Abstract
The p75 neurotrophin receptor, a member of the tumor necrosis factor superfamily of receptors, undergoes an α-secretase-mediated release of its extracellular domain, followed by a γ-secretase-mediated intramembrane cleavage. Like amyloid precursor protein and Notch, γ-secretase cleavage of the p75 receptor releases an intracellular domain (ICD). However, it has been experimentally challenging to determine the precise subcellular localization and functional consequences of the p75 ICD. Here, we utilized a nuclear translocation assay and biochemical fractionation approaches to follow the fate of the ICD. We found that the p75 ICD can translocate to the nucleus to activate a green fluorescent protein reporter gene. Furthermore, the p75 ICD was localized in nuclear fractions. Chromatin immunoprecipitation experiments indicated that nerve growth factor induced the association of endogenous p75 with the cyclin E1 promoter. Expression of the p75 ICD resulted in modulation of gene expression from this locus. These results suggest that the p75 ICD generated by γ-secretase cleavage is capable of modulating transcriptional events in the nucleus.
Keywords: Chromatin/Immunoprecipitation/ChIP, Proteases/Secretases, Protein/Nuclear Translocation, Protein/Processing, Receptors/Membrane, Neurotrophin, γ-Secretase, p75NTR
Introduction
The p75 neurotrophin receptor is the founding member of the tumor necrosis factor receptor superfamily that includes the Fas antigen, DR6, CD30, and CD40. This family of receptors is distinguished with multiple cysteine-rich domains for ligand binding, a single transmembrane sequence, and a noncatalytic cytoplasmic domain (1). The intracellular region of the p55 tumor necrosis factor receptor, Fas receptor, and p75 contains a death domain sequence (2). The death domain serves as a protein-protein docking site and is required for initiating tumor necrosis factor- and Fas-mediated apoptosis (3).
The p75 receptor is recognized by all the neurotrophins (NGF,3 brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4), which promote differentiation, growth, and survival of diverse cell types in the nervous system (4–6). Neurotrophins also initiate signaling through Trk tyrosine kinase receptors, which are capable of forming high affinity binding sites with p75 to potentiate responses at low concentrations of neurotrophins (7). The precursor form of neurotrophins (proneurotrophins) binds more avidly to p75 than the mature form (8, 9). In the absence of Trk receptors, p75 is capable of independent signaling that activates NF-κB, c-Jun N-terminal kinase (JNK), and the sphingomyelin cycle (10). In selected cell types, p75 can initiate cell death (11–13). Alternatively, p75 can serve as a co-receptor for several proteins that modulate axon outgrowth, such as Nogo, neuropilin-1, and plexin-A4 (14–16). Recently, p75 interaction with ephrin A has been shown to direct targeting of retinal ganglion cells during development (17).
Intramembrane cleavage events have been detected for p75 in many cell types (18, 19). Proteolysis through presenilin-dependent γ-secretase activity has emerged as a highly conserved and prevalent mechanism in receptor signaling responsible for the intramembrane cleavage of important proteins, such as Notch, ErbB4 tyrosine kinase receptors, CD44, low density lipoprotein, and β-amyloid precursor protein (20).
Inhibition of γ-secretase cleavage of p75 has been shown to prevent apoptosis (21). The receptor proteolysis was observed in vivo during naturally occurring cell death in the superior cervical ganglia. Moreover, overexpression of the p75 ICD resulted in apoptosis (22). These results indicate that p75-mediated apoptosis requires γ-secretase-dependent release of its ICD. Also, cleavage of p75 is required for inhibition of neurite outgrowth by myelin-associated glycoprotein with its receptor, Nogo receptor (23, 24). A complex between p75 and the Nogo receptor has been proposed to account for the ability of p75 to inhibit axonal regeneration through the action of the p75-interacting protein RhoA. Furthermore, myelin-associated glycoprotein binding to primary neurons induces proteolytic processing of p75 to produce the p75 ICD. Release of RhoA from p75 is involved in its activation, suggesting that the state of the p75 cytoplasmic domain is an important regulatory element.
Whether the p75 ICD is directed to the nucleus has been very difficult to determine due to the instability and the exceedingly low levels of the ICD fragment (19). Although several reports have indicated that the ICD of p75 can be found in the nucleus (25, 26), it is unclear what biological activities are displayed by the ICD. In this study, we have employed several biochemical approaches and a reporter gene assay to monitor the fate and potential role of the p75 ICD fragment.
EXPERIMENTAL PROCEDURES
Materials
Compound E, the γ-secretase inhibitor XVIII (catalog no. 565771), leptomycin B, and clasto-lactacystin β-lactone were obtained from Calbiochem. Thymidine was obtained from Sigma, and nocodazole from Tocris. Antibodies against acetylated histone H3K9 and RhoGDI were purchased from Upstate Biotechnology. A rabbit polyclonal antibody against the ICD of p75 (antiserum 9992) and its preimmune fraction were generated as described previously (27). Normal rabbit IgG (sc-2027) used as a negative control was from Santa Cruz Biotechnology. Anti-TAF135 antibody was purchased from BD Biosciences.
