Abstract
JNK pathway-associated phosphatase (JKAP, also named DUSP22) is expressed in various tissues, indicating that JKAP may have an important biological function. We showed that JKAP localized in the actin filament-enriched region. Expression of JKAP reduced cell migration, whereas a JKAP mutant lacking catalytic activity promoted cell motility. JKAP efficiently removed tyrosine phosphorylation of several proteins. We have identified focal adhesion kinase (FAK) as a substrate of JKAP. Overexpression of JKAP, but not JKAP mutant lacking catalytic activity, decreased FAK phosphorylation at tyrosines 397, 576, and 577 in H1299 cells. Consistent with these results, decreasing JKAP expression by RNA interference promoted cell migration and Src-induced FAK phosphorylation. Taken together, this study identified a new role for JKAP in the modulation of FAK phosphorylation and cell motility.
Keywords: Cell/Motility, Phosphorylation, Phosphorylation/Kinases/Tyrosine, Phosphorylation/Phosphatases/Tyrosine, Signal Transduction/Phosphoprotein Phosphatases/Dual Specificity, Tyrosine Kinase
Introduction
Dual specificity phosphatases (DUSPs),3 including MAPK phosphatases, can dephosphorylate both tyrosine and serine/threonine residues (1). Although DUSPs display limited sequence identity to the classical protein-tyrosine phosphatases (PTPs), they use the same cysteine-based catalytic mechanism and share striking structural similarity with the PTPs. Most MAPK phosphatases consist of a conserved catalytic region and an extended regulatory region, the cdc25 homology domain (2, 3). Some DUSPs lack this regulatory domain and, accordingly, have a low molecular weight; they have been classified as atypical DUSPs (4). JNK pathway-associated phosphatase (JKAP), an atypical DUSP, was originally identified from a differential display analysis of genes that are preferentially expressed in murine hematopoietic stem cells (5). JKAP is also named as VHR-related MKPX (VHX) (6), JNK stimulatory phosphatase-1 (JSP-1) (7), and LMW-DSP2 (8). This gene is now designated as DUSP22. JKAP has a canonical PTP signature motif, HCXXGXXR, at residues 87–94. JKAP is expressed in various types of tissues and cells (5), suggesting that JKAP may participate in essential biological processes. Previous studies have demonstrated that JKAP selectively activates JNK in human embryonic kidney 293T cells (5) and COS-1 cells (7). JKAP-deficient murine embryonic fibroblasts lack JNK activation in response to tumor necrosis factor α and transforming growth factor β (5). In contrast, JKAP was also shown to dephosphorylate and inactivate JNK and p38, but not ERK, in transfected COS-7 cells (8). JKAP expression has been found to suppress T cell antigen receptor-induced ERK2 activation in Jurkat T cells (6). The effects of JKAP on the activation of MAPKs are controversial, implying JKAP may exert distinct properties dependent on cell type and tissue specificity. Additionally, JKAP also acts as a negative regulator through decreased phosphorylation of estrogen receptor-α and STAT3 in estrogen- and interleukin-6/leukemia inhibitory factor-mediated signaling pathways, respectively (9, 10). Thus, JKAP may have multiple physiological substrates and participate in various signaling cascades.
The dynamic change of focal adhesions plays a central role in cell migration. Many proteins in focal adhesions, such as cytoskeletal proteins and signaling proteins, are regulated by phosphorylation (11). Although the roles of kinases in focal adhesions have been elucidated, the importance of phosphatases remains largely unknown. Focal adhesion kinase (FAK) is associated with the formation of focal contacts and is activated by tyrosine phosphorylation (12). Phosphorylation on Tyr-397, the autophosphorylated residue in FAK, creates an Src-binding site (13). Phosphorylation of FAK by Src on Tyr-576 and Tyr-577 within the catalytic domain, in turn, promotes the optimal activation of FAK (14). FAK could influence the cytoskeleton, structures of cell adhesion, and membrane protrusions to regulate cell motility (15). In this study, we show that FAK is a substrate of JKAP. Furthermore, JKAP co-localizes with actin filaments. By manipulating the expression and activity of JKAP, we have demonstrated a novel role of this phosphatase in coordination of cell motility through regulating FAK phosphorylation.
