Abstract
MicroRNA 183 (miR-183) has been reported to inhibit tumor invasiveness and is believed to be involved in the development and function of ciliated neurosensory organs. We have recently found that expression of miR-183 increased after the induction of cellular senescence by exposure to H2O2. To gain insight into the biological roles of miR-183 we investigated two potential novel targets: integrin β1 (ITGB1) and kinesin 2α (KIF2A). miR-183 significantly decreased the expression of ITGB1 and KIF2A measured by Western blot. Targeting of the 3′-untranslated region (3′-UTR) of ITGB1 and KIF2A by miR-183 was confirmed by luciferase assay. Transfection with miR-183 led to a significant decrease in cell invasion and migration capacities of HeLa cells that could be rescued by expression of ITGB1 lacking the 3′-UTR. Although miR-183 had no effects on cell adhesion in HeLa cells, it significantly decreased adhesion to laminin, gelatin, and collagen type I in normal human diploid fibroblasts and human trabecular meshwork cells. These effects were also rescued by expression of ITGB1 lacking the 3′-UTR. Targeting of KIF2A by miR-183 resulted in some increase in the formation of cells with monopolar spindles in HeLa cells but not in human diploid fibroblast or human trabecular meshwork cells. The regulation of ITGB1 expression by miR-183 provides a new mechanism for the anti-metastatic role of miR-183 and suggests that this miRNA could influence the development and function in neurosensory organs, and contribute to functional alterations associated with cellular senescence in human diploid fibroblasts and human trabecular meshwork cells.
Keywords: Aging, Cell/Adhesion, Diseases/Aging, Extracellular Matrix/Integrin, Gene/Regulation, RNA/MicroRNA
Introduction
MiR-1832 is predominantly expressed in ciliated ectodermal cells and tissues including retina and hair cells in the organ of Corti and has highly conserved orthologs in both deuterostomes and protostomes. MiR-183 has been found to be up-regulated in the retina of a mouse model of retinitis pigmentosa (1) as well as in colorectal cancer (2–4). Despite its up-regulation in colorectal carcinoma cells, it has been proposed that miR-183 may inhibit the invasiveness of certain cancer cells (5). This potential anti-metastatic role of miR-183 is supported by the observations that miR-183 expression is inversely correlated with the metastatic potential of lung cancer cells. Its ectopic expression in highly metastatic cells can inhibit cell migration and invasion. Furthermore, the expression of the VIL2 coding protein Ezrin, which is known to be functionally important in cancer progression, has been demonstrated to be post-transcriptionally regulated by miR-183 (5).
Based on the pattern of tissue expression of miR-183 it has been hypothesized that this miRNA may play some role in the development and function of ciliated neurosensory organs. Specifically, it has been proposed that miR-183 could contribute to reinforcing the post-mitotic differentiated state of hair cells (6). However, functional data for this miRNA is currently limited to the work by Wang et al. (5) on lung cancer cells.
We have recently found that, although miR-183 is normally expressed only at low levels in human trabecular meshwork (HTM) cells and human diploid fibroblasts (HDF), its expression increased significantly after the induction of cellular senescence by exposure to H2O2 in these two different cell types (7). Because cellular senescence is recognized as an anticancer mechanism (8), this observation has led us to hypothesize that miR-183 could play a role in the phenotypic changes characteristic of senescent cells, and, in particular, those involved in preventing malignant transformation of aging cells. To gain insight on the biological functions of miR-183, we investigated the effects of miR-183 expression in HeLa, HTM, and HDF cells and identified KIF2A and ITGB1 as targets post-transcriptionally regulated by this miRNA.
EXPERIMENTAL PROCEDURES
Cell Culture of Primary HTM, HDF, and HeLa Cells
Postmortem human eyes or cornea rings were obtained from the New York Eye Bank within 7 days postmortem according to the tenants of the Declaration of Helsinki. TM from a single individual was dissected out from surrounding tissue, digested in 10 mg of collagenase, 20 mg of bovine serum albumin, 5 ml of phosphate-buffered saline (PBS) solution. The cells were seeded on collagen-coated 3-cm Petri dishes and maintained at 37 °C in a humidified atmosphere of 5% CO2 in TM culture medium containing 20% fetal bovine serum. The TM culture medium was low glucose Dulbecco's modified Eagle's medium with l-glutamine and 110 mg/liter of sodium pyruvate, supplemented, with 100 μm nonessential amino acids, 100 units/ml of penicillin, and 100 μg/ml of streptomycin sulfate. All reagents were obtained from Invitrogen. Human dermal fibroblast cells were commercially obtained from Cell Applications, Inc. (San Diego, CA) and grown in Dulbecco's modified Eagle's medium with high glucose medium containing 10% fetal bovine serum. Human HeLa cells were kindly provided by Dr. Ping Yang and cultured in Dulbecco's modified Eagle's medium with high glucose medium containing 10% fetal bovine serum. HTM and HDF cells were used for transfection when they reached 95% confluence. HeLa cells were transfected during 50% confluence.
Luciferase Reporter Assay
The full-length (3′-untranslated region) 3′-UTR fragments of the ITGB1 and KIF2A genes were amplified by PCR from human cDNA using ITGB1 forward (5′-TGA CTC GAG CCG TGC AAA TCC CAC AAC ACT GAA) and reverse (5′-TGA GCG GCC GCA CAT CAG AGT CAA GAC ATC CGA TTT AAG); KIG2A forward (5′-TGA CTC GAG CCG TGC CCT TTA AAC CGG CAT TT) and reverse (5′-TAT GCG GCC GCC ATA TAA GAT GGA ATG GTC CAG CAG C) that contained XhoI and NotI sites, respectively, and were cloned into a pCR2.1 vector (Invitrogen). The XhoI-NotI-digested products were then transferred to a psiChECK2 vector (Promega Corp., Madison, WI). The H293A cells were co-transfected in 12-well plates using Effectene reagent (Qiagen) with 300 ng of the 3′-UTR-luciferase reporter vector and 7 ng of miRNA mimics or negative control mimic (Dharmacon, Inc., Chicago, IL). Twenty-four hours after transfection, firefly and Renilla luciferase activities were measured consecutively using dual-luciferase assays (Promega Corp., Madison, WI) according to the manufacturer's protocol. Negative control vectors were generated by cloning the same 3′-UTRs of ITGB1 or KIF2A in reverse orientation.
RNA Isolation and Quantitative-Polymerase Chain Reaction (Q-PCR)
Total RNA was isolated using an RNeasy mini kit (Qiagen Inc.). RNA yields were measured using RiboGreen fluorescent dye (Molecular Probes, Eugene, OR). First strand cDNA was synthesized from total RNA (1 μg) by reverse transcription using oligo(dT) and Superscript II reverse transcriptase (Invitrogen). Q-PCR assays were performed in a 20-μl mixture that contained 1 μl of the cDNA preparation and 1× iQ SYBR Green Supermix (Bio-Rad), using the following PCR parameters: 95 °C for 3 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s, plus melting curve 65 °C to 95 °C (increments 0.5 °C/5 s). The fluorescence threshold value (Ct) was calculated using the iCycle system software (Bio-Rad). The absence of nonspecific products was confirmed by both analysis of the melt curves and electrophoresis in 3% Super Acryl-Agarose gels. β-Actin was used as an internal standard of RNA expression for normalization. The specific primer pairs used were listed in Table 1.
TABLE 1.