Plasmids
The LexA-d1EGFP reporter gene was described previously (28). A PCR strategy was used to fuse the LexA DNA-binding domain (amino acids 1–87)-Gal4 transactivation domain (amino acids 768–881) cassette to the C terminus of the p75 and Fas receptors. The resulting constructs were subcloned into pcDNA3.1(+) using the KpnI and NotI sites. The plasmid encoding the rat p75 ICD was generated as described previously (35). All constructs were verified by automated DNA sequencing.
Cell Culture Treatments and Transfection
HEK293 and HeLa cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum or 5% fetal calf serum and penicillin/streptomycin (Invitrogen) at 37 °C in a humidified chamber. HEK293 cells were transfected with the LexA-d1EGFP plasmid using Lipofectamine 2000 (Invitrogen). Stably transfected clones were selected for 3 weeks in the presence of 500 μg/ml of Geneticin (Invitrogen). Resistant clones were expanded and tested for proper responses in the nuclear translocation assay with a Smad3 fusion protein in the presence of human transforming growth factor-β1 (R&D Systems). For transient transfection, HEK293 and HeLa cells were transferred to antibiotic-free Dulbecco's modified Eagle's medium with 10% fetal calf serum 12 h prior to transfection and then transfected with Lipofectamine 2000. Approximately 24 h after transfection, cells were harvested and/or processed as described below.
PC12 cells were maintained in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum and 10% horse serum supplemented with 2 mm glutamine. For experiments utilizing PC12-615 (29), cells were changed to serum-free Dulbecco's modified Eagle's medium containing 1 μm clasto-lactacystin β-lactone ∼16 h prior to NGF stimulation. NGF stimulation was performed by the addition of 50 ng of NGF/ml of medium (Harlan Bioproducts) for 3 h prior to harvesting.
Cells were harvested 48 h after transfection, washed once with cold PBS on ice, and lysed in radioimmune precipitation assay buffer (10 mm Tris, pH 8, 1 mm EDTA, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, 0.1% deoxycholate, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 25 μg/ml phenylmethylsulfonyl fluoride). Protein concentrations were determined by the Bradford assay.
Nuclear Translocation Assay
The LexA-d1EGFP-transfected HEK293 subclone R3 was used for these studies. An equal number of 293-R3 cells (4 × 105) were seeded in 6-well plates 1 day before transfection. Cells were transiently transfected with Lipofectamine 2000 using 4 μg of total DNA. A red fluorescent protein construct was cotransfected at a 1:50 ratio to normalize for transfection efficiency. Two days after transfection, cells were trypsinized and resuspended in 800 μl of FACS buffer (0.5% fetal bovine serum in 1× PBS) and stored on ice for FACS analysis.
Flow Cytometry
Transfected cells were analyzed with a FACScan flow cytometer (BD Biosciences). The data were analyzed using FlowJo software (Tree Star).
Western Blot Analysis
SDS-PAGE and immunoblot analysis were performed following electrophoresis on 4–20 or 15% SDS-polyacrylamide gels. After transfer to polyvinylidene difluoride membranes (Millipore), primary antibodies against specific proteins were used. They included rabbit polyclonal antiserum 9992 against the ICD of p75 (1:2000), the M20 antibody against the ICD of the Fas antigen (1:500; sc-716, Santa Cruz Biotechnology), and GFP (1:5000; Santa Cruz Biotechnology).
Subcellular Fractionation
Fractionation of PC12-615 cells into cytoplasmic or nuclear fractions was carried out using a well established protocol. Briefly, cells were collected and washed with PBS prior to resuspension in ice-cold buffer A (10 mm Hepes, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 1 μg/ml leupeptin). Cells were allowed to swell on ice for 5 min before Dounce homogenization. The homogenate was centrifuged at 218 × g to separate the cytoplasm from the nuclei. Nuclei were further purified on a 0.25–0.8 m discontinuous sucrose gradient. Nuclei were lysed, and nuclear proteins were released by sonication (6 × 10-s pulses with a Branson Model S-450D Sonifier, output of 30%) on ice. The total protein concentration of each fraction was determined by the Bradford assay, and equal amounts of protein were analyzed by Western blotting.
