPIC/H (full-length hGTSE-1 inducible H1299 monoclonal cell line) are stable monoclonal cell lines expressing inducible full-length hGTSE-1; JPIC/UP (full-length hGTSE-1 inducible U2OS polyclonal cell line) and EPIC/UP (hGTSE-1-(1–221) inducible U2OS polyclonal cell line) are stable polyclonal cell lines expressing the inducible full-length hGTSE-1 or hGTSE-1-(1–221) deletion, respectively, in a U2OS background. All inducible cell lines were generated by using the ecdysone-inducible system (Invitrogen) and were maintained in medium containing zeocin (Invitrogen) and G418 (Invitrogen). To induce hGTSE-1 expression, ponasterone A (Invitrogen) was added to the culture medium at a final concentration of 5 μm. Cycloheximide (50 μm/ml), MG132 (25 μm), 17-(allylamino)-17-emethoxygeldanamycin (17-AAG), and paclitaxel were purchased from Sigma.
Transfection and Vectors (DNA)
The cells were transfected using the calcium phosphate method, Lipofectamine 2000 reagent (Invitrogen), or FuGENE 6 (Roche Diagnostics) according to the manufacturer's instructions. Unless stated otherwise, the cells were analyzed 24 h after transfection. pcDNA3-hGTSE-1, pcDNA3-HA-hGTSE-1, and pEGFP-hGTSE-1 were described previously (15). GST-hGTSE-1 contains the full-length hGTSE-1 fused to GST (pGEX-4T1, GE Healthcare). To generate inducible cell lines, full-length pIND-hGTSE-1 and pIND-hGTSE-1-(1–221) were constructed by subcloning full-length hGTSE-1 or the EcoRI/XhoI fragment (1–221) respectively into the pIND vector (Invitrogen). pcDNA3-hGTSE-1-(1–221) was generated by subcloning the EcoRI/XhoI fragment of full-length hGTSE-1 into the pcDNA3 vector. GST-p21 was constructed by subcloning into pGEX-4T1 vector. pcDNA3-HA-WISp39 was generated by subcloning the PCR product in pcDNA3 vector.
siRNA
Cells were transfected with siRNAs using Oligofectamine reagent (Invitrogen), X-tremeGENE siRNA transfection reagent (Roche Diagnostics), or Lipofectamine RNAiMAX (Invitrogen) as recommended by the manufacturers. The mRNA-targeted sequences for hGTSE-1 (15), p21 (18; “p21-1”), and WISp39 (10) were described previously. All siRNA duplexes were synthesized by MWG Biotech. siRNAs referred as control are AACCUUUUUUUUUGGGGAAAA (15) or GUGACCAGCGAAUACCUGU (LacZ).
Cell Synchronization
U2OS cells were synchronized at the G1/S border by treating cells with 2.5 mm thymidine for 16 h followed by extensive wash and release into normal growth medium for 10 h to obtain cells in G2/M. The cell cycle stage was monitored by staining with propidium iodide (10 μg/ml), followed by cytometric analysis performed on a FACSCalibur (Becton-Dickinson) cytofluorimeter equipped with CellQuest software.
In Vitro Binding Assay
35S-labeled proteins were in vitro-translated (IVT) using TnT quick coupled transcription/translation system (Promega). 35S-labeled IVT proteins were incubated with similar amounts (as estimated by Coomassie Blue-stained SDS-PAGE gel) of purified GST or GST-tagged proteins (immobilized on glutathione-Sepharose 4B beads, Amersham Biosciences) in pulldown buffer (150 mm NaCl, 20 mm Hepes, pH 7.5, 0, 0.05% Nonidet P-40, 10% glycerol, 0.1 mm phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor mixture (Sigma)). Bound proteins were eluted, resolved on SDS-PAGE, and analyzed by autoradiography. For in vitro binding assays between GST, GST-hGTSE-1, and GST-p21 recombinant proteins, purified resin-bound GST-p21 was subjected to thrombin cleavage (1 unit/100 μg protein) for 6 h at room temperature to remove the GST tag. Digestion was stopped by adding 1 mm PMSF. Beads were pelleted at 3,000 rpm for 4 min, and supernatant (∼1 μg protein) was incubated with resin-bound GST-hGTSE-1 or GST. p21 presence was detected by immunoblot analysis using an antibody against p21.