Primer pairs used for real time Q-PCR
| Gene name | GenBankTM | Primersa |
|---|---|---|
| ITGB1 | NM_002211.3 | For: 5′-AATGAATGCCAAATGGGACACGGG |
| Rev: 5′-TTCAGTGTTGTGGGATTTGCACGG | ||
| ITGB2 | NM_000211.3 | For: 5′-AAACAACATCCAGCCCATCTTCGC |
| Rev: 5′-TGAGTTGGACCACATTGCTGGAGT | ||
| ITGB4 | NM_000213 | For: 5′-TGTCCTGGTGCACAAGAAGAAGGA |
| Rev: 5′-AGGCACAGTACTTCCAGCATAGCA | ||
| ITGB5 | NM_002213.3 | For: 5′-TCGGCAAGATCTATGGGCCTTTCT |
| Rev: 5′-TAACCTGCATGGCACTTGCATTCC | ||
| ITGA1 | NM_181501.1 | For: 5′-TGCCAATGAGACAGTCCCTGAAGT |
| Rev: 5′-TGGGTACAGCACAGGGTAACCATT | ||
| ITGA2 | NM_002203.3 | For: 5′-GCACTGGAAGCCACCAAATTAGCA |
| Rev: 5′-TGGGCCTTATCCCAATCTGACCAA | ||
| ITGA3 | NM_002204.2 | For: 5′-GCAAGTGGAGCTGTGCTTTGCTTA |
| Rev: 5′-ATGGAGAAGAAGCCGTGGAAGACA | ||
| ITGA5 | NM_002205.2 | For: 5′-AATTTGACAGCAAAGGCTCTCGGC |
| Rev: 5′-ACCACTGCAAGGACTTGTACTCCA | ||
| ITGA7 | NM_002206.2 | For: 5′-AACAACAGCCCTGTTTGCACTGAG |
| Rev: 5′-AGTGCAGTGAGTCAGGAAGCATGA | ||
| ITGAV | NM_002210.3 | For: 5′-TAGCAACTCGGACTGCACAAGCTA |
| Rev: 5′-AACCATTCCCAAAGTCCTTGCTGC | ||
| AURKA | NM_003600.2 | For: 5′-TTGGGTGGTCAGTACATGCTCCAT |
| Rev: 5′-AGCAAAGAACTCCAAGGCTCCAGA | ||
| AURKB | NM_004217 | For: 5′-AAGATTGCTGACTTCGGCTGGTCT |
| Rev: 5′-ATGCACCACAGATCCACCTTCTCA | ||
| KIF2A | NM_004520 | For: 5′-ACTCTCGTACCTGCATGATTGCCA |
| Rev: 5′-TCACCAGCAGCAGTTGGATCTACA | ||
| β-Actin | NM_001101.3 | For: 5′-CCTCGCCTTTGCCGATCCG |
| Rev: 5′-GCCGGAGCCGTTGTCGACG |
a For, forward; Rev, reverse.
RT2 ProfileTM PCR Array
Human extracellular matrix and adhesion molecular PCR array (ASABiosciences Corp., Frederick, MD) was performed following the manufacturer's instructions. Briefly, total RNA was extracted using an RNeasy mini kit followed by on-column DNase I treatment (Qiagen Inc.). The first strand cDNA was synthesized using RT2 first strand kit (C-03, ASABiosciences Corp.). Real time Q-PCR was performed by loading 25 μl of experimental mixture (1275 μl of 2× SABiosciences RT2 qPCR master mixture, 102 μl of diluted first strand cDNA synthesis, 1173 μl of ddH2O) to each well of a 96-well PCR plate using the following PCR parameters: 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min, plus melting curve at 65 to 95 °C (increments 0.5 °C/5 s). The fluorescence threshold value (Ct) was calculated using the iCycle system software (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase was used as an internal standard of RNA expression for normalization.
Cell Transfection
The plasmid used for expression of ITGB1 lacking the 3′-UTR (pITGB1) was the TrueORF clone RC203818 from OriGene Technologies (Rockville, MD), which contains an open reading frame of 2397 bp encoding the ITGB1 isoform 1A precursor with a C-terminal Myc-DDK tag in a pCMV6 vector. As a control plasmid (pCon), we used psiChECK2 from Promega Corp. MiR-183 mimic (183M) or control scramble miRNA (ConM) were purchased from Dharmacon Inc. (Chicago, IL). KIF2A siRNA (KIF2A-S, siRNA ID s229482), ITGB1 siRNA (ITGB1-S, siRNA ID s7575), and control scramble siRNA (ConS) were purchased from Applied Biosystems (Foster, CA). Transfections of HDF or HTM cells were performed with a Nucleofector system (Amaxa Inc., Gaithersburg, MD) using fibroblast or endothelial nucleofection solutions and programs U23 or T23, respectively, following the manufacturer's instructions. HeLa cells were transfected using an Effectene transfection kit (Qiagen Inc.).
Protein Extraction and Immunoblot
Cells were washed twice in cold PBS and proteins were extracted using RIPA buffer (150 mm NaCl, 10 mm Tris, pH 7.2, 0.1% SDS, 1.0% Triton X-100, 5 mm EDTA, pH 8.0) containing 1× protease inhibitor mixture (Roche Applied Science). Protein concentration was determined using a Micro-BCA Protein assay kit (Pierce). Total protein extracts (40 μg) were separated by 8% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked with 5% nonfat dry milk and incubated overnight with anti-ITGB1 (catalog number 600402, BioLegend, Inc., San Diego, CA) or KIF2A (catalog number A300-914A, Bethyl Laboratories, Inc., Montgomery, TX) primary antibodies, and then with a secondary antibody conjugated to horseradish peroxidase (Pierce). Immunoreactive proteins were visualized using chemiluminescence substrate (ECL Plus, GE Healthcare). For detection of an endogenous control of protein loading, the membrane was stripped with stripping buffer (25 mm glycine, pH 3.0, plus 1% SDS) and then incubated with anti-β-tubulin (SC-9935, Santa Cruz Biotechnology, Santa Cruz, CA).
Immunocytochemistry
Cells were fixed in methanol for 15 min at −20 °C for 3 days after transfection. For detection of KIF2A, cells were then rinsed in PBS and permeabilized in PBS, 0.5% Triton X-100 for 10 min and incubated with 10% blocking buffer (goat serum in PBS, 0.1% Triton X-100) for 30 min. After the blocking buffer was removed, cells were incubated for 90 min with rabbit polyclonal primary antibody anti-KIF2A (1:100) and mouse monoclonal anti-α-tubulin fluorescein isothiocyanate (FITC) (Invitrogen) (1:500), in 2% BSA, 0.1% Triton X-100, washed five times with PBS, 0.1% Triton X-100, and incubated under the same conditions with secondary antibody Alexa Fluor® 594 goat anti-rabbit IgG (H+L) (A-11012, Invitrogen). Nuclei were stained with 4′,6-diamidino-2-phenylindole (0.5 μg/ml) for 10 min. Cells were washed three more times and the glass coverslips were mounted to glass slides with Fluoramount-G (Southern Biotech, Birmingham, AL). Cell surface ITGB1 was analyzed using rabbit polyclonal anti-ITGB1 (1:50) following the same procedure as above except that Triton X-100 was removed in all steps and anti-α-tubulin-FITC was not added. The immunofluorescence images were recorded using a Nikon C90i confocal automated microscope with a ×60 lens, and visualized with EZ-C1.3.10 Nikon confocal software. The laser 1 wavelength 408 detecting blue nuclei was set to 6.15B with a large 100-μm pinhole. Laser 2 wavelength 488 detecting green fluorescence was set 5.45B with a medium 60-μm pinhole. Laser 3 wavelength 594 detecting red fluorescence was set 7.75B with a medium 60-μm pinhole. For evaluation of the percentage of cells with monopolar spindles, two independent observers analyzed 100 mitotic figures per slide in a masked fashion and the values were averaged.
