Abstract
Structural changes in channel proteins give critical insights required for understanding the gating transitions that underpin function. Tryptophan (Trp) is uniquely sensitive to its environment and can be used as a reporter of conformational changes. Here, we have used site-directed Trp insertion within the pore helices of the small mechanosensitive channel protein, MscS, to monitor conformational transitions. We show that Trp can be inserted in place of Leu at the two pore seal positions, Leu105 and Leu109, resulting in functional channels. Using Trp105 as a probe, we demonstrate that the A106V mutation causes a modified conformation in the purified channel protein consistent with a more open state in solution. Moreover, we show that solubilized MscS changes to a more open conformation in the presence of phospholipids or their lysoforms.
Keywords: Biophysics, Membrane/Channels, Methods/Fluorescence, Phospholipid, Protein Conformation, Escherichia Coli, Osmotic Stress
Introduction
Gating is a fundamental property of all ion and solute channels, and of some transporters, and involves structural transitions that convert the protein from a nonconducting to a conducting state. Channel gating is controlled by specific stimuli, such as transmembrane voltage, a chemical ligand, or a mechanical signal (1), and the coupling between the signal recognition and gating is a major focus of current channel research. Channel activation is frequently rapid, often complex, and may involve multiple states either as unstable intermediates or as well defined stable conformations (2). Channels may be required to respond to more than one signal and/or participate in adaptation processes, where they cease to conduct despite the continued exposure to the gating stimulus (3, 4). It is, therefore, necessary to detect gating under multiple conditions to understand channel mechanism and function. Patch clamp techniques, which measure the core properties of the channel signal sensitivity, conductance, open dwell time, selectivity, and adaptation, have become a major means to study channels in the membrane. However, this technology requires the channel to be ion-conducting and thus, cannot inform on structural conformations of the nonconducting states.
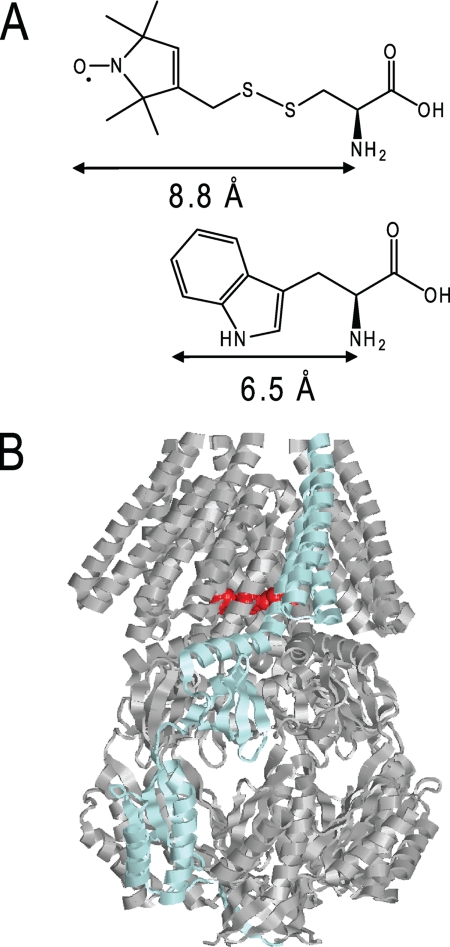
Our understanding of protein conformational changes has been greatly facilitated by biophysical measurements of both native and mutant proteins. In particular, fluorescent probes have proved invaluable complements to spin probes in EPR and related techniques. Tryptophan exhibits natural fluorescence that is very sensitive to environmental changes (5). The peak position in the emission spectrum reflects the polarity of the immediate environment of the Trp residue, quenching by small water-soluble agents is a measure of the accessibility of the residue to the bulk water phase and anisotropy indicates the mobility of the Trp residue. Trp is a rare amino acid in proteins, and its role is often structural. Introduction of Trp is potentially disruptive to the structure of the protein because of its bulky side chain, but although it is the largest natural amino acid, it is small for a fluorophore, particularly when compared with spin labels usually used for EPR spectroscopy. Importantly, the Trp aromatic group is more closely attached to the backbone than is either a spin probe or other fluorophores, which are connected through an engineered Cys residue (see Fig. 1a). The danger for modified residues to “snorkel” into environments moderately remote from their backbone attachment site is significant. Consequently, because Trp fluorescence reports local changes in conformation of the protein, an understanding of the potential of this reporter is important, which we have investigated here in the small mechanosensitive channel, MscS.
FIGURE 1.
A, structure of the fluorescence label tryptophan and for comparison, structure of the spin label MTSL. B, structure of MscS on the basis of the closed crystal structure (10). One subunit is shown in blue. Residue Leu105 in the pore that was changed against tryptophan is marked in red.