EXPERIMENTAL PROCEDURES
Plasmids
A human JKAP-encoding fragment was amplified using a cDNA pool of H1299 cells as a template. The fragment was inserted between BamHI and XhoI sites of pcDNA4/TO/myc-His B vector (Invitrogen). JKAP-C88S (JKAP-CS) mutant-expressing vector was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and was confirmed by DNA sequencing. The DNA fragment of wild type and mutant JKAP vectors was enzyme-digested, isolated, and inserted into pTriEX-GFP (−) vectors between the MluI and XhoI sites to form JKAP-GFP expression vector. The DNA fragments were also enzyme-digested and inserted into pGEX-4T-3 (GE Healthcare) vectors between the BamHI and XhoI sites to form glutathione S-transferase (GST)-JKAP and GST-JKAP-CS expression vectors. The interference RNA against human JKAP was generated using the pSUPER-retro.puro vector (OligoEngine, Seattle, WA) as a backbone. The 19-nucleotide sequences 5′-TACCTGTGCATCCCAGCAG-3′ (shJKAP-1) and 5′- ACACTGGTGATCGCATACA-3′ (shJKAP-2), corresponding to human JKAP coding sequence residues 148–166 and 289–307, respectively, were used for designing the inverted repeat insertion in pSUPER-retro.puro vector according to the manufacturer's instructions.
Establishment of Stably Transfected Cells
H1299 cells were transfected with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. To establish permanent tetracycline-inducible JKAP-expressing cells, H1299 cells were transfected with pcDNA4/TO-JKAP and pcDNA4/TO-JKAP-CS, respectively, and then transfected with pTriEX-TR-Neo vector, which encodes the tetracycline repressor (TR). Multiple clones (six to eight) from an individual transfection were then mixed to form H1299-JKAPTR and H1299-JKAP-CSTR cells. H1299-shJKAP-1, H1299-shJKAP-2, and H1299-pSUPER cells were generated by transfecting the pSUPER-shJKAP-1, pSUPER-shJKAP-2, or pSUPER-retro.puro plasmids, respectively.
Reagents and Antibodies
Tetracycline was purchased from Invitrogen. The other chemicals were purchased from Sigma. The anti-Myc monoclonal antibody was prepared from a culture of hybridoma clone 9E10 (American Type Culture Collection, Manassas, VA). The antibodies against phosphotyrosine 4G10 and FAK were from Upstate Biotechnology, Inc. (Waltham, MA). The anti-JKAP (DUSP22) and anti-FAK-pY397 antibodies were purchased from Abnova Corp. (Taipei, Taiwan) and Abcam Inc. (Cambridge, MA), respectively. The antibodies against Src, paxillin, and p130Cas (p130 Crk-associated substrate) were from Santa Cruz Biotechnology (Santa Cruz, CA). The antibody against β-actin was from Sigma. The other antibodies were purchased from Cell Signaling Technology (Beverly, MA). The peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG secondary antibodies were from Pierce.
Fluorescence Staining Analysis
Tested cells were rinsed with phosphate-buffered saline and fixed in 4% paraformaldehyde. After permeation with 0.5% Triton X-100, the cells were blocked with 5% bovine serum albumin. The treated cells were incubated with primary antibody or TRITC-conjugated phalloidin followed by the fluorescence-conjugated secondary antibody (Molecular Probes). Nuclei were detected with 4′,6-diamidino-2-phenylindole (Molecular Probes). The cover slides were mounted in mounting medium (Biomeda) and analyzed by Leica TCS SP2 confocal microscope (Mannheim, Germany).