Immunoprecipitation and Western Blot Analysis
Cells were harvested in ice-cold immunoprecipitation lysis buffer containing 50 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm Na3VO4, 1 mm dithiothreitol, 1 mm PMSF, 5 mm EDTA, and protease inhibitor mixture (Sigma) plus 1% Nonidet P-40 or in immunoprecipitation low stringency lysis buffer containing 50 mm Tris-HCl, pH 8, 50 mm NaCl, 0.1 mm Na3VO4, 2 mm dithiothreitol, 0.1 mm PMSF, 5 mm EDTA, and protease inhibitor mixture (Sigma) plus 0.1% Nonidet P-40, where stated. After centrifugation and preclearing, lysates were incubated at 4 °C for 2 h with 25 μl of protein A-Sepharose CL-4B (for anti-hGTSE-1, anti-HA, and anti-GFP immunoprecipitations) or GammaBind G Sepharose (for anti-FLAG immunoprecipitations) plus specific antibodies. Western blot analyses were performed with the following primary antibodies: affinity-purified LF1 anti-hGTSE-1 polyclonal antibody (14), affinity-purified anti-GFP polyclonal antibody, anti-p21 polyclonal, DO-1 anti-p53, anti-Hsp90 (Santa Cruz Biotechnology), anti-p21 monoclonal, anti-actin, anti-p27, anti-FLAG (Sigma), anti-p27/p57, anti-cyclin B1, anti-cleaved caspase-3 (Cell Signaling Technology), anti-cyclin A (BD Transduction Laboratories), anti-HA 12CA5 (Roche Applied Science), and anti-FKBPL (WISp39) (Proteintech Group). Bound primary antibodies were visualized by enhanced chemiluminescence (ECL; Amersham Biosciences) after addition of horseradish peroxidase-conjugated secondary antibodies.
Gel Filtration Chromatography
Subconfluent H1299 and U2OS cells plated in 8 dishes (500 cm2) were collected in gel filtration buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, 5 mm EDTA, 5% glycerol, 1 mm PMSF, and 0.5 mm NaF). Cell extracts were syringed and centrifuged at 13,000 rpm for 10 min, and the supernatant was resolved by gel filtration on a column of 1.6 cm inner diameter/70 cm length, filled with Superose 6 prep grade media. Flow rate was maintained constant at 1 ml/min, and collection of 2 ml fractions started after the void volume was reached. Fractions were analyzed by immunoblotting with anti-hGTSE-1, anti-p21, anti-WISp39, and anti-Hsp90 antibodies.
Immunofluorescence Analysis
These assays were performed as described previously (17). Glass slides were analyzed using a Leica DM4000B microscope.
Flow Cytometry
Cells were harvested by trypsinization, and cell suspensions were washed once in phosphate-buffered saline and then fixed in 70% ice-cold ethanol and stored at −20 °C until analysis. For FACS analysis, cell suspensions were treated with RNase (0.2 mg/ml) for 10 min at 4 °C and then stained with a propidium iodide solution (10 μg/ml) for at least 10 min in the dark. Cell cycle analysis was performed with a FACSCalibur (Becton-Dickinson) cytofluorimeter equipped with CellQuest software. The number of cells with a sub-G1 DNA content was calculated through normalization by biological background subtraction noise, i.e. the sub-G1 percentage of siCONT-transfected cells treated with paclitaxel was subtracted from the sub-G1 percentage of untreated siCONT-transfected cells. Experiments were performed in triplicate.
Colony Formation Assay
For each cell line, 3,000 cells were seeded on 60-mm dishes. After 24 h, cells were treated with the indicated doses of paclitaxel for 24 h. For knockdown experiments, cells were transfected with specific siRNAs 36 h before paclitaxel treatment, whereas for the inducible cell lines, ponasterone A was added to the culture media 16 h before paclitaxel treatment. Silencing/induction efficiency was controlled by immunoblotting. After 24 h of paclitaxel treatment, the cells were washed twice with phosphate-buffered saline, and fresh medium without drug was added. The cultures were microscopically monitored for colony formation, and when colonies were macroscopically visible, the cells were fixed in paraformaldehyde 3% for 20 min and stained with Crystal Violet 0.05% for 30 min in the dark. Finally, each plate was carefully washed twice with H2O and dried at 37 °C. The plates were scanned and counted for the colonies number, which is expressed as the mean ratio of scored colonies: paclitaxel-treated siCONT:sihGTSE-1 and ponasterone A:noninduced cells (-). Statistical significance was evaluated for each cell line set using Z-test method. Experiments were performed at least in triplicate.
RESULTS
hGTSE-1 Modulates p21 Turnover
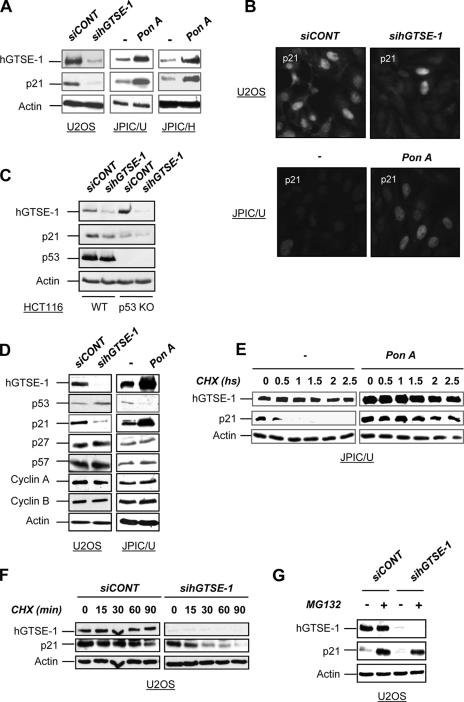
We found here that p21 protein levels change in parallel with hGTSE-1 expression in both hGTSE-1 knocked down cells and in hGTSE-1-inducible systems as evidenced by Western blot (Fig. 1A) and immunofluorescence (Fig. 1B) analyses. Regulation of p21 by hGTSE-1 occurs in U2OS (wild type p53), H1299 (p53-null) and in HCT116 wild type and p53-null (Fig. 1C) cells indicating a p53-independent mechanism. Importantly, such an effect was not seen with other members of the CIP/KIP family or cyclins (Fig. 1D), indicating that hGTSE-1 expression specifically targets p21. This is not a cell cycle related effect because no significant alteration in the cell cycle profile was seen in hGTSE-1-induced or -knocked down cells (supplemental Fig. S1, A and B, respectively). Similarly, regulation of p21 protein levels was reproduced using two different hGTSE-1 siRNAs (supplemental Fig. S1C) in different cell lines (data not shown), without significantly affecting p21 mRNA levels (supplemental Fig. S1D). In fact, through cycloheximide chase experiments, we found that hGTSE-1 is able to modulate p21 protein turnover because hGTSE-1 up-regulation increases (Fig. 1E) and hGTSE-1 knockdown (KD) decreases (Fig. 1F) p21 half-life. Treatment of hGTSE-1 knocked down cells with MG132 (Fig. 1G) or lactacystin (data not shown) abolished p21 degradation restoring its basal levels, suggesting that hGTSE-1 protein protects p21 from proteasome-dependent proteolysis.
