Abstract
The Tal1 transcription factor is essential for the development of the hematopoietic system and plays a role during definitive erythropoiesis in the adult. Despite the importance of Tal1 in erythropoiesis, only a small number of erythroid differentiation target genes are known. A chromatin precipitation and cloning approach was established to uncover novel Tal1 target genes in erythropoiesis. The BirA tag/BirA ligase biotinylation system in combination with streptavidin chromatin precipitation (Strep-CP) was used to co-precipitate genomic DNA bound to Tal1. Tal1 was found to bind in the vicinity of 31 genes including the E2-ubiquitin conjugase UBE2H gene. Binding of Tal1 to UBE2H was confirmed by chromatin immunoprecipitation. UBE2H expression is increased during erythroid differentiation of hCD34+ cells. Tal1 expression activated UBE2H expression, whereas Tal1 knock-down reduced UBE2H expression and ubiquitin transfer activity. This study identifies parts of the ubiquitinylation machinery as a cellular target downstream of the transcription factor Tal1 and provides novel insights into Tal1-regulated erythropoiesis.
Keywords: Blood Erythropoeisis, Chromatin/Immunoprecipitation/ChIP, Development Differentiation/Hematopoietic, Diseases/Cancer/Leukemia, Gene/Transcription, Proteases/Ubiquitination, Transcription
Introduction
Differentiation of hematopoietic stem cells (HSC)2 to mature blood cells involves lineage-specific activation and restriction of gene expression that is controlled by a network of transcription factors (1–4). Deregulated expression of these transcription factors in inappropriate lineages contributes to the development of leukemia (5, 6). The Tal1 (translocated in leukemia 1, or stem cell leukemia 1, Scl1) transcription factor is a critical regulator of hematopoiesis and vasculogenesis (for a review see, Lecuyer et al. (7) or Barton et al. (8)). Tal1-deficient mice fail to establish the hematopoietic system and exhibit defects in vasculogenesis and angiogenesis (9–12). Tal1 is also expressed in dividing cells that form new vessels and becomes down-regulated in differentiated endothelial cells (13, 14). The notion that Tal1 is involved in proliferation control is supported by the fact that it is found ectopically activated in more than 60% of all cases of pediatric T-cell acute lymphoblastic leukemia (T-ALL) (15–17). Deletion of the Tal1 gene in the mouse showed that Tal1 plays a role in definitive erythroid and megakaryocytic differentiation (18, 19). However, the conditional knock-out of Tal1 did not inhibit ongoing adult erythropoiesis. This leads to the speculation that Tal1 function is compensated to some degree by other bHLH transcription factors such as Lyl1 (20). Knock-down of Tal1 expression using shRNA in human or mouse CD34+ cells confirmed a role of Tal1 in adult myeloid progenitors and HSCs and leads to a decrease of erythroid and myeloid cell production. The same study revealed in transplantation experiments with human CD34+ Tal1 knock-down cells in NOD-SCID mice that Tal1 is important for the commitment of HSC (21). Furthermore, shRNA-mediated knock-down of Tal1 in hCD34+ cells leads to an 8-fold decrease of erythroid colonies in a colony-forming assay (22).
Tal1 binds to regulatory elements of target genes at so called E-box sites (CANNTG) with its basic helix-loop-helix (bHLH) domain. Tal1 has also functions that do not require direct DNA binding (23). Tal1 can be found in large protein complexes that include the transcriptional regulators E2A, Ldb1, LMO2, and GATA1 (24) and cofactors, such as CBP/p300, P/CAF, or Sin3a, providing histone acetylase and/or histone deacetylase activity, respectively (25, 26). Heterodimer formation with E-box-binding proteins like E2A in T-cells, may lead to an interference with the functions of these transcription factors in leukemia (27, 28). Besides a direct role in transcriptional activation or silencing, Tal1 may also keep a genetic locus poised for further regulation, as has been suggested for the α-globin locus (29). Moreover, Tal1 can act in a repressive complex containing the histone lysine methyltransferase Suv39h1 (30) or in conjunction with Brg1, the central component of the SWI/SNF complex, to modulate transcription (31). These observations argue for a coordinating role of Tal1 in chromatin and gene regulatory networks. The recent finding that Tal1 binds to regulatory elements of a number of transcription factor genes supports the notion that Tal1 has an important position within developmental transcriptional networks (32).
To identify novel Tal1 binding sites in the genome and new target genes in the erythrocytic lineage, a biotin/streptavidin chromatin precipitation (Strep-CP) cloning protocol was developed. In this study, 31 potential target genes of Tal1 were identified. It is shown that Tal1 regulates the E2 ubiquitin-conjugating enzyme, UBE2H, during erythrocyte differentiation. Ubiquitinylation is closely associated with many important cellular processes (33) and plays a crucial role during erythroid cell maturation (34). The identification of Tal1 as a positive regulator of the ubiquitinylation machinery is to our knowledge the first report of Tal1 as a regulator of this central enzymatic function.
EXPERIMENTAL PROCEDURES
Cell Culture
K562 cells were maintained in RPMI supplemented with 10% fetal calf serum and 1% penicillin/streptomycin, and induced for differentiation with 2 mm sodium butyrate for 2 days. GP2–293 retroviral packaging cells were transiently transfected with pMSCVneo-BirA ligase. After 48 h, K562 erythroblast cells were infected with the viral supernatants. Stable clones were selected for neomycin resistance. BirA ligase expression was monitored with immunoblot using anti-BirA ligase antibodies (GenWay Biotech). A second round of infection and subsequent double selection with puromycin and neomycin-generated BirA ligase/Tal1-BirA tag clones. Expression of Tal1 and biotinylation of the fusion protein were detected by immunoblot using anti-Tal1 antibody (Santa Cruz Biotechnology, sc 12984) and by streptavidin-horseradish peroxidase conjugate (BD Biosciences). Transfections were performed with MetafecteneTM according to the manufacturer's instructions.
Human CD34+ bone marrow cells were obtained from Stem Cell Technologies, expanded in vitro under serum-free conditions, and subjected to erythroid differentiation as described (35). Expansion took place in StemSpan medium (Stem cell Technologies) supplemented with Flt-1 (100 ng/ml), SCF (100 ng/ml), IL-3 (20 ng/ml), and IL-6 (20 ng/ml). At day 6, the cells were transferred into StemSpan with SCF (20 ng/ml), IL-3 (5 ng/ml), dexamethasone (2 μm, Sigma), estradiol (0.2 μm, Sigma), and erythropoietin (1 unit/ml, Applichem) for erythroid differentiation. Cells were harvested at day 9 of induction.