Cell Migration and Gap-closing Assay
Cell migration was performed in a standard Transwell assay (Costar). The Transwell-migrated cells were fixed in methanol, stained with Giemsa solution, and counted under a phase-contrast microscope. Results were presented as the number of cells per 10 fields. Gap-closing assay was performed using ibidi μ-dish with two-chamber inserts (ibidi GmbH, Munich, Germany). After cells became confluent, the cell-free gap of 500 μm was created by removing the two-chamber insert. Cells migrating into the gap region were monitored at 0, 15, and 24 h. Results presented the data from gap regions of three fields.
Cell Extract Preparation, GST Pulldown, and Immunoprecipitation
Cell extracts were prepared with lysis buffer (50 mm Tris (pH 8.0), 150 mm NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 2 μg/ml leupeptin, 5 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and 1 mm Na3VO4). GST fusion proteins were produced in the BL-21 strain of Escherichia coli and prepared according to the manufacturer's instructions. Cell extracts were pulled down with GST-Src-SH2 fusion proteins and GSH beads and precipitated by incubation with the anti-FAK antibody and protein G-agarose beads in lysis buffer with continuous rotation at 4 °C. The pulldown lysates and the precipitates were washed three times with lysis buffer before subjecting to phosphatase assays and Western blot analyses.
Phosphatase Assays and Western Blot Analyses
The pulldown complexes and immunoprecipitated complexes were washed once with phosphatase buffer (50 mm Tris (pH 7.0), 50 mm BisTris, 100 mm sodium acetate, and 10 mm dithiothreitol). The phosphatase reaction was performed at 37 °C for 30 min and then terminated by adding SDS sample buffer and heating at 95 °C for 5 min. The reaction mixtures and equal amount of cell extracts from each set of experiments were fractionated on SDS-PAGE. The protein bands were then transferred to polyvinylidene difluoride membranes and probed with primary antibody followed by the peroxidase-conjugated secondary antibody. Antibody reaction was performed using the SuperSignal reagent (Pierce) and exposed to x-ray films.
RESULTS
JKAP Regulates Cell Migration
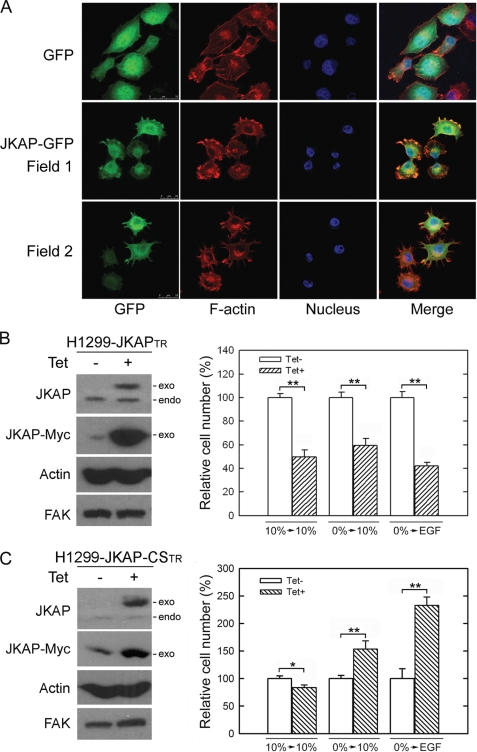
To investigate the intracellular localization of JKAP, we constructed a GFP-tagged JKAP expression vector and transfected it into H1299 cells. H1299-JKAP-GFP cells were stained with TRITC-labeled phalloidin for actin filaments. JKAP-GFP distributed throughout the whole cell but mainly in the cytoplasm (Fig. 1A). JKAP-GFP also co-localized with actin filaments in the cell periphery (Fig. 1A), indicating JKAP participation in cell motility. JKAP-CS was a mutant with a cysteine 88 to serine substitution, which caused the loss of phosphatase activity (5). The tetracycline-inducible cell lines for JKAP and JKAP-CS, H1299-JKAPTR and H1299-JKAP-CSTR, were established for examining the cellular function of JKAP (Fig. 1, B and C). The expression of Myc-tagged JKAP and JKAP-CS in H1299 cells was induced by the addition of tetracycline. The relative levels of endogenous and exogenous JKAP were determined by an anti-JKAP (DUSP22) antibody (Fig. 1, B and C). The JKAP expression reduced growth factor-induced and spontaneous cell migration (Fig. 1B, p < 0.01). In contrast, JKAP-CS expression enhanced growth factor-induced cell migration (Fig. 1C, p < 0.05), suggesting that the JKAP-CS mutant exhibits a dominant-negative function. These results indicate that JKAP could modulate cell migration and that this function is dependent on the phosphatase activity of JKAP.
