Abstract
The ubiquitin fold modifier 1 (Ufm1) is the most recently discovered ubiquitin-like modifier whose conjugation (ufmylation) system is conserved in multicellular organisms. Ufm1 is known to covalently attach with cellular protein(s) via a specific E1-activating enzyme (Uba5) and an E2-conjugating enzyme (Ufc1), but its E3-ligating enzyme(s) as well as the target protein(s) remain unknown. Herein, we report both a novel E3 ligase for Ufm1, designated Ufl1, and an Ufm1-specific substrate ligated by Ufl1, C20orf116. Ufm1 was covalently conjugated with C20orf116. Although Ufl1 has no obvious sequence homology to any other known E3s for ubiquitin and ubiquitin-like modifiers, the C20orf116·Ufm1 formation was greatly accelerated by Ufl1. The C20orf116·Ufm1 conjugate was cleaved by Ufm1-specific proteases, implying the reversibility of ufmylation. The conjugation was abundant in the liver and lungs of Ufm1-transgenic mice, fractionated into membrane fraction, and impaired in Uba5 knock-out cells. Intriguingly, immunological analysis revealed localizations of Ufl1 and C20orf116 mainly to the endoplasmic reticulum. Our results provide novel insights into the Ufm1 system involved in cellular regulation of multicellular organisms.
Keywords: E3 ubiquitin ligase, Post-translational modification, Ubiquitin, Ubiquitin ligase, Ubiquitylation, Ubiquitin-like modification, Ufm1, Ufmylation
Introduction
Protein-modifying systems contribute to functional changes in target proteins and/or to amplification of genetic information. Ubiquitylation is an example of the protein modification systems in which ubiquitin, composed of 76 amino acids, is covalently conjugated with target proteins (1). The covalent modification of cellular proteins with ubiquitin regulates a diverse array of biological processes, including protein degradation, cell cycle control, DNA repair, signal/transcriptional regulation, and stress response (1–3). Ubiquitylation is carried out by an elaborate enzymatic reaction consisting of three sequential steps. In the initial step, ubiquitin is activated by a ubiquitin-activating enzyme, E1, which forms a high energy thioester bond with ubiquitin via adenylation in an ATP-dependent manner. In the second step, the E1-activated ubiquitin is transferred to a ubiquitin-conjugating enzyme, E2, in a thioester linkage. In some cases, E2 can directly transfer the ubiquitin to substrate proteins in an isopeptide linkage; however, E2 mostly requires the participation of a ubiquitin-ligating enzyme, E3, to achieve substrate-specific ubiquitylation reaction in the cells. Although only one E1 had been thought to activate ubiquitin, another novel E1, such as Uba6/E1-L2, was discovered recently in higher eukaryotes (4, 5). Compared with the two E1s, E2 forms relatively large families. Whereas there are 13 E2s in Saccharomyces cerevisiae, the human genome encodes ∼30 E2s containing a core Ubc domain composed of about 150 amino acids in addition to several E2 variants (6), indicating evolutional variability. E3 has unexpectedly enormous diversity for strict attachment of ubiquitin to the target proteins, ensuring a variety of roles in ubiquitylation. Indeed, the number of E3s, which contain a core domain such as RING, HECT, and U-box domain, is estimated to exceed 1,000 in humans (7). Similar to E3s, deubiquitylating enzymes, in which about 100 genes are encoded in the human genome, also regulate the ubiquitylation because deubiquitylating enzymes remove ubiquitin from the target protein (8–10), ensuring the reversibility of ubiquitylation.
A set of novel molecules called ubiquitin-like proteins (UBLs)4 with structural similarities to ubiquitin were identified recently (11). In addition to ubiquitylation for protein degradation, it is generally considered that protein modification by UBLs serves many proteolysis-independent events, such as molecule assembly and functional conversion of proteins. The UBLs include SUMO, NEDD8, UCRP/ISG15, FAT10, Urm1, and Ufm1 as well as Atg8 and Atg12 proteins, which are involved in autophagy. These UBLs possess the C-terminal conserved glycine residue and are covalently attached to the target proteins or phospholipids through reaction cascades in a manner analogous to the ubiquitylation pathway. The E1 and E2 involved in UBL reactions have been identified. Although the E3s for small ubiquitin-related modifier, such as protein inhibitor of activated STATs, were also discovered recently (12–15), it remains elusive whether or not E3s for other UBLs exist.
Among the UBLs, the most recently identified, Ufm1 (ubiquitin fold modifier 1), conjugates to target protein(s) via unique E1- and E2-like enzymes (16–18). Ufm1 is synthesized as a pro-form and cleaved at the C terminus by the specific cysteine proteases, UfSP1 and UfSP2, to expose the conserved glycine residue (19). Thereafter, the mature Ufm1 is activated by a specific E1-like enzyme, Uba5, forming a high energy thioester bond. The activated Ufm1 is then transferred to an E2-like enzyme, Ufc1, in a similar thioester linkage. Although Ufm1 forms covalent complexes with cellular proteins in HEK293 cells and mouse tissues (16), to date, it is still puzzling whether a specific E3 is required for the Ufm1-conjugated reaction (ufmylation). In addition, the biological roles of the Ufm1-modifying system are largely unknown. To unravel these two issues, identification of the target protein(s) for ufmylation is essential. In the present study, we identified and characterized C20orf116 as the substrate for Ufm1. Subsequently, we found an E3-ligating enzyme for Ufm1, Ufl1, by proteomics using C20orf116 as bait and finally revealed the reversibility of the C20orf116·Ufm1 conjugate via UfSPs. Furthermore, we generated Ufm1-transgenic (tg) mice and examined the dynamic nature of Ufm1 conjugation profiles in different cell compartments and tissues.
EXPERIMENTAL PROCEDURES
DNA Construction
The cDNA encoding human C20orf116 was obtained by PCR from human liver cDNA with the C20orf116-s5′ primer (5′-TGGGATCCATGTGGCGCCTGTGTGGTA-3′) and the C20orf116-r3′ primer (5′-GTGCGGCCGCTCAGGCTGGGGCTTGGGCAG-3′). It was then subcloned into pcDNA3 vector (Invitrogen). The FLAG, Myc, and GFP tags were introduced at the C terminus of C20orf116. C20orf116ΔN50 (amino acids 51–314) was generated by PCR and then subcloned into pGEX-6p-1 vector (GE Healthcare) to express GST-fused C20orf116ΔN50 in Escherichia coli. The cDNA encoding human Ufl1 (KIAA0776) was purchased from the Kazusa DNA Research Institute and subcloned into pIRESpuro3 vector (Clontech). The 3× FLAG and GFP tags were introduced at the N terminus of Ufl1. The deletion mutants of Ufl1, named Ufl1 M1 (aa 1–212), Ufl1 M2 (aa 213–794), Ufl1 M3 (aa 453–794), Ufl1 M4 (aa 1–452), and Ufl1 M5 (aa 1–654), were generated by PCR. These cDNAs were subcloned into pIRESpuro3. To express maltose-binding protein (MBP)-fused Ufl1 and Ufl1 mutants in E. coli, these cDNAs were subcloned into pMAL vector (New England BioLabs, Ipswich, MA). All mutations mentioned above were confirmed by DNA sequencing. The cDNAs encoding Ufm1 and UfSPs have been described previously (16, 19).