FIGURE 1.
Regulation of p21 stability by hGTSE-1. A, U2OS cells were transiently transfected with a control siRNA (siCONT) or hGTSE-1 siRNA (sihGTSE-1) for 36 h. JPIC/U and JPIC/H cells were treated with ponasterone A (pon A) to induce hGTSE-1 expression or left untreated (−) for 24 h. B, an immunofluorescence analysis of endogenous p21 expression (visualized with an anti-p21 antibody) in U2OS cells transfected with siCONT or sihGTSE-1 (upper panels) or in JPIC/U cells treated with (pon A) or without (−) ponasterone A (lower panels) is shown. C, HCT 116 parental (WT) and p53-null (p53 KO) cells were transiently transfected with siCONT or sihGTSE-1 siRNA for 36 h. D, U2OS cells were transiently transfected with siCONT or sihGTSE-1 for 36 h. JPIC/U cells were treated (pon A) or not (−) with ponasterone A for 24 h. E, shown are cycloheximide (CHX) chase experiments of JPIC/U cells after the addition (pon A) or without addition (−) of ponasterone A for 24 h and then cycloheximide (50 μg/ml). Cells were lysed at the indicated time points. F, shown are cycloheximide chase experiments of U2OS cells after transfection of siCONT or sihGTSE-1 for 36 h and then treatment with cycloheximide (50 μg/ml) for the indicated time points. G, U2OS cells were transiently transfected with siCONT or sihGTSE-1 for 36 h, followed by addition of MG132 (25 μm) for 5 h. Immunoblots were performed with antibodies against hGTSE-1, p53, p21, p27, p57, cyclin A, cyclin B1, and actin.
hGTSE-1-dependent p21 Stabilization Requires a Functional Hsp90 Chaperone Complex
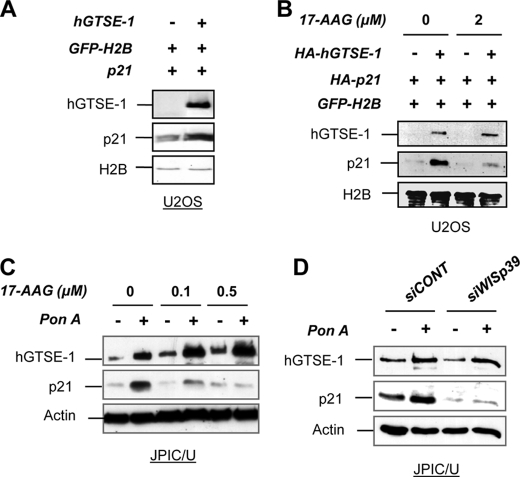
Co-transfection of p21 and hGTSE-1 results in increased p21 levels (Fig. 2A). The Hsp90 chaperone complex was previously shown to regulate the stability of newly synthesized p21 through the TPR co-chaperone protein WISp39 (10). We therefore explored the relevance of this complex on hGTSE-1-mediated p21 stabilization. Notably, we found that the specific Hsp90 inhibitor 17-AAG as well as the KD of WISp39 abrogates the effect of hGTSE-1 on ectopically expressed (Fig. 2B) and endogenous p21 levels (Fig. 2, C and D) thus indicating the requirement of a functional Hsp90-WISp39 complex for hGTSE-1-mediated p21 stabilization.
FIGURE 2.