Chromatin Immunoprecipitation
2 × 107 K562 cells were resuspended in 10 ml of fresh RPMI and cross-linked with 270 μl of 37% formaldehyde for 15 min. The cross-link was stopped by adding glycine to a final concentration of 0.125 m. The cells were washed with cold phosphate-buffered saline and with cell lysis buffer (10 mm Tris-Cl, pH 8, 10 mm NaCl, 0.2% Nonidet P-40, protease inhibitors). The cell pellet was resuspended in 400 μl of cell lysis buffer and incubated for 10 min on ice. The cells were spun down at 1000 rpm and resuspended in 1 ml of nucleic lysis buffer (50 mm NaCl, 10 mm EDTA, 1% SDS, protease inhibitors). Subsequently, the chromatin was sonicated using a bioruptor device (Diagenode, 15-ml tube, intensity H, interval 0.5) for 8 min resulting in chromatin fragments of 300–800 bp in length. The chromatin was centrifuged at full speed for 15 min, and the supernatant divided into different tubes in aliquots of 200 μl. 400 μl of dilution buffer (20 mm Tris-Cl pH8, 2 mm EDTA, 150 mm NaCl, 1% Triton X-100, protease inhibitors) was added to every tube. One sample was spared for the input control; to the others, 10 μl of magnetic protein-G beads (Dynabeads protein G-100, Invitrogen) were added, and the tubes were rotated at 4 °C for 2 h as a preclearing step. Afterward, the beads were spun down briefly and then subjected to a magnetic field. The supernatant was transferred to a fresh tube, 1 to 5 μg of antibody (Santa Cruz Biotechnology, sc 12984) was added, and the samples were incubated overnight at 4 °C on a rotating wheel. The next day, 10 μl of magnetic beads (for Strep-CP: Streptavidin Dynabeads M-280) were added to the lysates together with 100 μg/ml bovine serum albumin and 500 μg/ml tRNA and incubated for 3 h on a rotating wheel at 4 °C. The beads were captured using a magnetic rack, the supernatant was discarded, and the beads washed twice with IPwash1 (20 mm Tris-Cl, pH 8, 2 mm EDTA, 50 mm NaCl, 1% Triton-X 100, 0.1% SDS; in the case of Strep-CP, the beads were washed additionally twice with nucleic lysis buffer before IPwash1). Subsequently, the beads were washed twice with IPwash2 (10 mm Tris-Cl pH 8, 1 mm EDTA, 0.25 mm LiCl, 1% Nonidet P-40, 1% sodium deoxycholic acid) and twice with IPwash3 (20 mm Tris-Cl pH 7.6, 50 mm NaCl). Each wash includes incubation for 10 min on a rotational wheel; tubes were changed twice during the wash. The wash buffer was removed carefully, and the pellet resuspended in 150 μl of elution buffer (100 mm NaHCO3, 1% SDS) twice, the eluate was transferred to a new tube. For reversal of the cross-link, samples were adjusted to 300 mm NaCl and incubated at 67 °C overnight. (For Strep-CP the beads were suspended in elution buffer with 300 mm NaCl and incubated at 67 °C overnight with shaking.) Subsequently the DNA was precipitated, washed, and used for cloning or PCR.
Quantitative PCR
Quantitative PCR was performed using SYBR-green (Invitrogen) and a Light Cycler (Roche) according to the manufacturer's instructions. Synthesis of cDNA was performed with superscript reverse transcriptase (Invitrogen). Sequences of primer pairs used are available upon request.
Constructs
For knock-down experiments the small hairpin RNA expression vector psiRNA-h7SKGFPzeo (Invivogen) was employed. The following two regions in Tal1 were targeted by shRNA: 5′-GACAAGAAGCTCAGCAAGAAT-3′ and 5′-AAAGTTGTGCGGCGTATCTTC-3′. Transfected cells were isolated using fluorescence-activated cell sorting (FACS) and used for RNA isolation or selected by Zeocin (100 μg/ml) treatment to maintain stable expression of shRNAs. After selection, the cells were resorted by FACS to exclude cells with no or very low GFP signal and further cultivated in the presents of Zeocin.
The coding region of the Escherichia coli BirA biotin ligase gene (36) was PCR-amplified from genomic DNA. The fragment was cloned into the pMSCVneo retroviral expression vector (Clontech). The full-length human Tal1-coding region was cloned into the pBabe-puro retroviral expression vector. The stop codon was removed, and an oligonucleotide coding for the 23-amino acid biotinylation tag (BirA tag) (37) was inserted at the 3′-end of the Tal1-coding region.
Ubiquitin Transfer Assay
For measuring the cellular ubiquitin transfer, 20 μg of fresh cell extract was incubated with 1 μg of recombinant histone H2A (New England Biolabs) and 5 μg of purified Myc-tagged ubiquitin in a total volume of 20 μl of transfer buffer (20 mm Tris-Cl, pH 7.6, 20 mm KCl, 5 mm MgCl, 1 mm dithiothreitol, 10% glycerol, 5 mm ATP, protease inhibitors). The reaction was stopped by adding loading buffer and heating to 95 °C for 3 min. Ubiquitinylation was detected by immunoblot using anti-Myc antibody (9E10) and Alexa Fluor 680-coupled anti-mouse IgG (Molecular Probes). The membranes were scanned, and single lanes were quantified on a Odyssey system (LI-COR).
RESULTS
Identification of Potential Tal1 Target Genes
A stable K562 cell line harboring C-terminally BirA-tagged Tal1 and BirA ligase was established. The BirA ligase biotinylates the BirA-tagged Tal1 in vivo and permits the efficient isolation of the Tal1 protein by high affinity adsorption to streptavidin resin. These properties were employed in a chromatin precipitation assay analogous to the ChIP technique (Strep-CP). The Strep-CP-bound DNA was released, blunted, and amplified by ligation-mediated PCR. Amplified DNA was cloned, sequenced, and the genomic position of the Strep-CP DNA fragments was determined by data base analysis. From 151 analyzed clones, 43 hits were located within or adjacent to a gene. Some clones were found more than once, reducing the number of potential hits to 31 (Table 1). Most hits were located in the introns of genes; only UBE2H represented a gene with a hit in the 5′-region. In the case of CD7 and MAPK3K4, the cloned DNA was found 3′ of the gene. Almost all identified regions contained one or more E-box sequences (Table 1, lane E), except for C3AR, PTK2B, SDCCAG10, and STRAD. A large number of regions also contained GATA sites (Table 1, lane G), which sometimes can be found in conjunction with Tal1 binding sites. However, none of the regions was found to harbor an E-box/GATA combination, which was described as a typical Tal1 binding motif (24). Nevertheless, in a number of cases GATA and Tal1 sites were found within less than 50-bp distance from each other (Table 1, lane G/E), suggesting that Tal1 and GATA1 could be co-recruited at these sites as described for GATA1-activated genes (38).
TABLE 1.