MscS responds to increases in tension in the membrane bilayer arising from sudden osmotic transitions (hypoosmotic shock). It can also be triggered to open by the insertion of amphipaths or lysophospholipids (e.g. lysophosphatidyl choline, LPC)2 into the membrane bilayer (6–8). Gating is extremely rapid (2) and results in the closed channel transitioning to an open state that conducts the passage of small solutes, which releases the tension in the membrane by lowering the osmotic gradient that drives water entry (9). The structural organization of MscS is known through two complementary crystal structures: the wild type in a nonconducting state and a mutant, A106V, in an open state (10–12). MscS is a homoheptamer of 31-kDa subunits twisted around the central axis of the pore (Fig. 1b). Each subunit contributes three transmembrane helices (TM1–3). TM1 and TM2 form a paddle that is believed to sense the membrane tension; the TM3a helix lines the pore and TM3b, which lies along the membrane surface tangential to the axis of the pore, forms a resistor (12). The overall character of the pore is hydrophobic and residues Leu105 and Leu109 at the lower (cytoplasmic) end of the pore create a hydrophobic seal (10, 13). We have proposed that the structural transition from closed to open involves the rotation and radial movement of the TM3a helices, and Ala106 is a critical residue due to its location at the interhelix interface, where, in the closed state, it interacts with Gly108 on the adjacent pore helix (14). Ala106 was mutated to valine, and this protein variant was found to crystallize in an open state of MscS (12). In this structure, Val106 is positioned in an open pocket created by rotation of the TM helices and their radial movement. The mutated residue is not conformationally constrained in the crystal structure. The A106V mutant channel retains function but requires greater pressure to open, and at least two open states are detected by application of increased pressure in patch clamp analysis (14). The current study was undertaken to determine whether Trp substitutions in the pore region could be used to monitor structural transitions associated with either mutation (A106V) or perturbation of the MscS-containing detergent micelles by lipids or LPC.
EXPERIMENTAL PROCEDURES
Materials
1-Oleoyl-2-hydroxy-sn-glycero-3-phosphocholine (LPC); 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE); 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-sn-glycerol) (POPG); and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) were obtained from Avanti Polar Lipids Inc. (Alabaster, Alabama). n-Dodecyl-β-d-maltopyranoside (DDM) Sol grade was from Anatrace Inc., whereas all other chemicals were purchased from Sigma.
Mutants were made with the Stratagene QuikChangeTM protocol using the tryptophan-free MscS mutant, MscSΔW (W16Y/W240F/W251F) as template (15, 16). Primer sequences are available on request. All mutants were sequenced on both strands.
Physiological, Biochemical, and Electrophysiological Assays
The survival assay of osmotic down shock was described previously (17). Purification of MscS-H6 was performed as described (16), and Western blot analysis of whole-cell samples was essentially as carried out previously (15), except that detection was by Penta-His antibody (Qiagen). Patch clamp recordings were conducted on membrane patches derived from giant protoplasts using the strain MJF429 (18) transformed with MscS plasmids as described previously (14). Gene expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for up to 60 min before protoplast generation. Excised, inside-out patches were analyzed at a membrane potential of −20 mV with pipette and bath solutions containing: 200 mm KCl, 90 mm MgCl2, 10 mm CaCl2, and 5 mm HEPES buffer at pH 7. All data were acquired at a sampling rate of 50 kHz with 5-kHz filtration using an AxoPatch 200B amplifier and pClamp software (Molecular Devices). The pressure threshold for activation of the MscS channels was referenced against the activation threshold of MscL (PL:PS) to determine the pressure ratio for gating as previously (19–21). Measurements have been conducted on patches derived from a minimum of two protoplast preparations. Pressure ratios are given as mean ± S.E.
Fluorescence Spectroscopy
Steady-state and lifetime measurements were performed with a fluorescence spectrometer FLS920 (Edinburgh Instruments). For all steady-state measurements, excitation and emission slits were set to 2 nm, the excitation polarizer to 90°, and the emission polarizer was set to 0° (22). Emission spectra were corrected with a buffer spectrum and fitted to a skewed Gaussian curve to determine the emission maximum (23),
with fluorescence intensity I at wavelength λ and Imax at λmax, skew parameter of b, peak width at half-height of ωλ, and baseline offset of a.
Quantum yields (QYs) were determined relative to free Trp with a reference value of QYst = 0.14 (24) at 20 °C. The absorbance at 295 nm of the free Trp standard (ASt) was matched to the sample (A) and was kept below 0.05. Emission spectra were recorded with an excitation slit of 1.5 nm to match the slit width of our UV-visible spectrometer Varian Cary50. The emission spectra were fitted with Equation 1, and the fit was integrated over the whole peak range for both samples (I) and the standard (ISt). The quantum yields were then calculated according to Equation 2.
An excitation wavelength of 295 nm was taken for quenching experiments with acrylamide, and fluorescence intensities at 340 nm were recorded. Data were corrected for the inner filter effect of acrylamide (5) and were analyzed with the Stern-Volmer equation,
where I0 is the fluorescence intensity without quencher, I is in the presence of quencher, and KSV is the Stern-Volmer value.
Steady-state anisotropy was measured with an excitation at 295 nm and emission between 340 and 360 nm. The anisotropy in this range showed no wavelength dependence and was averaged. Measurements were also performed with a horizontally oriented excitation polarizer to correct with the instrumental factor G = Ihv/Ihh according to Equation 4,
where the fluorescence intensities I are labeled with a subscript indicating the orientation of the excitation and emission polarizers (v, vertical; h, horizontal), respectively. Apparent correlation times were calculated with the Perrin equation (Equation 5):
with anisotropy as r, mean lifetime as τav (see below), and fundamental anisotropy as r0, with a value of 0.232 for Trp at an excitation wavelength of 295 nm (25). Excitation anisotropy spectra were recorded with an emission wavelength of 360 nm to avoid Raman scattering in the recorded excitation range from 260 to 310 nm.