Cell Culture and Transfection
The media and reagents for cell culture were purchased from Invitrogen. HEK293 and MEFs were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 5 units/ml penicillin, and 50 μg/ml streptomycin. HEK293 cells at subconfluence were transfected with the indicated plasmids using the NeonTM transfection system (Invitrogen). Cells were analyzed at 48 h after transfection. Uba5+/+ and Uba5−/− MEFs prepared from day 10.5 embryos were transfected with pMESVts vector, a SV40 large T antigen expression vector (20), to establish immortalized MEFs. The immortalized cells were further transfected with C20orf116-FLAG and/or GFP-Ufm1 using the retrovirus vector system (21) and then cultured in medium containing 10 μg/ml puromycin and/or 5 μg/ml blasticidin to select stable transformants.
Short Hairpin RNA Interference against Ufl1
For plasmid-based RNA interference against HsUfl1, two oligonucleotides, siTop (5′-GATCCGAAAGTGGTCAGGTCACCATTCAAGAGATGGTGACCTGACCACTTTCTTTTTTGGAAA-3′) and siBottom (5′-AGCTTTTCCAAAAAAGAAAGTGGTCAGGTCACCATCTCTTGAATGGTGACCTGACCACTTTCG-3′), were synthesized. A fragment made by annealing these oligonucleotides was inserted into pSilencer H1 3.0 (Ambion, Austin, TX) via BamHI and HindIII restriction sites, and the resulting vector was transfected to knock down endogenous Ufl1.
Immunological Analysis
Immunoblot and immunoprecipitation analyses were performed as described previously (16). Immunoprecipitation analyses for Ufm1-conjugated proteins were carried out as described below. The cell lysates were denatured by boiling in 1% SDS-containing TNE buffer (10 mm Tris-Cl, pH 7.5, 1% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, and protease inhibitors) and then diluted by the addition of 10-fold volume TNE buffer without SDS followed by immunoprecipitation with anti-GFP or anti-FLAG antibody. 5% of total lysates used for immunoprecipitation and 20% of the immunoprecipitants were applied on each lane. The anti-C20orf116, anti-Ufl1, and anti-Uba5 polyclonal antibodies were raised in rabbits using the purified recombinant C20orf116ΔN50, MBP-Ufl1, and Uba5, respectively, as an antigen. The antibodies for Ufm1 and Ufc1 were described previously (16). The antibodies for lamin B (M-20; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), LDH (ab2101; Abcam, Inc., Cambridge, MA), cytochrome c (7H8.2C12; BD Biosciences), Bip (Affinity BioReagents, Inc., Golden, CO), actin (MAB1501R; Chemicon International, Inc., Temecula, CA), FLAG (M2; Sigma), and GFP (Invitrogen) were purchased.
Purified Recombinant Proteins
Recombinant GST-Ufm1ΔC2, GST-Ufm1ΔC3, GST-Uba5, GST-Ufc1, GST-C20orf116ΔN50, GST-UfSP1, GST-UfSP2, and GST-LC3 were produced in E. coli, and recombinant proteins were purified using glutathione-Sepharose 4B and PreScission protease according to the protocol provided by the manufacturer (Amersham Biosciences). Recombinant MBP-Ufl1 and the deletion mutants were expressed in E. coli, and these recombinant proteins were purified using amylose resin according to the protocol supplied by the manufacturer (New England BioLabs).
In Vitro Pull-down Assay
Recombinant purified MBP-Ufl1 or MBP-Ufl1 mutants and Ufc1 were mixed in TN buffer (10 mm Tris-HCl (pH 7.5) and 150 mm NaCl) for 3 h at 4 °C and subsequently precipitated with amylose resin. The mixtures were washed three times with ice-cold TNE buffer. The bound proteins were eluted using 10 mm maltose and analyzed by SDS-PAGE followed by immunoblot with anti-Ufc1 antibody and Coomassie Brilliant Blue staining.
In Vitro Ufm1 Conjugation and Deconjugation Assays
Purified recombinant proteins were dialyzed in 50 mm Tris (pH 8.5), 150 mm NaCl, and 1 mm dithiothreitol (reaction buffer). The purified Ufm1ΔC2 (0.1 μm), Uba5 (0.1 μm), Ufc1 (0.1 μm), C20orf116ΔN50 (0.1 μm), and MBP-Ufl1 (0.1 μm) were mixed in a reaction buffer containing 5 mm ATP and 10 mm MgCl2. The mixtures were incubated at 30 °C for 90 min, and the reaction was stopped by the addition of SDS-sample buffer containing 5% β-mercaptoethanol. For the deconjugation assay, the reconstitution reaction was stopped by the addition of 5 units apyrase (Sigma), followed by incubation at 30 °C for 30 min. UfSP1 (0.1–10 nm) or UfSP2 (0.1–10 nm) was added to the reaction mixture and then incubated at 30 °C for 15 min.
Generation of FLAGHis-Ufm1 tg Mice
FLAG- and His-tagged Ufm1 (FLAGHis-Ufm1) cDNA was introduced into the expression vector, pBsCAG2, which contains cytomegalovirus enhancer and chicken β-actin promoter, β-actin intron, and rabbit β-globin poly(A) signal (CAG) (22). The resulting construct (designated CAG-FLAGHis-Ufm1) was dissolved in 10 mm Tris-HCl (pH 7.6), 0.1 mm EDTA and then stored at 4 °C until use. Sperm suspension (1 × 106/29 μl) was mixed with 1 μl of the DNA solution (30 ng/μl) and incubated at ambient temperature for 2 min. Intracytoplasmic sperm injection (ICSI) was carried out according to the method of Kimura and Yanagimachi (23) with minor modifications. The oocytes were examined ∼6 h after ICSI for survival and activation. Oocytes with two well developed pronuclei were recorded at 7 h after ICSI to assess the fertilization ability. Two-cell stage embryos, cultured for ∼24 h and developed from oocytes fertilized by ICSI, were transferred to the oviducts of recipient females. 193 oocytes were microinjected, 157 (81.3%) survived injection, and 144 (91.7%) cleaved to the two-cell stage. Transfer of 144 embryos produced 49 offspring (34.0%). Of the 49 offspring generated by ICSI, four (8.2%) carried transgene DNA. To assess germ line transmission, four founder mice were mated with C57BL/6J mice. The transmission rate of the transgene to the F1 progeny, determined by PCR, was 46.7% (7 of 15), 47.4% (9 of 19), 54.5% (6 of 11), and 47.1% (8 of 17) in four founder mice (208, 503, 603, and 609 lines), respectively. These rates were within the variation range of the expected value, 50%, based on the Mendelian pattern. One of the tg lines, FLAGHis-Ufm1#503, was used in this study. Genotyping was carried out by PCR analysis. Mice were fed ad libitum a standard diet and maintained in an air-conditioned and light-controlled room (23 ± 1 °C, 55 ± 5%, 12/12 h light/dark). All animal experiments were performed in accordance with guidelines for the laboratory animal experimentation at Juntendo University School of Medicine.
Subcellular Fractionation
Fresh mouse livers were homogenized in the homogenate buffer (0.25 m sucrose, 10 mm HEPES (pH 7.4), and 1 mm dithiothreitol) using a Potter-Elvehjem homogenizer. The homogenate was centrifuged at 500 × g for 10 min. The supernatant was centrifuged at 10,000 × g for 30 min. The resultant precipitate was used as the mitochondria-lysosome fraction. The supernatant was further centrifuged at 100,000 × g for 60 min. The resulting supernatant and precipitate were used as the cytosol and microsome fractions, respectively. The nuclear fraction was prepared as described previously (24, 25).