Cell Invasion Assay
Cell invasion was evaluated using a QCMTM 96-well cell invasion assay kit (Millipore, Billerica, MA) following the manufacturer's protocol with slight modification. Briefly, 24 h post-transfection, 100 μl of pre-warmed serum-free medium was added to the inserts of the invasion assay plate coated with EC MatrixTM, a reconstituted basement membrane matrix of proteins derived from the Engelbreth Holm-Swarm mouse tumor. Following a 1-h rehydration, the medium from the inserts was carefully removed and 150 μl of medium containing 10% fetal bovine serum was added to the wells of the feeder tray. Transfected HeLa cells were then trypsinized and resuspended in culture medium at 5 × 105 cells/ml, and 100 μl of the cells were added to the invasion chamber. The plate was incubated for 24 h at 37 °C in a 5% CO2 incubator. At the end of the incubation period, cells were collected separately from both the top and bottom of the invasion chamber and lysed with 1:75 CyQuant GR dye. Cell lysates (150 μl) were transferred to a 96-well black plate and fluorescence was recorded with a plate reader using a 480/520 nm filter set (SpectraMax M5, Molecular Devices, Sunnyvale, CA).
Cell Migration Assay
Cell migration was analyzed using an OrisTM cell migration assay kit (Platypus Technologies, LLC., Madison, WI) following the manufacturer's instructions. Briefly, cells were trypsinized and resuspended in culture medium at 5 × 105 cells/ml 24 h post-transfection, 100 μl of the cell suspension was added to each collagen I-coated well of the Oris plate. After overnight incubation at 37 °C in a 5% CO2 incubator, the stoppers were removed from the Oris plate and each well washed with PBS to remove any unattached cells. Attached cells were incubated with complete culture medium for 30 h and stained with 5 μm Calcein AM (Invitrogen Corp.) for 30 min. The fluorescence was then recorded using a fluorescence filter set (excitation 494 and emission 517) with and without a Oris detection mask of the Oris plate (SpectraMax M5, Molecular Devices).
Cell Adhesion Assay
Cell adhesion was analyzed using a Vybrant Cell Adhesion Assay kit (Invitrogen Corp.) according to the manufacturer's instructions. Three days (HDF and HTM cells) or 1 day (HeLa cells) post-transfection, the cells were trypsinized and washed twice with PBS and resuspended in serum-free medium containing 5 μm Calcein AM at 5 × 106 cells/ml. After incubation at 37 °C for 30 min, the cells were washed twice with warm medium, resuspended in serum-free medium at 5 × 106 cells/ml, and 100 μl of the calcein-labeled cell suspension were added to laminin (1 mg/ml), gelatin (2% solution type B), collagen type I (0.1% solution), or fibronectin (0.5 mg/ml)-coated plates. The cells were incubated at 37 °C for 60 min and the fluorescence recorded using a plate reader (SpectraMax M5, Molecular Devices) with a fluorescence filter set (excitation 494 and emission 517). The cells were then washed five times with warm serum-free medium. After the last wash, 200 μl of PBS were added to each well and the fluorescence measured again using the same settings. The results were calculated as fluorescence after wash/fluorescence before wash.
Phagocytosis of pHRodo Escherichia coli and Collagen-coated Beads
HDF or HTM cells were transfected with 183M or ConM. Two days after transfection, collagen-coated beads (0.5 μl/ml to HDF cells) or pHRodo E. coli (10 μl/ml to HTM cells) were added to the culture medium and incubated overnight. HDF cells were trypsinized and treated with trypan blue (1:20) dilutions in PBS for 1 min at 37 °C. Cells were then washed and resuspended in PBS for fluorescence-activated cell sorter analysis in the FL1 (FITC) channel. HTM cells were trypsinized and washed for fluorescence-activated cell sorter analysis in the (561-nm laser) channel.
Electron Microscopy
HDF and HTM cells were transfected with 183M or ConM. Three days post-transfection, cells were fixed with 3% glutaraldehyde in 0.15 m sodium phosphate buffer, pH 7.4, for 2 h at room temperature. The cells were then processed for EM analysis in Microscopy Services Laboratory (University of North Carolina, NC). The thin sections of 65 nm were stained with uranyl acetate/lead citrate and examined with electron microscopy (JEM-1400; JEOL USA, Peabody, MA).
Statistical Analysis
The data were presented as the mean ± S.D. Statistical significance between groups was assessed by the Mann-Whitney U test. A value of p < 0.05 was considered statistically significant.
RESULTS
Targeting of ITGB1 and KIF2A by MiR-183
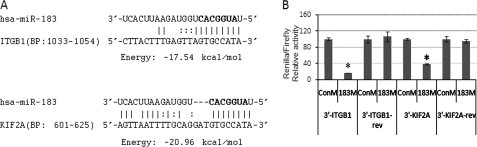
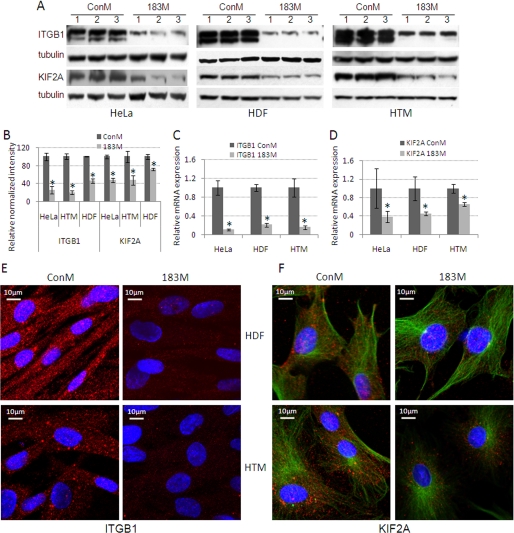
Two of the targets predicted by computational analysis for miR-183 are ITGB1 (Microcosm and PicTar-Vert) and KIF2A (Microcosm and TargetScan) (Fig. 1A). To investigate whether miR-183 was able to interact with the 3′-UTR of the ITGB1 and KIF2A mRNAs and inhibit the expression of these two genes, the complete 3′-UTRs of ITGB1 and KIF2A were cloned into a psiChECK2 dual-luciferase reporter vector. Human embryonic kidney 293 cells were co-transfected with 183M and psiChECK2 containing the 3′-UTRs of either ITGB1 or KIF2A. The results showed significantly lower expression of Renilla compared with cells transfected with the same reporter vectors and ConM. The effects of miR-183 on Renilla expression were eliminated when the 3′-UTRs of ITGB1 and KIF2A were cloned in reverse orientation (Fig. 1B). Consistent with these results, transfection of HeLa, HDF, and HTM cells with 183M led to a decrease in expression of ITGB1 and KIF2A analyzed by real time Q-PCR, Western blot, and immunocytochemistry (Fig. 2).
FIGURE 1.
Targeting of the 3′-UTRs of ITGB1 and KIF2A by miR-183. A, potential binding sites for miR-183 in the 3′-UTRs of ITGB1 and KIF2A predicted by computational analysis. BP indicates base pairs of the 3′-UTR of each gene where the sequences are located. The seed regions are highlighted in bold. B, to test for potential interactions with miR-183, the complete 3′-UTRs of ITGB1 (3′-ITGB1) and KIF2A (3′-KIF2A) were cloned into the psiChECK2 dual-luciferase reporter vector and co-transfected with either miR-183 mimic (183M) or scramble control mimic (ConM) into human embryonic kidney 293 cells. Negative controls were generated by cloning the same 3′-UTRs in reverse orientation (3′-ITGB1-rev and 3′-KIF2A-rev). The luciferase activities of Renilla (with inset) and firefly (endogenous) were analyzed 24 h post-transfection. The data represent the percentage of changes in the ratio of Renilla/firefly activities compared with the controls ± S.D. (n = 3; *, p < 0.05 by Mann-Whitney U test). Error bars are S.D.
