Abstract
Mitochondrial mRNA editing in trypanosomatid parasites involves several multiprotein assemblies, including three very similar complexes that contain the key enzymatic editing activities and sediment at ∼20S on glycerol gradients. These ∼20S editosomes have a common set of 12 proteins, including enzymes for uridylyl (U) removal and addition, 2 RNA ligases, 2 proteins with RNase III-like domains, and 6 proteins with predicted oligonucleotide binding (OB) folds. In addition, each of the 3 distinct ∼20S editosomes contains a different RNase III-type endonuclease, 1 of 3 related proteins and, in one case, an additional exonuclease. Here we present a protein-protein interaction map that was obtained through a combination of yeast two-hybrid analysis and subcomplex reconstitution with recombinant protein. This map interlinks ten of the proteins and in several cases localizes the protein region mediating the interaction, which often includes the predicted OB-fold domain. The results indicate that the OB-fold proteins form an extensive protein-protein interaction network that connects the two trimeric subcomplexes that catalyze U removal or addition and RNA ligation. One of these proteins, KREPA6, interacts with the OB-fold zinc finger protein in each subcomplex that interconnects their two catalytic proteins. Another OB-fold protein, KREPA3, appears to link to the putative endonuclease subcomplex. These results reveal a physical organization that underlies the coordination of the various catalytic and substrate binding activities within the ∼20S editosomes during the editing process.
Keywords: Organisms/Parasite, Organisms/Trypanosome, Protein/Protein-Protein Interactions, Protein/Domains, Protein/Protein-Protein Interactions, RNA/Editing, RNA/Processing, Subcellular Organelles/Mitochondria
Introduction
The post-transcriptional processing of mitochondrial mRNAs6 by RNA editing in trypanosomes is catalyzed by multiprotein complexes (1, 2). Central among these is the ∼20S complex (referred to as the 20S editosome hereafter) that contains the catalysts that cleave the mRNA, insert or remove uridylates (Us), and re-ligate the resultant products (reviewed in Refs. 3, 4). The sites of cleavage, the number of added or removed Us, and the alignment of the products to be ligated are specified by small (∼60 nucleotides) guide RNAs (gRNAs) (5). These gRNAs have partial complementarity to their pre-edited sequences but are fully complementary to the edited sequence. Each gRNA specifies the editing of numerous editing sites, and multiple rounds of these three coordinated reactions are required to edit each block of mRNA sequence that is specified by a single gRNA. Most gRNAs specify U insertion, which reflects the ∼10-fold greater number of U insertion sites compared with U deletion sites (reviewed in Ref. 6). Some gRNAs specify a combination of U insertion and U deletion, but no gRNAs have been identified that only specify U deletion. The gRNAs have 3′-oligonucleotide-U tails that are added post-transcriptionally and appear to be added by a homo-oligomeric complex of the kinetoplastid RNA editing 3′-terminal uridylyltransferase (TUTase) (7). The function of this gRNA U tail is unknown, but it may stabilize the interaction between gRNA, mRNA, and editosome (8).
Most mitochondrial mRNAs are extensively edited, which requires the use of numerous different gRNAs. The editing proceeds generally in the 3′ to 5′ direction, with respect to the mRNA, as a consequence of duplex formation between the 5′ “anchor” region of each gRNA and the 3′ end of the sequence block to be edited (9, 10). The anchor region of the first gRNA can form a duplex with pre-edited mRNA, but basepairing with successive gRNAs requires completed editing of the downstream region. This provides for the successive use of the gRNAs. Association of the mRNAs and gRNAs may be mediated by the MRP complex, which consists of two copies each of two related proteins (11). This complex catalyzes matchmaker RNA/RNA annealing, i.e. basepairing followed by dissociation of the RNA duplex from the proteins (11–13). RBP16, another trypanosomal mitochondrial RNA-binding protein, has also been shown to facilitate gRNA/pre-mRNA annealing in vitro (14, 15). It is uncertain how the gRNAs traffic to and from the 20S editosomes and how this process is controlled, especially between lifecycle stages that differentially edit mRNAs. Other complexes, including a novel multiprotein complex, may have this or other roles related to editing, because RNA interference knockdown of expression of their component proteins results in aberrant editing, including the loss of specific edited mRNAs (2, 16–18).
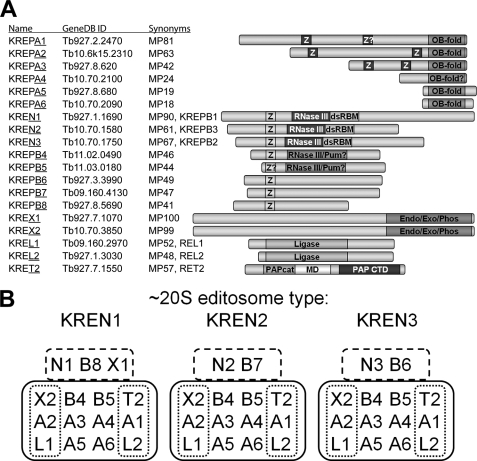
There are three very similar ∼20S editosomes, all of which have a common set of 12 proteins, but each has a different set of 2 or 3 related proteins (see Fig. 1) (19–23). The common set of proteins includes the four related KREPA3, KREPA4, KREPA5, and KREPA6 proteins, which have oligonucleotide/oligosaccharide binding (OB)-fold motifs, and two mutually related proteins KREPB4 and KREPB5, which each have a U1-like zinc finger motif and a degenerate RNase III motif (24). The common set also contains two heterotrimeric subcomplexes, one that can catalyze steps of insertion and the other steps of deletion editing (25). The insertion subcomplex consists of the 3′ TUTase KRET2 and the RNA ligase KREL2 linked by the KREPA1 protein, which contains two zinc finger motifs and an OB-fold motif. This subcomplex can accurately add Us to an editing-like mRNA/gRNA substrate and ligate the products as specified by the gRNA. Similarly, the deletion subcomplex consists of the U-specific 3′ exonuclease KREX2 and the RNA ligase KREL1 linked by the KREPA2 protein, which also has two zinc fingers and an OB-fold motif. It can catalyze U removal from an editing-like mRNA/gRNA substrate and ligate the products as specified by the gRNA. The KREPA1 and KREPA2 proteins are related to each other, and their C-terminal parts to the smaller KREPA3, KREPA4, KREPA5, and KREPA6 proteins (20, 22, 24, 25).
FIGURE 1.
Components and architecture of T. brucei 20S editosomes. A, protein components of T. brucei ∼20S editosomes identified to date. Each is designated according to the kinetoplastid RNA editing protein (KREP) nomenclature along with its ID in GeneDB, and synonyms. The identity and location of demonstrated and putative motifs are indicated in the diagram (3, 24). White Z on dark background, C2H2-type zinc finger; OB-fold, oligonucleotide binding fold; black Z on light background, U1-like C2H2 zinc finger; RNase III, RNase III catalytic-like domain; dsRBM, double-stranded RNA binding motif; RNase III-Pum, overlapping RNase III-like and Pumilio motifs; Endo-Exo-Phos, endonuclease-exonuclease-phosphatase family motif; ligase, polynucleotide ligase-mRNA capping catalytic domain; PAPcat, poly(A) polymerase catalytic domain; MD, middle domain; and PAP CTD, poly(A) polymerase C-terminal domain. Question marks indicate motifs with lower confidence values. B, architecture of the three identified 20S editosomes using a shorthand for the nomenclature as underlined in A (e.g. A1). Solid lines outline the proteins that are common to each complex and the trimeric U-deletion and U-insertion subcomplexes are outlined by the dotted lines. The dashed lines outline the endonuclease proteins along with their specific associated proteins that are unique to each of the three different types of 20S editosomes.