Fluorescence lifetimes were obtained in the time domain by single photon counting using a nanosecond-flashlamp with hydrogen filling. 10,000 counts were accumulated in the peak. Excitation and emission slits were set to 17 nm at 295 and 340 nm, respectively. The excitation polarizer was set to 0°, and the emission polarizer was set to the magic angle of 54.7°. Data were fitted to a sum of exponentials.
The average lifetime is calculated by the following equation.
To investigate the effect of LPC on the spectroscopic properties of MscS, a stock solution of 50 mg/ml LPC in methanol was prepared and added to a final concentration of 0.25 mg/ml. This corresponds to a final molar ratio of 1:2 LPC:DDM. Other lipids were dissolved in a buffer (50 mm phosphate, pH 7.5; 150 mm NaCl) containing 0.4% DDM and were added to a final concentration of 0.25 mg/ml with a final molar ratio of 1:5 lipid:DDM.
All measurements have been repeated at least in triplicate on at least two independent protein preparations, and mean values with standard deviations are given. The protein concentration in steady-state fluorescence experiments was 5 μm and 20 μm in lifetime experiments. Data were analyzed with the software Origin 8.0 (OriginLab) and F900 (Edinburgh Instruments).
RESULTS
MscS Tryptophan Mutants Are Stable and Functional
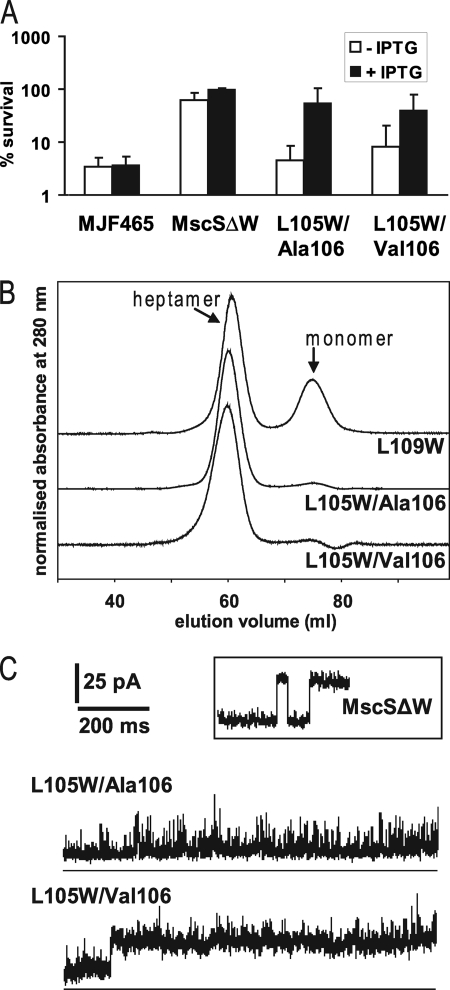
Previously, we constructed MscSΔW (W16Y/W240F/W251F), in which the natural Trp residues were replaced with phenylalanine or tyrosine, creating stable Trp-free channels. The Trp-free channels gated at a slightly higher tension than wild type MscS but exhibited normal electrophysiological profiles (16) and were found to protect cells against hypoosmotic shock (Fig. 2a). All subsequent mutants in this study used this MscSΔW as the framework into which new Trp residues were introduced. Unique Trp residues were substituted at the seal (L105W and L109W) in the presence and absence of an A106V mutation (12, 14). The L109W channel was incorporated into the membrane and was functional in protecting cells against osmotic downshock (data not shown). However, the protein was found to be unstable in detergent (Fig. 2b) and was not used further in this study. In contrast, both the L105W mutant channel (subsequently referred to as L105W/Ala106) and the L105W/Val106 mutant channel were expressed, incorporated into the membrane, and retained the heptameric structure in detergent (Fig. 2b). Protection against hypoosmotic shock was diminished relative to the parent protein. This effect was most marked when channel expression was not induced, and only low levels of channel proteins are produced from the plasmid borne genes. However, the channels carrying L105W mutations retained activity and protected when the expression was induced by addition of isopropyl 1-thio-β-d-galactopyranoside (Fig. 2a). We have previously shown that the A106V mutation alone provides slightly diminished protection against hypoosmotic shock (12), but this effect was not significantly enhanced by the presence of the L105W mutation (Fig. 2a).
FIGURE 2.
Functionality and stability of MscS mutants. A, osmotic down shock assay. Shown are the survival percentages relative to control samples. Samples that were induced with isopropyl 1-thio-β-d-galactopyranoside (IPTG) are shown as black bars, and samples without induction are shown as white bars. B, size exclusion profiles of the mutants MscS ΔW L109W, L105W/Ala106, and L105W/Val106. To determine the typical positions of heptameric and monomeric MscS in the elution profile (marked) fractions were taken for blue native electrophoresis and were observed to have masses typical of the heptamer and monomer, respectively (16). Intensities have been normalized and offset for clarity. C, examples of electrophysiological traces are shown. For comparison, a trace of the tryptophan-free mutant MscS ΔW is given in the inset.