FIGURE 2.
Decreased expression of ITGB1 and KIF2A mediated by miR-183. HeLa, HDF, and HTM cells were transfected with either 183M or ConM and the expression of ITGB1 and KIF2A was evaluated 1 (HeLa) or 3 days (HDF and HTM) post-transfection by Western blot, real time Q-PCR, and immunocytochemistry. A, Western blot analysis of ITGB1 and KIF2A protein expression from three independent transfection experiments. The two bands in the ITGB1 blots correspond to the precursor (upper band) and mature (lower band) forms of the protein. B, densitometry analysis of the Western blot data normalized with β-tubulin (n = 3; *, p < 0.05, Mann-Whitney U Test). Panels C and D represent the changes in expression of mRNA for ITGB1 and KIF2A measured by real time Q-PCR using β-ACTIN as normalization control (n = 3; *, p < 0.05, Mann-Whitney U test). Panels E and F show changes in expression of ITGB1 and KIF2A in HDF and HTM analyzed by immunocytochemistry staining with specific antibodies for ITGB1 or KIF2A (red). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue). Cells analyzed for KIF2A were also stained with specific antibodies for α-tubulin-FITC (green). The immunofluorescence images (×60) were recorded using a Nikon C90i confocal automated microscope under identical settings, and visualized with EZ-C1.3.10 Nikon confocal software. The figures are representative results from three independent experiments. Error bars are S.D.
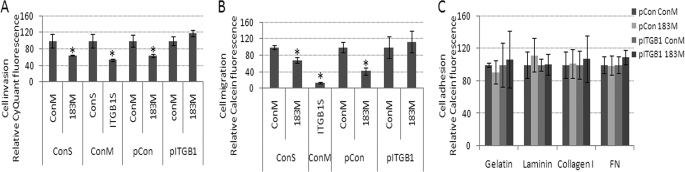
Effects of MiR-183 on Cell Migration, Invasion, and Adhesion Mediated by Inhibition of ITGB1
To test whether targeting of ITGB1 by miR-183 could result in functional effects in both cancer and normal cells, we analyzed the effects of miR-183 (183M) transfection on invasion, migration, and adhesion in HeLa cells, as well as the effects on migration and adhesion in normal HDF and HTM cells. Transfection of HeLa cells with 183M resulted in a significant decrease in their invasion and migration activities, and had no effects on their ability to adhere to laminin, gelatin, collagen type I, or fibronectin compared with ConM (Fig. 3). The effects on migration and invasion were mimicked by inhibition of ITGB1 expression with a specific siRNA (Fig. 3, A and B). Co-transfection of 183M with an expression plasmid for ITGB1 lacking 3′-UTR resulted in a significant increase on the effects of miR-183 on cell migration and invasion compared with HeLa cells transfected with 183M and control plasmid (Fig. 3, A and B).
FIGURE 3.
Decreased cell invasion and migration, but not cell adhesion in HeLa cells mediated by down-regulation of ITGB1 by miR-183. HeLa cells were co-transfected with ConM and ITGB1 siRNA (ITGB1S); ConM and control scramble siRNA (ConS); 183M and ConS; ConM and control plasmid (pCon); 183M and pCon; ConM and a plasmid expressing ITGB1 lacking the 3′-UTR (pITGB1); or 183M and pITGB1. Twenty-four hours post-transfection, cell invasion (A), cell migration (B), and cell adhesion (C) were assessed using the cell invasion assay kit (ECM555, Millipore), Oris Cell Migration Assay kit (CMA1.101, Platypus Technologies), and Vybrant Cell adhesion kit (Invitrogen) in plates coated with laminin (1 mg/ml), gelatin (2% solution type B), collagen type I (0.1% solution), or fibronectin (FN, 0.5 mg/ml). Data represent percentage of change compared with their individual controls (ConM plus ConV or ConM plus ConS) ± S.D. n = 4–6, *, p < 0.05 by Mann-Whitney U test. Error bars are S.D.
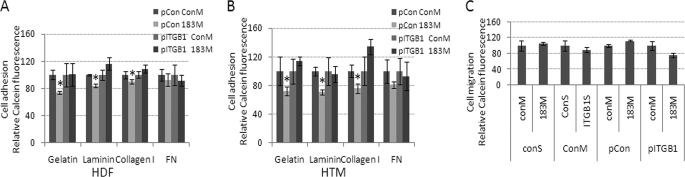
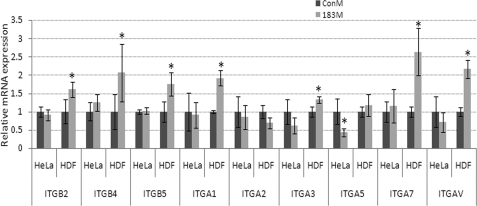
At a difference from what was observed in HeLa cells, transfection of 183M in normal HTM cells had no effects on cell migration (Fig. 4C), and led to a significant decrease in cell adhesion to laminin, gelatin, and collagen type I, in HDF and HTM cells, but not in HeLa cells compared with cells transfected with ConM plus control plasmid. Adhesion to fibronectin also showed a decrease but was not statistically significant. The effects on cell adhesion were rescued by co-transfection of a plasmid expressing ITGB1 lacking the 3′-UTR (Fig. 4, A and B). RT2 profileTM PCR array analysis of potential compensatory changes in expression of integrins associated with the inhibition of ITGB1 by miR-183 showed significant up-regulation of integrins ITGB2, ITGB4, ITGB5, and several α integrins in HTM cells (Table 2). Q-PCR analysis showed similar effects on these integrins by 183M in HDF cells. However, transfection with 183M in HeLa cells did not result in significant changes of expression of other β integrins different from ITGB1 and led to down-regulation of ITGA5 (Fig. 5).
FIGURE 4.
Decreased cell adhesion in HDF and HTM cells mediated by the down-regulation of ITGB1 by miR-183. pITGB1 or control (pCon) plasmids were co-transfected with either 183M or ConM into HDF (A) and HTM (B) cells. Three days post-transfection, cell adhesion ability was determined using the Vybrant cell adhesion kit (Invitrogen) in plates coated with laminin (1 mg/ml), gelatin (2% solution type B), collagen type I (0.1% solution), or fibronectin (FN, 0.5 mg/ml). Data represent percentage changes compared with control (ConM + pCon) ± S.D. n = 4–6, *, p < 0.05, compared with ConM plus pCon by Mann-Whitney U test. C, cell migration was not affected by miR183 in HTM cells. Assays were performed in HTM cells 3 days post-transfection using the Oris Cell Migration Assay kit. Data represent percentage changes compared with their individual controls (ConM + ConS or ConM + pCon) ± S.D. n = 4–6. Error bars are S.D.
TABLE 2.