Requirement of a functional Hsp90 machinery for hGTSE-1 mediated-p21 stabilization. A, U2OS cells were transfected with vectors encoding p21 with an empty vector or with hGTSE-1 for 24 h. A vector encoding GFP-tagged histone H2B (GFP-H2B) was used as transfection efficiency control. B, U2OS cells were transfected as in A and after 6 h were treated with 17-AAG for 16 h. C, JPIC/U cells were treated with the indicated doses of 17-AAG alone (−) or together with ponasterone A (pon A; +) for 16 h. D, JPIC/U cells were transfected with control (siCONT) or WISp39 (siWISp39) siRNAs for 72 h followed by the addition (+) or not (−) of ponasterone A for the last 16 h. Immunoblot analyses were performed using antibodies against hGTSE-1, p21, GFP, and actin as loading control.
hGTSE-1 Physically Interacts with p21 and Its Associated Chaperone Machinery in Vivo
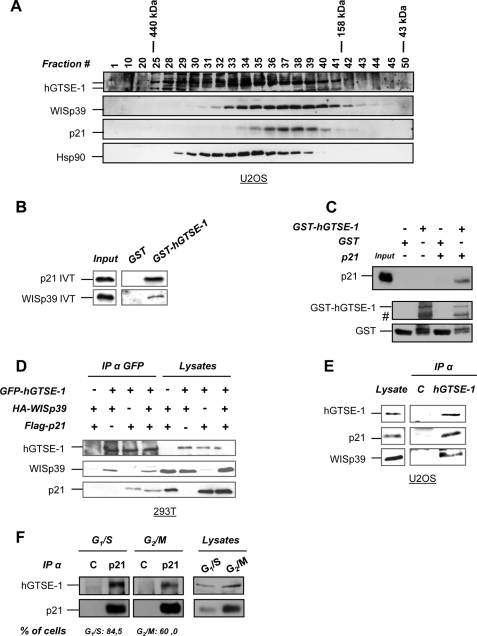
Gel filtration chromatography of U2OS (Fig. 3A) and H1299 (supplemental Fig. S2A) cell extracts followed by immunoblot analysis revealed that hGTSE-1 co-elutes with p21 and WISp39 in fractions resolved with a mass of ∼300 kDa, indicating that the three proteins could form a complex in vivo. First, we investigated whether hGTSE-1 could physically interact with p21 and/or WISp39 in vitro. To this purpose, in vitro pulldown assays using recombinant hGTSE-1 and 35S-labeled IVT p21 and WISp39 were performed. p21 and WISp39 were found to interact with hGTSE-1 (Fig. 3B) but not IVT p27 under the same experimental conditions (supplemental Fig. S2B), thus confirming the specificity of hGTSE-1 for p21 with respect to its closely related sibling. Such interaction was confirmed by incubating recombinant p21 and WISp39 with IVT hGTSE-1 (supplemental Fig. S2C). Interestingly, we observed that both hGTSE-1 and p21 recombinant proteins interact with each other in vitro demonstrating a direct association, with no bridging proteins mediating their binding (Fig. 3C). Moreover, overexpressed p21 and WISp39 were detected in hGTSE-1 immunoprecipitations (Fig. 3D), and the same complex was seen in reverse immunoprecipitations (data not shown) suggesting that hGTSE-1 is associated in vivo with p21 and its associated chaperone machinery. Finally, as expected, endogenous hGTSE-1 was detected in complex with endogenous WISp39 and p21 (Fig. 3E), confirming the existence of the protein complex in vivo.
FIGURE 3.
Interaction of hGTSE-1 with p21 and the co-chaperone WISp39. A, U2OS extracts were resolved by gel filtration on a Superose 6 column, and the fractions were analyzed by immunoblotting. B, in vitro binding assay using recombinant GST or GST-hGTSE-1 fusion protein incubated with 35S-labeled IVT p21 or WISp39. The left panel (input) shows 20% of the input of IVT. C, in vitro binding assay using recombinant resin-bound GST or GST-hGTSE-1 incubated with recombinant p21 after thrombin-mediated removal of the GST tag. #, degradation bands of hGTSE-1. The left panel (input) shows 10% of p21 input. D, 293T cells were transfected with FLAG-p21, HA-WISp39, and GFP-hGTSE-1 for 24 h, followed by immunoprecipitation (IP) using anti-GFP antibody. E, immunoprecipitation of endogenous hGTSE-1 from U2OS cells carried out in ”low stringency“ lysis buffer with anti-hGTSE-1 or -GFP (C) antibody as control. F, U2OS cells were synchronized at G1/S or G2/M phases of the cell cycle and subjected to immunoprecipitation with anti-p21 or -HA (C) as control. Bottom, FACS quantification of the percentage of cells in the specific phases of the cell cycle. Immunoblot analyses of A, C, D, E, and F, were performed using antibodies against hGTSE-1, p21, WISp39, Hsp90, GST, FLAG, HA, and GFP.