Immunofluorescence Microscopy
HeLa cells were transfected with C20orf116-GFP or GFP-Ufl1. At 24 h after transfection, the cells were fixed with ice-cold methanol and immunostained with anti-calreticulin (Affinity BioReagents) and anti-β-COP antibodies (Affinity BioReagents). All fluorescence images were obtained using a fluorescence microscope (Q550FV; Leica) equipped with a cooled charge-coupled device camera (CTR MIC; Leica). Pictures were taken using Leica Qfluoro software (Leica).
RESULTS
Identification of Ufm1- and Ufc1-interacting Protein, C20orf116
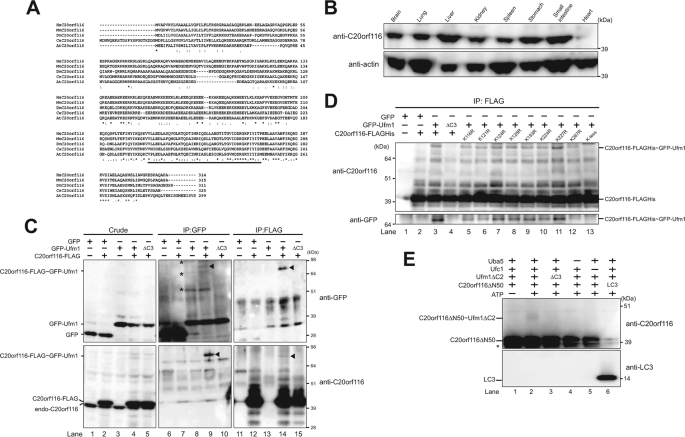
When the C-terminal conserved glycine residue in ubiquitin and Ufm1, essential for conjugation, is substituted with alanine residue, ubiquitin and Ufm1 form stable conjugates with the target proteins, which acquire resistance against attacks of deconjugating enzymes (16). Taking advantage of this property, the FLAG-tagged Ufm1G82A mutant was constructed. The mutant, in which the C-terminal glycine residue was substituted with an alanine residue, was expressed in HEK293 cells. Thereafter, the lysates were immunoprecipitated with anti-FLAG antibody, and the immunoprecipitants were then subjected to direct nanoflow LC-MS/MS analysis (26) in order to isolate the interacting protein(s), including target protein(s) (supplemental Table 1). A new uncharacterized protein, C20orf116, was identified as a Ufm1-interacting protein. C20orf116 is composed of 314 amino acids with a predicted molecular mass of ∼38 kDa and has a C-terminal proteasome-COP9-initiation factor (PCI) domain (Fig. 1A), which is found in proteins known to be involved in the formation of very large protein complexes like the proteasome and signalosome (27–29). This protein was also identified as a Ufc1-interacting protein by LC-MS/MS analysis using Ufc1 as bait (data not shown). This protein is conserved in multicellular organisms but not in yeasts, like Ufm1, Uba5, Ufc1, and UfSPs (16, 19) (supplemental Fig. 1). Immunoblot analysis with anti-C20orf116 antibody showed ubiquitous expression although with tissue-specific differences (e.g. specifically highly expressed in the liver) (Fig. 1B).
FIGURE 1.
C20orf116, a substrate for the Ufm1 modification system. A, sequence alignment of human C20orf116 (HsC20orf116) and its homologues of other species. The amino acid sequence of human C20orf116 was compared with those of other species by using the ClustalW program. *, indicate identical amino acids. Single and double dots indicate weakly and strongly similar amino acids, respectively. The underline indicates the PCI domain. B, tissue distribution of C20orf116. Homogenates prepared from various mouse tissues were analyzed by immunoblot with anti-C20orf116 and anti-actin antibodies. 20 μg of proteins were applied on each lane. C, C20orf116 is conjugated with Ufm1 in vivo. HEK293 cells were transfected with vectors at the indicated combinations. The cell lysates were immunoprecipitated (IP) with anti-GFP or anti-FLAG antibody, followed by immunoblot analysis with anti-GFP and anti-C20orf116 antibodies. The bands corresponding to GFP, GFP-Ufm1, endogenous C20orf116, C20orf116-FLAG, and C20orf116-FLAG·GFP-Ufm1 conjugate are indicated on the left. The arrowheads and asterisks indicate C20orf116-FLAG·GFP-Ufm1 conjugate and unknown proteins conjugated with GFP-Ufm1, respectively. D, determination of a lysine residue of C20orf116 for Ufm1 conjugation. GFP-Ufm1 was co-expressed with wild-type C20orf116 or the indicated mutants. The cell lysates were immunoprecipitated by anti-FLAG antibody and analyzed as described in C. E, in vitro reconstitution of C20orf116·Ufm1 conjugate. Purified recombinant C20orf116ΔN50 was incubated at 30 °C for 90 min with components of various combinations as indicated. The mixtures were boiled for 5 min with SDS-sample buffer containing 5% β-mercaptoethanol to stop the reaction. The samples were analyzed by immunoblot with anti-C20orf116 antibody. The bands corresponding to C20orf116ΔN50, LC3, and C20orf116ΔN50·Ufm1ΔC2 conjugate are indicated. *, degradative product of C20orf116ΔN50. The data shown in B–E are representative of three experiments with similar results.
C20orf116 Is a Target for Ufm1
Because C20orf116 was identified as both a Ufm1- and Ufc1-interacting protein, we first examined whether C20orf116 is a substrate for Ufm1 in vivo. To do this, GFP-tagged Ufm1 (GFP-Ufm1) was expressed alone and co-expressed with C-terminal FLAG-tagged C20orf116 (C20orf116-FLAG) in HEK293 cells. Each cell lysate was prepared and subjected to immunoprecipitation analysis with anti-GFP antibody followed by immunoblotting with anti-GFP and anti-C20of116 antibodies. Expression of GFP-Ufm1 resulted in the appearance of several bands that shifted to higher molecular weight (see the band shown by asterisks in Fig. 1C, top, lane 8). Because we did not detect any higher bands when Ufm1ΔC3 lacking the C-terminal glycine 83 of mature Ufm1 (GFP-Ufm1ΔC3) was expressed (Fig. 1C, lane 10), these higher size bands were thought to be conjugates between GFP-Ufm1 and endogenous proteins. By co-expression of GFP-Ufm1 with C20orf116-FLAG, an additional band with a molecular mass of 80 kDa (see the band shown by an arrowhead) was detected (Fig. 1C, top, lane 9). The band was also clearly recognized by anti-C20orf116 antibody (Fig. 1C, bottom, lane 9), suggesting the presence of C20orf116-FLAG·GFP-Ufm1 conjugate. To further confirm this result, C20orf116-FLAG was immunoprecipitated by anti-FLAG antibody and subsequently immunoblotted with anti-GFP and anti-C20orf116 antibodies. Consistent with the above data, a higher sized band (see the band shown by an arrowhead) was observed when C20orf116-FLAG was co-expressed with GFP-Ufm1 but not alone or with GFP-Ufm1ΔC3 (Fig. 1C, bottom, lanes 12, 14, and 15). The band was also recognized by anti-GFP antibody (Fig. 1C, top, lane 14). These results indicate the existence of the C20orf116-FLAG·GFP-Ufm1 complex. Because the conjugate had an apparent molecular mass of 80 kDa, it could be composed of one GFP-Ufm1 (40 kDa) and one C20orf116-FLAG (40 kDa).