PCR array data showed that significant alteration of genes involved extracellular matrix and adhesion molecules by miR-183 in HTM cells
| Symbol | Unigene | GeneBank | Description | Fold of 183 m vs ConM |
|---|---|---|---|---|
| ADAMTS1 | Hs.643357 | NM_006988 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | −1.66 ± 0.069 |
| COL11A1 | Hs.523446 | NM_080629 | Collagen, type XI, α 1 | 2.03 ± 0.36 |
| COL12A1 | Hs.101302 | NM_004370 | Collagen, type XII, α 1 | −1.57 ± 0.17 |
| COL15A1 | Hs.409034 | NM_001855 | Collagen, type XV, α 1 | −2.66 ± 0.18 |
| COL16A1 | Hs.368921 | NM_001856 | Collagen, type XVI, α 1 | −2.03 ± −0.15 |
| COL4A2 | Hs.508716 | NM_001846 | Collagen, type IV, α 2 | 1.15 ± 0.046 |
| COL6A1 | Hs.474053 | NM_001848 | Collagen, type VI, α 1 | −1.26 ± 0.04 |
| COL8A1 | Hs.654548 | NM_001850 | Collagen, type VIII, α 1 | 1.33 ± 0.18 |
| CTNNA1 | Hs.534797 | NM_001903 | Catenin (cadherin-associated protein), α 1, 102 kDa | 1.35 ± 0.14 |
| CTNNB1 | Hs.476018 | NM_001904 | Catenin (cadherin-associated protein), β 1, 88 kDa | 1.35 ± 0.17 |
| CTNND1 | Hs.166011 | NM_001331 | Catenin (cadherin-associated protein), δ1 | 1.29 ± 0.16 |
| ECM1 | Hs.81071 | NM_004425 | Extracellular matrix protein 1 | 1.31 ± 0.05 |
| HAS1 | Hs.57697 | NM_001523 | Hyaluronan synthase 1 | −1.74 ± 0.23 |
| ICAM1 | Hs.643447 | NM_000201 | Intercellular adhesion molecule 1 | 3.46 ± 0.24 |
| ITGA1 | Hs.644352 | NM_181501 | Integrin, α 1 | 1.86 ± 0.13 |
| ITGA2 | Hs.482077 | NM_002203 | Integrin, alpha 2 (CD49B, α 2 subunit of VLA-2 receptor) | 1.84 ± 0.07 |
| ITGA3 | Hs.265829 | NM_002204 | Integrin, alpha 3 (antigen CD49C, α 3 subunit of VLA-3 receptor) | 1.99 ± 0.5 |
| ITGA5 | Hs.505654 | NM_002205 | Integrin, α 5 (fibronectin receptor, α -polypeptide) | 1.42 ± 0.18 |
| ITGA7 | Hs.524484 | NM_002206 | Integrin, α 7 | 1.48 ± 0.26 |
| ITGAV | Hs.436873 | NM_002210 | Integrin, α V (vitronectin receptor, alpha polypeptide, antigen CD51) | 1.3 ± 0.19 |
| ITGB1 | Hs.643813 | NM_002211 | Integrin, beta 1 (fibronectin receptor, β -polypeptide, antigen CD29 includes MDF2, MSK12) | −6.47 ± 0.23 |
| ITGB2 | Hs.375957 | NM_000211 | Integrin, β 2 (complement component 3 receptor 3 and 4 subunit) | 2.57 ± 0.71 |
| ITGB4 | Hs.632226 | NM_000213 | Integrin, β 4 | 2.59 ± 0.28 |
| ITGB5 | Hs.536663 | NM_002213 | Integrin, β 5 | 1.69 ± 0.18 |
| KAL1 | Hs.521869 | NM_000216 | Kallmann syndrome 1 sequence | 1.9 ± 0.08 |
| LAMA1 | Hs.270364 | NM_005559 | Laminin, α 1 | 3.07 ± 0.92 |
| LAMA2 | Hs.200841 | NM_000426 | Laminin, α 2 | |
| LAMA3 | Hs.436367 | NM_000227 | Laminin, α 3 | 3.22 ± 0.22 |
| LAMC1 | Hs.609663 | NM_002293 | Laminin, γ1 (formerly LAMB2) | 1.33 ± 0.09 |
| MMP11 | Hs.143751 | NM_005940 | Matrix metallopeptidase 11 (stromelysin 3) | 1.59 ± 0.11 |
| MMP12 | Hs.1695 | NM_002426 | Matrix metallopeptidase 12 (macrophage elastase) | 1.81 ± 0.38 |
| MMP14 | Hs.2399 | NM_004995 | Matrix metallopeptidase 14 (membrane-inserted) | 2.01 ± 0.22 |
| MMP15 | Hs.80343 | NM_002428 | Matrix metallopeptidase 15 (membrane-inserted) | 2.23 ± 0.15 |
| MMP16 | Hs.546267 | NM_005941 | Matrix metallopeptidase 16 (membrane-inserted) | 2.18 ± 0.09 |
| MMP2 | Hs.513617 | NM_004530 | Matrix metallopeptidase 2 (gelatinase A, 72-kDa gelatinase, 72-kDa type IV collagenase) | 1.33 ± 0.22 |
| MMP9 | Hs.297413 | NM_004994 | Matrix metallopeptidase 9 (gelatinase B, 92-kDa gelatinase, 92-kDa type IV collagenase) | −1.57 ± 0.08 |
| NCAM1 | Hs.503878 | NM_000615 | Neural cell adhesion molecule 1 | 2.1 ± 0.37 |
| SPARC | Hs.111779 | NM_003118 | Secreted protein, acidic, cysteine-rich (osteonectin) | 1.61 ± 0.13 |
| SPG7 | Hs.185597 | NM_003119 | Spastic paraplegia 7 (pure and complicated autosomal recessive) | 1.41 ± 0.06 |
| SPP1 | Hs.313 | NM_000582 | Secreted phosphoprotein 1 | 3.56 ± 0.14 |
| THBS2 | Hs.371147 | NM_003247 | Thrombospondin 2 | 1.56 ± 0.32 |
| TIMP1 | Hs.522632 | NM_003254 | TIMP metallopeptidase inhibitor 1 | 1.28 ± 0.09 |
| TIMP2 | Hs.633514 | NM_003255 | TIMP metallopeptidase inhibitor 2 | 1.52 ± 0.36 |
| TNC | Hs.143250 | NM_002160 | Tenascin C | −1.71 ± 0.08 |
| VCAM1 | Hs.109225 | NM_001078 | Vascular cell adhesion molecule 1 | 1.44 ± 0.15 |
| VTN | Hs.2257 | NM_000638 | Vitronectin | 1.79 ± 0.49 |
FIGURE 5.
Effects of miR-183 on the expression of α and β integrins (ITGAs and ITGBs) in HeLa and HDF cells. HeLa and HDF cells were transfected with either 183M or ConM and the expression of ITGBs and ITGAs was evaluated 1 (HeLa) or 3 days (HDF) post-transfection by real time Q-PCR and normalized by β-actin. Specific primers of ITGBs and ITGAs are indicated in Table 1. Data represent fold-changes in cells transfected with 183M compared with control cultures transfected with ConM ± S.D. (n = 3; *, p < 0.05, Mann-Whitney U test). Error bars are S.D.