FIGURE 1.
JKAP co-localizes with actin filaments and reduces cell migration. A, H1299 cells were stably transfected with GFP (upper panel) or GFP-tagged JKAP (lower panel, Field 1 and Field 2) and stained with TRITC-conjugated phalloidin for F-actin and 4′,6-diamidino-2-phenylindole for nucleus. B and C, H1299-JKAPTR cells and H1299-JKAP-CSTR cells were cultured with or without 2 μg/ml tetracycline (Tet) for 15 h. The protein levels and cell migration abilities were examined by Western blotting (left panel) or by Transwell assays (right panel), respectively. EGF, epidermal growth factor; exo, exogenous JKAP; endo, endogenous JKAP. Quantitative data were means ± S.E. of one representative result from three independent experiments. *, p < 0.05; **, p < 0.01, compared with the Tet− group.
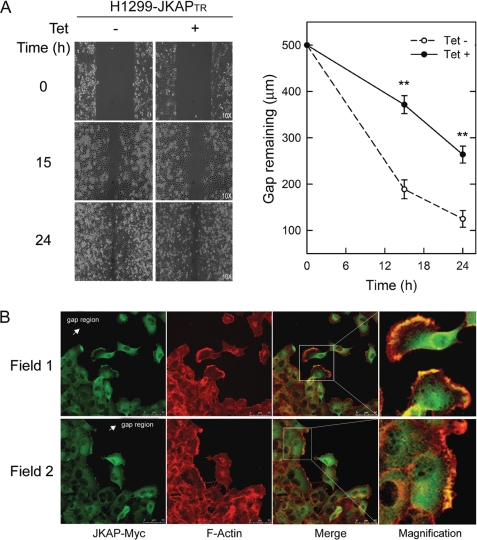
Furthermore, we used a gap-closing assay to monitor cell migration ability. Fifteen to 24 h after the gap creation, the remaining gap regions of the JKAP-expressing cells were wider (372 and 264 μm at 15 and 24 h, respectively) than those of control cells (189 and 125 μm at 15 and 24 h, respectively). The results demonstrated that the JKAP-expressing cells migrated significantly slower than control cells (Fig. 2A, p < 0.01). To examine JKAP distribution in migrating cells, the assayed cells were also fixed and then immunostained with anti-Myc antibody and TRITC-labeled phalloidin. Similar to the localization of JKAP-GFP, Myc-tagged JKAP resided mostly in the cytoplasm, although nuclear staining was also observed (Fig. 2B). Interestingly, JKAP co-localized with actin filaments in the lamellipodia of the leading edges of migratory cells (Fig. 2B). These data indicate that JKAP participates in the regulation of cell movement.
FIGURE 2.