To find out whether the interaction of hGTSE-1 with p21 is cell cycle-regulated, we synchronized U2OS cells (which express low levels of hGTSE-1 in G1 phase (15)) at the G1/S boundary or at G2/M phases of the cell cycle through thymidine block and release, respectively, followed by immunoprecipitation of endogenous p21 protein. The p21-hGTSE-1 complex was observed both in G1/S and G2/M phases of the cell cycle (Fig. 3F), indicating a constitutive interaction of these proteins throughout the cell cycle in U2OS cells.
hGTSE-1 N-terminal Portion Is Sufficient for Binding and Stabilizing p21
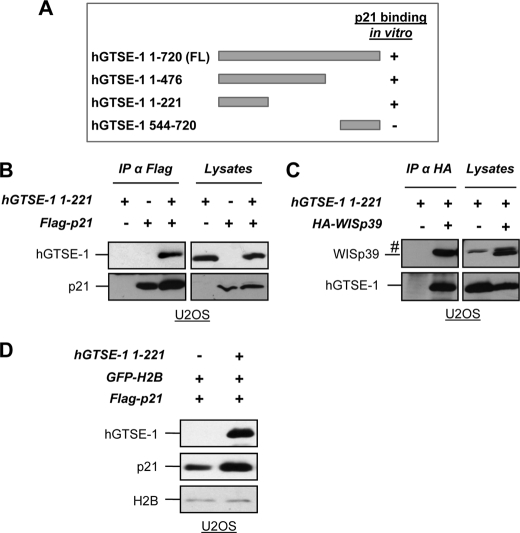
Interaction mapping through in vitro pulldown (Fig. 4A and supplemental Fig. S2, D and E) and immunoprecipitation assays (Fig. 4B) indicated that residues 1–221 of hGTSE-1 (hGTSE-1-(1–221)) are responsible for binding with p21 as well as with WISp39 (Fig. 4C). Importantly, the hGTSE-1-(1–221) deletion preserves the ability of the full-length in stabilizing p21 as demonstrated by co-transfection experiments (Fig. 4D).
FIGURE 4.
Mapping of hGTSE-1 region involved in p21 binding and stability. A, schematic representation of hGTSE-1 deletions and their in vitro binding with p21. B, U2OS cells were transfected with vectors encoding hGTSE-1-(1–221) and FLAG-p21 and subjected to immunoprecipitation with an anti-FLAG antibody. C, U2OS cells were transfected with vectors encoding hGTSE-1-(1–221) and HA-WISp39 and subjected to immunoprecipitation (IP) with an anti-HA antibody. #, unspecific band. D, U2OS cells were transfected with vectors encoding p21 with an empty vector or with hGTSE-1-(1–221) for 36 h. A vector encoding GFP-tagged histone H2B (GFP-H2B) was used as transfection efficiency control. FL, full-length.
hGTSE-1 Modulates the Cellular Response to Paclitaxel-induced Apoptosis by Regulating p21 Levels
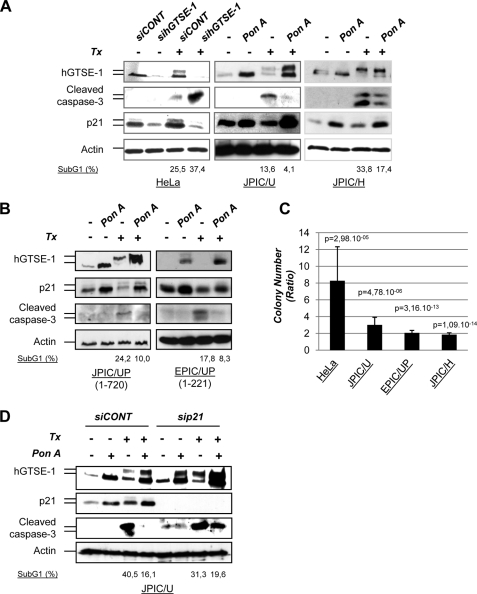
The biological consequence of p21 regulation by hGTSE-1 is reflected by the well known activity of p21 in hampering paclitaxel-induced apoptosis (19). Indeed, siRNA-mediated hGTSE-1 KD sensitizes cells to paclitaxel, whereas cells overexpressing hGTSE-1 display resistance to such treatment, as detected by caspase-3 cleavage, sub-G1 DNA content (Fig. 5A) and colony formation assays (Fig. 5C and supplemental Fig. S3A). This effect occurring in HeLa, U2OS (wild type p53), and H1299 (p53-null) cells suggested again a p53-independent mechanism. In fact, similar experiments performed upon transient or stable down-regulation of p53, indicated that such an effect is p53-independent (supplemental Fig. S3, B and C).
FIGURE 5.
Modulation of the cellular response to paclitaxel (Tx) treatment by hGTSE-1, in a p21-dependent manner. A, HeLa cells transfected with a control (siCONT) or hGTSE-1 (sihGTSE-1) siRNA for 36 h were treated with paclitaxel (0.5 μm) for an additional 24 h (left panel). JPIC/U cells (middle panel) or JPIC/H cells (right panel) were treated or without ponasterone A (pon A and −, respectively) for 16 h followed by addition of paclitaxel (0,5 μm) for another 24 h. The upper bands of hGTSE-1 and p21 detected in paclitaxel-treated cells correspond to phosphorylated forms seen in mitotic cells (5, 13). B, JPIC/UP and EPIC/UP cells were treated with (pon A) or without (−) ponasterone A for 16 h, followed by addition of paclitaxel (1 μm) for another 24 h. C, colony formation assays of cells treated as in A and B. JPIC/U and EPIC/UP cells were treated with 1 μm paclitaxel for 24 h. JPIC/UP cells behave as JPIC/U and are not shown. JPIC/H cells were treated with 0.2 μm paclitaxel for 24 h. HeLa cells were treated with 0,05 μm paclitaxel for 24 h. Histograms represent the mean ratio between paclitaxel-treated siCONT:sihGTSE-1 (HeLa) and ponasterone A:noninduced cells (−) (JPIC/U, EPIC/UP, and JPIC/H). D, JPIC/U cells were treated as in A but after transfecting a control (siCONT) or p21 (sip21) siRNAs 36 h before ponasterone A treatment. All immunoblots were performed with antibodies against hGTSE-1, p21, cleaved caspase-3, and actin as loading control. Numbers at the bottom of the immunoblots indicate representative values of the sub-G1 DNA content calculated trough normalization by biological background subtraction noise (see ”Experimental Procedures“).