Next, to determine the lysine residue of C20orf116 for Ufm1 conjugation, we prepared several point mutants in which each lysine residue of C20orf116 conserved across species was substituted with an arginine residue (K116R, K121R, K124R, K128R, K193R, K224R, K227R, and K267R) and a lysineless mutant in which all conserved lysine residues were substituted with arginine residues (K-less in Fig. 1D). Then we examined the effect of each mutant on the conjugate formation. As shown in Fig. 1D, immunoprecipitation experiments revealed that the K267R mutant was less likely to form a conjugate with GFP-Ufm1 than the wild-type C20orf116 and other mutants (Fig. 1D, lane 12), although some conjugate formation was still observed. In contrast, the lysineless mutant completely abolished the conjugation formation (Fig. 1D, lane 13). These results suggest that Lys-267 is the main lysine residue for Ufm1 conjugation, although other lysine residue(s) could become the site when Lys-267 was mutated. There is also a possibility that K267R mutation causes a conformational change of the PCI domain and subsequently disrupts the protein-protein interaction, leading to loss of the ufmylation.
In the next step, we tested whether Ufm1 can conjugate with C20orf116 in vitro. The in vitro reconstitution assay was performed by using recombinant proteins expressed in E. coli. Due to insolubilization of the full-length C20orf116, we used C20orf116 with the N-terminal 50 amino acids deleted, whose region is rich in hydrophobic residues (C20orf116ΔN50). Recombinant Uba5, Ufc1, mature Ufm1 (i.e. Ufm1ΔC2) with an exposed C-terminal glycine 83 residue, and C20orf116ΔN50 were purified, mixed, and incubated in the presence or absence of ATP, and then they were subjected to SDS-PAGE under reducing conditions. Ufm1ΔC3 was used as a negative control. A ∼40 kDa band, corresponding to the C20orf116ΔN50·Ufm1ΔC2 complex, was observed, which was dependent on Uba5 and Ufc1 (Fig. 1E, lanes 2, 4, and 5). This complex was not observed when the reaction mixture was ATP-free (Fig. 1E, lane 1). Moreover, Ufm1ΔC3 could not form the complex in this reaction (Fig. 1E, lane 3). Furthermore, Ufm1 did not form a conjugate with the unrelated protein LC3 in these assay conditions (Fig. 1E, lane 6), implying substrate specificity of Ufm1. Taken together, these results indicate that C20orf116 is a real substrate for Ufm1.
Identification of C20orf116- and Ufc1-interacting Protein, Ufl1
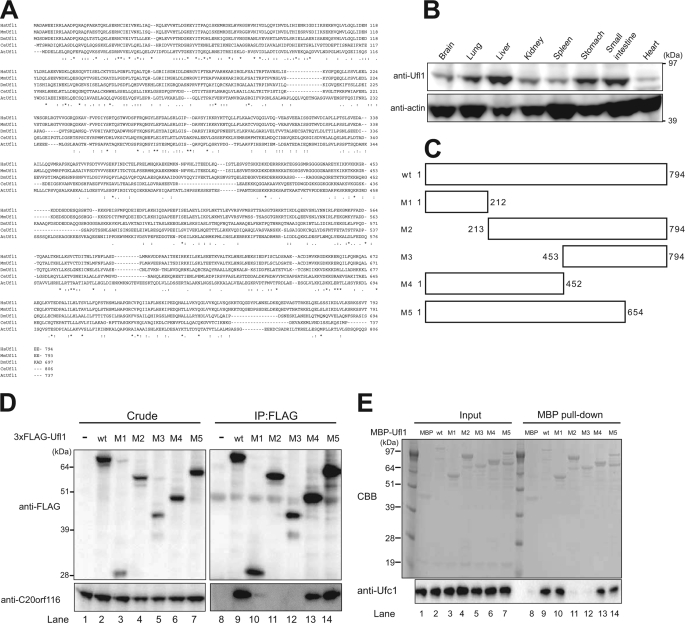
To identify the proteins that regulate C20orf116 conjugation with Ufm1, we carried out direct nanoflow LC-MS/MS analysis using C20orf116-FLAG as bait. The analysis identified an uncharacterized protein, KIAA0776, that was named Ufl1 (Ufm1-ligase 1) (supplemental Table 1; for explanation, see below). Ufl1 is composed of 794 amino acids with a predicted molecular mass of ∼90 kDa (Fig. 2A) and has no typical functional domain or homology with other known proteins. This protein was also identified as an Ufc1-interacting protein by the LC-MS/MS analysis using Ufc1 as bait (data not shown; see Fig. 2E). Like C20orf116, this protein is conserved in multicellular organisms but not in yeasts (supplemental Fig. 1). Immunoblot analysis with anti-Ufl1 antibody showed ubiquitous expression in tissues, with exceptionally high expression in the liver (Fig. 2B).
FIGURE 2.
Identification of Ufc1- and C20orf116-interacting protein, Ufl1. A, sequence alignment of human Ufl1 (HsUfl1) and its homologues. Alignment was performed as described in the legend to Fig. 1A. B, tissue distribution of Ufl1. Homogenates prepared as described in the legend to Fig. 1B were analyzed by immunoblot with anti-Ufl1 and anti-actin antibodies. C, schematic representation of constructs for Ufl1 (wt) and its deletion mutants (M1–M5). D, Ufl1 interacts with C20orf116. HEK293 cells were transfected with 3×FLAG-tagged Ufl1 or its mutants (M1–M5). The cell lysates were immunoprecipitated (IP) with anti-FLAG antibody, followed by immunoblot analysis with anti-FLAG and anti-C20orf116 antibodies. E, Ufl1 interacts with Ufc1 in vitro. Each purified recombinant MBP, MBP-Ufl1, and MBP-Ufl1 mutant (M1–M5) was incubated with recombinant Ufc1 at 4 °C for 3 h in TN buffer. The mixtures were pulled down with amyrose resin and then subjected to SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining or immunoblot with anti-Ufc1 antibody. The data shown in B, D, and E) are representative of three experiments with similar results.
N-terminal Region of Ufl1 Is Essential for Interaction with both C20orf116 and Ufc1
Next, to determine the C20orf116-interacting domain of Ufl1, we constructed a series of FLAG-tagged deletion mutants of Ufl1 (Fig. 2C), and they were expressed in HEK293 cells. Each cell lysate was prepared and then immunoprecipitated with anti-FLAG antibody followed by immunoblot analysis with anti- FLAG and anti-C20orf116 antibodies. As predicted, the wild-type Ufl1 (aa 1–794) efficiently interacted with endogenous C20orf116 (Fig. 2D, lane 9). Similarly, the C-terminal deleted mutants of Ufl1, such as M4 (aa 1–452) and M5 (aa 1–654), also interacted with C20orf116 (Fig. 2D, lanes 13 and 14). The interaction with C20orf116 was still noted in the N-terminal region of Ufl1 named M1 (aa 1–212), although the immunoblot signal was very weak (Fig. 2D, lane 10). In contrast, the N-terminal deleted mutants of Ufl1, including M2 (aa 213–794) and M3 (aa 453–794), did not interact with C20orf116 (Fig. 2D, lanes 11 and 12). We also confirmed the interaction between recombinant C20orf116 and Ufl1 M1 (supplemental Fig. 2), indicating direct interaction via the N-terminal region of Ufl1.
In contrast to C20orf116 interaction with Ufl1, there was hardly any in vivo interaction between Ufl1 and Ufc1 (data not shown). However, an in vitro pull-down assay with recombinant proteins expressed in E. coli revealed a direct interaction between Ufl1 and Ufc1 (Fig. 2E, lane 9). Furthermore, the pull-down assay with mutant recombinant Ufl1 proteins showed clearly that the N-terminal region (aa 1–212) is sufficient for the interaction with Ufc1 (Fig. 2E, lane 10). Taken together, these results suggest that in addition to the presence of the intracellular stable complex of C20orf116 and Ufl1, the interaction between Ufc1 and Ufl1 is transient in vivo, or there exists certain protein(s) that hinders the physical interaction in the cell.