Expression of JKAP decreases cell motility. H1299-JKAPTR cells were cultured with or without 2 μg/ml tetracycline (Tet) overnight before the gap-closing assay. A, after a gap was created, cells migrating into the gap region were monitored at 0, 15, and 24 h. Quantitative data were means ± S.E. of one representative result from three independent experiments. **, p < 0.01, compared with the Tet− group. B, migrating H1299-JKAPTR cells that were cultured with 2 μg/ml tetracycline were fixed at 15 h (Field 1 and Field 2) and subjected to double immunostaining with anti-Myc antibody for JKAP-Myc and TRITC-conjugated phalloidin for F-actin. Magnifications of the insets are shown in the right panels.
JKAP Is a FAK Phosphatase
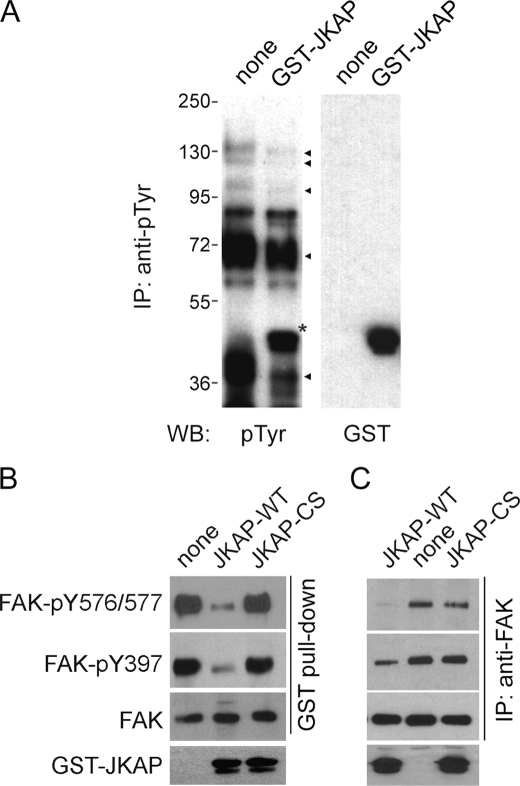
An in vitro JKAP phosphatase reaction was used to determine whether JKAP could dephosphorylate cellular proteins. Through Western blot analysis with an anti-phosphotyrosine antibody (4G10), we found that JKAP was capable of removing tyrosine phosphorylation from immunoprecipitated tyrosine-phosphorylated proteins with apparent molecular masses of 130, 100, 72, and 36 kDa (Fig. 3A). Through immunoprecipitation-coupled Western blot analyses of H1299-JKAPTR cell extracts, we also found the basal and Src-induced tyrosine phosphorylation levels of several proteins (with molecular masses of 130, 70, 35, and 25 kDa) were decreased by JKAP expression (supplemental Fig. 1). These data suggest that JKAP may serve as a protein-tyrosine phosphatase in mammalian cells.
FIGURE 3.
JKAP decreases FAK phosphorylation in vitro. A, tyrosine-phosphorylated proteins were immunoprecipitated (IP) from pervanadate (5 μm, 1 h)-treated cell extracts and subjected to JKAP phosphatase assay. B and C, cellular FAKs were pulled down with GST-Src-SH2 fusion protein (B) or immunoprecipitated with the anti-FAK antibody (C) as substrates and then left untreated or treated with GST-JKAP-WT or GST-JKAP-CS proteins to perform phosphatase reactions. Equal amounts of reaction mixtures were subjected to Western blot (WB) analyses using specific antibodies as indicated. Results shown are one representative result of three independent experiments.
Based on the molecular mass and its role in regulating cell adhesion and migration, we speculated that FAK (125 kDa) could be a substrate of JKAP. To prove that FAK was a substrate of JKAP, FAK complexes were isolated from cell extracts using the GST-Src-SH2 protein and subjected to an in vitro JKAP assay. We found that JKAP efficiently dephosphorylated FAK at Tyr-397, Tyr-576, and Tyr-577 (Fig. 3B). The same result was obtained using immunoprecipitated FAK as a substrate (Fig. 3C). In contrast, JKAP-CS mutant failed to reduce the tyrosine phosphorylation level of FAK (Fig. 3, B and C). These data indicate that FAK is a substrate of JKAP.