Fractionation of HEK293 cells was carried out in accordance with the protocol published by Méndez and Stillman (30). Briefly, cells were collected and washed once with ice-cold PBS before resuspension in ice-cold buffer B (10 mm Hepes, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 340 mm sucrose, 10% glycerol, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 1 μg/ml leupeptin). Following an 8-min incubation on ice, nuclei were collected by centrifugation (P1) at 1300 × g for 5 min. A clarified supernatant fraction (S2) was obtained by centrifugation of the supernatant fraction (S1) at 16,000 rpm at for 15 min 4 °C. The P1 fraction was washed with buffer B, resuspended in buffer C (3 mm EDTA, 0.2 mm EGTA, 1 mm dithiothreitol, and protease inhibitors as listed above), and incubated on an orbital shaker at 4 °C for 30 min. Soluble (S3) and insoluble (P3) fractions were obtained by centrifugation at 1700 rpm for 5 min at 4 °C, and P3 was resuspended in buffer C and sonicated with a Branson Model S-450D Sonifier fitted with a microtip to solubilize the chromatin fraction. Equal proportions of each fraction based on volume were analyzed by SDS-PAGE, followed by immunoblotting. Densitometry was performed by high resolution scanning of autoradiography film, followed by quantification with ImageJ.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assays were performed as described previously (31) with further modifications. A formaldehyde solution (11% formaldehyde, 0.1 m NaCl, 1 mm EDTA, 0.5 mm EGTA, and 50 mm Hepes, pH 8) was added directly to the culture medium to a final concentration of 1%, and cells were incubated with gentle agitation at room temperature for 5 min. Cross-linking was inhibited by the addition of glycine to a final concentration of 0.125 m and incubation at room temperature for 5 min. Cells were collected by centrifugation, and the pellet was resuspended in ice-cold lysis buffer 1 (1 ml/10-cm plate; 50 mm Hepes, pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Triton X-100, and protease inhibitors), followed by a 10-min incubation at 4 °C. Nuclei were collected by centrifugation, and the nuclear pellet was resuspended in 8 ml of lysis buffer 2 (200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 10 mm Tris, pH 8.0, and protease inhibitors) and rocked at room temperature for 10 min. Following centrifugation, chromatin pellets were resuspended in 3 ml of lysis buffer 3 (1 mm EDTA, 0.5 mm EGTA, and 10 mm Tris, pH 8) and sonicated with a Branson Model S-450D Sonifier fitted with a microtip continuously for 10 min in a dry ice/ethanol bath to an average length of 500 bp. Sheared chromatin (25 μg/immunoprecipitation) was precleared with a 50% slurry of a 1:1 mixture of protein A/G beads (Roche Applied Science) that had been blocked in 1 mg/ml IgG-free bovine serum albumin (Jackson ImmunoResearch Laboratories) for 3 h at 4 °C. Specific immunoprecipitations were performed at 4 °C overnight with rocking. Immune complexes were collected by the addition of 20 μl of protein A/G beads for 3 h. The beads were then washed three times with 140 mm NaCl, twice with 500 mm NaCl, twice with 250 mm LiCl, and three times with 1 mm Tris and 10 mm EDTA. After the addition of Tris/EDTA containing 0.5% SDS, 200 μg/ml RNase A (Sigma), and 200 μg/ml proteinase K (Roche Applied Science), samples were incubated at 55 °C for 3 h, followed by an overnight incubation at 65 °C. The samples were then extracted with phenol/chloroform, followed by ethanol precipitation in the presence of 20 μg of glycogen (Roche Applied Science). The precipitated DNA was resuspended in Tris/EDTA buffer for analysis by PCR.
Chromatin Immunoprecipitation PCR Primers and Conditions
PCR was performed using standard reactions with 5 μl of immunoprecipitated material as a template. PCR conditions (40 total cycles) were used, which allowed products to remain within a linear range for comparison. Following amplification, PCR products were run on a 1% agarose gel and stained with ethidium bromide. For rat (PC12) amplicons, the following sequences were used: CycE1 upstream control region, 5′-TGGGAAGGCATTCTGAAGCAC-3′ and 5′-GGTTGGGAGAGGATTGAGATTGC-3′ (T = 57 °C); and CycE1 promoter, 5′-GCAGGACACGCCCATATTAG-3′ and 5′-CTACACCGCGCTAGCTGTC-3′ (T = 61 °C).
Cell Cycle Synchronization and Quantitative Reverse Transcription- PCR
HeLa cells transfected with either control or p75 ICD-expressing plasmids were allowed to recover for 4 h after transfection, followed by the addition of 2 mm thymidine. Cells were incubated with thymidine for 16 h, washed extensively with PBS, and allowed to cycle for 4 h in complete Dulbecco's modified Eagle's medium before the addition of nocodazole (10 μg/ml). After 12 h of additional incubation, mitotic cells were collected and extensively washed before replating. Samples were collected at 0, 3, and 6 h after cell cycle block release, and total RNA was extracted using TRIzol (Invitrogen). The total RNA was treated to remove contaminating DNA (Turbo DNase removal kit, Ambion) before reverse transcription of 1 μg of total RNA (Transcriptor cDNA synthesis kit, Roche Applied Science). Quantitative PCR was performed using the Opticon 2 quantitative thermal cycler (MJ Research) and iQ SYBR Green Supermix (Bio-Rad). The following primers were used for PCR amplification: human CycE1, 5′-GTGGTGCGACATAGAGAACTG-3′ and 5′-CGCTGCTCTGCTTCTTACC-3′; and human β-actin, 5′-GAGGCCCAGAGCAAGAGAGG-3′ and 5′- GTACTTGCGCTCAGGAGGAGC-3′.