Potential target genes of Tal1
| Gene ID | Symbol | Position ChIP | G | E | G/E | ||
|---|---|---|---|---|---|---|---|
| 1 | 109 | ADCY3 | chr2:24,930,644 | Intron | 2 | 1 | |
| 2 | 421 | ARVCF | chr22:18,350,757 | Intron | 2 | 4 | 1 |
| 3 | 2972 | BRF1 | chr14:104,763,411 | Intron | 2 | 4 | 1 |
| 4 | 50515 | CHST11 | chr12:103,670,754 | Intron | 1 | ||
| 5 | 924 | CD7 | chr17:77,864,557 | 3′ | 6 | ||
| 6 | 64072 | CDH23 | chr10:72,928,029 | Intron | 2 | 4 | |
| 7 | 719 | C3AR1 | chr12:8,105,009 | Intron | 2 | ||
| 8 | 1825 | DSC3 | chr18:26,872,574 | Intron | 1 | ||
| 9 | 115350 | FCRL1 | chr1:156,050,473 | Intron | 2 | ||
| 10 | 2200 | FBN1 | chr15:46,618,525 | Intron | 4 | 1 | 1 |
| 11 | 146713 | HRNBP3 | chr17:74,601,985 | Intron | 1 | ||
| 12 | 23185 | LARP5 | chr10:900,483 | Intron | 1 | ||
| 13 | 4580 | MTX1 | chr1:153,470,433 | Intron | 2 | ||
| 14 | 4216 | MAP3K4 | chr6:161,461,859 | 3′ | 1 | 2 | |
| 15 | 4643 | MYO1E | chr15:57,269,664 | Intron | 1 | 2 | |
| 16 | 84628 | NTNG2 | chr9:134,074,489 | Intron | 3 | ||
| 17 | 51594 | NAG | chr2:15,349,191 | Intron | 3 | 2 | 2 |
| 18 | 4842 | NOS1 | chr12:116,181,400 | Intron | 1 | 2 | 1 |
| 19 | 353238 | PADI6 | chr1:17,583,042 | Intron | 4 | 4 | 1 |
| 20 | 192111 | PGAM5 | chr12:131,803,298 | Intron | 2 | 5 | 1 |
| 21 | 5335 | PLCG1 | chr20:39,222,294 | Intron | 2 | 1 | 1 |
| 22 | 310674 | Plekho1 | chr1:148,394,131 | Intron | 1 | 1 | |
| 23 | 5530 | PPP3CA | chr4:102,283,984 | Intron | 1 | ||
| 24 | 2185 | PTK2B | chr8:27,305,895 | Intron | 1 | ||
| 25 | 10283 | SDCCAG10 | chr5:64,273,606 | Intron | 1 | ||
| 26 | 85464 | SSH2 | chr17:25,015,577 | Intron | 3 | 1 | |
| 27 | 92335 | STRAD | chr17:59,151,570 | Intron | 2 | ||
| 28 | 6814 | STXBP3 | chr1:109,154,566 | 3′ | 1 | 1 | |
| 29 | 7187 | TRAF3 | chr14:102,427,027 | Intron | 4 | 4 | 2 |
| 30 | 7328 | UBE2H | chr7:129,380,792 | 5′ | 3 | 2 | 1 |
| 31 | 9736 | USP34 | chr2:61,464,749 | Intron | 1 | 4 |
Candidate Tal1 Target Genes Are Mostly Membrane-associated or Cytoplasmic
The potential Tal1 target genes were grouped according to the localization of their products in the cell. Most of the Tal1 targets are described to be membrane-associated or present in the cytoplasm (Fig. 1A). This is in contrast to a recent study that found a large number of Tal1 target genes encoding for nuclear proteins (32). However, our results are in agreement with this study in the observation that a large number of Tal1 candidate target genes have catalytic activity or mediate protein modifications (Fig. 1B, DAVID Data Bank analysis (39)) and thus are involved in signaling. Several genes are implicated in regulation of the cytoskeleton or in calcium signaling, UBE2H and USP34 are involved in ubiquitinylation.
FIGURE 1.
Candidate target genes of Tal1. A, localization of the proteins within the cell. The largest number of Tal1 target genes encode for membrane-bound or membrane-associated proteins. Another fraction is cytoplasmatic, and only three genes are predominantly nuclear. For five genes, no data could be obtained concerning their intracellular localization. B, functional categorization of potential target genes. Most potential target genes exhibit catalytic activity, some may modify proteins, and a subset is involved in regulation of the cytoskeleton or calcium signaling. Two genes are involved in ubiquitinylation.
Influence of Tal1 on Candidate Target Gene Expression
To investigate which of the identified genes depend directly or indirectly on Tal1 for their expression, we stably knocked down Tal1 in K562 cells. The knock-down resulted in a reduction of Tal1 mRNA to less than 20% of its normal level and to a concomitant down-regulation of the known Tal1 target gene glycophorin A (GPA) (Fig. 2A, see Fig. 5C for Tal1 Western blot). Next, quantitative RT-PCR on all 31 genes was performed, and expression levels were compared with control shRNA-K562 cells. The expression of seven genes was down-regulated (Fig. 2A), and nine genes were up-regulated (Fig. 2B) in shTal1-K562 cells. The other potential Tal1 target genes did not display altered expression in shTal1-K562 cells or expression could not be detected (data not shown).
FIGURE 2.
Tal1 knock-down by shRNA reveals effects of Tal1 on candidate target genes. Stable shTal1-K562 cells were established, and the expression of candidate target genes was compared with shRNA control cells. Upon Tal1 knock-down genes were down-regulated (A) and up-regulated (B), respectively. Expression was determined by quantitative RT-PCR and calculated as fold induction (expression level of the shRNA-control-K562 cells was set as 1.0). Values were normalized to the expression level of the housekeeping gene GAPDH. Error bars represent the deviation from four evaluations.
FIGURE 5.
Tal1 regulates UBE2H expression. A, transient ectopic expression of Tal1 increases endogenous UBE2H expression. A Tal1-IRES-GFP vector was transfected transiently into K562 cells, GFP-positive cells were isolated, and the relative expression levels of GPA and UBE2H were determined by quantitative RT-PCR. Expression levels are shown as -fold induction compared with cells transfected with empty IRES-GPF-vector. B, transient down-regulation of Tal1 results in a decrease of endogenous UBE2H expression. A vector expressing GFP and shRNAs interfering with Tal1 expression were transfected into K562 cells. Transfected cells were isolated by GFP sorting 2 days after transfection. Expression levels were determined by quantitative RT-PCR. Two independent shRNAs reduced Tal1 expression. Tal1 knock-down also diminished GPA and UBE2H expression, but not G6PDH expression. The relative amount of transcripts is shown as fold induction compared with cells transfected with an sh-control vector only. All values were normalized to the GAPDH level and represent values from three independent determinations performed in duplicate. C, stable shTal1-K562 knock-down cells showed a reduced amount of Tal1 protein. D, chromatin immunoprecipitation shows diminished binding of Tal1 to the UBE2H locus in shTal1-K562 knock-down cells. ChIP with a sh-control cell line and with the Tal1 target gene GPA served as positive controls. ChIP with IgG and an αGFP antibody were used as negative controls. The experiments shown represent one of three independent ChIP assays performed. E, expression of the E2 ubiquitin conjugase UBE2H is reduced in the stable shTal1 knock-down cells. Expression of the E1 ligase UBE1 and the ubiquitin-specific peptidase USP5 remains unchanged. F, expression of E2 ubiquitin conjugases in stable shTal1-K562 knock-down cells. The expression of UBE2H, UBE2D2, and UBE2L3 was reduced by more than 50% in the Tal1 knock-down cells. Relative expression was determined by real-time PCR. Error bars represent variation from three independent determinations performed in duplicate.