In addition to the set of twelve common proteins each different editosome also contains related sets of paired proteins. These are the RNase III type endonucleases KREN1, KREN2, and KREN3 that occur in combination with KREPB8, KREPB7, and KREPB6, respectively (20, 26).7 The KREN1 editosome also contains the U-specific exonuclease KREX1 (Fig. 1B). Some editosome preparations also contained the KREH1 helicase (20).7 These KREN1·KREPB8·KREX1, KREN2·KREPB7, and KREN3·KREPB6 protein sets might be considered endonuclease subcomplexes, although, unlike the insertion and deletion heterotrimeric subcomplexes, they have not been observed as separate physical and functional entities. The KREN1 editosomes specifically cleave substrates that undergo deletion editing in vitro while KREN2 and KREN3 each specifically cleave different insertion editing sites in vitro (20, 26–28). Hence the KREN1, KREN2, and KREN3 editosomes must collaborate in some way for the editing of blocks where Us are both inserted and deleted.
The composition of the 20S editosomes along with what is known of their physical and functional associations and their organization into subcomplexes indicates that their structural organization is important to their function (25). The KREPA3, KREPA4, and KREPA6 proteins, as well as the KREPB4 and KREPB5 proteins, are essential for editosome integrity. Knockdown of the expression of any one of these results not only in the loss of the specific protein but also essentially complete loss of all of the ∼20S editosomes and degradation of their constituent proteins (29–35). An earlier yeast two-hybrid study using full-length proteins identified a number of pairwise interactions in the editosome (25). Using a fragment-based approach, herein we report the characterization of several additional pairwise interactions among editosome proteins and the localization of interaction domains of newly as well as previously identified pairs. We also report the co-expression in Escherichia coli of multiple recombinant proteins that form stable associations. The results indicate that the KREPA3, KREPA4, and KREPA6 proteins form a network of interactions, among themselves as well as with the KREPA1 and KREPA2 proteins in the heterotrimeric insertion and deletion subcomplexes, that is mediated by their predicted OB-fold domains. The KREPA3 protein also interacts with the KREPB5 protein implying linkage of the network with the endonuclease subcomplexes.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Analysis
A yeast two-hybrid strategy described by James et al. (36) based on fragment libraries was adopted. The fragment libraries were constructed from pools of plasmids that contained genes for 16 components of the Trypanosoma brucei ∼20S editosome, namely KREPA1, KREPA2, KREPA3, KREPA4, KREPA6, KREN1, KREN2, KREN3, KREPB4, KREPB5, KREPB8, KREL1, KREL2, KRET2, KREX1, and KREX2. The KREPA5, KREPB6, and KREPB7 components had not yet been identified at the time of library construction. Plasmids encoding other genes suggested to be involved in trypanosome RNA editing were also included. These are MRP1 and MRP2 (37), mHel61 (38), RBP16 (39), REAP-1 (40), TbRGG1 (16, 41), and KRET1 (7, 42). The plasmid pools were partially digested with five restriction enzymes that recognize 4-bp sequences, all generating 5′-CG overhangs (AciI, HinPII, MaeII, MspI, and TaqI), and fragments ranging up to 2 kb were gel-isolated and ligated into ClaI-digested yeast two-hybrid plasmids pGAD-C1, -C2, and -C3 and pGBD-C1, -C2, and -C3, which are identical except for the translational reading frame of the polylinker region (36). Thus, a total of 30 ligation reactions were carried out (6 vectors × 5 digests), theoretically resulting in C-terminal fusions of Gal4 activation and DNA-binding domains with any given fragment in all three possible reading frames. For each ligation reaction, 0.5 μg of fragments were mixed with 0.5 μg of plasmid. Ligations were transformed into library-efficient DH5α cells (Invitrogen). After incubation in 1 ml of SOC (Invitrogen) medium at 37 °C for 1 h, cells were grown in 50 ml of LB medium (10 g of Bacto-tryptone, 5 g of Bacto-yeast extract, and 10 g of NaCl in 1000 ml of H2O) with 100 μg/ml ampicillin to a density of A600 1.5, and libraries were obtained by plasmid isolation.
The following genes were prepared as baits by generating C-terminal fusions of Gal4 activation (pGAD-C1) or DNA binding domain (pGBD-C1) with their entire open reading frame: KREPA2, KREPA3, KREPA6, KREPB4, KREPB5, KREPB8, and KREX1. Bait plasmids were introduced into yeast strain PJ69-4A using the lithium acetate method (43). Screens were carried out by transforming bait strains with the libraries using the same protocol, scaled up 30-fold. Transformation reactions were plated on drop-out medium, i.e. lacking tryptophan, leucine, and adenine, and incubated for up to 1 week at 30 °C. Colonies were replated on the same medium for confirmation and then subjected to plasmid isolation. Inserts were identified by sequencing. Bait and putative prey plasmids were retransformed into strain PJ69-4A, and interactions were considered as confirmed when the resulting yeast clones gave rise to colonies within 7 days on selective drop-out medium.
Expression and Purification of Recombinant Proteins in E. coli
KREPA1196–762 (superscript numbers and asterisks throughout this report refer to amino acid positions in the precursor proteins, i.e. before removal of the N-terminal mitochondrial targeting signal upon import, and the C-terminal residue, respectively), KREPA1369–762*, KREPA1369–569, KREPA1396–482, KREPA1416–569, KREP1A1-OB626–762*, KREPA256–587*, KREPA2-OB474–587*, KREPA322–393*, KREPA347–393*, KREPA3-OB245–393*, KREPA619–164*, KREPA6-OB19–148, KREPB540–253, KREL151–469*, KREL221–416*, and KRET220–487* were amplified by PCR using primers designed to place vector-specific restriction sites at the 5′ and 3′ ends of the gene and cloned into expression vectors, pSKB3, pRSF, pCDF, and pACYC (Novagen) and pProEX (Invitrogen). The pair KREPA6·KREPA3-OB was cloned into the bi-cistronic expression vector pRSF, and the pairs KREL1·KREPA2 and KREPA1·KRET2 both were cloned into pET21d for bi-cistronic expression.
KREPA6 with an N-terminal cleavable His tag was co-transformed with other proteins in compatible vectors without any tag and co-expressed in E. coli BL21-Gold(DE3) (Stratagene). Cells were grown to an A600 of ∼0.6–0.8 at 37 °C in Luria broth containing vector-specific antibiotics and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C for 20 h. Cells were harvested by centrifugation and resuspended in buffer A (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 20 mm imidazole, and 10% glycerol). Cells were lysed by three cycles in a French Press, and insoluble material was removed by centrifugation. Subsequent purification steps were carried out at 4 °C. The supernatant was passed through an Ni-NTA column (Qiagen), washed with buffer A, and eluted with buffer A containing 250 mm imidazole. Recombinant TEV protease was added to the collected fractions after Ni-NTA purification, and the mixture was dialyzed against buffer B (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 10% glycerol) at 4 °C for 24 h. The protein solution was passed through a second Ni-NTA column to remove the cleaved His tag, His-tagged recombinant TEV protease, and any uncleaved KREPA6. The flow-through was concentrated to 5–10 mg/ml using a Centricon spin-column and applied onto a Superdex 200 column (Amersham Biosciences) for a size-exclusion chromatography step in buffer C (20 mm HEPES, pH 7.5, 500 mm NaCl, 2 mm dithiothreitol, and 10% glycerol). Each step of protein purification was monitored by SDS-PAGE.
For the preparation of KREPA1·KRET2 complexes, KREPA1369–569, KREPA1369–482, KREPA1396–482, KREPA1416–569, and KRET220–487* were expressed and purified separately. An equimolar mixture of purified KREPA1, after removal of the His tag and KRET2 with His tag, was passed through an Ni-NTA column, washed with buffer A, and eluted subsequently with buffer A containing 250 mm imidazole. The purification steps were monitored by SDS-PAGE.
Western Analysis
Recombinant proteins expressed as described above were fractionated on precast 10% SDS-PAGE gels (Bio-Rad) and transferred to a polyvinylidene difluoride membrane by Western blotting as described previously (25). Blots were probed with monoclonal antibodies against KREPA1, KREPA2, and KREL1 (22) or a polyclonal antibody against KREPA6 (25) and developed with the ECL system (Amersham Biosciences).