An explanation for the poor protective ability of the L105W channel became evident from the electrophysiology. The L105W single pore mutation shortens the open dwell time leading to a rapidly transitioning channels (Fig. 2c, upper panel versus inset) but the pressure required to open the channel, expressed as the PL:PS ratio (19, 21, 26) was not changed in comparison to that for MscSΔW (1.34; n = 4 versus 1.33; n = 6 for MscSΔW/L105W and MscSΔW, respectively). Introduction of the A106V mutation into MscSΔW/L105W resulted in a PL:PS ratio of 1.29 (n = 7) and sometimes a distinct separate state with a longer dwell time (Fig. 2c, lower panel). In ∼30% of patches (3/10) the sustained open state observed in L105W/Val106 channels was superimposed on the unstable openings initially observed with L105W/Ala106. Two open states were previously described for the A106V mutant, that differed in their open dwell time; however, stable openings were observed for both states, compared with the unstable openings seen here for L105W/Val106 (14). The L105W mutation destabilizes an open state, whereas A106V can cause an additional stable open state; both these properties are found in the double mutant. The combined effect of the two mutations was a mild gain-of-function phenotype. Wild type MscS channels can be expressed from plasmids without changing the growth behavior of cells but gain-of-function mutations cause immediate growth inhibition upon induction (27). In contrast, cells induced for L105W/Val106 expression exhibited normal growth for up to ∼45–60 min (∼ two generations) and then exhibited severe inhibition (supplemental Fig. S1a). Such behavior would be consistent with the combination of L105W and Val106 causing infrequent but persistent transitions to an open state, such that growth inhibition only becomes evident after accumulation of higher levels of the channel protein. Western blot analysis of whole-cell samples suggests that growth inhibition correlates with the maximum level of MscS protein, suggesting that lower levels are tolerated, but high levels allow for rare stable openings (supplemental Fig. S1b). Although the mutations in L105W/Val106 are adjacent in the linear sequence of MscS and both produce bigger side chains, this does not disrupt packing, as this mutant is functional and forms a stable complex. The Leu side chains, and therefore the presumed location of the Trp groups, are oriented into the lumen of the pore, even in the closed structure. Although Trp side chains are bigger than Leu, the lumen location reduces the expected effect on packing caused by the mutation. Given the structure of the α-helix, Val106 and Trp105 side chains cannot interact directly with each other as they are exposed on different sides of the helix.
Fluorescence Analysis Is Consistent with Different Conformations Caused by the A106V Mutation
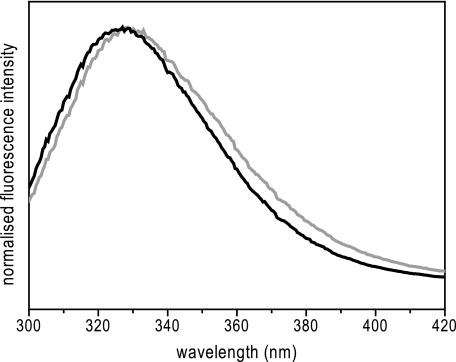
During purification, L105W/Ala106 and L105W/Val106 proteins behaved almost identically. Both proteins were purified to homogeneity by a two-step process involving a metal chelating column and size exclusion chromatography. The peak position during size exclusion chromatography indicated that both proteins retain the heptameric state in detergent micelles (Fig. 2b). This simple purification regime provided reproducible samples of stable and functional MscS proteins with the Trp probe introduced into the pore for fluorescence spectroscopy. A summary of fluorescence spectroscopic properties is given in Table 1. A small but significant red shift in the peak position was observed for the L105W/Val106 protein (λmax, 329.5 nm) relative to L105W/Ala106 (λmax, 327.7 nm) as shown in Fig. 3. In comparison to Trp fully accessible to water (λmax, 347 nm), both mutants seem to retain the Trp in an apolar environment, which may reflect the close packing of the Trp residues expected in both the closed and open states. However, the quantum yield for L105W/Val106 was increased 1.5-fold relative to L105W/Ala106, suggesting a significant environmental change (Table 1).
TABLE 1.
Comparison of fluorescence spectroscopic properties of L105W/Ala106 and L105W/Val106
| L105W/Ala106 | L105W/Val106 | |
|---|---|---|
| λmax in emission spectrum | 327.7 ± 0.6 nm | 329.5 ± 0.7 nm |
| Quantum yield | 0.10 ± 0.01 | 0.16 ± 0.01 |
| Lifetime τ1 | 1.44 ± 0.26 ns | 1.81 ± 0.73 ns |
| Amplitude factor α1 | 0.48 ± 0.11 | 0.38 ± 0.02 |
| Lifetime τ2 | 3.74 ± 0.61 ns | 4.73 ± 1.11 ns |
| Amplitude factor α2 | 0.39 ± 0.05 | 0.53 ± 0.07 |
| Lifetime τ3 | 7.05 ± 0.75 ns | 7.84 ± 1.12 ns |
| Amplitude factor α3 | 0.13 ± 0.08 | 0.09 ± 0.06 |
| Average lifetime τav | 4.11 ± 0.20 ns | 4.53 ± 0.29 ns |
| Anisotropy r | 0.135 ± 0.012 | 0.106 ± 0.017 |
| Apparent correlation time ϴapp | 5.7 ns | 3.8 ns |
| Stern-Volmer quenching constant KSV | 2.49 ± 0.30 m−1 | 3.13 ± 0.45 m−1 |
| Bimolecular quenching constant kq | 6.1·108m−1 s−1 | 6.9·108m−1 s−1 |
FIGURE 3.