Because the hGTSE-1-(1–221) deletion stabilizes p21 as full-length hGTSE-1 does, we tested the effect of its expression in a U2OS-inducible pool of cells (EPIC/UP) as compared with a control full-length hGTSE-1 inducible-pool (JPIC/UP). We observed that, similar to JPIC/UP, cells with induced expression of hGTSE-1-(1–221) display resistance to paclitaxel-induced apoptosis (Fig. 5, B and C, and supplemental Fig. S3A), indicating that this region of hGTSE-1 is sufficient to up-regulate p21 levels and to protect cells from paclitaxel-mediated cytotoxicity. Importantly, hGTSE-1 mediated chemoresistance to paclitaxel-induced cell death is dependent on p21 expression as siRNA-mediated p21 KD restores the sensitivity to apoptosis (Fig. 5D).
Signaling pathways involved in paclitaxel-induced cell death are as yet poorly understood, but it is known that paclitaxel is able to induce cell death independent of p53 status (20) and to abnormally activate G2/M-specific Cdk1, preferentially affecting cells crossing the G2/M boundary (19). In fact, we observed a significant reduction of paclitaxel-induced Cdk1 kinase activity in hGTSE-1-induced cells, which is relieved by p21 KD (supplemental Fig. S4, A and B, respectively). These findings mark p21 as the effector of hGTSE-1-induced resistance to paclitaxel treatment and support the observation that hGTSE-1 levels increase in SKOV-3 ovarian cancer cells during the acquisition of a paclitaxel resistance phenotype in vitro (supplemental Fig. S4C). In addition, the reported effect of hGTSE-1 on cell death could be extended to other type of cellular stresses besides microtubule disruption such as DNA damage, as an effect of hGTSE-1 on doxorubicin-induced cell death is observed in p53-null tumor cell lines (supplemental Fig. S5). Collectively, the data presented here indicate a novel function of hGTSE-1 in protecting p21 from proteasome-dependent degradation through its association with the Hsp90-WISp39 chaperone machinery, thus giving a further level of modulation of p21-specific interactions with its multiple partners (21), with the final outcome of mediating cellular chemoresistance to the microtubule disrupting agent paclitaxel.
DISCUSSION
Here, we present data describing a p53-independent effect of hGTSE-1 in modulating p21 stability, as part of a protein complex that controls p21 turnover. Importantly, such regulation of p21 levels does not rely on significant alterations in the cell cycle and is specific for p21, because no other members of the CIP/KIP family of Cdk inhibitors are affected by hGTSE-1 expression.
p21 abundance must be finely regulated to act in accordance to the requested outcome, as variations in its levels may determine whether it acts as an inhibitor or as an assembly factor of cyclin-Cdk complexes (4, 5), thereby hindering or stimulating proliferation, respectively. In fact, timed regulation of p21 levels and function is essential for cell cycle and apoptosis control (22); therefore, it is important to precisely understand the molecular mechanisms underlying these events. Regulation of p21 intracellular levels is mainly achieved by well established transcriptional mechanisms (23) in combination with post-translational processes, both acting in concert to carry out a coordinated regulation of p21 levels within the established cellular context (10).
We found here that hGTSE-1 protects p21 from a proteasome-dependent degradation and for this activity hGTSE-1 requires a functional Hsp90 complex containing the co-chaperone WISp39. WISp39 binds the N terminus of newly synthesized p21, concomitantly recruiting it to the Hsp90 machinery by means of its TPR domain; likewise, treatment of cells with the Hsp90 inhibitor 17-AAG abrogates WISp39-dependent stabilization of p21 (10). hGTSE-1-dependent regulation of p21 levels is blocked by 17-AAG or WISp39 KD, and, in addition, the three endogenous proteins co-fraction in gel filtration assays and are found in the same complex in vivo (as demonstrated by endogenous co-immunoprecipitation studies). Such results provide evidence that hGTSE-1 could be a component of such chaperone machinery that controls p21 stability.
p21 is an intrinsically unstructured and highly unstable protein (6, 24), and its interaction with molecular partners has been proposed to shield it from degradation. In fact, the capacity of p21 to be directly recognized by the proteasome complex, when it is free (25), highlights that binding partners of p21, such as hGTSE-1, could display an essential role in maintaining p21 steady state levels. For instance, bound Cdk2 induces a stable conformation of the N terminus of p21 (6), and association with proliferating cell nuclear antigen has been proposed to mask the region of p21 involved in C8-α subunit binding and recognition by the proteasome (24). Here, we demonstrate that hGTSE-1 is physically associated with p21 and critically involved in keeping p21 protein levels as a component of the Hsp90-WISp39 chaperone complex.