RESULTS
Mapping of Direct Protein-protein Interactions with the Yeast Two-hybrid System
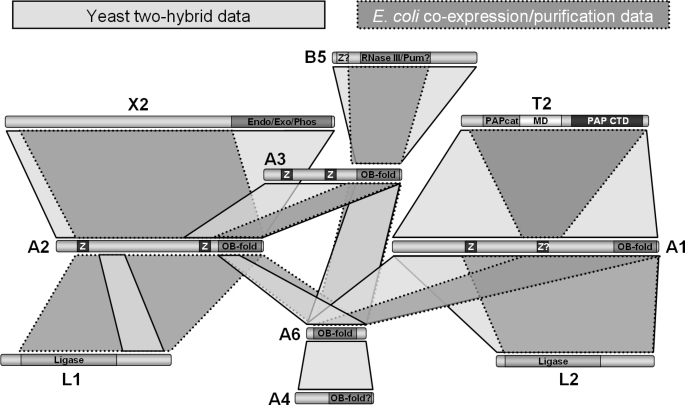
A yeast two-hybrid strategy was used to identify direct interactions between T. brucei 20S editosome proteins. In summary, screens with five full-length editosome proteins as baits (KREPA2, KREPA3, KREPA6, KREPB5, and KREL1) against a library containing fragments from sixteen editosome components as well as from seven other editing-associated proteins (see “Experimental Procedures” for details) identified protein domains involved in six pairwise interactions (summarized in Fig. 10). Five of these interactions involved previously unrecognized binding partners. Genes encoding the following ∼20S editosome components had been included in the prey libraries but were neither found among the clones selected for further analysis nor used as baits: KREPA1, KREN1, KREN2, KREN3, KREL2, KRET2, and KREX2. Screens with editosome proteins KREPB4, KREPB8, or KREX1 as baits did not identify any reproducible interactions. None of the editing-associated proteins included in the libraries (MRP1, MRP2, mHel61, RBP16, REAP-1, TbRGG1, or KRET1) were identified as direct interactors.
FIGURE 10.
Summary of identified interactions. Direct interactions are shown in light and dark gray as identified by yeast two-hybrid analysis (either in this study or, in case of KREPA2-KREX2, and the KREPA1 interactions, in Ref. 25) and bacterial co-expression/purification, respectively. Confirmed and predicted motifs are indicated as in Fig. 1. See schematics of individual interactions (Figs. 2–9) for identification of amino acid residues involved in the interactions.
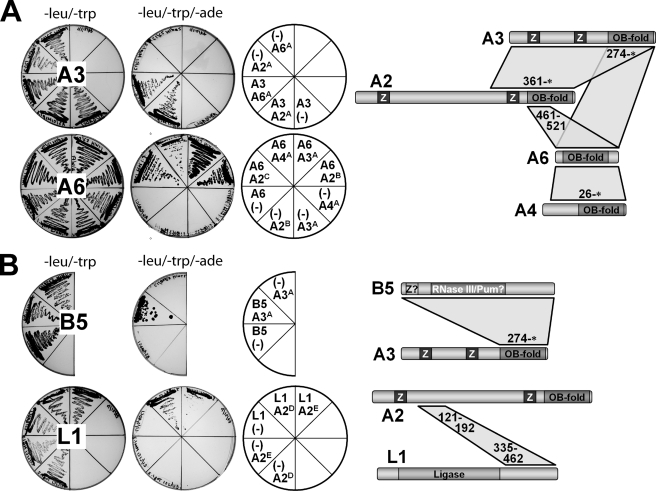
A screen using a full-length KREPA3 fused to GBD as bait against the GAD libraries resulted in the identification of multiple KREPA2 and KREPA6 fragments that supported growth on selective medium. In each case, the prey plasmids that contained the shortest fragments were identified (KREPA2361–587* and KREPA68–164*, respectively), and their direct interaction was confirmed by co-transformation of the isolated plasmid and the KREPA3 bait plasmid (Fig. 2A, top row). Similarly, a screen using full-length KREPA6 as bait identified fragments of KREPA2, KREPA3, and KREPA4 that indicate direct interaction with this protein (Fig. 2A, bottom row). These are KREPA2461–587* and KREPA2217–521 (which localize KREPA2 interaction between residues 461 and 521), KREPA3274–393*, and KREPA426–218*. A screen with full-length KREPB5 as bait identified direct interaction with KREPA3274–393* (Fig. 2B, top row). Full-length KREPA2 was screened as bait in fusion with GAD, because a previous study had indicated that full-length KREPA2 in fusion with GBD is self-activating (25). The screen confirmed interaction with KREPA3 and identified a fragment of the C-terminal domain of KREL1 (residues 335–462) as responsible for mediating interaction with KREPA2 (data not shown). A secondary screen using KREL1335–462 fused to GBD as bait and the GAD libraries revealed that KREPA2121–201 and KREPA2102–192 fragments mediated direct interaction (Fig. 2B, bottom row), suggesting that the overlapping KREPA2 region 121–192 is sufficient for interaction with KREL1. These data indicate that these four KREPA-group proteins interact with each other via their OB-fold domain regions and that this region of KREPA3 also mediates interaction with the KREPB5 protein.
FIGURE 2.
Identification of binary interactions with the yeast two-hybrid system. Retests of gene fragments identified in screens with full-length baits KREPA3 and KREPA6 (A), or full-length KREPB5 and the C-terminal domain of KREL1 (amino acid residues 335–462) (B) by co-transforming bait (DNA binding domain) and prey (activation domain) plasmids into yeast cells. Left column: leucine/tryptophan drop-out plates selecting for cells harboring bait and prey plasmids. Middle column: leucine/tryptophan/adenine drop-out plates selecting for bait-prey protein-protein interaction. Right column: schematic showing bait-prey combinations in the respective plate sectors with bait on top and prey on bottom. “(-)” indicates controls with empty pGAD or pGBD plasmids. Letters in superscript identify protein fragments encoded in prey plasmids; an asterisk indicates the natural C terminus of the protein (A2A = 361-*, A2B = 217–521, A2C = 461-*, A2D = 102–192, A2E = 121–201; A3A = 274-*; A4A = 26-*; and A6A = 8-*). The diagrams to the right of the plate images illustrate the location of the interacting regions within the proteins. See Fig. 1 for motif symbols.
Reconstitution of Binary Complexes with Recombinant Protein Expressed in E. coli
In a parallel approach, selected T. brucei editosome proteins were either co-expressed in E. coli, with only one of the proteins containing a His6 tag, or expressed and purified separately and then mixed. Pairs and groups for reconstitution of binary and higher order complexes, respectively, were selected based on yeast two-hybrid data published previously (25) or described above. In many cases, expression of full-length T. brucei proteins in E. coli as soluble protein was not successful. However, soluble recombinant proteins were successfully obtained for numerous fragments, and progressively smaller fragments were produced in attempts to identify interacting domains. Family sequence alignments and secondary structure predictions were used to avoid cutting in predicted helices and to avoid start and end points in hydrophobic stretches. As a result, several protein fragments chosen to express in vitro are not exactly the same as the fragments that were identified by yeast two-hybrid. Interactions that were detected were confirmed by co-elution in the form of complexes during Ni-NTA and/or size-exclusion chromatography.