RESULTS
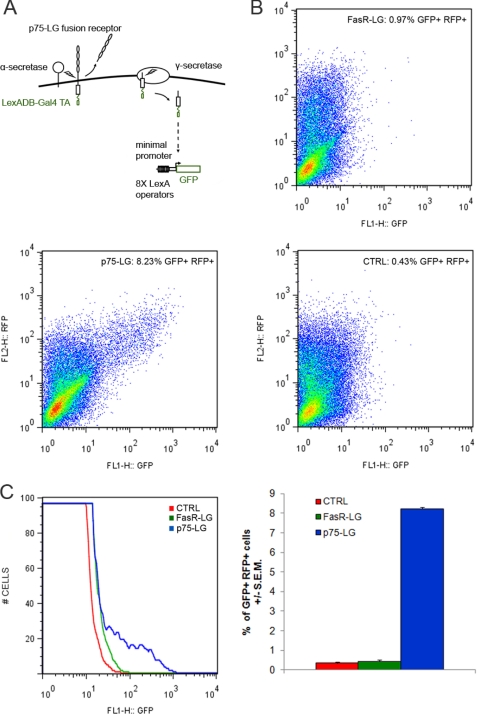
Cleavage of the full-length p75 neurotrophin receptor releases the extracellular domain from a membrane-bound C-terminal fragment. The C-terminal fragment is subsequently cleaved by γ-secretase to give rise to the soluble p75 ICD (18, 19). To determine whether p75 is directly involved in nuclear signaling, we used a reporter gene assay to detect nuclear translocation events. A protein sequence containing the LexA DNA-binding domain with a Gal4 transactivation domain (referred to as LG) was fused to the p75 C-terminal domain. The LG sequences are capable of directly activating a GFP reporter gene controlled by eight LexA operator sequences (28, 32). Nuclear translocation is required to transactivate the LexA operator sites of the reporter gene. The translocation and transactivation events were documented previously for several nuclear proteins (28, 33). Activation of the GFP reporter gene can be monitored by FACS analysis, fluorescent microscopy, and Western blot analysis.
We constructed LG fusion proteins for the p75 receptor (p75-LG) and also the Fas receptor (FasR-LG). A HEK293 cell line stably transfected with the GFP reporter gene was generated (293-R3) and used to analyze these proteins (Fig. 1A). Previous studies indicated that this assay can efficiently detect the nuclear translocation of Smad3 after transforming growth factor-β stimulation (28). Accordingly, when we transiently transfected a Smad3-LG fusion protein in 293-R3 cells, we found a significant increase in the number of GFP-expressing cells after treatment with transforming growth factor-β (data not shown). Moreover, transfection of the LG cassette did not induce GFP expression. Nuclear translocation of the LG fusion protein is therefore necessary for activation of the reporter gene in this system (28, 33).
FIGURE 1.
FACS analysis of the nuclear translocation of p75 and FasR and fusion proteins. A, schematic representation of the reporter assay. B, FACS analysis for nuclear translocation of FasR and p75 fusion proteins. C, quantification of GFP-expressing cells. DB, DNA-binding domain; TA, transactivation domain; RFP, red fluorescent protein; CTRL, control.
p75 Nuclear Translocation
The p75-LG fusion protein was transiently transfected in 293-R3 cells to detect nuclear translocation events. The FasR-LG construct was used as a negative control because FasR is structurally related to p75 but does not undergo proteolytic processing, such as α- and γ-secretase cleavages (34). After transfection of p75-LG, GFP expression was induced in a significant population of cells detected by FACS analysis. In contrast, transfection of FasR-LG did not promote GFP expression (Fig. 1, B and C).
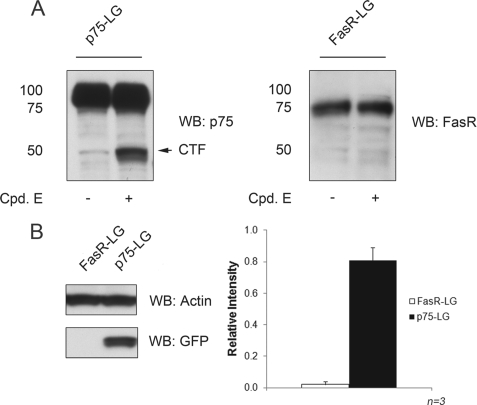
Western blot analysis indicated that both the FasR-LG and p75-LG proteins were appropriately expressed in 293-R3 cells (Fig. 2A). Fusion with the LG sequences did not interfere with p75 biosynthesis and proteolytic processing. In fact, generation of the p75 ICD from full-length p75-LG was sensitive to the γ-secretase inhibitor compound E. Upon γ-secretase inhibition, we promptly observed the accumulation of an ∼50-kDa band corresponding to the membrane-bound C-terminal fragment-LG fusion protein (Fig. 2A). No cleavage product was observed for the FasR-LG construct (Fig. 2A). Analysis of GFP protein expression by Western blot analysis confirmed that p75-LG induced high levels of GFP expression in 293-R3 cells (Fig. 2B), whereas FasR-LG-transfected cells did not express GFP.
FIGURE 2.
Western blot analysis of FasR and p75 fusion proteins and GFP protein expression. A, expression of FasR and p75 fused to the reporter cassette (LG) assessed by immunoblotting with antibodies against each receptor. B, Western blot (WB) analysis of GFP protein in FasR-LG- and p75-LG-transfected 293-R3 cells and quantitation of analysis. Error bars represent means ± S.E. (n = 3). Cpd. E, compound E; CTF, C-terminal fragment.