Candidate Gene Expression upon Erythroid Differentiation of hCD34+ Cells
The expression of Tal1-dependent genes was analyzed in differentiating erythroid cells established from primary hCD34+ cells. To this aim, we employed a serum-free in vitro erythrocytic differentiation protocol of primary hCD34+ bone marrow cells established by the Weissman group (35). Human CD34+ bone marrow cells were expanded for 6 days and subjected to erythrocyte differentiation for 9 days. The harvested cell pellet showed a red color, and the cells were benzidine-positive indicating hemoglobinization. Expression of Tal1 was increased by five times in the differentiated cells (Fig. 3A). Concomitantly, the erythrocyte marker GPA was expressed more than 180 times higher and expression of the adult type α- and β-globin genes were increased 350 times compared with the uninduced expanded hCD34+ cells (Fig. 3B). We then analyzed the expression of Tal1-regulated genes upon erythroid differentiation. Five of seven genes that are potentially positively regulated by Tal1 in K562 cells are up-regulated in the differentiated primary CD34+ cells (Fig. 3C, left side). Additionally, four of nine genes that are negatively regulated by Tal1 in K562 cells, are down-regulated in the erythroid cells (Fig. 3C, right side). The strongest increase of expression in erythrocytic cells compared with uninduced cells was observed in the case of the UBE2H gene.
FIGURE 3.
Tal1 target genes expression during hCD34+ erythroid differentiation. Human bone marrow CD34+ cells were expanded for 6 days and subjected to erythroid differentiation in serum-free medium by erythropoietin treatment for 9 days. A, Tal1 expression was five times higher in the Epo-treated cells than in untreated controls. B, erythroid differentiation was accompanied by an increase of GPA, α-globin, and β-globin transcription. C, Tal1 target gene expression in differentiated erythroid cells. Relative expression values are given as fold induction compared with the expression level in uninduced control cells. Values were normalized to the expression level of the housekeeping gene GAPDH. Error bars represent the deviation from four evaluations. Nd, expression not detectable.
Analysis of the Tal1 Target UBE2H
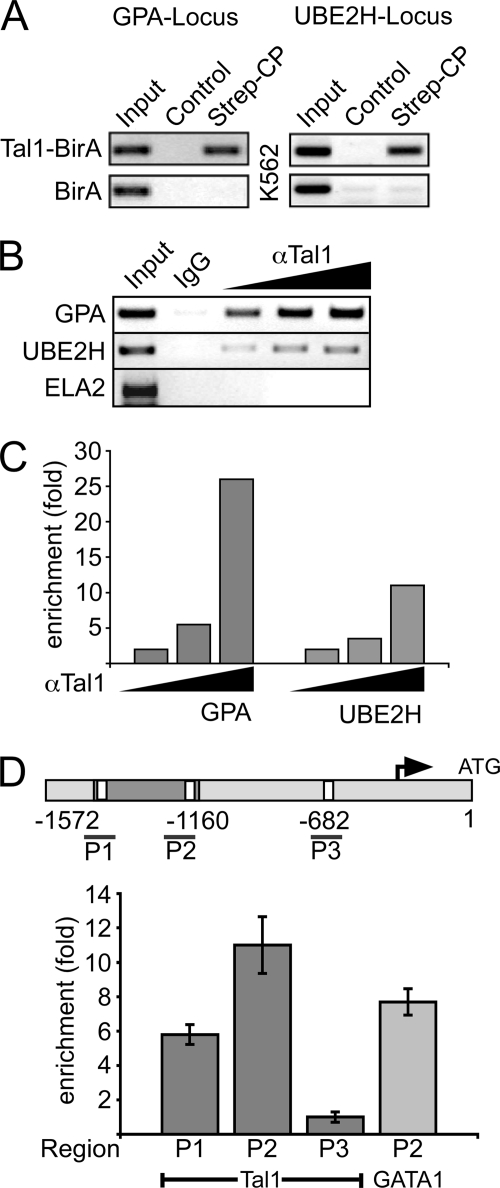
The UBE2H-associated ChIP-DNA was in the 5′ region (−1572 to −1160 relative to the ATG translational start codon) of the E2-ubiquitin conjugase gene UBE2H. Binding of Tal1 to this sequence could be verified by an independent Strep-CP using specific primers for the cloned UBE2H region (Fig. 4A). Strep-CP for the established Tal1 target gene glycophorin A (GPA) was performed as a positive control, and a K562 cell subclone that expresses only the BirA ligase but not the BirA-tagged Tal1 construct served as a negative control (Fig. 4A). Subsequently, binding of Tal1 to the UBE2H promoter was ascertained in wild-type K562 cells using a standard ChIP protocol with increasing amounts of Tal1-specific antibody (Fig. 4B). ChIP of GPA and neutrophile elastase 2 (ELA2) promoters served as positive and negative controls, respectively (Fig. 4B). ChIP-DNA was quantified by real-time PCR, confirming enrichment of the UBE2H promoter DNA in an antibody concentration-dependent manner (Fig. 4C).
FIGURE 4.
Tal1 binding to the UBE2H locus. A, Strep-CP using specific primers for the GPA locus (left) or the UBE2H locus (right) was performed. Cells that only expressed the BirA ligase but not the BirA-tagged Tal1 construct were used as negative controls (BirA). B, conventional ChIP assay and PCR of the UBE2H locus using wild-type K562 cells and increasing amounts of an anti-Tal1 antibody (1, 2.5, and 5 μg). ChIP-PCR reactions targeting the GPA locus and the neutrophile elastase 2 (ELA2) locus served as positive and negative controls, respectively. C, quantitative anti-Tal1 ChIP-PCR showing the enrichment (fold) compared with signals obtained with the negative control. D, Tal1 and GATA1 binding to the cloned region. Upper part, scheme of the 5′ region of the UBE2H gene. The cloned region is between −1572 and −1160 with respect to translational initiation. One primer pair (P1) covering the −1543 E-box site, a primer pair (P2) covering the −1171 E-box site, and third primer pair (P3) further downstream was used. Dark gray, UBE2H 5′ region cloned by Strep-CP. White boxes, E-box sites. Lower part, real-time PCR ChIP quantification. Best enrichment upon Tal1 ChIP was obtained with the primer pair P2 within the cloned region. GATA1 binding could also be detected at the 5′-UBE2H region. Values were obtained from two independent experiments, normalized to the input DNA (to correct for PCR efficiencies), and expressed as fold enrichment compared with the signal obtained with the isotype control.
Inspection of the cloned 5′ UBE2H region using TESS (transcription element search system) (40) revealed the presence of two potential Tal1 binding sites at position −1543 and position −1171 (Fig. 4D and supplemental Fig. S2). One additional E-box is located at position −682, between the cloned region and the ATG translational start codon. The E-box at position −1171 has an overlapping binding site for the transcription factor GATA, similar to a composite TAL1/GATA site that is present in the promoter of the GPA gene (41). The proximity of E-boxes and GATA sites is a feature frequently found in Tal1-regulated genes (42). Using the UCSC genome browser and the BLAST alignment tool (43, 44), it was found that the region with the E-boxes and the GATA site is evolutionary conserved in primates, but not in mice (supplemental Fig. S2).