Effects of MiR-183 on Mitotic Spindle Organization
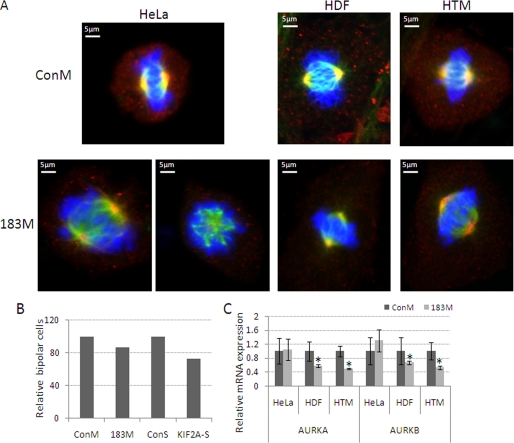
Given the reported role of KIF2A on organization of the mitotic spindle (9, 10), we investigated whether the down-regulation of this protein by miR-183 could result in an increase in the presence of cells with monopolar spindles. Transfections with 183M resulted in a 13% increase in the number of cells with monopolar spindles in HeLa cells. Similarly, inhibition of KIF2A with a specific siRNA in HeLa cells resulted in an increase of 27% in the formation of monopolar spindles (Fig. 6, A and B). However, neither 183M nor KIF2A siRNA led to a significant increase in cells with monopolar spindles in either HDF or HTM cells. Because the activity of KIF2A is known to be negatively modulated by the levels of phosphorylation mediated by aurora kinases A and B (AURKA and AURKB), we tested whether miR-183 affected the expression of these two genes by Q-PCR. The mRNA levels for both genes were substantially decreased in HDF and HTM cells, but not HeLa cells, transfected with 183M compared with those transfected with ConM (Fig. 6C).
FIGURE 6.
Effects of KIF2A and miR-183 on bipolar spindle formation. A, HeLa, HDF, and HTM cells were transfected with 183M, KIF2A siRNA (KIF2A-S), or their controls (ConM or ConS). One (HeLa) or 3 days (HDF and HTM) post-transfection cells were stained with specific antibodies for KIF2A (red), α-tubulin-FITC (green), and 4′,6-diamidino-2-phenylindole (blue). Immunofluorescence images were recorded using a Nikon C90i confocal automated microscope, and visualized with EZ-C1.3.10 Nikon confocal software. The images presented in panel A are representative results from three independent experiments. B, quantitative data of bipolar cells in HeLa cells. Data represent percentage changes of bipolar cells compared with their individual controls. C, changes in expression of mRNA of aurora kinases AURKA and AURKB measured by real time Q-PCR using specific primers and β-ACTIN was used as internal normalization control. Data represent fold-changes between cells transfected with 183M and ConM ± S.D. (n = 3, *, p < 0.05, Mann-Whitney U test). Error bars are S.D.
Effects of MiR-183 on Phagocytosis and Vacuolization in HDF and HTM Cells
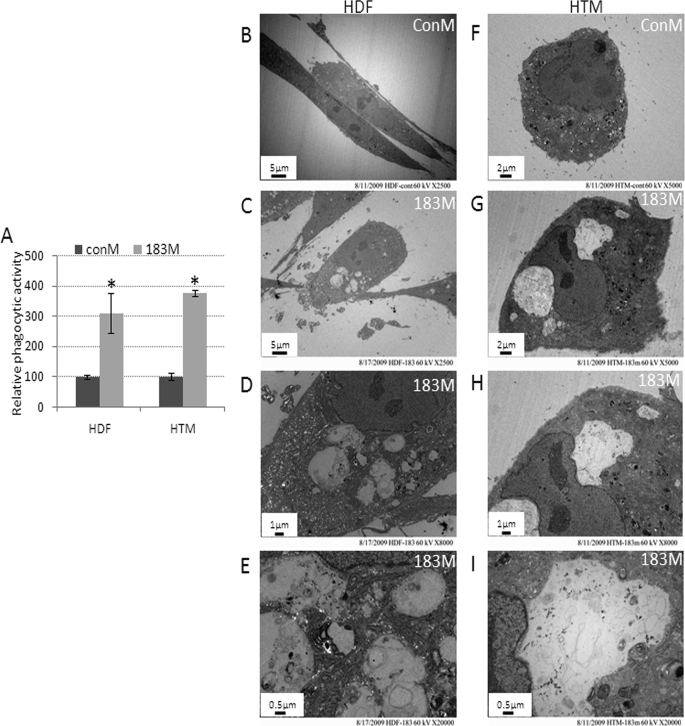
Given the known role of ITGB1 in phagocytosis, we tested whether miR-183 could affect the phagocytic activity of HDF and HTM cells. Surprisingly, transfections with 183M led to a large and statistically significant increase in the ability to phagocyte fluorescence-labeled collagen-coated beads by HDF cells (311% increase compared with ConM) as well as in the ability to phagocyte pHRodo E. coli by HTM cells (376% increase compared with ConM, Fig. 7A). This increase in phagocytic activity was associated with increased vacuolization. Analysis by electron microscopy confirmed the presence of relatively large vacuoles containing amorphous material in both HDF and HTM cells transfected with 183M compared with cells transfected with ConM (Fig. 7, B–I).
FIGURE 7.
Panel A, increase in phagocytic activity and cell vacuolization induced by miR-183 in HDF and HTM cells. Normal HDF and HTM cells were transfected with 183M or ConM. Two days post-transfection, HDF cells were incubated with collagen-coated fluorescence beads and HTM cells with pHRodo E. coli overnight. Cells were then collected and analyzed by flow cytometry in the FL1 (fluorescence isothiocyante) channel (collagen-coated fluorescence beads) or FL2 (561 nm-laser) channel (pHRodo E. coli). Data represent the change in phagocytic activity in cells transfected with 183M compared with control cultures transfected with ConM ± S.D. (n = 3–5, *, p < 0.05, Mann-Whitney U test). Panels B–I show representative electron microscopy images of increased vacuolization in HDF and HTM cells 3 days after transfection with 183M compared with cells transfected with ConM. The images (×2,500) were recorded in both ConM- (B) and 183M (C)-transfected cells. The images (×5,000) were recorded in both ConM- (F) and 183M (G)-transfected cells. Higher magnification images (×8,000 and 20,000) of vesicles present in cells transfected with 183M are also shown (D and E, HDF; H and I, HTM). The figures are representative results from three independent experiments. Error bars are S.D.
DISCUSSION
To gain a better understanding of the biological role of miR-183 we identified two novel targets genes that are post-transcriptionally regulated by this miRNA: ITGB1 and KIF2A.
The inhibition of ITGB1 by miR-183 resulted in some clear functional alterations typically associated with loss of expression of this protein. Interestingly, these effects were clearly different in HeLa cells compared with normal HDF and HTM cells.
In HeLa cells miR-183 exerted a significant inhibitory effect on migration and invasion, but not in cell adhesion. These effects on migration and invasion were prevented by expression of ITGB1 lacking the 3′-UTR, indicating that they were mediated, at least in part, by inhibition of ITGB1. The inhibition of migration and invasion of HeLa cells through direct targeting of ITGB1 is consistent with the anti-metastatic role proposed for miR-183 by Wang et al. (5) and provides an additional mechanism that might contribute to the regulation of cancer invasiveness through the inhibition of ITGB1. A large body of experimental evidence supports an essential role for ITGB1 during tumor induction and invasiveness (11–15). Elevated expression of ITGB1 and activation of ITGB1-coupled signaling have been implicated in the induction and propagation of a wide variety of human cancers (16–21). In addition, blocking ITGB1 binding activity has been shown to revert the transformed phenotype of human breast cancer cells (22, 23), and targeted disruption of ITGB1 in a transgenic mouse model of human breast cancer inhibited both the initiation and maintenance of mammary tumor growth in vivo (24).