The p75 ICD Localizes to the Nuclear Compartment
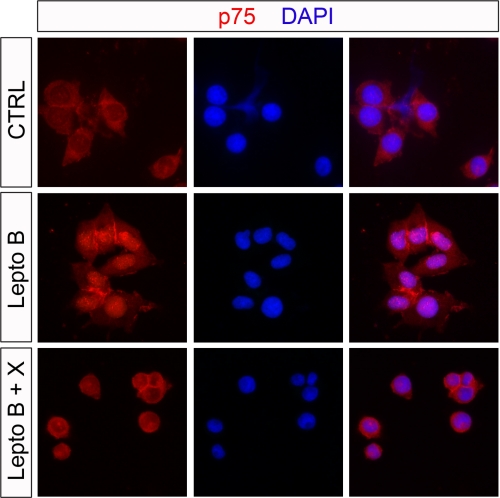
The nuclear translocation assay provides evidence that p75 sequences can be directed to the nuclear compartment. To confirm these findings, we analyzed the localization of endogenous p75 in PC12 cells by immunofluorescence microscopy. Under control conditions, we found, as expected, that p75 was localized to the cell surface and in the cytosol (Fig. 3). Upon treatment with leptomycin B, a potent nuclear protein export inhibitor (35), a detectable amount of p75 could be observed in the nucleus. Interestingly, nuclear accumulation could be blocked by treatment with inhibitor X, a γ-secretase inhibitor, suggesting that p75 proteolytic processing and subsequent p75 ICD production are necessary for nuclear translocation.
FIGURE 3.
Nuclear accumulation of p75 in PC12 cells. PC12 cells were treated for 1 h with the nuclear export inhibitor leptomycin B (Lepto B; 20 ng/ml), vehicle control solution (CTRL), or a combination of leptomycin B and γ-secretase inhibitor XVIII (Lepto B + X; 1 μm final concentration) and then fixed and immunostained with anti-p75 antibody. Inhibition of the nuclear export machinery promoted an accumulation of p75 immunoreactivity in the nucleus as shown by co-localization with 4′,6-diamidino-2-phenylindole (DAPI), which is blocked by the inhibition of γ-secretase.
To further investigate p75 ICD nuclear-cytoplasmic shuttling, we conducted cell fractionation experiments in HEK293 cells transiently transfected with a p75 ICD construct. The p75 ICD was found in both the cytoplasmic and nuclear fractions as assessed by the localization of the nuclear specific protein TAF135 and the cytoplasmic restricted protein RhoGDI (36). Interestingly, we observed a significant proportion of the nuclear p75 ICD to be located with a chromatin-enriched fraction (Fig. 4, A and B). Cell fractionation experiments performed in PC12 cells confirmed these results for the endogenous p75 ICD protein. Endogenously generated p75 ICD was present in both the cytoplasmic and nuclear fractions (Fig. 4C). Localization of a nuclear protein (H3K9) and a cytoplasmic protein (RhoGDI) was assessed to control for the specificity of the fractionation procedure. The results of this set of experiments show that the p75 ICD can translocate into the nucleus, specifically within a chromatin-enriched fraction, and that nuclear-cytoplasmic shuttling is dependent upon p75 cleavage by γ-secretase.
FIGURE 4.
The p75 ICD can be detected in nuclear fractions. A, HEK293 cells transiently transfected with a plasmid expressing the p75 ICD were fractionated into total cell extract (TCE) or cytoplasmic (S2), nucleoplasmic (S3), or chromatin-enriched (P3) fractions and probed with antibody against p75, cytoplasmically restricted RhoGDI, or the chromatin-associated protein TAF135. B, the p75 ICD signal shown in A was quantified by densitometry. The levels were normalized to the signal from the total cell extract band. Error bars represent means ± S.E. (n = 3). C, PC12 cells were fractionated into cytoplasmic and nuclear fractions, and proteins were analyzed by Western blotting. Whole cell extract from HEK293 cells transiently transfected with a plasmid expressing the p75 ICD (Trans. Ctrl.) was loaded to ensure proper identification of the band corresponding to the p75 ICD fragment (band marked with an asterisk). The membrane was probed with antibody against p75, RhoGDI, or nuclear restricted acetylated histone H3 lysine 9 (Ac H3-K9). Boxed areas represent different regions of the same membrane.
Endogenous p75 Binds to the CycE1 Promoter upon NGF Stimulation
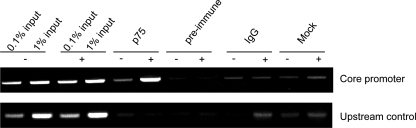
A number of proteins have been found to bind to the cytoplasmic domain of p75. They include SC-1 (36), RhoA (37), and TRAF6 (38, 39). In addition, several p75-interacting proteins, such as NRIF (neurotrophin receptor-interacting factor) (21), NADE (p75NTR-associated cell death executor) (40), and NRAGE (neurotrophin receptor-interacting MAGE homolog) (41), contribute to apoptosis in immortalized cell lines. Each protein binds to separate domains in the cytoplasmic domain of the p75 receptor. Of particular interest is SC-1, a zinc finger protein that has been localized to both the nucleus and cytoplasm. A principal function of SC-1 is to regulate growth arrest though modulation of cyclin E gene expression (42). To determine whether endogenously expressed p75 can also be detected at the CycE1 promoter region, we performed chromatin immunoprecipitation experiments in PC12 cells. After treatment with NGF for 3 h and immunoprecipitation with an antibody (9992) against the ICD of p75 (27), we could detect p75 bound to the CycE1 promoter region (−213 to +23 of the transcriptional start site). In contrast, we were unable to detect p75 either in untreated cells or at an upstream control region (−6058 to −5855 from the transcriptional start site of CycE1) (Fig. 5). In addition, no signal could be detected from material precipitated with a preimmune fraction of the anti-p75 antibody (9992), rabbit IgG, or protein A/G beads alone, showing that the detected signal was specific to the anti-p75 antibody. Taken together, these results and the biochemical fractionation (Fig. 4) indicate that p75 can be found in a nuclear fraction and specifically interacts with a known transcriptional target of a p75-interacting protein (42).