Quantitative ChIP revealed that Tal1 binding can be detected with primers located at the −1543 site (P1), the strongest enrichment was achieved with primers covering the −1171 site (P2), whereas little if any specific binding of Tal1 to the E-box at −682 (P3 fragment) was found (Fig. 4D). Furthermore, GATA1 binding to the cloned region could be detected by ChIP using a GATA1-specific antibody and the P2 primer pair (Fig. 4D). We conclude that both Tal1 and GATA1 localize within the cloned region upstream of the endogenous UBE2H gene.
Tal1 Is an Activator of UBE2H Gene Expression
Both, Tal1 and the ubiquitin conjugase UBE2H are highly expressed in CD71+ early erythroid cells (supplemental Fig. S1) (45). Furthermore, a mass spectrometric approach recently identified a number of proteins of the ubiquitin pathway that are abundantly expressed in the erythrocyte proteome. Among these proteins were the E2-ubiquitin conjugase UBE2H, the E1 ligase UBE1 and the ubiquitin-specific protease USP5. In this study, the erythrocyte marker and Tal1 target gene GPA was also found (46). Because of their abundance in erythrocytes, UBE1 and USP5 were included as controls in further experiments. To examine if the Tal1 level directly affects UBE2H expression, the protein was transiently expressed from a Tal1-IRES-GFP vector in K562 cells. Tal1 expressing GFP-positive cells were enriched by fluorescence-activated cell sorting (FACS). Subsequently, endogenous UBE2H gene expression was determined by quantitative RT-PCR. Ectopic expression of Tal1 resulted in a 3-fold increase of the endogenous UBE2H transcript and a 7-fold increase of GPA transcript (Fig. 5A). Next, Tal1 expression was transiently knocked down in K562 cells by transfection of a vector expressing a Tal1-specific shRNA. An efficient knock-down of Tal1 was achieved with two different shRNAs, and a vector expressing a control shRNA did not change Tal1 expression (Fig. 5B). Expression of UBE2H and GPA was markedly reduced in transient Tal1 knock-down cells (Fig. 5B). Expression of glucose-6-phosphate dehydrogenase (G6PDH) remained unchanged. These results verify the data obtained from the stable Tal1 knock-down in K562 cells and confirmed that the observed UBE2H decrease was not due to establishing of the shTal1-K562 cells. In the stable knock-down the Tal1 protein was significantly reduced (Fig. 5C). Consequently, Tal1 was no longer detected by ChIP on the UBE2H or the GPA locus (Fig. 5D). The absence of Tal1 on the UBE2H and GPA promoters is in accordance with down-regulation of GPA and UBE2H expression (Figs. 5E and 2A). Expression of the ubiquitin-specific protease USP5 and the ubiquitin E1 ligase UBE1 remained unchanged. To examine if UBE2H was the only affected E2 ubiquitin conjugase in the stable shTal1 knock-down cells, we determined the expression of eight other E2 conjugases, which are present in CD71+ cells and K562 cells (UBE2R1, UBE2D1, UBE2D2, UBE2D3, UBE2E1, UBE2L3, UBE2E2, UBE2E3). In addition to UBE2H, the expression levels of UBE2D2 and UBE2L3 were diminished by more than 50% in the stable shTal1-K562 cell line (Fig. 5F). We also identified an E-box site within the UBE2L3 promoter and could detect Tal1 binding in this region by ChIP (supplemental Fig. S3) suggesting an involvement of Tal1 in expression regulation of UBE2L3. Taken together, these data show that Tal1 is a positive regulator of the E2-ubiquitin conjugase UBE2H in K562 cells and might also influence other E2 conjugases including UBE2L3.
UBE2H Expression and Overall Ubiquitinylation Activity Are Increased during Erythroid Differentiation in a Tal1-dependent Manner
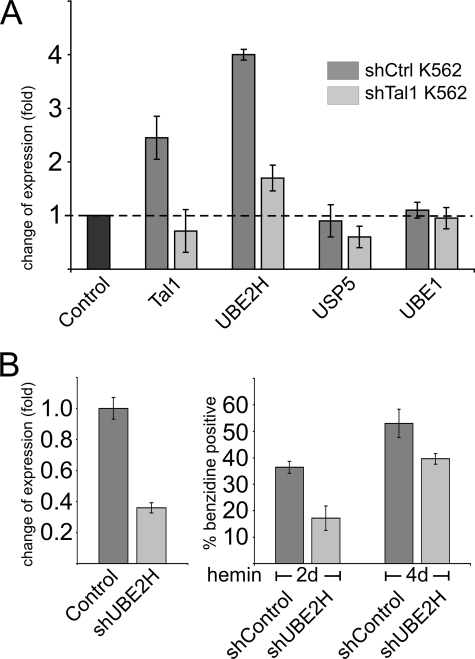
K562 cells can be differentiated toward the erythrocyte lineage by sodium butyrate or hemin treatment. Erythroid differentiation of K562 cells is accompanied by an increase in expression of the erythrocytic marker gene GPA (47) and an increase of hemoglobinized cells, which can be visualized by benzidine staining. To investigate a link between the expression level of Tal1 and UBE2H during erythrocyte differentiation, K562 were induced toward erythroid differentiation, and UBE2H expression was monitored by quantitative RT-PCR. Butyrate treatment up-regulated expression of Tal1 and UBE2H, whereas UBE1 and USP5 expression remained unchanged (Fig. 6A, dark gray). Up-regulation of Tal1 and UBE2H expression was substantially decreased in induced shTal1-K562 cells (Fig. 6A, light gray), suggesting a role of Tal1 in the induction of E2-conjugase expression in K562 cells. Hemin-induced erythropoiesis of K562 cells increases UBE2H expression ∼2.5-fold and gave rise to about 60% of benzidine-positive cells (not shown).
FIGURE 6.
Tal1-dependent up-regulation of UBE2H upon erythroid differentiation. A, induction of Tal1 and UBE2H in butyrate-induced control K562 cells and unaltered expression of Tal1 and UBE2H in shTal1-K562 knock-down cells. Cells were treated with 2 mm sodium butyrate for 2 days to induce erythroid differentiation. Expression levels were determined by quantitative RT-PCR. Values are expressed in fold induction compared with the expression level before induction (independent triplicates). Note that the absolute expression levels differ as shown in Fig. 5. B, influence of UBE2H on hemoglobin synthesis. Stable shUBE2H-K562 cells show a reduced UBE2H transcript level (left side). The shUBE2H-K562 cells show a lower number of benzidine-positive cells upon hemin (30 μm) induction (right side).
We generated stable shUBE2H-K562 cells with reduced UBE2H expression (Fig. 6B, left side). Upon hemin induction toward the erythrocyte lineage, the shUBE2H-K562 cells generated a smaller number of benzidine-positive cells at day 2 of induction, a difference that was less pronounced after 4 days of induction (Fig. 6B, right side). These data indicate that UBE2H may contribute to but does not determine hemin-induced hemoglobin synthesis.