Ufl1 Is an E3 Ligase for Ufm1·C20orf116 Conjugation
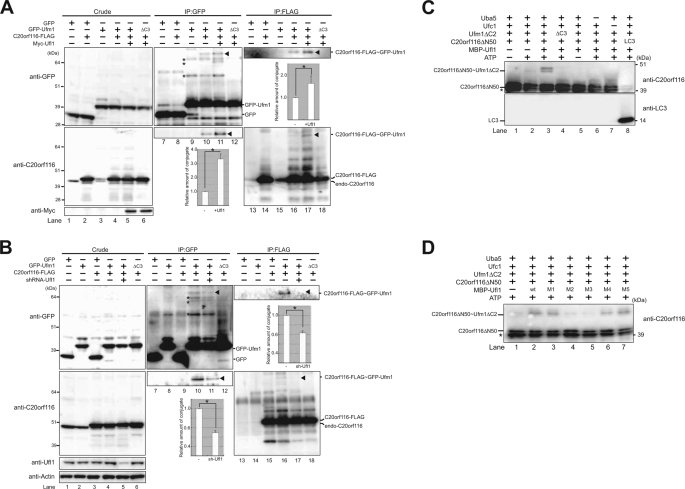
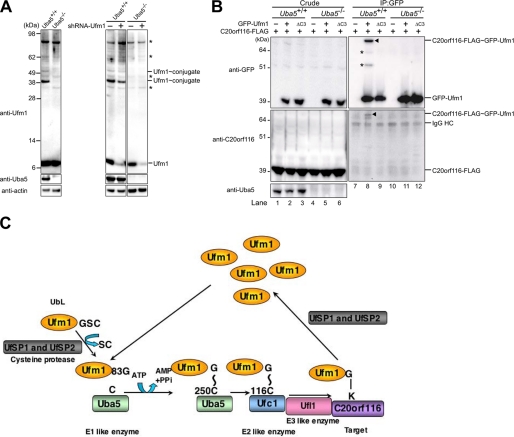
Because Ufl1 interacted with both Ufc1 and C20orf116 (Fig. 2), we postulated that Ufl1 is a ligase for C20orf116 modification by Ufm1. To test this, we examined the effect of Ufl1 overexpression on the formation of C20orf116·Ufm1 conjugate. After co-expression of proteins at the indicated combinations (Fig. 3A), the lysates were immunoprecipitated with anti-GFP or anti-FLAG antibody and then analyzed by immunoblotting with anti-GFP and anti-C20orf116 antibodies. The formation of C20orf116-FLAG·GFP-Ufm1 conjugation (see the band shown by the arrowhead) was clearly enhanced by the expression of Myc-Ufl1 (compare with lanes 10 and 11 or lanes 16 and 17 in Fig. 3A). To further verify the effects of Ufl1 on the conjugate formation, we performed knockdown analysis against Ufl1. After knockdown of endogenous Ufl1 and subsequent co-expression of proteins at the indicated combinations (Fig. 3B), the lysates were immunoprecipitated with anti-GFP or anti-FLAG antibody and then analyzed by immunoblotting with anti-GFP and anti-C20orf116 antibodies. As expected, short hairpin RNA-mediated down-regulation of Ufl1 was accompanied by a significant reduction of C20orf116-FLAG·GFP-Ufm1 conjugate (compare with lanes 10 and 11 or lanes 16 and 17 in Fig. 3B). The in vitro constitution assay clearly showed enhancement of the C20orf116 conjugation with Ufm1 following the addition of Ufl1 to the reaction (Fig. 3C, lane 3, and supplemental Fig. 3). Taken together, these results indicate that Ufl1 is an E3 ligase to ufmylate C20orf116.
FIGURE 3.
Ufl1, a novel E3-like enzyme for the Ufm1 conjugation system. A, formation of C20orf116·Ufm1 conjugate is significantly enhanced by Ufl1 in vivo. HEK293 cells were transfected with vectors at the indicated combinations, and then the cell lysates were immunoprecipitated (IP) with anti-GFP or anti-FLAG antibody. The cell lysates (Crude) and immunoprecipitants (IP) were subjected to SDS-PAGE, followed by immunoblot analysis with anti-GFP and anti-C20orf116 antibodies. The bands corresponding to GFP, GFP-Ufm1, endogenous C20orf116, C20orf116-FLAG, and C20orf116-FLAG·GFP-Ufm1 conjugate are indicated. The arrowheads and asterisks indicate C20orf116-FLAG·GFP-Ufm1 conjugate and unknown proteins conjugated with GFP-Ufm1, respectively. Quantitative densitometry of immunoblotting data from three individual experiments was performed, and the ratios of C20orf116-FLAG·GFP-Ufm1 relative to GFP-Ufm1 (middle bottom graph) and C20orf116-FLAG·GFP-Ufm1 relative to FLAG-C20orf116 (right top graph) were plotted. Data are means ± S.D. of three experiments. *, p < 0.05. B, formation of C20orf116·Ufm1 conjugate is markedly reduced by Ufl1 knockdown in vivo. At 24 h after introduction of control or Ufl1 short hairpin RNA vector, HEK293 cells were transfected with vectors at the indicated combinations, and then the cell lysates were analyzed as described in A. Quantitative densitometry of immunoblotting data from three individual experiments was performed, and the ratios of C20orf116-FLAG·GFP-Ufm1 relative to GFP-Ufm1 (middle bottom graph) and C20orf116-FLAG·GFP-Ufm1 relative to FLAG-C20orf116 (right upper graph) were plotted. Data are mean ± S.D. of three experiments. *, p < 0.05. C, Ufl1 enhances the formation of C20orf116·Ufm1 conjugate in vitro. The conjugation reactions described in Fig. 1E were performed in the presence or absence of MBP-Ufl1. The bands corresponding to C20orf116ΔN50 and C20orf116ΔN50·Ufm1ΔC2 conjugates are indicated. *, degradative form of C20orf116ΔN50. D, N-terminal region of Ufl1 is sufficient for enhancing C20orf116·Ufm1 in vitro. The conjugation reactions described in C were performed in the presence of MBP-Ufl1 and its mutants. The bands corresponding to C20orf116ΔN50 and C20orf116ΔN50·Ufm1ΔC2 conjugate are indicated. *, degradative form of C20orf116ΔN50. The data shown in A–D are representative of three experiments with similar results.
To determine the essential domain of Ufl1 necessary for the ligase activity, we carried out the in vitro reconstitution analysis using a series of Ufl1 mutants. The analysis revealed facilitation of C20orf116 conjugation with Ufm1 following the addition of C-terminal deleted mutants of Ufl1, such as M4 (aa 1–452) and M5 (aa 1–654) as well as wild-type Ufl1 (Fig. 3D, lanes 2, 6, and 7). However, the N-terminal deleted mutants of Ufl1, including M2 (aa 213–794) and M3 (aa 453–794) had no effect (Fig. 3D, lanes 4 and 5). Interestingly, the conjugation was also effectively accelerated by the addition of Ufl1 M1 (aa 1–212) (Fig. 3D, lane 3), suggesting the essential roles of the N-terminal region of Ufl1 in C20orf116 conjugation with Ufm1.