FIGURE 5.
Endogenous p75 binds to the CycE1 promoter upon NGF stimulation. PC12 cells were either left untreated (−) or treated with NGF for 3 h (+) prior to cross-linking, and chromatin immunoprecipitation was performed with the indicated antibodies. The precipitated material was amplified by PCR using primers flanking the CycE1 transcriptional start site (core promoter, −213 to +23). A region 6 kb upstream of the rat CycE1 transcriptional start site (upstream control, −6058 to −5855) was used as a PCR control. input lanes represent 0.1–1% (first four lanes) of the total chromatin used for each immunoprecipitation.
p75 ICD Expression Can Modulate CycE1 mRNA Levels
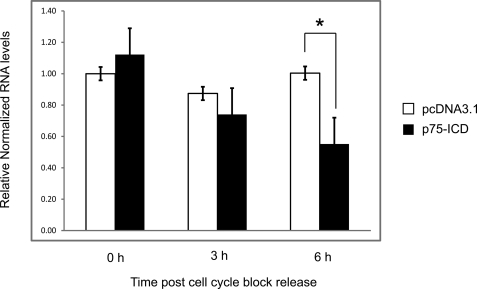
Because we were able to detect p75 at the CycE1 promoter, we next asked whether the p75 ICD might have any effect on CycE1 gene expression. We used HeLa cells for these experiments because they can be easily and reproducibly synchronized by thymidine/nocodazole block. HeLa cells were transfected either with empty vector or with a plasmid expressing the p75 ICD and synchronized by thymidine/nocodazole-induced cell cycle arrest. Quantitative reverse transcription-PCR analysis of total RNA indicated that the levels of CycE1 mRNA were significantly decreased in p75 ICD-expressing cells 6 h after cell cycle block release (Fig. 6). This finding is consistent with a previous study showing that expression of the p75 interaction partner SC-1 represses transcription of CycE1 in serum withdrawal-synchronized 3T3 cells (42).
FIGURE 6.
Expression of the p75 ICD in cell cycle-synchronized HeLa cells leads to decreased levels of CycE1 mRNA. HeLa cells transfected with either the pcDNA3.1 empty vector or a p75 ICD-expressing plasmid were synchronized by thymidine/nocodazole block. After release from cell cycle block, total cellular RNA was collected at 0, 3, or 6 h and subjected to quantitative reverse transcription-PCR. CycE1 levels were normalized to β-actin RNA levels. RNA levels are displayed relative to pcDNA3.1-transfected cells at the 0-h time point. Error bars represent means ± S.E. *, p < 0.05, Student's t test (n = 3).
DISCUSSION
Regulated intramembrane proteolysis is a conserved mechanism that has been proposed to regulate intracellular signaling events. Because many transmembrane proteins undergo γ-secretase cleavage, one prevailing explanation is that these proteolytic events are designed to remove and degrade proteins from the cell surface. An alternative view is that the ICD sequences, once released from the plasma membrane, may have important functions in a different subcellular compartment, such as the cytoplasm and/or the nucleus. In this work, we have demonstrated that the p75 ICD is capable of translocating into the nucleus, where it can activate gene expression of a heterologous gene encoding GFP. Moreover, the p75 ICD can be detected in nuclear fractions and is capable of interacting with the CycE1 promoter and modulating its gene expression. Our data suggest that the p75 receptor may employ regulated intramembrane proteolysis to transmit an intracellular signal. Similar to the cleavage of Notch, the p75 ICD may function as a nuclear transcriptional modulator. The ICDs that result from γ-secretase cleavage of Notch appear to act as nuclear signaling proteins (43, 44). For example, the p75 ICD may activate a set of genes that are involved in cell death or alternatively modulate genes that are associated with neuronal differentiation.
Cleavage of p75 by regulated intramembrane proteolysis is of particular importance for a number of reasons. The ICD of p75 has the potential of binding many intracellular proteins, including TRAF6, SC-1, NADE, NRAGE, and RhoA (6). The p75 cytoplasmic domain may bring these proteins to function in different cellular compartments. Alternatively, p75 may interact with proteins possessing defined nuclear localization signals, such as importin-β, that might act to shuttle the p75 ICD into the nucleus (45). Cleavage of the C-terminal fragment also gives rise to a small peptide whose significance is unknown, but it is analogous to the amyloid-β peptides generated from amyloid precursor protein. Another compelling reason that p75 cleavage is important is that many cell types up-regulate p75 receptors under pathological or inflammatory conditions. For example, after seizures of adult rats, there is prominent expression of p75 in cortical neurons (46), which normally do not express this receptor. Moreover, induction of p75 has been observed in many cell types, including oligodendrocytes, Schwann cells, microglia, macrophages, and smooth muscle cells (5, 6).