JKAP Regulates FAK Phosphorylation
To further examine whether JKAP modulated the FAK phosphorylation level in a cellular context, H1299-JKAPTR and H1299-JKAP-CSTR cells were transiently transfected with Src, a FAK kinase, to enhance FAK phosphorylation and then left untreated or treated with tetracycline for 0–12 h. Src-transfected cells exhibited stronger FAK phosphorylation at Tyr-576/577 and Tyr-397 sites in comparison with untransfected cells (Fig. 4A). Quantitative analysis showed that 12-h induction of JKAP expression decreased the FAK phosphorylation at residues Tyr-576/577 (to 0.13-fold) and Tyr-397 (to 0.39-fold); conversely, JKAP-CS expression increased Tyr-576/577 and Tyr-397 phosphorylation to 2.69- and 1.32-fold, respectively, in comparison with those derived from the control cells at the 0-h time point (Fig. 4A). The collective results from three independent experiments demonstrate that increasing JKAP expression significantly decreased the FAK phosphorylation, whereas JKAP-CS had the opposite effect (Fig. 4B). Other focal adhesion proteins, such as paxillin and p130Cas, also undergo regulated phosphorylation (16). Expression of JKAP reduced paxillin phosphorylation at the Tyr-118 residue; conversely, JKAP-CS increased this phosphorylation (Fig. 4, A and B, p < 0.05). JKAP did not affect p130Cas phosphorylation at Tyr-165 in the same experimental condition (Fig. 4A). Overexpression of a paxillin mutant that cannot be phosphorylated at Tyr-118 affected the turnover of focal contacts and cell motility (16), indicating that paxillin Tyr-118 phosphorylation influences the dynamics of cell adhesion. Using immunostaining analysis, we showed that induction of JKAP expression was accompanied by a decrease of paxillin-rich focal adhesions (Fig. 4C, arrows). This result was consistent with the finding that JKAP expression decreased paxillin phosphorylation and disturbed cell motility.
FIGURE 4.
JKAP dephosphorylates FAK and decreases paxillin-rich focal adhesions. A, H1299-JKAPTR cells and H1299-JKAP-CSTR cells were transfected with Src and then incubated with 2 μg/ml tetracycline (Tet) for 6 and 12 h. The cell extracts were subjected to Western blotting using specific antibodies as indicated. Results shown are one representative result from three independent experiments. The relative band intensities on Western blot assay shown under blot were determined using a computing densitometer equipped with the Gel-Pro analyzer program, which were normalized by arbitrarily setting the densitometry of control to 1. B, bar graphs depicted the results from A (open bars, H1299-JKAPTR; shaded bars, H1299-JKAP-CSTR cells). Quantitative data were means ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01, compared with Src-only control. C, H1299-JKAPTR cells were cultured with or without 2 μg/ml tetracycline for 12 h and subjected to double immunostaining with an anti-paxillin antibody (red) as well as an anti-Myc antibody (green). Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Arrows indicate those cells with higher JKAP expression and less paxillin-rich focal adhesions.