C20orf116·Ufm1 Conjugation Is Reversible
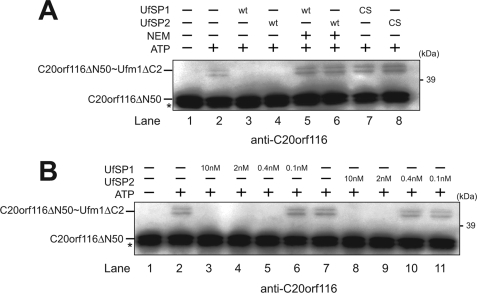
Recently, our group identified the Ufm1-specific cysteine proteases, UfSP1 and UfSP2 (19). Both enzymes can cleave out not only the C terminus of pro-Ufm1 but also Ufm1 conjugates. However, it is not clear whether or not the enzymes can cleave the C20orf116·Ufm1 conjugation. To determine the in vitro deconjugation activity of UfSPs against the C20orf116·Ufm1 conjugate, we prepared the C20orf116ΔN50·Ufm1 complex in vitro (see Fig. 3C) and then mixed it with recombinant UfSP1 or UfSP2. The complex was completely deconjugated by both UfSP1 and UfSP2 (Fig. 4A, lanes 3 and 4), indicating the reversibility of C20orf116 conjugation with Ufm1. Moreover, the cleavage was suppressed by the addition of N-ethylmaleimide (NEM), a sulfhydryl blocking agent (Fig. 4A, lanes 5 and 6), suggesting the dependence of their active site cysteine residues in the deconjugation. Indeed, substitution of the active site cysteine residue of UfSPs with serine residue (UfSP1/C53S and UfSP2/C294S) (19, 30) resulted in loss of the deconjugating activity (Fig. 4A, lanes 7 and 8). Notably, UfSP2 was much less efficient in the isopeptidase activity than UfSP1. Although 0.4 nm UfSP1 completely cleaved the C20orf116ΔN50·Ufm1 complex, 2 nm UfSP2 was required to digest it to a similar extent (Fig. 4B). Collectively, these results suggest that UfSP1 and UfSP2 catalyze the deconjugation of the C20orf116·Ufm1 complex.
FIGURE 4.
Ufm1·C20orf116 conjugation is reversible. A, deconjugation of C20orf116·Ufm1 by UfSP1 and UfSP2. The in vitro conjugation reaction described in Fig. 3C was stopped by the addition of 5 units of apyrase (lane 2) and then incubated with purified UfSP1 (10 nm) (lanes 3 and 5), UfSP1C53S (10 nm) (lane 7), UfSP2 (10 nm) (lanes 4 and 6), and UfSP2C249S (10 nm) (lane 8) at 30 °C for 15 min. UfSP1 and UfSP2 were pretreated with 1 mm N-ethylmaleimide (NEM) (lanes 5 and 6). The in vitro conjugation reaction without ATP was used as a negative control (lane 1). The mixtures were boiled for 5 min in SDS-sample buffer containing 5% β-mercaptoethanol and then subjected to SDS-PAGE, followed by immunoblot with anti-C20orf116 antibody. B, comparison of isopeptidase activity among UfSPs. The in vitro conjugation and its stop reactions were performed as described in A. The reaction mixtures were incubated with UfSP1 (lanes 3–6) or UfSP2 (lanes 8–11) at various concentrations and then analyzed as described in A. The in vitro conjugation reactions in the presence and absence of ATP were used as a positive and negative control, respectively (lanes 1 and 2). The bands corresponding to C20orf116ΔN50 and C20orf116ΔN50·Ufm1ΔC2 are indicated. *, degradative form of C20orf116ΔN50. The data shown in A and B are representative of three experiments with similar results.
Ufm1 Conjugation System in Mice
Next, to investigate the C20orf116 conjugation with Ufm1 and ufmylation in general in mice, we generated tg mice that expressed FLAGHis-Ufm1 under the control of the constitutive CAG promoter (FLAGHis-Ufm1 tg). Four tg mouse lines were obtained, and one line, FLAGHis-Ufm1#503, was used in this study. This tg line showed no gross abnormalities, such as morphology, growth, or fertilization (data not shown).
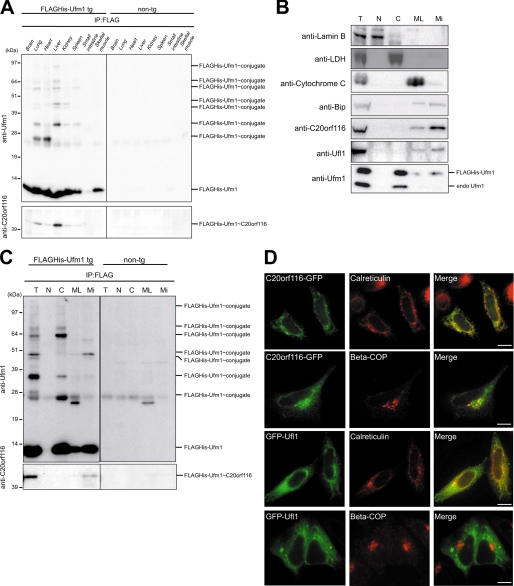
Non-tg and FLAGHis-Ufm1 tg mice were dissected, and then the lysates from each genotype tissue were immunoprecipitated with anti-FLAG antibody followed by immunoblot analysis with anti-Ufm1 and anti-C20orf116 antibodies. In addition to the 10-kDa free FLAGHis-Ufm1, several FLAGHis-Ufm1-conjugated proteins with sizes of about 25, 35, 45, 47, 55, 64, and 90 kDa in FLAGHis-Ufm1 tg, but not non-tg mice, were detected by immunoblot with anti-Ufm1 antibody (Fig. 5A, top). It is worth noting that both the number and amount of these conjugates differed among tissues, suggesting the tissue-specific roles of the Ufm1 conjugation system (Fig. 5A, top). A 45-kDa protein included in the immunoprecipitants was recognized by anti-C20orf116 antibody, implying the presence of the FLAGHis-Ufm1·C20orf116 complex (Fig. 5A, bottom). The complex was abundant in the liver and lung (Fig. 5A, bottom).
FIGURE 5.
The Ufm1 system in mice. A, differences in Ufm1 conjugates among tissues. Lysates from the indicated tissues of non-tg or FLAGHis-Ufm1 tg were immunoprecipitated (IP) with anti-FLAG antibody, and the immunoprecipitants were analyzed by immunoblot with anti-Ufm1 and anti-C20orf116 antibodies. 20 μg of proteins were applied on each lane. B, subcellular fractionation of liver of FLAGHis-Ufm1 tg. Subcellular fractionation was performed as described under “Experimental Procedures.” Each fraction was subjected to SDS-PAGE and then analyzed by immunoblot with the indicated antibodies. T, total homogenate; N, nuclear fraction; C, cytosol fraction; ML, mitochondria and lysosome fraction; Mi, microsome fraction. The total amounts of proteins in each fraction prepared from 20 μg of total homogenate were applied on each lane. C, subcellular distribution of Ufm1-conjugated proteins. Each fraction prepared as described in B was immunoprecipitated with anti-FLAG antibody, and the immunoprecipitants were analyzed by immunoblot with anti-Ufm1 and anti-C20orf116 antibodies. The data shown in A–C are representative of three experiments with similar results. D, subcellular localization of Ufl1 and C20orf116. HeLa cells were transfected with C20orf116-GFP or GFP-Ufl1 and then immunostained with anti-calreticulin or anti-β-COP antibody. The right-hand panels show merged images. Bar, 10 μm.