In the experiments reported here, we have used immortalized cell lines rather than primary neurons because of the difficulty in obtaining sufficient materials for analysis and the inherent problems in detecting the fate of the cleaved ICD fragment. Despite these reservations, the results indicate that the p75 receptor is capable of nuclear localization and modulation of transcriptional activity. The increase in binding of endogenous p75 to the CycE1 promoter after NGF treatment (Fig. 5) implies that other target genes may also be affected by p75.
A number of experiments point to potentially important biological implications of p75 cleavage. Recent studies of malignant gliomas found that inhibition of γ-secretase cleavage of p75 results in a decrease in glioma invasion and an increase in survival (47). In this case, generation of the ICD in glioma cells produces a more invasive phenotype, suggesting that there are changes in gene expression as a result of p75 cleavage. The p75 receptor has also been implicated in amyloid-β-induced neurodegeneration (48). The involvement of p75 cleavage has not yet been assessed in this case or in the interaction with ephrin A reverse signaling (17). It is notable that the induction and cleavage of p75 occur in selective neurons that are destined to undergo intramembranous γ-secretase cleavage of amyloid precursor protein (49), which suggests that p75 cleavage may also represent an important early event during neurodegeneration. Further experiments to investigate the downstream effects of p75 cleavage will yield insight into these and other diverse possibilities.
Acknowledgments
We thank Naoko Tanese, Hodaka Fujii, Jeffrey N. Savas, Lauren M. Young, and Sophie Restituito for kindly providing regents and advice during the course of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant AG025970 (to M. V. C.).
This article was selected as a Paper of the Week.
- NGF
- nerve growth factor
- ICD
- intracellular domain
- PBS
- phosphate-buffered saline
- FACS
- fluorescence-activated cell sorter
- GFP
- green fluorescent protein
- CycE1
- cyclin E1
- FasR
- Fas receptor.
REFERENCES
- 1.Locksley R. M., Killeen N., Lenardo M. J. (2001) Cell 104, 487–501 [DOI] [PubMed] [Google Scholar]
- 2.Feinstein E., Kimchi A., Wallach D., Boldin M., Varfolomeev E. (1995) Trends Biochem. Sci. 20, 342–344 [DOI] [PubMed] [Google Scholar]
- 3.Haase G., Pettmann B., Raoul C., Henderson C. E. (2008) Curr. Opin. Neurobiol. 18, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang E. J., Reichardt L. F. (2001) Annu. Rev. Neurosci. 24, 677–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao M. V. (2003) Nat. Rev. Neurosci. 4, 299–309 [DOI] [PubMed] [Google Scholar]
- 6.Barker P. A. (2004) Neuron 42, 529–533 [DOI] [PubMed] [Google Scholar]
- 7.Hempstead B. L., Martin-Zanca D., Kaplan D. R., Parada L. F., Chao M. V. (1991) Nature 350, 678–683 [DOI] [PubMed] [Google Scholar]
- 8.Lee R., Kermani P., Teng K. K., Hempstead B. L. (2001) Science 294, 1945–1948 [DOI] [PubMed] [Google Scholar]
- 9.Chao M. V., Bothwell M. (2002) Neuron 33, 9–12 [DOI] [PubMed] [Google Scholar]
- 10.Dobrowsky R. T., Werner M. H., Castellino A. M., Chao M. V., Hannun Y. A. (1994) Science 265, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 11.Rabizadeh S., Oh J., Zhong L. T., Yang J., Bitler C. M., Butcher L. L., Bredesen D. E. (1993) Science 261, 345–348 [DOI] [PubMed] [Google Scholar]
- 12.Frade J. M., Rodríguez-Tébar A., Barde Y. A. (1996) Nature 383, 166–168 [DOI] [PubMed] [Google Scholar]
- 13.Casaccia-Bonnefil P., Carter B. D., Dobrowsky R. T., Chao M. V. (1996) Nature 383, 716–719 [DOI] [PubMed] [Google Scholar]
- 14.Wang K. C., Kim J. A., Sivasankaran R., Segal R., He Z. (2002) Nature 420, 74–78 [DOI] [PubMed] [Google Scholar]
- 15.Wong S. T., Henley J. R., Kanning K. C., Huang K. H., Bothwell M., Poo M. M. (2002) Nat. Neurosci. 5, 1302–1308 [DOI] [PubMed] [Google Scholar]
- 16.Ben-Zvi A., Ben-Gigi L., Klein H., Behar O. (2007) J. Neurosci. 27, 13000–13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim Y. S., McLaughlin T., Sung T. C., Santiago A., Lee K. F., O'Leary D. D. (2008) Neuron 59, 746–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanning K. C., Hudson M., Amieux P. S., Wiley J. C., Bothwell M., Schecterson L. C. (2003) J. Neurosci. 23, 5425–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung K. M., Tan S., Landman N., Petrova K., Murray S., Lewis R., Kim P. K., Kim D. S., Ryu S. H., Chao M. V., Kim T. W. (2003) J. Biol. Chem. 278, 42161–42169 [DOI] [PubMed] [Google Scholar]