As shown in Fig. 7A, ubiquitin-conjugating enzyme activity is up-regulated 1.7-fold in Tal1-positive cells during butyrate-stimulated erythroid differentiation, as measured by the amount of high molecular weight ubiquitin conjugates. Therefore, we tested the possibility that Tal1 modulates overall cellular ubiquitin transfer activity. A ubiquitin transfer assay was performed using K562 sh-control cells and shTal1-K562 knock-down cells. As shown in Fig. 7B, ubiquitin transfer activity in Tal1 knock-down cells is decreased. We speculate that this decrease is caused by down-regulation of E2 conjugases in the Tal1 knock-down, including UBE2H. Thus, we conclude that Tal1 is required for the increase of ubiquitin transfer activity during erythroid differentiation.
FIGURE 7.
Tal1-dependent up-regulation of ubiquitinylation upon erythroid differentiation. A, ubiquitin transfer activity after sodium butyrate-induced erythroid differentiation of K562 cells. Extracts from cells were incubated with histone H2A and Myc-tagged ubiquitin for 1 h. Immunoblotting with anti-Myc antibody was performed to detect ubiquitin conjugates and immunoblotting against tubulin was included as a loading control (not shown). B, decreased ubiquitin transfer activity of shTal1-K562 cells. Immunoblots were quantified on an Odyssey scanner system (LI-COR). Error bars represent the standard deviation from at least two independent determinations. Stars (*) indicate free ubiquitin molecules.
DISCUSSION
This study identifies potential target genes of the hematopoietic transcription factor Tal1 in erythroid cells. We show that UBE2H, a member of the ubiquitinylation machinery, is a downstream target of the transcription factor Tal1. To the best of our knowledge, this is the first report linking Tal1 to the regulation of ubiquitinylation in erythropoiesis.
Identification of Tal1 Target Genes
The identification of Tal1 target genes is essential for the understanding of how Tal1 functions during differentiation and how it interferes with normal development in leukemia. Despite the important properties of Tal1 in development, differentiation, and leukemic transformation, little is known about the downstream cellular mechanisms controlled by Tal1 during erythropoiesis. A recent advance in the understanding of Tal1 biology is the finding that transcription factors including Runx1 are Tal1 target genes (32, 48). Runx1 is a central player in hematopoiesis and acts together with Tal1 in a transcriptional network that determines development of the embryonic mesoderm into hemangioblast tissue (49). It is important to identify and to examine Tal1 target genes to unravel its developmental and leukemogenic functions.
In this study, we identified novel target sites of Tal1 in the genome by precipitation of tagged Tal1 and direct cloning of bound DNA. Most of the identified sequences have intergenic locations without obvious connections to distinct genes. 31 sequences were found associated with coding regions, mostly within introns. This observation is in accordance with other studies, which show that a large fraction of transcription factor binding sites is not located in the immediate 5′ region of genes (50).
Sixteen of the identified genes are directly or indirectly regulated by Tal1 in K562 cells. Of these sixteen genes, fourteen genes changed their expression during in vitro erythrocyte differentiation of primary hCD34+ and thus are candidate Tal1 target genes in erythropoiesis. Nine genes changed their expression during differentiation as expected from the Tal1 knock-down in K562 cells. CDH23 was also identified as a Tal1 target gene in early embryonic hematopoietic development (32). The same study reported members of the focal adhesion kinase (fak) pathway as targets of Tal1. In our study we identified PTK2B (fak2, an important member of the fak kinase family) as a Tal1 target gene, which highlights the notion that Tal1 regulates this pathway. Another potential Tal1 target that was also identified in T-cells by others is TRAF3 (51), a TNF receptor-associated factor. However, we could not verify a dependence of TRAF3 expression on the presence of Tal1 in our system.
BRF1, CHST11, FBN1, and Plekho1 were markedly down-regulated in erythroid cells compared with the expanded hCD34+ control. These genes may have a function in progenitor cells and may not be required in more mature cells. This makes them interesting candidates for stem cell or progenitor-specific Tal1-regulated genes.
In the group of up-regulated genes during differentiation, the peak expression of the E2-ubiquitin-conjugating enzyme UBE2H coincides with high Tal1 expression in CD71+ early erythroid cells (supplemental Fig. S1). Our observation that UBE2H is strongly induced during erythrocyte differentiation of human CD34+ primary cells is also in line with previous results (52). It was also demonstrated that UBE2H expression is up-regulated during differentiation of erythroblasts to reticulocytes and reduced during terminal differentiation stages (53, 54). A similar expression pattern was described for Tal1, which is highly expressed in early erythrocyte stages and down-regulated during later stages of maturation (55, 56). These observations may suggest a role of E2-conjugases and specifically UBE2H during late erythrocyte differentiation and a regulation by the hematopoietic transcription factor Tal1.
Ubiquitinylation as a Tal1-regulated Process
Up to now, more than 50 E2-ubiquitin-conjugating enzymes and hundreds of E3 ligases have been identified (33, 57). Ubiquitinylation is involved in multiple cellular functions and is required for enhanced degradation of proteins during erythrocyte maturation (58). The importance of ubiquitinylation-mediated protein degradation during development is underlined by the observation that proteasome inhibitors decrease the ability of erythroid precursor cells to undergo late differentiation processes such as enucleation (59). The idea that Tal1 modulates erythroid maturation via ubiquitinylation is supported by our observation that Tal1 knock-down cells exhibit reduced ubiquitin transfer activity. Furthermore, butyrate-induced erythroid differentiation leads to Tal1-dependent up-regulation of UBE2H and a concomitant increase in overall ubiquitin transfer activity. Thus, Tal1 expression, erythroid differentiation, and up-regulation of cellular ubiquitin transfer activity are linked. UBE2H is also not the only E2-conjugase with decreased expression in shTal1 knock-down cells. The observation that Tal1 also binds to the UBE2L3 promoter hints to a direct involvement of Tal1 in the expression control of ubiquitinylation components. However, if Tal1 knock-down influences other E2-conjugases directly or if the ubiquitinylation machinery is altered due to secondary effects remains to be determined.
In the hematopoietic stem cell and in hematopoietic lineage commitment ubiquitinylation plays a role in the regulation of several important stem cell factors and commitment factors, including c-kit, cyclin D, Bcl-2, NFκB, Runx1, GATA1, C/EBPα, HoxA9, Gfi-1, STAT3, p21 (for a review, see Marteijn et al. (60)), and also Tal1 (61). UBE2H was first identified as a homologue to yeast Ubc8 with 54% amino acid sequence identity (62). It has been reported that UBE2H expression is a limiting factor for proteosomal degradation activity in the muscle (63). UBE2H is able to transfer ubiquitin moieties to histones in vitro (64) and UBE2H exhibits a nuclear localization when overexpressed in HEK293 cells (not shown). Interestingly, histone H2A ubiquitinylation is increased during erythrocyte development (65). These observations may suggest that UBE2H plays a role in the regulation of chromatin during late stages of erythrocyte differentiation. Our finding that the increase of benzidine-positive K562 cells during erythroid differentiation is delayed in UBE2H knock-down cells also supports the notion that UBE2H is involved in erythrocyte maturation. The specific role of UBE2H in the hematopoietic system should be further analyzed.