Knockdown of JKAP Enhances Cell Migration and FAK Phosphorylation
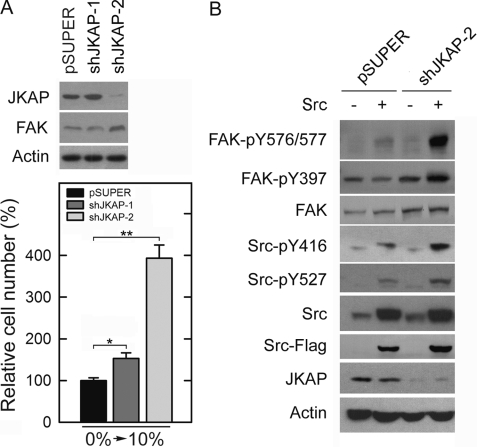
To further confirm the role of JKAP in the regulation of cell migration, we constructed expression vectors that encode two shRNAs targeting distinct segments of human JKAP mRNA. Endogenous JKAPs were efficiently knocked down in the cells stably transfected with shJKAP-2 but not shJKAP-1 (Fig. 5A). As expected, shJKAP-2 caused the cells to migrate significantly faster than the control cells (Fig. 5A, p < 0.01). In contrast, shJKAP-1 only showed a marginal, although statistically significant, effect on cell migration. This result confirmed that endogenous JKAP modulated cell motility. To examine the effects of shJKAP-2 on FAK phosphorylation, H1299-shJKAP-2 cells were stably transfected with Src to activate FAK. Down-regulating JKAP expression markedly enhanced Src-induced phosphorylation of FAK at Tyr-397, Tyr-576, and Tyr-577 residues (Fig. 5B), further confirming that JKAP is indeed a physiological modulator of FAK signaling.
FIGURE 5.
Knockdown of JKAP expression increases cell migration and FAK phosphorylation. A, levels of indicated proteins and cell migration abilities of H1299-pSUPER, H1299-shJKAP-1, and H1299-shJKAP-2 cells were examined by Western blotting (upper panel) or by Transwell assays (lower panel), respectively. Quantitative data were means ± S.E. of one representative result from three independent experiments. *, p < 0.05; **, p < 0.01, compared with H1299-pSUPER cells. B, H1299-pSUPER and H1299-shJKAP-2 cells were stably transfected with Src. The levels of indicated proteins in cell extracts were examined by Western blot analyses. Results shown are one representative of three independent experiments.
DISCUSSION
DUSPs were first found to be involved in the down-regulation of MAPK signaling cascades (reviewed in Refs. 3, 17). Recently, many newly identified DUSPs were shown to have little or no phosphatase activity against MAPKs and to have different substrate specificities and physiological roles from those of typical MAPK phosphatases (18–21). Here, we demonstrated that JKAP, an atypical DUSP, suppressed Src-induced FAK phosphorylation and reduced cell migration.
The movement of cells is controlled by the dynamic assembly and disassembly of actin filaments underlying the plasma membrane (22). Results presented here showed that JKAP co-localized with actin filaments in the lamellipodia of the leading edges of migratory cells, suggesting that JKAP could regulate cell motility. Furthermore, JKAP-expressing cells migrated significantly slower. Conversely, expression of JKAP-CS and knockdown of endogenous JKAP markedly enhanced growth factor-induced cell migration. These results further indicate that JKAP could modulate cell migration and that this function is dependent on phosphatase activity of JKAP.
FAK was first identified as a tyrosine-phosphorylated, Src-associated protein in v-Src-transformed cells (23). Phosphorylation of FAK at Tyr-397 in response to integrin or growth factor signaling leads to the recruitment and activation of Src (13); subsequently, the activated Src phosphorylates other tyrosine residues of FAK to induce maximal kinase activity (14). In addition to regulation by Src, phosphorylation of FAK may be regulated by tyrosine phosphatases, given that vanadate treatment of fibroblasts leads to sustained phosphorylation of FAK (24). Several PTPs have been implicated in regulation of FAK phosphorylation status and function. Both PTP1B and PTPD1 positively regulate FAK phosphorylation (25, 26). Expression of PTP1B induces FAK phosphorylation by activating Src through dephosphorylation of Src at residue Tyr-527 (27). Conversely, PTP-SHP2 and PTP-PEST are negative regulators of FAK phosphorylation (28, 29). Our study indicates that JKAP is another phosphatase negatively regulating FAK phosphorylation.