p21 levels in nontransformed fibroblasts oscillate during the cell cycle, displaying peaks in G1 and G2 (26), whereas in the S phase, it is efficiently targeted for degradation mainly by the SCF-Skp2 ubiquitin ligase complex (27) and by the anaphase-promoting complex-Cdc20 complex in mitosis (28). Synchronization of nontransformed fibroblasts at the G1/S boundary shows that p21 levels increase concomitantly with those of hGTSE-1 as cells exit the S phase (data not shown), suggesting that hGTSE-1 could have a physiological function in maintaining p21 steady state levels, possibly during these specific phases of the cell cycle that may require additional and more stringent regulation of p21 expression. However, in the U2OS tumor cell line, low levels of hGTSE-1 are detected in G1 (15), and the hGTSE-1-p21 complex is observed both in G1/S and G2/M phases of the cell cycle, indicating that hGTSE-1 could constitutively regulate p21 levels throughout the cell cycle in transformed cells.
Importantly, p21 malfunction in tumor cells often derives from variation in its levels and/or subcellular localization, mainly induced by oncogene-activated pathways (29–31). High levels of p21 in breast cancer (as associated with ErbB2 overexpression (32)), as well as in other type of cancers (22), have been linked to poor prognosis.
As a functional consequence, we show that up-regulation of hGTSE-1 expression with the concomitant stabilization of p21 results in chemoresistance to paclitaxel-induced cell death. Moreover, we demonstrate that the region of hGTSE-1 involved in p21 binding, corresponding to amino acids 1–221, results to be sufficient both for stabilizing p21 as well as for inhibiting paclitaxel-induced cell death.
p21 is known to regulate cellular responses to stress such as microtubule damage (33). We found here that hGTSE-1 overexpression protects cells from paclitaxel-triggered apoptosis and that such an effect depends on p21. Hampering paclitaxel-induced cell death has previously been attributed to the ability of p21 to inhibit Cdk1 activity (19). In fact, we observed that paclitaxel-induced Cdk1 kinase activity diminishes upon induction of hGTSE-1 in a p21-dependent way. These results point to p21 as a mediator of hGTSE-1-induced resistance to paclitaxel treatment.
We have previously demonstrated a role of hGTSE-1 in attenuating p53-mediated apoptotic response (15, 17). We now add that the opposite regulation of p53 and p21 stability by hGTSE-1 supports its combined role in promoting cell survival by shifting the equilibrium of the p53 response from apoptosis to survival. This protective effect could also be extended to other types of stress-induced apoptosis such as DNA damage or reactive oxygen species, in which p21 plays an important role in influencing cell fate decision (34). Thus, cancer cells expressing higher and constitutive levels of hGTSE-1 protein could display an unbalanced behavior against proapoptotic signaling pathways.
Acknowledgments
We thank Stefania Marzinotto for collaboration with microscopy; Stefania Gobessi, Loredana Alison, and Ramiro Mendoza-Maldonado for help and technical support; and Silvano Piazza for statistical analysis. We are indebted to Mauro Giacca and Marco Bestagno (International Centre for Genetic Engineering and Biotechnology) for access to FACS and to Alessandro Vindigni for helpful advice on gel filtration chromatography. We are grateful to R. Zhang for pcDNA3-HA-p21, to M. C. Hung for pcDNA3-FLAG-p21, to G. Whal for pBOS-H2B-GFP-N1 (GFP-H2B), to G. Del Sal for pcDNA3-p27, to R. Agami for pSUPER-Retro p53 (pSR-p53) vectors, and to B. Vogelstein for HCT-116 p53 knock-out and parental cells.
This work was supported by Associazione Italiana per la Ricerca sul Cancro, Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) Fondo per gli Investimenti della Ricerca di Base RBIN04N4-003, TRANSlational and Functional Onco-Genomics (TRANSFOG) FP6 503438 (to C. S.), and PICT07-00283 (to M. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental ”Experimental Procedures,“ Figs. S1–S5, and an additional reference.
- Cdk
- cyclin-dependent kinase
- hGTSE-1
- human GTSE-1
- siRNA
- small interfering RNA
- siCONT
- control siRNA
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- Hsp90
- heat shock protein 90
- IVT
- in vitro-translated
- KD
- knockdown
- 17-AAG
- 17-allylamino geldanamycin
- HA
- hemagglutinin
- FACS
- fluorescent-activated cell sorting.