Emission spectra of MscS mutants. The emission spectrum of L105W/Ala106 (black) is compared with L105W/Val106 (gray). An excitation at 295 nm was used. The protein concentration was 5 μm.
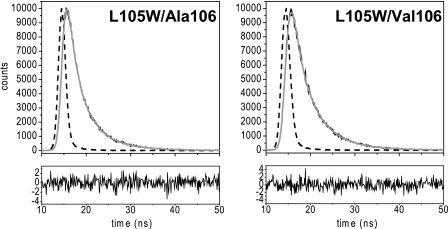
When fluorescence decay was analyzed, three exponential contributions were required to explain the data for both mutants (Fig. 4). The calculated weighted mean lifetime was slightly longer for L105W/Val106 (4.53 ns) than for L105W/Ala106 (4.11 ns). Side chains of certain amino acids like tyrosine or histidine are known to quench Trp fluorescence intrinsically (28). However, the only His residues in MscS are in the carboxy-terminal tag, and no Tyr residue is close to L105W in MscS, as judged from the crystal structures. However, in recent years, it has become clear that the carbonyl group of the peptide bond is also an important relaxation pathway for Trp by excited state electron transfer (29). The fluorescence lifetime and quantum yield strongly depend on the protein conformation (30, 31). Therefore, the difference in lifetimes between L105W/Ala106 and L105W/Val106 indicates a changed orientation of the Trp side chain relative to the backbone or a change in backbone conformation itself.
FIGURE 4.
Fluorescence lifetime traces for L105W/Ala106 and L105W/Val106. Experimental decays are shown in black, instrumental response functions are shown as dashed lines, and fits are shown as gray lines. Residuals are shown below. The protein concentration was 20 μm.
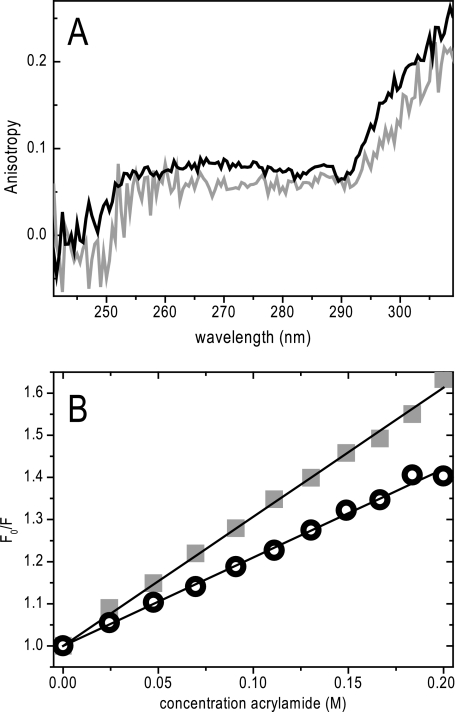
Anisotropy can also give an indication of the packing around the fluorophore that may restrict the free rotation of the fluorophore around its covalent bond. The anisotropy changes with the mobility of the fluorophore caused by the overall rotation of the protein, segmental movements, and the side chain mobility of the fluorophore itself. The steady-state anisotropy for L105W/Val106 was lower (r = 0.106) than for L105W/Ala106 (r = 0.135), which is partly explained by the longer fluorescence lifetime for L105W/Val106. However, apparent correlation times calculated from the anisotropies take the lifetime into account and still show a difference. For L105W/Val106 and L105W/Ala106, much higher anisotropies are expected only if the mobility is considered. The large complex of 210 kDa, plus the attached detergent and lipid molecules, will cause a slow overall tumbling of the protein and side chain mobility within the pore will be restricted. The low anisotropies can also be explained by a second effective mechanism to lower anisotropies, Förster resonance energy transfer (FRET). Because the distance between the Trp residues contributed from each subunit at position 105 will be small, an efficient homoFRET can be expected. The Förster distance R0, the characteristic distance with 50% efficiency of energy transfer, for a Trp couple is 6–12 Å, which is close to the predicted separation expected in the MscS open state and greater than that predicted for the closed state (32). To explain the difference of the anisotropy for both mutants, either homoFRET in L105W/Val106 is more efficient, or the side chain mobility is increased. A more efficient homoFRET is expected if the distance between the Trp side chains in L105W/Val106 is smaller in comparison to L105W/Ala106 or the orientation between the Trp residues is more favorable for FRET. We recorded excitation anisotropy spectra to distinguish between the possible causes, mobility or homoFRET. Weber reported (33, 34) that no homoFRET is observed at the red edge of the excitation spectrum. Because the excitation anisotropy spectra of L105W/Ala106 and L105W/Val106 do not approach each other at high wavelengths (Fig. 5a), we conclude that the difference in anisotropy is caused by a change of the mobility of the Trp side chain. Thus, the mobility is increased for L105W/Val106, consistent with a widened pore. Preliminary time resolved anisotropy experiments show a fast and a very slow decay (data not shown). The fast contribution is presumably due to homoFRET and side chain movements. This is faster and not resolved in the experiment for L105W/Val106 in comparison to L105W/Ala106, which can be explained by the change in side chain movements as suggested above. The slow contribution is caused by overall tumbling of the complex.