Next, to investigate the localization of Ufm1, C20orf116, Ufl1, and C20orf116·Ufm1 conjugate, we performed cell fractionation classified into nuclear, cytosol, mitochondria, and lysosome (ML in Fig. 5), and microsomal fractions from the livers of control and FLAGHis-Ufm1 tg mice by differential centrifugation. The fractions were analyzed quantitatively by immunoblot with the indicated antibodies (Fig. 5B). The majority of endogenously free Ufm1 and FLAGHis-Ufm1 was fractionated into the cytoplasm, whereas a small population of free Ufm1 was detected in microsomal fractions (Fig. 5B). Interestingly, C20orf116 and Ufl1 were mainly collected into the microsome fraction (Fig. 5B). Immunoprecipitation of each fraction with anti-FLAG antibody revealed Ufm1-conjugated proteins of about 25, 35, and 64 kDa in the cytosol fraction and about 45, 47, and 55 kDa in the microsomal fraction (Fig. 5C). The C20orf116·FLAGHis-Ufm1 conjugate was detected only in the microsomal fraction (Fig. 5C). We also examined the subcellular distribution of C20orf116-GFP and GFP-Ufl1 in HeLa cells. Consistent with the biochemical analysis mentioned above, immunocytochemical analysis revealed predominant localization of Ufl1 in the endoplasmic reticulum, based on co-staining with anti-calreticulin (endoplasmic reticulum marker) antibody (Fig. 5D). Similarly, C20orf1116 was mainly localized in the endoplasmic reticulum, although a small fraction was detected in the Golgi apparatus based on staining with anti-COP1 (Golgi marker) antibody (Fig. 5D).
Uba5 Is Essential for C20orf116 Conjugation with Ufm1
Finally, we investigated C20orf116 conjugation with Ufm1 in Uba5 knock-out mice. Because Uba5 knock-out mice died at embryonic day 12.5, we prepared wild type and Uba5−/− MEFs. The phenotypes of Uba5−/− mice will be described in detail elsewhere.5 We confirmed the loss of several conjugates with Ufm1, such as proteins of 39 and 51 kDa, which were otherwise observed in control MEFs, in Uba5−/− MEFs (Fig. 6A), indicating that Uba5 is really the activating enzyme for Ufm1. Next, to verify the loss of C20orf116 with Ufm1 in Uba5 knock-out mice, we stably expressed C20orf116-FLAG together with GFP-Ufm1 or GFP-Ufm1ΔC3 into wild type (Uba5+/+) or Uba5 knock-out (Uba5−/−) MEFs using the retrovirus system. The lysates were immunoprecipitated with anti-GFP antibody, and the precipitants were analyzed by immunoblot with anti-GFP and anti-C20orf116 antibodies. Contrary to conjugation of C20orf116-FLAG with GFP-Ufm1 in Uba5+/+ MEFs, such conjugation was not recognized in Uba5−/− MEFs (Fig. 6B), indicating that Uba5 is essential for C20orf116 conjugation with Ufm1.
FIGURE 6.
Impairment of Ufm1 conjugation in Uba5 knock-out mice. A, Ufm1 conjugation in Uba5 knock-out MEFs. MEFs prepared from embryonic day 10.5 Uba5+/+ and Uba5−/− were lysed, and the lysates were analyzed by immunoblot with anti-Ufm1, anti-Uba5, and anti-actin antibodies. A knockdown of Ufm1 was not accompanied by any change of the ∼35-, ∼40-, ∼62-, and ∼90-kDa Ufm1-reactive proteins, whereas levels of free Ufm1 and the 39- and 51-kDa proteins (the latter two proteins were not recognized in Uba5 knock-out MEFs) were decreased by the knockdown (right-hand panels). Thus, we concluded that the ∼35, ∼40, ∼62, and ∼90 kDa bands indicated by asterisks are nonspecific. B, Uba5 is indispensable for C20orf116 conjugation with Ufm1. Uba5+/+ (lanes 1–3) and Uba5−/− (lanes 4–6) MEFs were stably transfected with C20orf116-FLAG (lanes 1–6) together with GFP-Ufm1 (lanes 2 and 5) or GFP-Ufm1ΔC3 (lanes 3 and 6), and then the cell lysates were immunoprecipitated with anti-GFP antibody (lanes 7–12). The lysates (Crude) and immunoprecipitants (IP) were subjected to SDS-PAGE, followed by immunoblot analysis with the indicated antibodies. The bands corresponding to GFP-Ufm1, C20orf116-FLAG, C20orf116-FLAG·GFP-Ufm1, and IgG HC are indicated. *, unknown proteins conjugated with GFP-Ufm1. The data shown in A and B are representative of three experiments with similar results. C, overall features of the Ufm1 conjugation pathway. G∼C, thioester linkage; G-K, isopeptide bond.
DISCUSSION
We have reported the existence of the Ufm1-conjugating system by discovering the specific E1 (Uba5), E2 (Ufc1), and deconjugating enzymes (UfSPs) for Ufm1 (16, 19, 30). However, in addition to the lack of identification of the target protein(s), it also remains unknown whether or not an E3 ligase(s) for ufmylation is present in the cell. Herein we identified by sensitive proteomics analysis the E3 ligase for ufmylation named Ufl1, together with C20orf116 as the target protein. Based on this finding, we propose the following overall features of the Ufm1 conjugation pathway (Fig. 6C). Initially, the newly synthesized pro-Ufm1 is cleaved at the C terminus by UfSP1 and UfSP2 to expose a conserved glycine residue essential for the conjugation. The mature Ufm1 is activated by Uba5 in an ATP-dependent manner and then transferred to Ufc1. Finally, Ufm1 is covalently conjugated with the target protein (e.g. C20orf116) via Ufl1 (E3-ligating enzyme). The resulting C20orf116 conjugate with Ufm1 is cleaved by UfSPs (UfSP1 and UfSP2).
E3s for ubiquitylation are defined by three criteria: they bind the substrate (either directly or indirectly); they bind the E2; and most importantly, they stimulate the transfer of ubiquitin from the E2-ubiquitin thioester intermediate to an amide bond either with the substrate or with another ubiquitin moiety. According to this scenario, Ufl1 fulfilled all three criteria (i.e. Ufl1 interacted with C20of116 (substrate) and Ufc1 (E2) (Fig. 2) and stimulated the transfer of Ufm1 to C20orf116 both in vivo and in vitro) (Fig. 3). This conjugation reaction was not observed when ATP, E1 (Uba5), or E2 (Ufc1) was excluded, demonstrating the dependence on the E2 intermediate. Surprisingly, Ufl1 did not have any domain(s) conserved in known E3 ligases for ubiquitylation, such as RING finger, HECT, and U-box, whereas the N-terminal region highly conserved across species was basically sufficient for enhancement of C20orf116 conjugation with Ufm1 in vitro. Because the active site cysteine residue, which should be conventionally found in HECT type E3 ligase, for a thioester linkage with ubiquitin (31), was not detected in the N-terminal region of Ufl1, Ufl1 might be a scaffold type E3 ligase like RING finger E3s rather than a HECT type E3 ligase. Further characterization and structural analysis of the N-terminal region are needed for clarification of the molecular mechanism catalyzed by Ufl1. It is worth considering that besides C20orf116-Ufm1 conjugate, a conjugate (∼60 kDa) between GFP-Ufm1 and an unknown endogenous protein seemed to be promoted by forced expression of Myc-Ufl1 (Fig. 3A), suggesting that Ufl1 might have an E3 ligase activity for another unknown protein.