- 20.Ebinu J. O., Yankner B. A. (2002) Neuron 34, 499–502 [DOI] [PubMed] [Google Scholar]
- 21.Kenchappa R. S., Zampieri N., Chao M. V., Barker P. A., Teng H. K., Hempstead B. L., Carter B. D. (2006) Neuron 50, 219–232 [DOI] [PubMed] [Google Scholar]
- 22.Majdan M., Lachance C., Gloster A., Aloyz R., Zeindler C., Bamji S., Bhakar A., Belliveau D., Fawcett J., Miller F. D., Barker P. A. (1997) J. Neurosci. 17, 6988–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domeniconi M., Cao Z., Spencer T., Sivasankaran R., Wang K., Nikulina E., Kimura N., Cai H., Deng K., Gao Y., He Z., Filbin M. (2002) Neuron 35, 283–290 [DOI] [PubMed] [Google Scholar]
- 24.Domeniconi M., Zampieri N., Spencer T., Hilaire M., Mellado W., Chao M. V., Filbin M. T. (2005) Neuron 46, 849–855 [DOI] [PubMed] [Google Scholar]
- 25.Frade J. M. (2005) J. Neurosci. 25, 1407–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Podlesniy P., Kichev A., Pedraza C., Saurat J., Encinas M., Perez B., Ferrer I., Espinet C. (2006) Am. J. Pathol. 169, 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber L. J., Chao M. V. (1995) Dev. Biol. 167, 227–238 [DOI] [PubMed] [Google Scholar]
- 28.Hoshino A., Matsumura S., Kondo K., Hirst J. A., Fujii H. (2004) Mol. Cell 15, 153–159 [DOI] [PubMed] [Google Scholar]
- 29.Hempstead B. L., Rabin S. J., Kaplan L., Reid S., Parada L. F., Kaplan D. R. (1992) Neuron 9, 883–896 [DOI] [PubMed] [Google Scholar]
- 30.Méndez J., Stillman B. (2000) Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi Y., Rayman J. B., Dynlacht B. D. (2000) Genes Dev. 14, 804–816 [PMC free article] [PubMed] [Google Scholar]
- 32.Hoshino A., Fujii H. (2007) Cell Stress Chaperones 12, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoshino A., Hirst J. A., Fujii H. (2007) J. Biol. Chem. 282, 17706–17711 [DOI] [PubMed] [Google Scholar]
- 34.Zampieri N., Xu C. F., Neubert T. A., Chao M. V. (2005) J. Biol. Chem. 280, 14563–14571 [DOI] [PubMed] [Google Scholar]
- 35.Fornerod M., Ohno M., Yoshida M., Mattaj I. (1997) Cell 90, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 36.Chittka A., Chao M. V. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10705–10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita T., Tucker K. L., Barde Y. A. (1999) Neuron 24, 585–593 [DOI] [PubMed] [Google Scholar]
- 38.Khursigara G., Orlinick J. R., Chao M. V. (1999) J. Biol. Chem. 274, 2597–2600 [DOI] [PubMed] [Google Scholar]
- 39.Powell J. C., Twomey C., Jain R., McCarthy J. V. (2009) J. Neurochem. 108, 216–230 [DOI] [PubMed] [Google Scholar]
- 40.Mukai J., Hachiya T., Shoji-Hoshino S., Kimura M. T., Nadano D., Suvanto P., Hanaoka T., Li Y., Ire S., Greene L. A., Sato T. A. (2000) J. Biol. Chem. 275, 17566–17570 [DOI] [PubMed] [Google Scholar]
- 41.Salehi A. H., Xanthoudakis S., Barker P. A. (2002) J. Biol. Chem. 277, 48043–48050 [DOI] [PubMed] [Google Scholar]
- 42.Chittka A., Arevalo J. C., Rodriguez-Guzman M., Pérez P., Chao M. V., Sendtner M. (2004) J. Cell Biol. 164, 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortini M. E. (2002) Nat. Rev. Mol. Cell Biol. 3, 673–684 [DOI] [PubMed] [Google Scholar]
- 44.Weinmaster G. (2000) Curr. Opin. Genet. Dev. 10, 363–369 [DOI] [PubMed] [Google Scholar]
- 45.Hanz S., Perlson E., Willis D., Zheng J. Q., Massarwa R., Huerta J. J., Koltzenburg M., Kohler M., van Minnen J., Twiss J. L., Fainzilber M. (2003) Neuron 40, 1095–1104 [DOI] [PubMed] [Google Scholar]
- 46.Roux P., Colicos M. A., Barker P. A., Kennedy T. E. (1999) J. Neurosci. 19, 6887–6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L., Rahn J. J., Lun X., Sun B., Kelly J. J., Weiss S., Robbins S. M., Forsyth P. A., Senger D. L. (2008) PLoS Biol. 6, e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bengoechea T. G., Chen Z., O'Leary D., Masliah E., Lee K. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7870–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller T., Meyer H. E., Egensperger R., Marcus K. (2008) Prog. Neurobiol. 85, 393–406 [DOI] [PubMed] [Google Scholar]