Tal1, Ubiquitinylation, and Leukemia
A role of E3-ubiquitin ligases within large protein complexes such as in SCF (SKP1-Cul1-F-box-protein) or in APC/C (anaphase-promoting complex/cyclosome) in cell cycle control and cancer is well documented (66). Recently, it was shown that the ubiquitin E3 ligase Cul4A is important for stem cell engraftment and self-renewal (67). Interestingly, Cul4A was also identified as a potential target gene for Tal1 in T-ALL (51). A link between an E2-conjugase and cancer has been demonstrated for UBCH10, which plays a role in cell cycle control and can serve as a tumor marker (68, 69). This raises the question whether deregulation of ubiquitinylation by Tal1-UBE2H could also be important for cancer. Recently, UBE2H has been identified as a strong candidate oncogene for hepatocellular carcinoma. It was shown that UBE2H-expressing cells have an increased proliferation rate (70). However, analysis of the dataset collected by Ferrando et al. (16) using the Oncomine data base (71) revealed that UBE2H is down-regulated in 10 of 14 Tal1-expressing T-ALL samples. It is therefore possible that an influence of Tal1 on UBE2H expression might have different outcomes, depending on the cell type. Examination of a link between misexpression of Tal1 and deregulation of components of the ubiquitinylation machinery, such as UBE2H, may help to elucidate mechanisms underlying leukemic conversion and to uncover novel therapeutic routes.
Acknowledgments
We thank Hans-Peter Rahn for assisting with FACS analysis. Thomas Sommer (MDC) kindly provided expression vectors for Myc-tagged ubiquitin.
This work was supported by Grant LA 1389/3-1 from the Deutsche Forschungsgemeinschaft.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- HSC
- hematopoietic stem cell
- shRNA
- short hairpin RNA
- ChIP
- chromatin immunoprecipitation assay
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GPA
- glycophorin A.
REFERENCES
- 1.Orkin S. H. (2000) Nat. Rev. Genet. 1, 57–64 [DOI] [PubMed] [Google Scholar]
- 2.Cantor A. B., Orkin S. H. (2002) Oncogene 21, 3368–3376 [DOI] [PubMed] [Google Scholar]
- 3.Gottgens B. (2004) Vox Sang 87, Suppl. 1, 15–19 [DOI] [PubMed] [Google Scholar]
- 4.Smale S. T. (2003) Nat. Immunol. 4, 607–615 [DOI] [PubMed] [Google Scholar]
- 5.Passegué E., Jamieson C. H., Ailles L. E., Weissman I. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, Suppl. 1, 11842–11849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hope K. J., Jin L., Dick J. E. (2004) Nat. Immunol. 5, 738–743 [DOI] [PubMed] [Google Scholar]
- 7.Lécuyer E., Hoang T. (2004) Exp. Hematol. 32, 11–24 [DOI] [PubMed] [Google Scholar]
- 8.Barton L. M., Gottgens B., Green A. R. (1999) Int. J. Biochem. Cell Biol. 31, 1193. [DOI] [PubMed] [Google Scholar]
- 9.Shivdasani R. A., Mayer E. L., Orkin S. H. (1995) Nature 373, 432–434 [DOI] [PubMed] [Google Scholar]
- 10.Robb L., Elwood N. J., Elefanty A. G., Köntgen F., Li R., Barnett L. D., Begley C. G. (1996) EMBO J. 15, 4123–4129 [PMC free article] [PubMed] [Google Scholar]
- 11.Visvader J. E., Fujiwara Y., Orkin S. H. (1998) Genes Dev. 12, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porcher C., Liao E. C., Fujiwara Y., Zon L. I., Orkin S. H. (1999) Development 126, 4603–4615 [DOI] [PubMed] [Google Scholar]
- 13.Kallianpur A. R., Jordan J. E., Brandt S. J. (1994) Blood 83, 1200–1208 [PubMed] [Google Scholar]
- 14.Pulford K., Lecointe N., Leroy-Viard K., Jones M., Mathieu-Mahul D., Mason D. Y. (1995) Blood 85, 675–684 [PubMed] [Google Scholar]
- 15.Bash R. O., Hall S., Timmons C. F., Crist W. M., Amylon M., Smith R. G., Baer R. (1995) Blood 86, 666–676 [PubMed] [Google Scholar]
- 16.Ferrando A. A., Herblot S., Palomero T., Hansen M., Hoang T., Fox E. A., Look A. T. (2004) Blood 103, 1909–1911 [DOI] [PubMed] [Google Scholar]
- 17.Look A. T. (1997) Science 278, 1059–1064 [DOI] [PubMed] [Google Scholar]
- 18.Hall M. A., Curtis D. J., Metcalf D., Elefanty A. G., Sourris K., Robb L., Gothert J. R., Jane S. M., Begley C. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikkola H. K., Klintman J., Yang H., Hock H., Schlaeger T. M., Fujiwara Y., Orkin S. H. (2003) Nature 421, 547–551 [DOI] [PubMed] [Google Scholar]
- 20.Hall M. A., Slater N. J., Begley C. G., Salmon J. M., Van Stekelenburg L. J., McCormack M. P., Jane S. M., Curtis D. J. (2005) Mol. Cell Biol. 25, 6355–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunet de la Grange P., Armstrong F., Duval V., Rouyez M. C., Goardon N., Romeo P. H., Pflumio F. (2006) Blood 108, 2998–3004 [DOI] [PubMed] [Google Scholar]
- 22.Brunet de la Grange P., Zink E., Armstrong F., Rouyez M. C., Pflumio F. (2008) Stem Cells 26, 1658–1662 [DOI] [PubMed] [Google Scholar]
- 23.Kassouf M. T., Chagraoui H., Vyas P., Porcher C. (2008) Blood 112, 1056–1067 [DOI] [PubMed] [Google Scholar]
- 24.Wadman I. A., Osada H., Grütz G. G., Agulnick A. D., Westphal H., Forster A., Rabbitts T. H. (1997) EMBO J. 16, 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S., Qiu Y., Shi Y., Xu Z., Brandt S. J. (2000) EMBO J. 19, 6792–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S., Brandt S. J. (2000) Mol. Cell Biol. 20, 2248–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S. T., Sun X. H. (1998) J. Biol. Chem. 273, 7030–7037 [DOI] [PubMed] [Google Scholar]
- 28.O'Neil J., Shank J., Cusson N., Murre C., Kelliher M. (2004) Cancer Cell 5, 587–596 [DOI] [PubMed] [Google Scholar]
- 29.Anguita E., Hughes J., Heyworth C., Blobel G. A., Wood W. G., Higgs D. R. (2004) EMBO J. 23, 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen J., Huang S., Pack S. D., Yu X., Brandt S. J., Noguchi C. T. (2005) J. Biol. Chem. 280, 12956–12966 [DOI] [PubMed] [Google Scholar]