FIGURE 5.
A, excitation anisotropy spectra of L105W/Ala106 (black) and L105W/Val106 (gray). B, Stern-Volmer plot of acrylamide quenching. Quenching experiments for L105W/Ala106 (open circles) and for L105W/Val106 (closed squares) are shown. Linear fits to these data are shown as lines. The protein concentration was 5 μm.
Fluorescence quenching was used to determine the accessibility of the pore Trp residue in both mutants (Fig. 5b). Acrylamide, which has no charge and therefore shows no misleading contributions from electrostatic interactions, was used as a small, water soluble quencher. The Stern-Volmer quenching constant is increased for L105W/Val106 (KSV = 3.13 m−1) in comparison to L105W/Ala106 (KSV = 2.49 m−1). This is also reflected in the bimolecular quenching constants (kq = 6.9 108 m−1s−1 versus 6.1 108 m−1s−1 for L105W/Val106 and L105W/Ala106, respectively). These data indicate increased accessibility of Trp105 from the bulk water phase in L105W/Val106 in comparison with L105W/Ala106, consistent with a modified protein conformation.
Structural Modifications Caused by Lipids and LPC
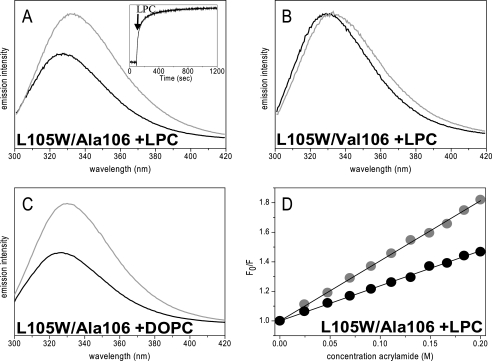
Spectroscopic properties change if solubilized lipids or their lysoforms are added to solubilized MscS, indicating a more open conformation. LPC caused a red shift of the emission spectrum of Trp in both L105W/Ala106 and L105W/Val106 by 6 and 4 nm, respectively (Fig. 6, a and b), resulting in the Trp residue in both proteins approaching the same λmax of 333 nm. A similar red shift is observed when lipids are added to L105W/Ala106, azolectin as well as single synthetic lipids DOPC, DOPE, or POPG all have a comparable effect to LPC (Fig. 6c and data not shown). In addition, the fluorescence intensity for L105W/Ala106 increased in the presence of LPC and lipids, such that its quantum yield approached that of L105W/Val106. In both incubations, the MscS channel retained the homoheptameric structure as evidenced by blue native gel electrophoresis (data not shown). Finally, quenching by acrylamide increased for the L105W/Ala106 mutant in the presence of LPC consistent with greater access to the aqueous phase of the Trp residue (Fig. 6d). As controls, fluorescence of free Trp does not change after addition of LPC nor does the fluorescence of L105W/Ala106 or L105W/Val106 change upon addition of methanol or concentrated DDM solutions, which were used to make up the stock solutions of LPC or lipids, respectively (data not shown). These data are consistent with lipids and LPC causing conformational changes in MscS even when the protein is dissolved in detergent micelles.
FIGURE 6.
Effect of LPC and DOPC on emission properties. Emission spectra of L105W/Ala106 (A) and L105W/Val106 (B) before (black) and after (gray) addition of 0.25 mg/ml LPC. The inset in A shows the emission intensity change at 350 nm over time. C, addition of 0.25 mg/ml DOPC to L105W/Ala106. D, Stern-Volmer plots of acrylamide quenching experiments with L105W/Ala106 in the absence (black) or presence (gray) of 0.25 mg/ml LPC. The protein concentration was 5 μm.
DISCUSSION
Crystal structures of membrane proteins are often difficult to obtain and are almost exclusively not embedded into the natural lipid bilayer. This leads to unanswered mechanistic questions, even when multiple structures are available, as is the case for MscS. Ultimately, dynamic information is required that provides insights not only into the relevance of the crystal states but also into the potential conformational pathways undertaken during channel gating. Clues emerge from electrophysiology, but these are restricted to open and subconducting states. We show here that a Trp probe introduced within the pore of a channel can be used as a gating probe applying fluorescence spectroscopy.
Replacing Leu105, which is one of the key seal residues in the pore of MscS, with Trp resulted in a functional and stable MscS complex. In the nonconducting (Ala106) crystal structure, the Leu residues are tightly packed, and it was a surprise that the protein tolerated the Trp substitution at this position, as introduction of Trp increases the volume by 72 Å3 per subunit (35). The L105W protein was clearly able to pack the Trp residues and attain a stable conformation as it could be solubilized by detergent and retained its heptameric structure. This is in contrast to L109W, which was expressed, integrated into the membrane and was functional but was unstable after detergent solubilization. The L105W substitution causes disruption of the open state, because stable openings were not observed in patch-clamp analysis. Steric hindrance caused by the Trp side chains may explain this observation, as in the Val106 (i.e. more open) structure the Leu105 side chains are separated by only ∼6 Å. Addition of the A106V mutation could produce stabilized channel openings in the membrane patch, consistent with the ability of MscS A106V to attain a long lived stable open state (14).