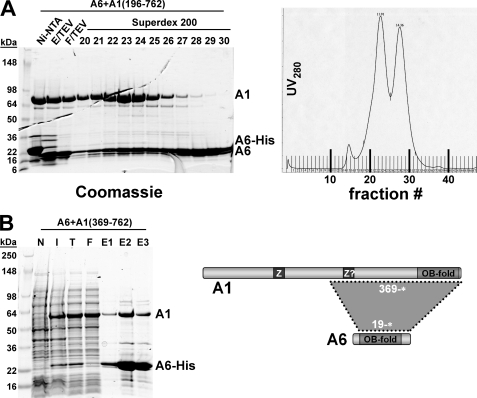
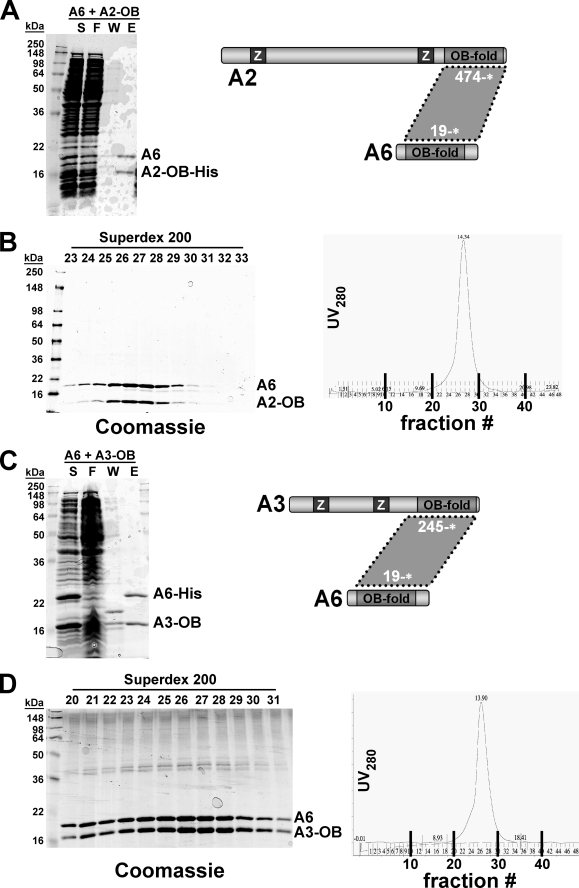
The OB-fold proteins KREPA1, KREPA2, and KREPA3 were all found to form binary complexes with the smallest OB-fold protein, KREPA6. When co-expressed with His6-tagged KREPA619–164*, untagged KREPA1196–762* and KREPA1369–762* co-eluted with KREPA6 from the Ni-NTA column, indicating direct interaction (Fig. 3, A (Ni-NTA) and B (E1–3), respectively). Fractionation of the KREPA619–164*·KREPA1196–762* Ni-NTA co-eluate on a Superdex 200 size-exclusion column resulted in separation of two peaks (Fig. 3A, right panel). Analysis of individual fractions by SDS-PAGE and Coomassie staining identified the first peak as the KREPA6·KREPA1 complex (Fig. 3A, left panel, fractions 20–25). The second peak, corresponding to an estimated molecular mass of 58 kDa, appeared to be a KREPA6 homotetramer (fractions 27–30). Fractionation by size-exclusion chromatography of a mixture of individually purified KREPA6 and KREPA1 OB-fold domains (fragments 19–148 and 626–762*, respectively) suggested that these domains mediate the interaction: a shoulder migrating ahead of a major peak (the latter representing individual fragments) was only detectable in the presence of both fragments, indicative of formation of a heteromeric complex (supplemental Fig. S1). Similarly, co-expressing KREPA619–164* and the predicted OB-fold domains of KREPA2 (residues 474–587*) or KREPA3 (residues 245–393*), with only one of the respective partners being His-tagged, resulted in co-elution of the respective polypeptide pairs from Ni-NTA columns (Figs. 4, A and C, lanes E). Fractionation of the eluates by size-exclusion chromatography revealed heteromeric complexes that eluted as a single, well defined peak (Fig. 4, B and D).
FIGURE 3.
Reconstitution of the KREPA6·KREPA1 complex. T. brucei KREPA6 and KREPA1 were co-expressed in E. coli, and complexes were purified by Ni-NTA chromatography via a His6 tag on KREPA6 (A and B) and gel filtration over a Superdex 200 sizing column (A). A, purification of the KREPA619–164·KREPA1196–762 complex by Ni-NTA chromatography, followed by cleavage of the His tag and fractionation on a Superdex 200 sizing column. Left panel, Coomassie staining of loading material and peak fractions; right panel, UV280 profile of sizing column eluates. B, purification of the KREPA619–164·KREPA1369–762 complex. The diagram here and in the following figures illustrates the locations of the interacting regions within the proteins. Ni-NTA, eluate from first Ni-NTA column; E/TEV, eluate from first Ni-NTA after treatment with TEV protease; F/TEV, flow-through from second Ni-NTA to remove His tag, TEV, and uncleaved protein, which remain bound to the column; N, lysate from uninduced cells; I, lysate from induced cells; T, total lysate; F, flow-through Ni-NTA; and E1–3, Ni-NTA elution fractions.
FIGURE 4.
Reconstitution of KREPA6·KREPA2 and KREPA6·KREPA3 complexes. A, T. brucei KREPA619–164 and KREPA2474–587 were co-expressed in E. coli and co-purified by Ni-NTA chromatography via a His6 tag on KREPA2. B, His tag, TEV protease and uncleaved KREPA2 were removed by TEV cleavage and Ni-NTA chromatography, and the flow-through was collected, concentrated, and fractionated on a Superdex 200 sizing column. Left panel, Coomassie staining of peak eluate fractions; right panel, UV280 profile of eluates. C, T. brucei KREPA619–164 and KREPA3245–393 were co-expressed in E. coli and co-purified by Ni-NTA chromatography via a His6 tag on KREPA6. D, after TEV cleavage and Ni-NTA chromatography to remove His tag, TEV protease and uncleaved KREPA6, the flow-through was collected, concentrated, and fractionated on a Superdex 200 sizing column. Left panel, Coomassie staining of peak eluate fractions; right panel, UV280 profile of eluates. S, soluble fraction; F, flow-through Ni-NTA; W, final wash Ni-NTA; and E, Ni-NTA elution fractions.
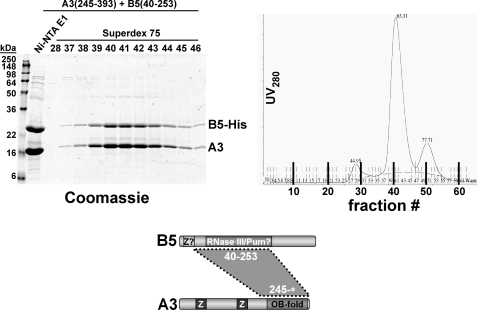
The OB-fold domain of KREPA3 also mediated interaction with the RNase III-like protein KREPB5 (Fig. 5). After co- expression, KREPA3245–393* co-eluted with His6-tagged KREPB540–253 from the Ni-NTA column (Fig. 5, left panel, lane E1) and the KREPA3·KREPB5 complex fractionated as a major, well defined peak on a Superdex 75 column (Fig. 5, right panel).
FIGURE 5.
Reconstitution of the KREPA3·KREPB5 complex. T. brucei KREPA3245–393 and KREPB540–253 were co-expressed in E. coli and co-purified by Ni-NTA chromatography via a His6 tag on KREPB5, followed by gel filtration over a Superdex 75 sizing column. Left panel, Coomassie staining of loading material and peak eluate fractions; right panel, UV280 profile of eluates. Ni-NTA E1, elution fraction 1.
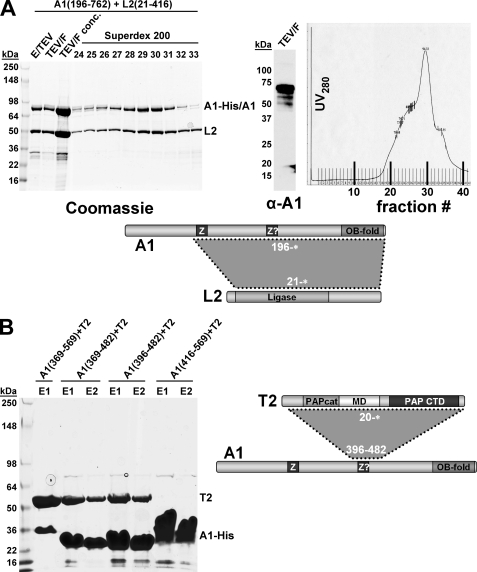
Recombinant KREPA1, in addition to its interaction with KREPA6, formed binary complexes with two editosome enzymes, the RNA ligase KREL2 and the TUTase KRET2, respectively (Fig. 6). After co-expression of His6-tagged KREPA1196–762* with untagged KREL221–416* both proteins co-eluted from the Ni-NTA column (Fig. 6A, lane E/TEV). Fractionation of the eluate by size-exclusion chromatography resulted in identification of a broad peak, likely representing the binary KREPA1·KREL2 complex overlapping with dissociated, free proteins (Fig. 6A, right panel). Mixing of purified KREPA1369–569, 369–482, 396–482 or KREPA1416–569 with individually purified KRET220–487* (after removal of KRET2 His tag with TEV protease), and subsequent analysis on a second Ni-NTA column indicated that KREPA1 region 396–482 is sufficient for interaction with KRET2 and that residues 396–416 are critical because KRET2 did not co-elute with KREPA1416–569 (Fig. 6B).