- 31.Xu Z., Meng X., Cai Y., Koury M. J., Brandt S. J. (2006) Biochem. J. 399, 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson N. K., Miranda-Saavedra D., Kinston S., Bonadies N., Foster S. D., Calero-Nieto F., Dawson M. A., Donaldson I. J., Dumon S., Frampton J., Janky R., Sun X. H., Teichmann S. A., Bannister A. J., Gottgens B. (2009) Blood 113, 5456–5465 [DOI] [PubMed] [Google Scholar]
- 33.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 34.Haas A. L. (1991) Adv. Exp. Med. Biol. 307, 191–205 [DOI] [PubMed] [Google Scholar]
- 35.Mahajan M. C., Karmakar S., Krause D., Weissman S. M. (2009) Exp. Hematol. 37, 1143–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard P. K., Shaw J., Otsuka A. J. (1985) Gene 35, 321–331 [DOI] [PubMed] [Google Scholar]
- 37.Schatz P. J. (1993) Biotechnology 11, 1138–1143 [DOI] [PubMed] [Google Scholar]
- 38.Tripic T., Deng W., Cheng Y., Zhang Y., Vakoc C. R., Gregory G. D., Hardison R. C., Blobel G. A. (2009) Blood 113, 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Gen. Biol. 4, P3. [PubMed] [Google Scholar]
- 40.Schug J. (2008) in Current Protocols in Bioinformatics (Baxevanis A. D. ed) Chapter 2, unit 2.6–2.6.15 [Google Scholar]
- 41.Lahlil R., Lécuyer E., Herblot S., Hoang T. (2004) Mol. Cell Biol. 24, 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swiers G., Patient R., Loose M. (2006) Dev. Biol. 294, 525–540 [DOI] [PubMed] [Google Scholar]
- 43.Kent W. J. (2002) Genome Res. 12, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhn R. M., Karolchik D., Zweig A. S., Wang T., Smith K. E., Rosenbloom K. R., Rhead B., Raney B. J., Pohl A., Pheasant M., Meyer L., Hsu F., Hinrichs A. S., Harte R. A., Giardine B., Fujita P., Diekhans M., Dreszer T., Clawson H., Barber G. P., Haussler D., Kent W. J. (2009) Nucleic Acids Res. 37, D755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su A. I., Cooke M. P., Ching K. A., Hakak Y., Walker J. R., Wiltshire T., Orth A. P., Vega R. G., Sapinoso L. M., Moqrich A., Patapoutian A., Hampton G. M., Schultz P. G., Hogenesch J. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kakhniashvili D. G., Bulla L. A., Jr., Goodman S. R. (2004) Mol. Cell Proteomics 3, 501–509 [DOI] [PubMed] [Google Scholar]
- 47.Grebenová D., Kuzelová K., Pluskalová M., Peslová G., Halada P., Hrkal Z. (2006) Blood Cells Mol. Dis. 37, 210–217 [DOI] [PubMed] [Google Scholar]
- 48.Landry J. R., Kinston S., Knezevic K., de Bruijn M. F., Wilson N., Nottingham W. T., Peitz M., Edenhofer F., Pimanda J. E., Ottersbach K., Göttgens B. (2008) Blood 111, 3005–3014 [DOI] [PubMed] [Google Scholar]
- 49.Ema M., Rossant J. (2003) Trends Cardiovasc. Med. 13, 254–259 [DOI] [PubMed] [Google Scholar]
- 50.Jothi R., Cuddapah S., Barski A., Cui K., Zhao K. (2008) Nucleic Acids Res. 36, 5221–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palomero T., Odom D. T., O'Neil J., Ferrando A. A., Margolin A., Neuberg D. S., Winter S. S., Larson R. S., Li W., Liu X. S., Young R. A., Look A. T. (2006) Blood 108, 986–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keller M. A., Addya S., Vadigepalli R., Banini B., Delgrosso K., Huang H., Surrey S. (2006) Physiol. Genom. 28, 114–128 [DOI] [PubMed] [Google Scholar]
- 53.Wefes I., Mastrandrea L. D., Haldeman M., Koury S. T., Tamburlin J., Pickart C. M., Finley D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4982–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haldeman M. T., Finley D., Pickart C. M. (1995) J. Biol. Chem. 270, 9507–9516 [DOI] [PubMed] [Google Scholar]
- 55.Hoang T., Paradis E., Brady G., Billia F., Nakahara K., Iscove N. N., Kirsch I. R. (1996) Blood 87, 102–111 [PubMed] [Google Scholar]
- 56.Brady G., Billia F., Knox J., Hoang T., Kirsch I. R., Voura E. B., Hawley R. G., Cumming R., Buchwald M., Siminovitch K. (1995) Curr. Biol. 5, 909–922 [DOI] [PubMed] [Google Scholar]
- 57.Willis M. S., Patterson C. (2006) J. Mol. Cell Cardiol. 41, 567–579 [DOI] [PubMed] [Google Scholar]
- 58.Boches F. S., Goldberg A. L. (1982) Science 215, 978–980 [DOI] [PubMed] [Google Scholar]
- 59.Chen C. Y., Pajak L., Tamburlin J., Bofinger D., Koury S. T. (2002) Exp. Hematol. 30, 634–639 [DOI] [PubMed] [Google Scholar]
- 60.Marteijn J. A., Jansen J. H., van der Reijden B. A. (2006) Leukemia 20, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 61.Tang T., Arbiser J. L., Brandt S. J. (2002) J. Biol. Chem. 277, 18365–18372 [DOI] [PubMed] [Google Scholar]
- 62.Kaiser P., Seufert W., Höfferer L., Kofler B., Sachsenmaier C., Herzog H., Jentsch S., Schweiger M., Schneider R. (1994) J. Biol. Chem. 269, 8797–8802 [PubMed] [Google Scholar]
- 63.Li Y. P., Lecker S. H., Chen Y., Waddell I. D., Goldberg A. L., Reid M. B. (2003) Faseb J. 17, 1048–1057 [DOI] [PubMed] [Google Scholar]
- 64.Kaiser P., Mandl S., Schweiger M., Schneider R. (1995) FEBS Lett. 377, 193–196 [DOI] [PubMed] [Google Scholar]
- 65.Hensold J. O., Swerdlow P. S., Housman D. E. (1988) Blood 71, 1153–1156 [PubMed] [Google Scholar]
- 66.Nakayama K. I., Nakayama K. (2006) Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 67.Li B., Jia N., Waning D. L., Yang F. C., Haneline L. S., Chun K. T. (2007) Blood 110, 2704–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okamoto Y., Ozaki T., Miyazaki K., Aoyama M., Miyazaki M., Nakagawara A. (2003) Cancer Res. 63, 4167–4173 [PubMed] [Google Scholar]
- 69.Rape M., Kirschner M. W. (2004) Nature 432, 588–595 [DOI] [PubMed] [Google Scholar]
- 70.Keng V. W., Villanueva A., Chiang D. Y., Dupuy A. J., Ryan B. J., Matise I., Silverstein K. A., Sarver A., Starr T. K., Akagi K., Tessarollo L., Collier L. S., Powers S., Lowe S. W., Jenkins N. A., Copeland N. G., Llovet J. M., Largaespada D. A. (2009) Nat. Biotech. 27, 264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. (2004) Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]