Ufm1-specific proteases, UfSP1 and UfSP2, can cleave both the C terminus of pro-Ufm1 and several Ufm1 conjugates, although the apparent efficiency of these two reactions seems different (19). It is generally accepted that whereas deubiquitylating enzymes containing the motif extending the core domain are responsible for deconjugation of the modifier from the conjugates, deubiquitylating enzymes composed of only the core region, such as cysteine, aspartic acid, and histidine boxes, digest the C-terminal peptide of ubiquitin and UBLs. UfSP2 has an N-terminal elongated region, which is not found in UfSP1, suggesting isopeptidase activity of UfSP2. However, UfSP1, rather than UfSP2, efficiently deconjugated Ufm1 from the C20orf116 conjugate (Fig. 4B). Because UfSP1 is conserved in higher organisms compared with UfSP2 (supplemental Fig. 1), the high efficiency of isopeptidase activity of UfSP1 might have been acquired in the late stages of evolution. However, it is possible that the difference between UfSP1 and UfSP2 in our deconjugation assay was also due to the quality of the different preparations and different protein stabilities.
Analysis of the FLAGHis-Ufm1 tg mice showed several bands corresponding to Ufm1-conjugated proteins in various mouse tissues, and both the amounts and numbers were different among tissues. These results strongly suggest strict regulation of each Ufm1 conjugate formation in a tissue-specific manner. Although we could not exclude the possibility that some of these bands correspond to multi- or poly-Ufm1 conjugates, the 45 kDa band is probably a Ufm1-conjugated C20orf116. The conjugation was recognized in many tissues but was abundant in the lung and liver, suggesting the specific role of C20orf116 modification by Ufm1 in such tissues. Intriguingly, several conjugates including C20orf116·Ufm1 were fractionated into the microsome fraction. Consistently, some populations of Ufm1 and the majority of both Ufl1 and C20orf116 were also fractionated into the microsome fraction. Moreover, Ufl1 and C20orf116 were mainly localized to the endoplasmic reticulum, suggesting that Ufm1 conjugation could regulate events closely associated with the membranes (e.g. membrane transport).
C20orf116 has a C-terminal PCI domain frequently found in proteins involved in three protein complexes: the proteasome, COP9/signalosome, and eIF3 translational initiation complex. The common presence of PCI domains in these protein complexes supports a model in which the PCI domain can play a role in assembling multiple subunits (29, 32). Importantly, the site of modification with Ufm1 in C20orf116 is most likely the Lys-267 residue within the PCI domain (Fig. 1D). Considering this issue, the conjugation might be related to the formation of a certain protein complex(es). Further analysis of our tg and knock-out mice should clarify the biological roles of Ufm1 conjugation in cells.
Acknowledgments
We thank T. Kouno and A. Sakamoto for excellent technical assistance, Dr. M. Hagiwara (Tokyo Medical and Dental University) for providing the pLRT-X retrovirus vectors, and Dr. T. Kitamura (Tokyo University) for providing the pMXs-puro retrovirus vectors.
This work was supported by grants from the Japan Science and Technology Agency (to M. K.); the Ministry of Education, Science and Culture of Japan (to M. K. and K. T.); and New Energy and Industrial Technology Development Organization (to T. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
K. Tatsumi, H. Y. Mukai, Y. Sou, T. Hara, C. H. Chung, K. Tanaka, M. Yamamoto, and M. Komatsu, manuscript in preparation.
- UBL
- ubiquitin-like protein
- UfSP
- Ufm1-specific protease
- tg
- transgenic
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- aa
- amino acids
- MEF
- mouse embryonic fibroblast
- ICSI
- intracytoplasmic sperm injection
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- PCI
- proteasome-COP9-initiation factor
- MBP
- maltose-binding protein.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 3.Pickart C. M., Eddins M. J. (2004) Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 4.Groettrup M., Pelzer C., Schmidtke G., Hofmann K. (2008) Trends Biochem. Sci. 33, 230–237 [DOI] [PubMed] [Google Scholar]
- 5.Jin J., Li X., Gygi S. P., Harper J. W. (2007) Nature 447, 1135–1138 [DOI] [PubMed] [Google Scholar]
- 6.Scheffner M., Nuber U., Huibregtse J. M. (1995) Nature 373, 81–83 [DOI] [PubMed] [Google Scholar]
- 7.Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 8.Amerik A. Y., Hochstrasser M. (2004) Biochim. Biophys. Acta 1695, 189–207 [DOI] [PubMed] [Google Scholar]
- 9.D'Andrea A., Pellman D. (1998) Crit. Rev. Biochem. Mol. Biol. 33, 337–352 [DOI] [PubMed] [Google Scholar]
- 10.Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 11.Schulman B. A., Harper J. W. (2009) Nat. Rev. Mol. Cell Biol. 10, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung C. D., Liao J., Liu B., Rao X., Jay P., Berta P., Shuai K. (1997) Science 278, 1803–1805 [DOI] [PubMed] [Google Scholar]
- 13.Liu B., Liao J., Rao X., Kushner S. A., Chung C. D., Chang D. D., Shuai K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt D., Müller S. (2003) Cell Mol. Life Sci. 60, 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuai K., Liu B. (2005) Nat. Rev. Immunol. 5, 593–605 [DOI] [PubMed] [Google Scholar]
- 16.Komatsu M., Chiba T., Tatsumi K., Iemura S., Tanida I., Okazaki N., Ueno T., Kominami E., Natsume T., Tanaka K. (2004) EMBO J. 23, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima T., Tatsumi K., Ozaki Y., Kawakami T., Suzuki A., Ogasahara K., Komatsu M., Kominami E., Tanaka K., Yamane T. (2007) Biochem. Biophys. Res. Commun. 362, 1079–1084 [DOI] [PubMed] [Google Scholar]
- 18.Sasakawa H., Sakata E., Yamaguchi Y., Komatsu M., Tatsumi K., Kominami E., Tanaka K., Kato K. (2006) Biochem. Biophys. Res. Commun. 343, 21–26 [DOI] [PubMed] [Google Scholar]
- 19.Kang S. H., Kim G. R., Seong M., Baek S. H., Seol J. H., Bang O. S., Ovaa H., Tatsumi K., Komatsu M., Tanaka K., Chung C. H. (2007) J. Biol. Chem. 282, 5256–5262 [DOI] [PubMed] [Google Scholar]
- 20.Lee G. H., Ogawa K., Drinkwater N. R. (1995) Am. J. Pathol. 147, 1811–1822 [PMC free article] [PubMed] [Google Scholar]
- 21.Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. (2003) Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 22.Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 23.Kimura Y., Yanagimachi R. (1995) Biol. Reprod. 52, 709–720 [DOI] [PubMed] [Google Scholar]
- 24.Blobel G., Potter V. R. (1966) Science 154, 1662–1665 [DOI] [PubMed] [Google Scholar]
- 25.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 26.Natsume T., Yamauchi Y., Nakayama H., Shinkawa T., Yanagida M., Takahashi N., Isobe T. (2002) Anal. Chem. 74, 4725–4733 [DOI] [PubMed] [Google Scholar]
- 27.Aravind L., Ponting C. P. (1998) Protein Sci. 7, 1250–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann K., Bucher P. (1998) Trends Biochem. Sci. 23, 204–205 [DOI] [PubMed] [Google Scholar]
- 29.Wei N., Deng X. W. (2003) Annu. Rev. Cell Dev. Biol. 19, 261–286 [DOI] [PubMed] [Google Scholar]
- 30.Ha B. H., Ahn H. C., Kang S. H., Tanaka K., Chung C. H., Kim E. E. (2008) J. Biol. Chem. 283, 14893–14900 [DOI] [PubMed] [Google Scholar]
- 31.Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayeur G. L., Fraser C. S., Peiretti F., Block K. L., Hershey J. W. (2003) Eur. J. Biochem. 270, 4133–4139 [DOI] [PubMed] [Google Scholar]