FIGURE 6.
Reconstitution of KREPA1·KREL2 and KREPA1·KRET2 complexes. A, T. brucei KREPA1196–762 and KREL221–416 were co-expressed in E. coli and co-purified by Ni-NTA chromatography via a His6 tag on KREPA1, followed by gel filtration over a Superdex 200 sizing column. Left panel, Coomassie staining of loading material and peak eluate fractions; middle panel, immunoblot analysis of fraction TEV/F with anti-KREPA1 monoclonal antibody; right panel, UV280 profile of eluates. B, co-purification of various KREPA1 fragments with KRET220–487. The His6-tagged KREPA1 and KRET2 proteins were individually purified via Ni-NTA chromatography. Prior to mixing and final Ni-NTA chromatography, the His6 tag on KRET2 was removed with TEV protease. E/TEV, eluate from first Ni-NTA after treatment with TEV protease; TEV/F, flow-through from second Ni-NTA to remove His tag, TEV, and uncleaved protein, which remain bound to the column; and E1–2, Ni-NTA elution fractions.
Reconstitution of Ternary and Higher Order Complexes with Recombinant Editosome Proteins
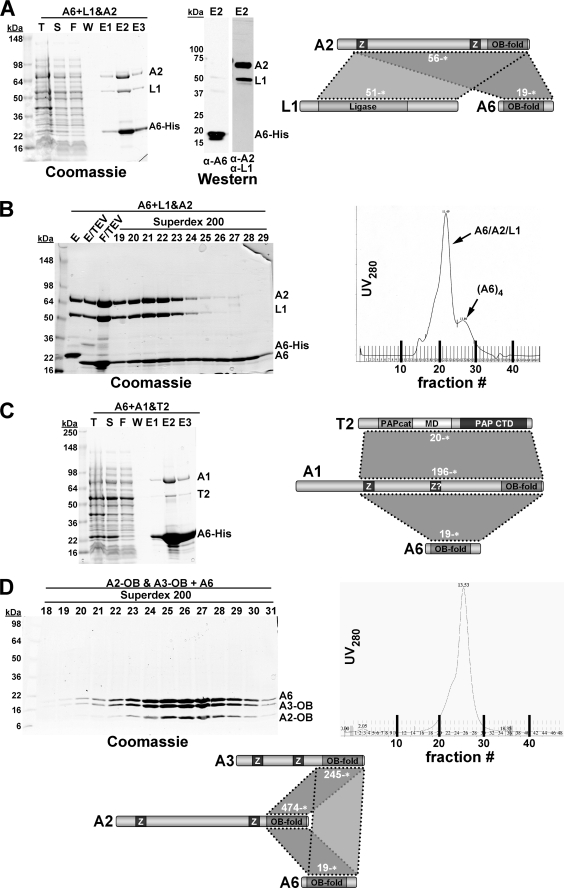
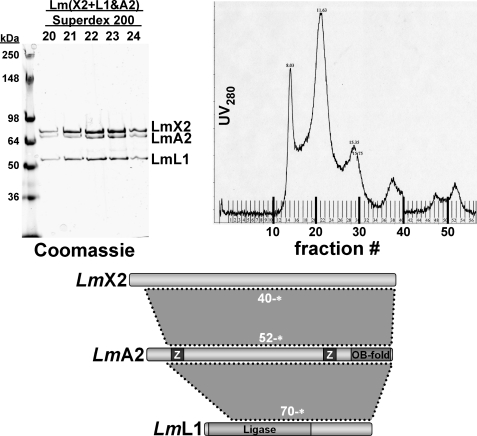
Ternary complexes of T. brucei editosome protein KREPA6 in combination with KREPA2·KREL1, KREPA1·KRET2, and KREPA3·KREPA2 were produced using the co-expression strategy described above and purified via His6 tags on KREPA6 (Fig. 7). As with some of the binary KREPA6 complexes above, the original Ni-NTA eluates contained KREPA6 in more than stoichiometric amounts, indicating the formation of homo-oligomeric KREPA6 complexes in addition to the heteromeric complexes (Fig. 7, A and C, fractions E2). During fractionation of the KREPA619–164*· KREPA256–587*·KREL151–469* co-eluate by size-exclusion chromatography, these KREPA6 complexes eluted as a separate but overlapping peak (Fig. 7B, fractions 25–29). To reconstitute a ternary Leishmania major KREX2·KREPA2·KREL1 complex, corresponding to the aforementioned T. brucei U deletion subcomplex (25), LmKREX240–675* with an N-terminal His6 tag was expressed in E. coli and purified by Ni-NTA chromatography (not shown). LmKREL170–424* and LmKREPA252–623* were co-expressed in E. coli, and the cell lysate was mixed with the purified KREX2. KREX2-associated complexes were purified by a second Ni-NTA chromatography step and fractionated by gel filtration over a Superdex 200 size-exclusion column (Fig. 8). Like other size-fractionated binary and ternary complexes, the Leishmania major KREX2·KREPA1·KREL1 complex appeared to contain components in approximately equimolar amounts. The apparent molecular mass of this complex was ∼350 kDa, consistent with a (KREX2·KREPA2· KREL1)2 heterohexamer. Reconstitution of the T. brucei KREX2·KREPA2·KREL1 ternary complex was not successful because full-length TbKREX2 could not be expressed in soluble form in E. coli.
FIGURE 7.
Reconstitution of ternary KREPA6 complexes. Combinations of T. brucei editosome proteins were co-expressed in E. coli, and complexes were purified by Ni-NTA chromatography via a His6 tag on KREPA6 (A–C) or on KREPA2 (D). A, purification of the KREPA619–164·KREPA256–588·KREL151–469 complex. Left panel, Coomassie staining; right panel, immunoblot analysis of the peak eluate fraction with a polyclonal antibody against KREPA6 or monoclonal antibodies against KREPA2 and KREL1. B, fractionation of the KREPA2·KREL1·KREPA6 complex from A on a Superdex 200 column after removal of the His tag from KREPA6. Left panel, Coomassie staining of loading material and peak eluate fractions; right panel, UV280 profile of eluates. C, purification of the KREPA619–164· KREPA1196–762·KRET220–487 complex. D, purification of the KREPA619–164·KREPA3245–393·KREPA2474–587 complex. Following initial Ni-NTA chromatography, TEV cleavage, and second Ni-NTA chromatography (to remove His6 tag, TEV protease, and uncleaved KREPA2; not shown), the flow-through was collected, concentrated, and fractionated on a Superdex 200 sizing column. Left panel, Coomassie staining of peak eluate fractions; right panel, UV280 profile of eluates. Plus sign (“+”) and ampersand (“&”) indicate co-expression from the same plasmid or from co-transformed individual plasmids, respectively. T, total lysate; S, soluble fraction; F, flow-through Ni-NTA; W, final wash Ni-NTA; E, Ni-NTA elution fractions; E/TEV, eluate from first Ni-NTA after treatment with TEV protease; and F/TEV, flow-through from second Ni-NTA to remove His tag, TEV, and uncleaved protein, which remain bound to the column.
FIGURE 8.
Reconstitution of the Leishmania KREPA2·KREX1·KREL1 complex. L. major KREX240–675 with an N-terminal His6 tag was expressed in E. coli and purified by Ni-NTA chromatography. L. major KREL170–490 and KREPA252–623 were co-expressed in E. coli, and the cell lysate was mixed with the purified KREX2. KREX2-associated complexes were purified by Ni-NTA chromatography (not shown) and fractionated by gel filtration over a Superdex 200 sizing column. Left panel, Coomassie staining of the Superdex 200 fractions corresponding to the major peak; right panel, full UV280 profile of column eluate.