At a difference from what was observed in HeLa cells, normal HDF and HTM cells transfected with miR-183 showed significant alterations in cell adhesion to collagen, gelatin, and laminin, but no changes in cell migration. The decrease in cell adhesion induced by miR-183 in HDF and HTM cells was dependent on the inhibition of ITGB1 because it could be prevented by expression of ITGB1 lacking the 3′-UTR. Similar effects on cell adhesion mediated by the loss of ITGB1 expression have been previously reported in endothelial cells from ITGB1-null mice (25), and can be explained because ITGB1 is a component of the main receptors for collagen (a1b1 and a2b1) and laminin (a3b1 and a6b1). Although ITGB1 is also part of the main fibronectin (a3b1, a4b1, and a5b1), transfection of miR-183 did not have significant effects on adhesion to fibronectin. These results were also consistent with those from Carlson et al. (25) in endothelial cells from ITGB1-null mice but differ from data obtained with ITGB1−/− embryonic cells (26, 27), which do not adhere to fibronectin. An explanation for these discrepancies proposed by Carlson et al. (25) is that in some cells there may be a compensatory up-regulation of other β integrins that might support fibronectin interactions in the absence of ITGB1 in vitro. Our results support this concept because transfection with miR-183 resulted in significant up-regulation of β integrins ITGB2, ITGB4, and ITGB5, as well as several α integrins.
The observed alterations in the expression of other integrins different from ITGB1 after transfection with miR-183 may also be relevant to understand the lack of effects of miR-183 on cell migration in HTM cells as well as the different responses elicited by this miRNA in HeLa cells compared with normal HDF and HTM cells. In HeLa cells, where miR-183 led to an ITGB1-dependent inhibition of cell migration, no significant compensatory up-regulation of other integrins was observed.
Together with its proposed role in cancer metastasis, miR-183 has been hypothesized to be involved in the development and function of ciliated neurosensory organs due to its preferential expression in these tissues. ITGB1 is known to play a major role in normal development of many tissues and organs (28–30), including ciliated neurosensory organs like the retina (31–35), the inner ear (36–38), and the olfactory system (39, 40). This suggests that miR-183 could influence tissue development through modulation of ITGB1. There is also increasing experimental evidence supporting an essential role for ITGB1-containing cell-matrix contacts in mechanotransduction (41, 42) Therefore, regulation of ITGB1 by miR-183 could be relevant to the specific sensorial function of some ciliated neurosensory cells, such as those in the inner ear.
Similarly, the up-regulation of miR-183 observed in senescent HDF and HTM cells and the subsequent effects on ITGB1 expression could lead to alterations in organization of actin cytoskeleton (43) and alter the ability of these cells to sense and respond to mechanical stimuli. Responses elicited by mechanical stress in the TM are believed to play an important role in the maintenance of normal levels of intraocular pressure (44–46). An increase in senescent cells has also been hypothesized to contribute to the loss in the ability of this tissue to maintain normal levels of intraocular pressure in glaucoma (47, 48). Therefore, alterations in the ITGB1 expression mediated by miR-183 in senescent cells could be a contributing factor affecting the functionality of the TM during aging and glaucoma.
In addition to ITGB1, our data showed that miR-183 also targeted KIF2A, a kinesin essential for both bipolar spindle assembly and chromosome movement (10). Cells lacking KIF2A have been shown to form monopolar spindles instead of bipolar spindles in mitosis. Although our results showed clear targeting to the 3′-UTR of the KIF2A mRNA, transfection with miR-183 resulted in a significant increase in the presence of cells with monopolar spindles only in HeLa cells, but not in HDF or HTM cells. Formation of bipolar spindles has been reported under conditions of partial knockdown of KIF2A in other cell types (9). The difference on the effects of miR-183 on the formation of monopolar spindles observed between HeLa cells and normal HDF and HTM cells are not likely to depend on differences in targeting the 3′-UTR of KIF2A because, in all cases, miR-183 induced a similar decrease in expression of both the KIF2A transcript and protein. A possible explanation for these differences may be related to the different effects of miR-183 on the expression of AURKA and AURKB in HeLa cells compared with normal HDF and HTM cells. These two kinases were down-regulated by miR-183 in HDF and HTM cells, but no expression changes were observed in HeLa cells. These two kinases negatively regulate the activity of KIF2A through phosphorylation. Therefore, a decline in AURKA and AURKB expression mediated by miR-183 in HDF and HTM cells should result in increased activity of KIF2A that could potentially compensate for the decrease in expression of this protein. The effects of miR-183 on the expression of AURKA and AURKB in HDF and HTM cells are unlikely to be mediated by direct targeting of their mRNAs because these two genes lack predicted target sites for this miRNA. Therefore, these results suggest that other still unknown targets of miR-183 may be involved in the regulation of aurora kinases.
MiR-183 induced a surprising increase on phagocytosis of collagen in HDF and E. coli HTM cells that were associated in both cases with increased vacuolization. These effects of miR-183 are unlikely to be mediated by down-regulation of ITGB1 (49), Ezrin (50, 51), or KIF2A (10). Although the observed compensatory up-regulation of other integrins may explain why inhibition of ITGB1 by miR-183 may not result in a decline in phagocytic activity, such compensatory up-regulation of integrins is not known to result by itself in an increase in phagocytic activity similar to that induced by miR-183. Therefore, this observation highlights the complexity of the biological effects mediated by miR-183 and strongly suggests the existence of additional unknown targets that remain to be identified.
In conclusion, the regulation of ITGB1 expression by miR-183 provides a new mechanism for the anti-metastatic role of miR-183 and suggests that this miRNA could influence tissue development and function in neurosensory organs and contribute to functional alterations associated with cellular senescence in HDF and HTM cells. Additional biological effects of miR-183 such as the observed increase in phagocytic activity appear to be independent from the regulation of ITGB1, KIF2A, and Ezrin, and suggest the presence of important additional targets for this miRNA.
Acknowledgments
We thank Nomingerel Tserentsoodol and Ying Hao, Duke Cancer Center, for technical support of the confocal and electron microscopes.
This work was supported, in whole or in part, by National Institutes of Health Grants EY01894, EY016228, EY019137, and EY05722 from the NEI, and Research to Prevent Blindness.
- miR-183
- microRNA 183
- HTM
- human trabecular meshwork
- HDF
- human diploid fibroblasts
- KIF2A
- kinesin 2α
- ITGB1
- integrin β1
- 183M
- miR-183 mimic
- ConM
- miRNA control mimic
- pITGB1
- expressing ITGB1 lacking 3′-UTR vector
- KIF2A-S
- KIF2A siRNA
- 3′-UTR
- 3′-untranslated region
- PBS
- phosphate-buffered saline
- Q-PCR
- quantitative-PCR
- siRNA
- small interfering RNA
- FITC
- fluorescein isothiocyanate
- AURK
- aurora kinase.