A significant difference between the two mutants L105W/Ala106 and L105W/Val106 was detectable by fluorescence spectroscopy. Introduction of the A106V mutation causes the reporting Trp residue to reside in a more polar environment, to be more accessible from the bulk water phase, and less restricted in its side chain motions. These data are consistent with solubilization, allowing some relaxation of the subunit packing, causing the pore in the A106V mutant to be open more frequently than in the wild type. A fully and stable open state might be expected to cause more drastic effects on the fluorescence properties of the protein. However, only a small portion of the L105W/Val106 channels might be in the open state in solution. Our approach looking at averaged properties cannot resolve populations.
In contrast to the finding that the MscS A106V mutation is found in an open conformation in crystals and when solubilized in solution, electrophysiological experiments indicate that it is more difficult to open when embedded in the membrane than wild type MscS. However, for two reasons, the conformer distribution in solution and the required membrane tension in electrophysiological experiments cannot be directly compared. The tension in the membrane in comparison to the micelle and direct interaction with detergent or lipid molecules is different, which, in turn, will change the energetically favorable conformations. Furthermore, the population of the open state depends on the required tension but also on the open dwell time of the channel. The observed growth inhibition suggests that occasional spontaneous gating leads to the formation of stable open channels. The most surprising observation was that lipids and LPC modified the behavior of the Trp residues in both L105W/Ala106 and L105W/Val106. The greatest change was observed for the former, and the observed properties of the Trp fluorescence in L105W/Ala106 were close to those observed for the effect of introducing the A106V mutation. The simplest interpretation of these data is that lipids and LPC cause structural modifications in both proteins, but that their effects on the Ala106 protein are to generate a more open state. Two immediate mechanisms may account for these changes. First, lipids and LPC could bind directly to MscS channels and thereby cause the structural change. The binding could influence the position of the sensor paddle and thereby cause an opening. Second, the detergent may exert some tension on the protein, which would account for the observation that the first crystallized forms of MscS and MscL are in closed states (10, 36). Lipids or LPC may then modify the tension resulting in a transition to a more open state. This interpretation would be consistent with the observation that when A106V was crystallized, it was found to be in an open state (12). A titration of LPC to L105W/Ala106 (supplemental Fig. S2) exhibited a half-saturation at ∼0.25 mm, which indicates that rather high amounts are required to observe the effect. The required concentration is also dependent on the detergent concentration. Both observations would support the second explanation that the effect is through the bulk phase rather than specific lipid-binding sites (37). Furthermore, the purified MscS still has about three phospholipid molecules associated with each subunit, which corresponds to 30–40% of the boundary layer of lipid (supplemental Fig. S3) (38, 39), consistent with phospholipids bound to MscS that are not removed under our purification regime.
The Trp fluorescence data are all consistent with detergent solubilization, allowing the L105W/Val106 protein to move into a conformation closer to the open state. Given that this channel is predominantly in the closed state in cells and in membrane patches, the data are consistent with the channel in detergent experiencing a modified tension that is insufficient to generate the completely open state unless augmented by mutations that stabilize it. Thus, it is possible that the properties of the MscS channel proteins in detergent reflect an intermediate state, close to the closed state, that can be relaxed by lipids or LPC. This observation opens a novel pathway to analysis of MscS channels outside of membrane patches. It has to be stressed, to avoid confusion, that the observed effect of LPC is not related to experiments described earlier where LPC was introduced asymmetrically to one side of the lipid bilayer in liposomes to change the bilayer tension and cause an opening of the reconstituted MscS channel (8, 40). This is particularly clear as “normal” lipids in our experiments have the same effect as LPC.
Finally, we have demonstrated that Trp, one of the smallest spectroscopic probes, is extremely useful as a conformational probe within the pore of a channel. It will allow researchers to detect channel gating independent from the patch clamp technique, for example, in reconstituted systems under changing conditions or in the living cell. These spectroscopic approaches will help to elucidate structural properties of unstable yet mechanistically important states of channels that are generally too unstable to crystallize.
Acknowledgments
We thank Andrea Raab and Jörg Feldmann (Chemistry Department, University of Aberdeen) for inductively coupled plasma mass spectrometry measurements and Peter Kapusta (PicoQuant GmbH, Berlin) for time-resolved anisotropy measurements.
This work was supported by The Wellcome Trust (Grant 040174 and 086903).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- LPC
- lysophosphatidylcholine
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DDM
- n-dodecyl-β-d-maltopyranoside
- FRET
- Förster resonance energy transfer
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-sn-glycerol)
- QY
- quantum yield
- TM
- transmembrane helix.