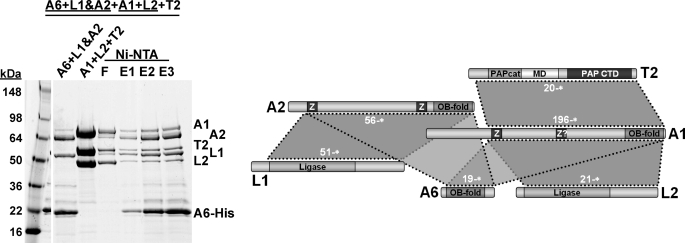
Because the yeast two-hybrid data and the reconstitution experiments above indicated that KREPA6 can interact with both KREPA2 and KREPA1, it could potentially provide a physical linkage between the trimeric U deletion and U insertion complexes in the 20S editosome. To investigate this possibility we attempted to reconstitute from recombinant protein a heterohexameric complex containing components of both subcomplexes. First, a ternary KREPA6·KREPA2·KREL1 complex was produced as described for Fig. 7 using His6-tagged KREPA6 for purification (Fig. 9, first lane). A KREPA1·KREL2 complex and KRET2 were purified separately and the His tags removed by TEV cleavage (not shown). The ternary KREPA1·KREL2·KRET2 complex was obtained by gel filtration of a mixture of the three proteins (Fig. 9, second lane). After mixing the two ternary complexes, a complex containing all six proteins could be obtained by Ni-NTA purification via the His6 tag on KREPA6 (Fig. 9, Ni-NTA lanes). This suggests that KREPA6 indeed is capable of engaging in multiple simultaneous interactions with partner proteins as well as with itself. It cannot be completely ruled out that some KREPA6 dissociated from its complex with KREPA2 and KREL1 before becoming available for interaction with, and purification of, the KREPA·KREL2·KRET2 complex. However, as discussed in more detail below, a propensity of KREPA6 for multiple simultaneous interactions is consistent with data presented here and elsewhere.
FIGURE 9.
Reconstitution of a higher order editosome protein complex. A complex containing T. brucei editosome proteins KREPA1, KREPA2, KREPA6, KREL1, KREL2, and KRET2 was reconstituted as follows. KREPA619–164, KREL151–469, and KREPA256–587 were co-expressed and co-purified by Ni-NTA via a His6 tag on KREPA6 (lane 1). KREPA1196–762 and KREL221–416 were co-expressed and co-purified by Ni-NTA chromatography via a His6 tag on KREPA1, followed by TEV cleavage and Ni-NTA chromatography to remove His tag, TEV protease, and uncleaved KREPA1. The flow-through was collected, concentrated, and fractionated on a Superdex 200 gel-filtration column to obtain the pure binary KREPA1·KREL2 protein complex (not shown). Editosome protein KRET220–487 was purified by Ni-NTA chromatography, followed by TEV cleavage, a second Ni-NTA chromatography step to remove TEV protease, His tag, and uncleaved protein, and a final gel-filtration step (Superdex 75 column) to obtain pure protein (not shown). The ternary KREPA1·KREL2·KRET2 complex was obtained by mixing KREPA1·KREL2 and KRET2, followed by gel filtration on a Superdex 200 column (lane 2). The full complex was obtained by mixing the pure ternary complexes of KREL1·KREPA2·KREPA6 and KREL2·KREPA1·KRET2, followed by Ni-NTA purification via the His6 tag on KREPA6 (lanes 4–6). F, flow-through Ni-NTA; E1–3, Ni-NTA elution fractions.
As summarized in Fig. 10, the E. coli co-expression/co-purification approach confirmed four of the five new interactions identified in the present yeast two-hybrid screen (KREPA2-KREPA3, KREPA6-KREPA2, KREPA6-KREPA3, and KREPA3-KREPB5), along with five that had been reported previously (KREPA2-KREX2, KREPA2-KREL1, KREPA6-KREPA1, KREPA1-KRET2, and KREPA1-KREL2) (25). In six cases, this approach localized the interacting domains more precisely.
DISCUSSION
A combination of yeast two-hybrid screening and co-expression of recombinant proteins in E. coli was used to map protein-protein interactions among ten proteins in the T. brucei 20S editosome and to reconstitute binary and higher order complexes of editosome proteins. These data enabled the development of a map of the network of interactions among these proteins and, combined with data from an earlier yeast two-hybrid study that had used full-length proteins (25), are summarized in Fig. 10. New interactions were identified between five proteins, and numerous protein domains that are responsible for the interactions among the ten proteins were localized. The interactions identified among the KREPA family of proteins reveal a network of interactions that is essential for the structural integrity of the 20S editosome. Interestingly, the interactions in the “core” of the 20S editosome all appear to be mediated via OB-fold containing proteins, with the OB-folds from different KREPA proteins interacting with each other. Using the methods employed here none of the four enzymes nor KREPB4 and KREPB5 appear to interact mutually (Fig. 10).
KREPA3 and KREPA6 are key components in the interaction network and for 20S editosome structural integrity. Both can interact with multiple other partners in the complex and, in doing so, provide a physical linkage between the heterotrimeric deletion and insertion subcomplexes that were previously identified (25). KREPA6, the smallest of the KREPA proteins, can directly interact with the KREPA1 and KREPA2 zinc finger-OB-fold proteins that are the coordinators of the insertion and deletion subcomplex, respectively. KREPA6 can also directly interact with the KREPA3 and KREPA4 OB-fold proteins. This capacity for multiple interactions suggests a role for KREPA6 in bridging the deletion and insertion subcomplexes. This is consistent with functional studies of this protein in which knockdown of KREPA6 expression by RNA interference resulted in disruption of the 20S editosome and initial accumulation of ∼10S editosome fragments, followed by virtually complete disappearance of editosome proteins and cell death (31, 34). Similarly, KREPA3 also appears to have a central role in this interaction network. It can directly interact with KREPA2 and KREPA6, and again this crucial structural role is consistent with functional studies. Knockdown of KREPA3 in procyclic form T. brucei resulted in accumulation of smaller complexes that appeared to be associations of the deletion and insertion subcomplexes with other proteins (30), and a more extensive knockdown in bloodstream forms also resulted in the virtual disappearance of ∼20S editosomes (29). Furthermore, KREPA3 was shown here to also be able to directly interact with the RNase III-like protein KREPB5, which along with the related KREPB4 protein, has been suggested to function in association of the editosome endonucleases, KREN1, KREN2, and KREN3 (26). Hence, KREPA3 may therefore have a role in binding the endonuclease component to the editosome. Interestingly, knockdown of KREPA3 expression in procyclic T. brucei did not affect U-deletion or U-insertion activities but led to complete loss of endonucleolytic activities that could be partially restored by adding recombinant KREPA3 to cell extracts (29, 30). Thus, KREPA3 and KREPA6 together appear to provide key links for the integration of the three enzymatic subcomplexes responsible for endonucleolytic cleavage, U addition/ligation, and U removal/ligation in the core of the editosome.
KREPA4 has been shown to be essential for integrity of the ∼20S editosome (32), although it does not have as central a position in the interaction map as does KREPA6 with which it can interact (Fig. 10). One possibility is that KREPA4 may only form stable interactions with other editosome proteins in the presence of additional components and such interactions would have escaped detection in our yeast two-hybrid screen (see below). Another KREPA family protein, KREPA5, was only recently discovered and therefore not included in the library. Based on our findings, however, we would predict that KREPA5 also participates in the OB-fold interaction network and might be essential for editosome integrity.