REFERENCES
- 1.Loscher C. J., Hokamp K., Kenna P. F., Ivens A. C., Humphries P., Palfi A., Farrar G. J. (2007) Genome Biol 8, R248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandrés E., Cubedo E., Agirre X., Malumbres R., Zárate R., Ramirez N., Abajo A., Navarro A., Moreno I., Monzó M., García-Foncillas J. (2006) Mol. Cancer 5, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yantiss R. K., Goodarzi M., Zhou X. K., Rennert H., Pirog E. C., Banner B. F., Chen Y. T. (2009) Am. J. Surg. Pathol. 33, 572–582 [DOI] [PubMed] [Google Scholar]
- 4.Motoyama K., Inoue H., Takatsuno Y., Tanaka F., Mimori K., Uetake H., Sugihara K., Mori M. (2009) Int. J. Oncol. 34, 1069–1075 [DOI] [PubMed] [Google Scholar]
- 5.Wang G., Mao W., Zheng S. (2008) FEBS Lett. 582, 3663–3668 [DOI] [PubMed] [Google Scholar]
- 6.Soukup G. A. (2009) Brain Res. 1277, 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G., Luna C., Qiu J., Epstein D. L., Gonzalez P. (2009) Mech. Ageing Dev. 130, 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohtani N., Mann D. J., Hara E. (2009) Cancer Sci. 100, 792–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang C. Y., Wong J., Coppinger J. A., Seki A., Yates J. R., 3rd, Fang G. (2008) J. Cell Biol. 181, 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem N. J., Compton D. A. (2004) J. Cell Biol. 166, 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsay A. G., Marshall J. F., Hart I. R. (2007) Cancer Metastasis Rev. 26, 567–578 [DOI] [PubMed] [Google Scholar]
- 12.Felding-Habermann B. (2003) Clin. Exp. Metastasis 20, 203–213 [DOI] [PubMed] [Google Scholar]
- 13.Kemperman H., Driessens M., La Rivière G., Meijne A. M., Roos E. (1994) Invasion Metastasis 14, 98–108 [PubMed] [Google Scholar]
- 14.White D. E., Muller W. J. (2007) J. Mammary Gland Biol. Neoplasia 12, 135–142 [DOI] [PubMed] [Google Scholar]
- 15.Hood J. D., Cheresh D. A. (2002) Nat. Rev. Cancer 2, 91–100 [DOI] [PubMed] [Google Scholar]
- 16.Ahmed N., Riley C., Oliva K., Stutt E., Rice G. E., Quinn M. A. (2003) J. Pathol. 201, 229–237 [DOI] [PubMed] [Google Scholar]
- 17.Casey R. C., Skubitz A. P. (2000) Clin. Exp. Metastasis 18, 67–75 [DOI] [PubMed] [Google Scholar]
- 18.Graff J. R., Deddens J. A., Konicek B. W., Colligan B. M., Hurst B. M., Carter H. W., Carter J. H. (2001) Clin. Cancer Res. 7, 1987–1991 [PubMed] [Google Scholar]
- 19.Marotta A., Tan C., Gray V., Malik S., Gallinger S., Sanghera J., Dupuis B., Owen D., Dedhar S., Salh B. (2001) Oncogene 20, 6250–6257 [DOI] [PubMed] [Google Scholar]
- 20.Oktay M. H., Oktay K., Hamele-Bena D., Buyuk A., Koss L. G. (2003) Hum. Pathol. 34, 240–245 [DOI] [PubMed] [Google Scholar]
- 21.Cannistra S. A., Ottensmeier C., Niloff J., Orta B., DiCarlo J. (1995) Gynecol. Oncol. 58, 216–225 [DOI] [PubMed] [Google Scholar]
- 22.Wang F., Hansen R. K., Radisky D., Yoneda T., Barcellos-Hoff M. H., Petersen O. W., Turley E. A., Bissell M. J. (2002) J. Natl. Cancer Inst. 94, 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weaver V. M., Petersen O. W., Wang F., Larabell C. A., Briand P., Damsky C., Bissell M. J. (1997) J. Cell Biol. 137, 231–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White D. E., Kurpios N. A., Zuo D., Hassell J. A., Blaess S., Mueller U., Muller W. J. (2004) Cancer Cell 6, 159–170 [DOI] [PubMed] [Google Scholar]
- 25.Carlson T. R., Hu H., Braren R., Kim Y. H., Wang R. A. (2008) Development 135, 2193–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens L. E., Sonne J. E., Fitzgerald M. L., Damsky C. H. (1993) J. Cell Biol. 123, 1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fässler R., Pfaff M., Murphy J., Noegel A. A., Johansson S., Timpl R., Albrecht R. (1995) J. Cell Biol. 128, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanasaki K., Kanda Y., Palmsten K., Tanjore H., Lee S. B., Lebleu V. S., Gattone V. H., Jr., Kalluri R. (2008) Dev. Biol. 313, 584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wederell E. D., de Iongh R. U. (2006) Semin. Cell Dev. Biol. 17, 759–776 [DOI] [PubMed] [Google Scholar]
- 30.De Arcangelis A., Georges-Labouesse E. (2000) Trends Genet. 16, 389–395 [DOI] [PubMed] [Google Scholar]
- 31.Skeith A., Dunlop L., Galileo D. S., Linser P. J. (1999) Brain Res. Dev. Brain Res. 116, 123–126 [DOI] [PubMed] [Google Scholar]
- 32.Clegg D. O., Mullick L. H., Wingerd K. L., Lin H., Atienza J. W., Bradshaw A. D., Gervin D. B., Cann G. M. (2000) Results Probl. Cell Differ. 31, 141–156 [DOI] [PubMed] [Google Scholar]
- 33.Marrs G. S., Honda T., Fuller L., Thangavel R., Balsamo J., Lilien J., Dailey M. E., Arregui C. (2006) Mol. Cell. Neurosci. 32, 230–241 [DOI] [PubMed] [Google Scholar]
- 34.Leu S. T., Jacques S. A., Wingerd K. L., Hikita S. T., Tolhurst E. C., Pring J. L., Wiswell D., Kinney L., Goodman N. L., Jackson D. Y., Clegg D. O. (2004) Dev. Biol. 276, 416–430 [DOI] [PubMed] [Google Scholar]
- 35.Cann G. M., Bradshaw A. D., Gervin D. B., Hunter A. W., Clegg D. O. (1996) Dev. Biol. 180, 82–96 [DOI] [PubMed] [Google Scholar]
- 36.Littlewood Evans A., Müller U. (2000) Nat. Genet. 24, 424–428 [DOI] [PubMed] [Google Scholar]
- 37.Davies D., Holley M. C. (2002) J. Comp. Neurol. 445, 122–132 [DOI] [PubMed] [Google Scholar]
- 38.Toyama K., Ozeki M., Hamajima Y., Lin J. (2005) Hear Res. 201, 21–26 [DOI] [PubMed] [Google Scholar]
- 39.Emsley J. G., Hagg T. (2003) Exp. Neurol. 183, 273–285 [DOI] [PubMed] [Google Scholar]
- 40.Belvindrah R., Hankel S., Walker J., Patton B. L., Müller U. (2007) J. Neurosci. 27, 2704–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puklin-Faucher E., Sheetz M. P. (2009) J. Cell Sci. 122, 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedland J. C., Lee M. H., Boettiger D. (2009) Science 323, 642–644 [DOI] [PubMed] [Google Scholar]
- 43.Filla M. S., Woods A., Kaufman P. L., Peters D. M. (2006) Invest. Ophthalmol. Vis. Sci. 47, 1956–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnstone M. A. (2004) J. Glaucoma 13, 421–438 [DOI] [PubMed] [Google Scholar]
- 45.Bradley J. M., Kelley M. J., Rose A., Acott T. S. (2003) Invest. Ophthalmol. Vis. Sci. 44, 5174–5181 [DOI] [PubMed] [Google Scholar]
- 46.Borrás T. (2003) Prog. Retin. Eye Res. 22, 435–463 [DOI] [PubMed] [Google Scholar]
- 47.Luna C., Li G., Liton P. B., Epstein D. L., Gonzalez P. (2009) Mol. Vis. 15, 534–544 [PMC free article] [PubMed] [Google Scholar]
- 48.Chow J., Liton P. B., Luna C., Wong F., Gonzalez P. (2007) Mol. Vis. 13, 1926–1933 [PMC free article] [PubMed] [Google Scholar]
- 49.Dupuy A. G., Caron E. (2008) J. Cell Sci. 121, 1773–1783 [DOI] [PubMed] [Google Scholar]
- 50.Lugini L., Lozupone F., Matarrese P., Funaro C., Luciani F., Malorni W., Rivoltini L., Castelli C., Tinari A., Piris A., Parmiani G., Fais S. (2003) Lab. Invest. 83, 1555–1567 [DOI] [PubMed] [Google Scholar]
- 51.Fais S. (2007) Cancer Lett. 258, 155–164 [DOI] [PubMed] [Google Scholar]