REFERENCES
- 1.Kung C. (2005) Nature 436, 647–654 [DOI] [PubMed] [Google Scholar]
- 2.Shapovalov G., Lester H. A. (2004) J. Gen. Physiol. 124, 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akitake B., Anishkin A., Sukharev S. (2005) J. Gen. Physiol. 125, 143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinac B., Buechner M., Delcour A. H., Adler J., Kung C. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2297–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, 3rd Ed., pp. 530–577, Springer, New York [Google Scholar]
- 6.Martinac B., Adler J., Kung C. (1990) Nature 348, 261–263 [DOI] [PubMed] [Google Scholar]
- 7.Perozo E., Kloda A., Cortes D. M., Martinac B. (2002) Nat. Struct. Biol. 9, 696–703 [DOI] [PubMed] [Google Scholar]
- 8.Vásquez V., Sotomayor M., Cordero-Morales J., Schulten K., Perozo E. (2008) Science 321, 1210–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berrier C., Coulombe A., Szabo I., Zoratti M., Ghazi A. (1992) Eur. J. Biochem. 206, 559–565 [DOI] [PubMed] [Google Scholar]
- 10.Bass R. B., Strop P., Barclay M., Rees D. C. (2002) Science 298, 1582–1587 [DOI] [PubMed] [Google Scholar]
- 11.Steinbacher S., Bass R., Strop P., Rees D. C. (2007) Curr. Top. Membr. 58, 1–24 [Google Scholar]
- 12.Wang W., Black S. S., Edwards M. D., Miller S., Morrison E. L., Bartlett W., Dong C., Naismith J. H., Booth I. R. (2008) Science 321, 1179–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anishkin A., Sukharev S. (2004) Biophys. J. 86, 2883–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards M. D., Li Y., Kim S., Miller S., Bartlett W., Black S., Dennison S., Iscla I., Blount P., Bowie J. U., Booth I. R. (2005) Nat. Struct. Mol. Biol. 12, 113–119 [DOI] [PubMed] [Google Scholar]
- 15.Miller S., Edwards M. D., Ozdemir C., Booth I. R. (2003) J. Biol. Chem. 278, 32246–32250 [DOI] [PubMed] [Google Scholar]
- 16.Rasmussen A., Rasmussen T., Edwards M. D., Schauer D., Schumann U., Miller S., Booth I. R. (2007) Biochemistry 46, 10899–10908 [DOI] [PubMed] [Google Scholar]
- 17.Booth I. R., Edwards M. D., Black S., Schumann U., Bartlett W., Rasmussen T., Rasmussen A., Miller S. (2007) in Methods in Enzymology (Sies M., Haeussinger D. eds.) pp. 47–61, Elsevier, Amsterdam, Netherlands: [DOI] [PubMed] [Google Scholar]
- 18.Levina N., Tötemeyer S., Stokes N. R., Louis P., Jones M. A., Booth I. R. (1999) EMBO J. 18, 1730–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blount P., Schroeder M. J., Kung C. (1997) J. Biol. Chem. 272, 32150–32157 [DOI] [PubMed] [Google Scholar]
- 20.Blount P., Sukharev S. I., Moe P. C., Nagle S. K., Kung C. (1996) Biol. Cell 87, 1–8 [PubMed] [Google Scholar]
- 21.Ou X., Blount P., Hoffman R. J., Kung C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11471–11475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladokhin A. S., Jayasinghe S., White S. H. (2000) Anal. Biochem. 285, 235–245 [DOI] [PubMed] [Google Scholar]
- 23.Clark E. H., East J. M., Lee A. G. (2003) Biochemistry 42, 11065–11073 [DOI] [PubMed] [Google Scholar]
- 24.Kirby E. P., Steiner R. F. (1970) J. Biol. Chem. 245, 6300–6306 [PubMed] [Google Scholar]
- 25.Valeur B., Weber G. (1977) Photochem. Photobiol. 25, 441–444 [DOI] [PubMed] [Google Scholar]
- 26.Blount P., Sukharev S. I., Schroeder M. J., Nagle S. K., Kung C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11652–11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada K., Moe P. C., Blount P. (2002) J. Biol. Chem. 277, 27682–27688 [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Barkley M. D. (1998) Biochemistry 37, 9976–9982 [DOI] [PubMed] [Google Scholar]
- 29.Adams P. D., Chen Y., Ma K., Zagorski M. G., Sönnichsen F. D., McLaughlin M. L., Barkley M. D. (2002) J. Am. Chem. Soc. 124, 9278–9286 [DOI] [PubMed] [Google Scholar]
- 30.Alston R. W., Lasagna M., Grimsley G. R., Scholtz J. M., Reinhart G. D., Pace C. N. (2008) Biophys. J. 94, 2280–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan C. P., Barkley M. D. (2004) Biophys. J. 86, 3828–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Meer B. W., Coker G., III, Chen Y. (1994) Resonance Energy Transfer: Theory and Data, p. 148, Wiley & Sons, New York [Google Scholar]
- 33.Weber G. (1960) Biochem. J. 75, 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber G. (1960) Biochem. J. 75, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards F. M. (1977) Annu. Rev. Biophys. Bioeng. 6, 151–176 [DOI] [PubMed] [Google Scholar]
- 36.Chang G., Spencer R. H., Lee A. T., Barclay M. T., Rees D. C. (1998) Science 282, 2220–2226 [DOI] [PubMed] [Google Scholar]
- 37.Wilson M. L., Dahlquist F. W. (1985) Biochemistry 24, 1920–1928 [DOI] [PubMed] [Google Scholar]
- 38.Deitrich C. L., Raab A., Pioselli B., Thomas-Oates J. E., Feldmann J. (2007) Anal. Chem. 79, 8381–8390 [DOI] [PubMed] [Google Scholar]
- 39.Marsh D., Horváth L. I. (1998) Biochim. Biophys. Acta 1376, 267–296 [DOI] [PubMed] [Google Scholar]
- 40.Vásquez V., Sotomayor M., Cortes D. M., Roux B., Schulten K., Perozo E. (2008) J. Mol. Biol. 378, 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]