A striking aspect of the interaction network formed by the KREPA family proteins is that these interactions are mediated by the putative OB-fold domains that are common to these proteins. OB-folds are defined by multiple β-strands that are coiled to form approximately a closed β-barrel and are often capped by an α-helix (44). OB-folds were originally identified as binding sites for oligonucleotides and oligosaccharides but have since been shown to be involved in various DNA and RNA processing pathways in which they function not only as nucleic acid-binding modules but also as mediators of protein-protein interactions (45). The KREPA family OB-folds appear to be most closely related to the single strand DNA-binding protein (SSB) OB-fold (24, 25). SSB proteins fall into two major groups. Eubacterial SSBs form homo-tetramers or -dimers, in which separate parts of the same OB-fold domain are involved in oligomerization and nucleic acid binding, respectively. Eukaryotic SSBs typically form hetero-oligomers with multiple OB-fold domains in which some OB-folds are responsible for nucleic acid binding while others mediate protein-protein interactions, often with multiple partners (45, 46). Recombinant KREPA6, KREPA3, and KREPA4 all have been shown to be able to bind RNA in vitro, with a preference for gRNA and oligo(U) sequences (32, 34, 47). The RNA-binding activity of KREPA3 was mapped to the OB-fold domain (47). In addition, there is evidence that the OB-fold domains of KREPA1 and KREPA2 provide substrate-binding platforms for the enzymatic activities in the deletion and insertion subcomplexes, respectively (25, 48). Thus, the multiple OB-fold domains in the editosome may perform the dual roles of interaction with RNA substrates and with other proteins.
Not all interactions among editosome proteins must necessarily be simultaneous. Indeed, the orderly processing of the mRNA/gRNA duplex could be orchestrated by a handing off of the RNA substrate from one OB-fold domain to another, similar to sequential DNA binding by the replication protein A complex (49). It could also entail a sequential series of interactions of one protein with multiple partner proteins that affect the overall structure of the 20S editosomes and perhaps the conformations of its component proteins. However, reconstitution experiments presented here suggest that editosome OB-fold domains have the capacity to simultaneously interact with more than one protein partner. KREPA6, KREPA3, and tagged KREPA2 OB-fold can form a trimeric complex (Fig. 7D). In addition KREPA6 was capable of linking the KREPA2·KREL1 and KREPA1· KRET2·KREL2 complexes into a heterohexameric complex (Fig. 9). The stoichiometries of the trimeric and hexameric complexes are uncertain but appear to contain higher molar amounts of A6. Hence it is unclear whether the same KREPA6 molecule interacted with both KREPA1 and KREPA2 or whether multiple KREPA6 molecules interacted with each other and with other proteins. Indeed, the protein stoichiometry of the 20S editosome itself is not yet elucidated, and a full understanding of the architecture and dynamics of this complex will depend on determining the precise number of each of its components. The estimated molecular weight of the ∼20S editosome as well as analysis of complexes that had been purified via tagged components are consistent with the presence of only a single molecule of each of the larger proteins (3, 20, 21, 25). It is conceivable, however, that smaller components are present in more than one copy. Indeed, several lines of evidence suggest that this might be the case at least for KREPA6: (i) the KREPA6 band in silver-stained 20S editosomes is usually more intense than its molecular weight would suggest (e.g. Fig. 1 in Ref. 20), (ii) recombinant KREPA6 tends to form homotetramers (Figs. 3A and 7B) reminiscent of E. coli SSB (50), and (iii) KREPA6 RNA-binding activity showed strong cooperativity (34).
In addition to providing an explanation how the deletion and insertion subcomplexes might be physically linked in the 20S editosome, we have also mapped interactions within these two subcomplexes more precisely. The deletion subcomplex contains two enzymes, the KREX2 U-specific 3′ exonuclease and the KREL1 RNA ligase, which interact with the KREPA2 zinc finger-OB-fold protein. Likewise, the insertion subcomplex contains the KRET2 TUTase and the KREL2 RNA ligase, both of which interact with the KREPA1 zinc finger-OB-fold protein. The yeast two-hybrid data have identified a region within the C-terminal domain of KREL1 (residues 335–463) as responsible for mediating interaction with KREPA2 (Fig. 2B). This was not unexpected, because the N-terminal two-thirds of KREL1 have been identified as the catalytic domain of the protein (Fig. 1A) (51, 52), and recombinant KREL1 with a C-terminal truncation failed to bind to KREL1-depleted editosomes (53). The corresponding interacting region in KREPA2 is located just C-terminal to the first zinc finger. This is consistent with co- immunoprecipitation experiments using recombinant protein, which showed that mutating the KREPA2 zinc fingers did not affect pulldown of KREL1 (data not shown). Interestingly, expression of LtKREPA2 with disrupted zinc fingers in Leishmania resulted in substantial breakdown of the 20S editosome (54), suggesting that these motifs do have a functional role, possibly in interaction with KREX2 or perhaps in proper folding of KREPA2 itself. The latter would be consistent with the finding that correct folding of recombinant KREPA3, which also contains two zinc finger motifs, required zinc ions (47). Expression of KREPA3 with mutated zinc fingers in T. brucei still resulted in incorporation into editosomes but failed to rescue the lethal effect of depleting endogenous KREPA3 (29). The corresponding protein in the insertion subcomplex, KREPA1, also contains two predicted zinc finger motifs (22, 24). Using truncated versions of recombinant KREPA1 we have identified a region of <100 amino acids surrounding the second zinc finger motif as sufficient for interaction with the KRET2 TUTase (Fig. 6B). Interestingly, this zinc finger motif has an unusual 22-amino acid insertion between the cysteine and histidine residues (105 amino acids in L. major), and its significance had therefore been pondered (22, 24). Mutational analyses may be useful for determining the significance of this C2H2 motif more definitively. We have not mapped the KREPA1-KREL2 interaction in more detail but, based on the analogous KREPA2-KREL1 interaction, it seems very likely that it involves the C-terminal domain of KREL2.
It remains to be investigated how the architecture of the core of the editosome described in this work can be integrated with the recently reported three-dimensional structures of 20S editosomes (55, 56). Golas et al. (56), using complexes purified via tagged KREPA3, reported a bipartite appearance in the majority of their structures, which might reflect the division into deletion and insertion subcomplexes. Similarly, Li et al. (55), studying Leishmania editosomes purified via tagged KREPB5, reported a quasi-4-fold symmetry in the central region of their structure. It is tempting to speculate that the open channels in the middle of this region might be surrounded by OB-fold domains, which orchestrate the movements of RNA molecules through the complex. Detailed mapping of the locations of individual components within these structures, combined with crystal structures as they become available (52, 57), will be useful. In the present analysis, eight of the 19 known protein components of 20S editosomes have been used as baits to screen the generated fragment libraries. Additional yeast two-hybrid studies should therefore prove useful. The reconstitution strategy described here represents an informative complementary approach.
The architecture of the 20S editosome is now beginning to emerge. The structural organization reported here nicely mirrors the functions of the component parts of the editosome and provides a prelude to detailed structure/function analysis of this fascinating complex. While elegant in its relative simplicity, it is uncertain if this static picture completely reflects the in vivo reality or if proteins and subunits of the complex dynamically associate and dissociate with each other during the editing process. Further studies on the structure and function of the 20S editosome complexes will be needed to unravel the architecture and dynamics of this multiprotein assembly.
Acknowledgments
We thank Troy Gilliam for excellent technical support and Elizabeth Craig (University of Wisconsin) for the gift of the pGAD and pGBD plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants GM042188 (to K. S.) and GM077418 (to W. H.). Research was conducted using equipment made possible by support from the Economic Development Administration, United States Department of Commerce, and the M. J. Murdock Charitable Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
C. Zelaya Soares and K. Stuart, unpublished observation.
- mRNA
- messenger RNA
- U
- uridylate
- gRNA
- guide RNA
- TUTase
- terminal uridylyltransferase
- OB
- oligonucleotide binding
- GAD
- Gal4 activation domain
- GBD
- Gal4 DNA-binding domain
- TEV
- tobacco etch virus
- Ni-NTA
- nickel-nitrilotriacetic acid
- SSB
- single strand DNA-binding protein.