REFERENCES
- 1.Brugarolas J., Chandrasekaran C., Gordon J. I., Beach D., Jacks T., Hannon G. J. (1995) Nature 377, 552–557 [DOI] [PubMed] [Google Scholar]
- 2.Parker S. B., Eichele G., Zhang P., Rawls A., Sands A. T., Bradley A., Olson E. N., Harper J. W., Elledge S. J. (1995) Science 267, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 3.Kagawa S., Fujiwara T., Kadowaki Y., Fukazawa T., Sok-Joo R., Roth J. A., Tanaka N. (1999) Cell Death Differ. 6, 765–772 [DOI] [PubMed] [Google Scholar]
- 4.LaBaer J., Garrett M. D., Stevenson L. F., Slingerland J. M., Sandhu C., Chou H. S., Fattaey A., Harlow E. (1997) Genes Dev. 11, 847–862 [DOI] [PubMed] [Google Scholar]
- 5.Dash B. C., El-Deiry W. S. (2005) Mol. Cell. Biol. 25, 3364–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriwacki R. W., Hengst L., Tennant L., Reed S. I., Wright P. E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11504–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., Clurman B. E. (2000) Mol. Cell 5, 403–410 [DOI] [PubMed] [Google Scholar]
- 8.Bloom J., Amador V., Bartolini F., DeMartino G., Pagano M. (2003) Cell 115, 71–82 [DOI] [PubMed] [Google Scholar]
- 9.Child E. S., Mann D. J. (2006) Cell Cycle 5, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 10.Jascur T., Brickner H., Salles-Passador I., Barbier V., El Khissiin A., Smith B., Fotedar R., Fotedar A. (2005) Mol. Cell 17, 237–249 [DOI] [PubMed] [Google Scholar]
- 11.Coqueret O. (2003) Trends Cell Biol. 13, 65–70 [DOI] [PubMed] [Google Scholar]
- 12.Utrera R., Collavin L., Lazarević D., Delia D., Schneider C. (1998) EMBO J. 17, 5015–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collavin L., Monte M., Verardo R., Pfleger C., Schneider C. (2000) FEBS Lett. 481, 57–62 [DOI] [PubMed] [Google Scholar]
- 14.Monte M., Collavin L., Lazarevic D., Utrera R., Dragani T. A., Schneider C. (2000) Gene 254, 229–236 [DOI] [PubMed] [Google Scholar]
- 15.Monte M., Benetti R., Buscemi G., Sandy P., Del Sal G., Schneider C. (2003) J. Biol. Chem. 278, 30356–30364 [DOI] [PubMed] [Google Scholar]
- 16.Pfleger C. M., Kirschner M. W. (2000) Genes Dev. 14, 655–665 [PMC free article] [PubMed] [Google Scholar]
- 17.Monte M., Benetti R., Collavin L., Marchionni L., Del Sal G., Schneider C. (2004) J. Biol. Chem. 279, 11744–11752 [DOI] [PubMed] [Google Scholar]
- 18.Saxena S., Jónsson Z. O., Dutta A. (2003) J. Biol. Chem. 278, 44312–44319 [DOI] [PubMed] [Google Scholar]
- 19.Yu D., Jing T., Liu B., Yao J., Tan M., McDonnell T. J., Hung M. C. (1998) Mol. Cell 2, 581–591 [DOI] [PubMed] [Google Scholar]
- 20.Debernardis D., Siré E. G., De Feudis P., Vikhanskaya F., Valenti M., Russo P., Parodi S., D'Incalci M., Broggini M. (1997) Cancer Res. 57, 870–874 [PubMed] [Google Scholar]
- 21.Pratt W. B., Morishima Y., Osawa Y. (2008) J. Biol. Chem. 283, 22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbas T., Dutta A. (2009) Nat. Rev. Cancer [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gartel A. L., Tyner A. L. (1999) Exp. Cell Res. 246, 280–289 [DOI] [PubMed] [Google Scholar]
- 24.Cayrol C., Ducommun B. (1998) Oncogene 17, 2437–2444 [DOI] [PubMed] [Google Scholar]
- 25.Touitou R., Richardson J., Bose S., Nakanishi M., Rivett J., Allday M. J. (2001) EMBO J. 20, 2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Jenkins C. W., Nichols M. A., Xiong Y. (1994) Oncogene 9, 2261–2268 [PubMed] [Google Scholar]
- 27.Bornstein G., Bloom J., Sitry-Shevah D., Nakayama K., Pagano M., Hershko A. (2003) J. Biol. Chem. 278, 25752–25757 [DOI] [PubMed] [Google Scholar]
- 28.Amador V., Ge S., Santamaría P. G., Guardavaccaro D., Pagano M. (2007) Mol. Cell 27, 462–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou B. P., Liao Y., Xia W., Spohn B., Lee M. H., Hung M. C. (2001) Nat. Cell Biol. 3, 245–252 [DOI] [PubMed] [Google Scholar]
- 30.Li Y., Dowbenko D., Lasky L. A. (2002) J. Biol. Chem. 277, 11352–11361 [DOI] [PubMed] [Google Scholar]
- 31.Westbrook T. F., Nguyen D. X., Thrash B. R., McCance D. J. (2002) Mol. Cell. Biol. 22, 7041–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W., Klos K. S., Zhou X., Yao J., Yang Y., Smith T. L., Shi D., Yu D. (2003) Cancer 98, 1123–1130 [DOI] [PubMed] [Google Scholar]
- 33.Li W., Fan J., Banerjee D., Bertino J. R. (1999) Mol. Pharmacol. 55, 1088–1093 [DOI] [PubMed] [Google Scholar]
- 34.Chen W., Sun Z., Wang X. J., Jiang T., Huang Z., Fang D., Zhang D. D. (2009) Mol. Cell 34, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]