REFERENCES
- 1.Lukes J., Hashimi H., Zíková A. (2005) Curr. Genet. 48, 277–299 [DOI] [PubMed] [Google Scholar]
- 2.Weng J., Aphasizheva I., Etheridge R. D., Huang L., Wang X., Falick A. M., Aphasizhev R. (2008) Mol. Cell 32, 198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stuart K. D., Schnaufer A., Ernst N. L., Panigrahi A. K. (2005) Trends Biochem. Sci. 30, 97–105 [DOI] [PubMed] [Google Scholar]
- 4.Simpson L., Aphasizhev R., Gao G., Kang X. (2004) RNA 10, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum B., Bakalara N., Simpson L. (1990) Cell 60, 189–198 [DOI] [PubMed] [Google Scholar]
- 6.Stuart K., Allen T. E., Heidmann S., Seiwert S. D. (1997) Microbiol. Mol. Biol. Rev. 61, 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aphasizhev R., Sbicego S., Peris M., Jang S. H., Aphasizheva I., Simpson A. M., Rivlin A., Simpson L. (2002) Cell 108, 637–648 [DOI] [PubMed] [Google Scholar]
- 8.Koslowsky D. J., Reifur L., Yu L. E., Chen W. (2004) RNA Biol. 1, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abraham J. M., Feagin J. E., Stuart K. (1988) Cell 55, 267–272 [DOI] [PubMed] [Google Scholar]
- 10.Maslov D. A., Simpson L. (1992) Cell 70, 459–467 [DOI] [PubMed] [Google Scholar]
- 11.Schumacher M. A., Karamooz E., Zíková A., Trantírek L., Lukes J. (2006) Cell 126, 701–711 [DOI] [PubMed] [Google Scholar]
- 12.Zíková A., Kopecná J., Schumacher M. A., Stuart K., Trantírek L., Lukes J. (2008) Int. J. Parasitol. 38, 901–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller U. F., Göringer H. U. (2002) Nucleic Acids Res. 30, 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ammerman M. L., Fisk J. C., Read L. K. (2008) RNA 14, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisk J. C., Presnyak V., Ammerman M. L., Read L. K. (2009) Mol. Cell. Biol. 29, 5214–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimi H., Zíková A., Panigrahi A. K., Stuart K. D., Lukes J. (2008) RNA 14, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acestor N., Panigrahi A. K., Carnes J., Zíková A., Stuart K. D. (2009) RNA 15, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisk J. C., Ammerman M. L., Presnyak V., Read L. K. (2008) J. Biol. Chem. 283, 23016–23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rusché L. N., Cruz-Reyes J., Piller K. J., Sollner-Webb B. (1997) EMBO J. 16, 4069–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panigrahi A. K., Ernst N. L., Domingo G. J., Fleck M., Salavati R., Stuart K. D. (2006) RNA 12, 1038–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aphasizhev R., Aphasizheva I., Nelson R. E., Gao G., Simpson A. M., Kang X., Falick A. M., Sbicego S., Simpson L. (2003) EMBO J. 22, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panigrahi A. K., Schnaufer A., Carmean N., Igo R. P., Jr., Gygi S. P., Ernst N. L., Palazzo S. S., Weston D. S., Aebersold R., Salavati R., Stuart K. D. (2001) Mol. Cell. Biol. 21, 6833–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panigrahi A. K., Gygi S. P., Ernst N. L., Igo R. P., Jr., Palazzo S. S., Schnaufer A., Weston D. S., Carmean N., Salavati R., Aebersold R., Stuart K. D. (2001) Mol. Cell. Biol. 21, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worthey E. A., Schnaufer A., Mian I. S., Stuart K., Salavati R. (2003) Nucleic Acids Res. 31, 6392–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnaufer A., Ernst N. L., Palazzo S. S., O'Rear J., Salavati R., Stuart K. (2003) Mol. Cell 12, 307–319 [DOI] [PubMed] [Google Scholar]
- 26.Carnes J., Trotter J. R., Peltan A., Fleck M., Stuart K. (2008) Mol. Cell. Biol. 28, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carnes J., Trotter J. R., Ernst N. L., Steinberg A., Stuart K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16614–16619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trotter J. R., Ernst N. L., Carnes J., Panicucci B., Stuart K. (2005) Mol. Cell 20, 403–412 [DOI] [PubMed] [Google Scholar]
- 29.Guo X., Ernst N. L., Stuart K. D. (2008) Mol. Cell. Biol. 28, 6939–6953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law J. A., O'Hearn S. F., Sollner-Webb B. (2008) RNA 14, 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law J. A., O'Hearn S., Sollner-Webb B. (2007) Mol. Cell. Biol. 27, 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salavati R., Ernst N. L., O'Rear J., Gilliam T., Tarun S., Jr., Stuart K. (2006) RNA 12, 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B., Ernst N. L., Palazzo S. S., Panigrahi A. K., Salavati R., Stuart K. (2003) Eukaryotic Cell 2, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarun S. Z., Jr., Schnaufer A., Ernst N. L., Proff R., Deng J., Hol W., Stuart K. (2008) RNA 14, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babbarwal V. K., Fleck M., Ernst N. L., Schnaufer A., Stuart K. (2007) RNA 13, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vondrusková E., van den Burg. J., Zíková A., Ernst N. L., Stuart K., Benne R., Lukes J. (2005) J. Biol. Chem. 280, 2429–2438 [DOI] [PubMed] [Google Scholar]
- 38.Missel A., Souza A. E., Nörskau G., Göringer H. U. (1997) Mol. Cell. Biol. 17, 4895–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelletier M., Read L. K. (2003) RNA 9, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madison-Antenucci S., Hajduk S. L. (2001) Mol. Cell 7, 879–886 [DOI] [PubMed] [Google Scholar]
- 41.Vanhamme L., Perez-Morga D., Marchal C., Speijer D., Lambert L., Geuskens M., Alexandre S., Ismaïli N., Göringer U., Benne R., Pays E. (1998) J. Biol. Chem. 273, 21825–21833 [DOI] [PubMed] [Google Scholar]
- 42.Aphasizhev R., Aphasizheva I., Simpson L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10617–10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gietz D., St. Jean A., Woods R. A., Schiestl R. H. (1992) Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bochkarev A., Bochkareva E. (2004) Curr. Opin. Struct. Biol. 14, 36–42 [DOI] [PubMed] [Google Scholar]
- 45.Richard D. J., Bolderson E., Khanna K. K. (2009) Crit. Rev. Biochem. Mol. Biol. 44, 98–116 [DOI] [PubMed] [Google Scholar]
- 46.Xu D., Guo R., Sobeck A., Bachrati C. Z., Yang J., Enomoto T., Brown G. W., Hoatlin M. E., Hickson I. D., Wang W. (2008) Genes Dev. 22, 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brecht M., Niemann M., Schlüter E., Müller U. F., Stuart K., Göringer H. U. (2005) Mol. Cell. 17, 621–630 [DOI] [PubMed] [Google Scholar]
- 48.Ernst N. L., Panicucci B., Carnes J., Stuart K. (2009) RNA 15, 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bochkareva E., Belegu V., Korolev S., Bochkarev A. (2001) EMBO J. 20, 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., Keck J. L. (2008) Crit. Rev. Biochem. Mol. Biol. 43, 289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnaufer A., Panigrahi A. K., Panicucci B., Igo R. P., Jr., Wirtz E., Salavati R., Stuart K. (2001) Science 291, 2159–2162 [DOI] [PubMed] [Google Scholar]
- 52.Deng J., Schnaufer A., Salavati R., Stuart K. D., Hol W. G. (2004) J. Mol. Biol. 343, 601–613 [DOI] [PubMed] [Google Scholar]
- 53.Gao G., Simpson A. M., Kang X., Rogers K., Nebohacova M., Li F., Simpson L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang X., Falick A. M., Nelson R. E., Gao G., Rogers K., Aphasizhev R., Simpson L. (2004) J. Biol. Chem. 279, 3893–3899 [DOI] [PubMed] [Google Scholar]
- 55.Li F., Ge P., Hui W. H., Atanasov I., Rogers K., Guo Q., Osato D., Falick A. M., Zhou Z. H., Simpson L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12306–12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Golas M. M., Böhm C., Sander B., Effenberger K., Brecht M., Stark H., Göringer H. U. (2009) EMBO J. 28, 766–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng J., Ernst N. L., Turley S., Stuart K. D., Hol W. G. (2005) EMBO J. 24, 4007–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]