Different from PTPs, DUSPs have shallow but broader catalytic pockets, which would allow them to dephosphorylate both tyrosine and serine/threonine residues. Interestingly, the structure around the catalytic site in several atypical DUSPs, including JKAP, VHR, and VHY, is identical to PTPs, although little sequence homology is shown (31–33). In fact, VHR has been shown to dephosphorylate the tyrosine residue of STAT5 in the context of interferon signaling (30). In this study, we demonstrate that JKAP can dephosphorylate FAK at tyrosine residues. A previous study shows that JKAP also dephosphorylates serine 118 of estrogen receptor-α, which in turn attenuates estrogen-mediated signaling (9). These studies suggest that atypical DUSPs can have various substrate specificities and efficiently exert dephosphorylation activity. It deserves further exploration how these atypical DUSPs, which lack apparent substrate-binding domain, exhibit selective activity toward their substrates.
The results of previous reports (9, 10) and this study showed that JKAP can regulate cellular functions through targeting molecules other than MAPKs. Furthermore, in addition to FAK, JKAP may also regulate other proteins (e.g. paxillin) in focal adhesions directly or indirectly (Fig. 4). Further studies can elucidate the mechanisms by which JKAP regulates cell migration through other signaling proteins in coordination with the suppression of FAK phosphorylation.
Acknowledgment
We thank the Optical Biology Core, National Health Research Institute, for confocal microscopy assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01-AI066895 (to T.-H. T.). This work was also supported by National Health Research Institutes, Taiwan, Grants MG-097-PP-03 (to Y.-R. C.) and 98A1-IMPP01-014 (to T.-H. T.) and Department of Health, Taiwan, Grant DOH97-TD-G-111-0037 (to Y.-R. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- DUSP
- dual specificity phosphatase
- FAK
- focal adhesion kinase
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- JNK
- c-Jun N-terminal kinase
- JKAP
- JNK pathway-associated phosphatase
- JKAP-CS
- JKAP mutant lacking catalytic activity
- MAPK
- mitogen-activated protein kinase
- PTP
- protein-tyrosine phosphatase
- TRITC
- tetramethylrhodamine isothiocyanate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TR
- tetracycline repressor
- sh
- short hairpin.
REFERENCES
- 1.Patterson K. I., Brummer T., O'Brien P. M., Daly R. J. (2009) Biochem. J. 418, 475–489 [DOI] [PubMed] [Google Scholar]
- 2.Barford D., Flint A. J., Tonks N. K. (1994) Science 263, 1397–1404 [PubMed] [Google Scholar]
- 3.Neel B. G., Tonks N. K. (1997) Curr. Opin. Cell Biol. 9, 193–204 [DOI] [PubMed] [Google Scholar]
- 4.Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. (2004) Cell 117, 699–711 [DOI] [PubMed] [Google Scholar]
- 5.Chen A. J., Zhou G., Juan T., Colicos S. M., Cannon J. P., Cabriera-Hansen M., Meyer C. F., Jurecic R., Copeland N. G., Gilbert D. J., Jenkins N. A., Fletcher F., Tan T. H., Belmont J. W. (2002) J. Biol. Chem. 277, 36592–36601 [DOI] [PubMed] [Google Scholar]
- 6.Alonso A., Merlo J. J., Na S., Kholod N., Jaroszewski L., Kharitonenkov A., Williams S., Godzik A., Posada J. D., Mustelin T. (2002) J. Biol. Chem. 277, 5524–5528 [DOI] [PubMed] [Google Scholar]
- 7.Shen Y., Luche R., Wei B., Gordon M. L., Diltz C. D., Tonks N. K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13613–13618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoyama K., Nagata M., Oshima K., Matsuda T., Aoki N. (2001) J. Biol. Chem. 276, 27575–27583 [DOI] [PubMed] [Google Scholar]
- 9.Sekine Y., Ikeda O., Hayakawa Y., Tsuji S., Imoto S., Aoki N., Sugiyama K., Matsuda T. (2007) Oncogene 26, 6038–6049 [DOI] [PubMed] [Google Scholar]
- 10.Sekine Y., Tsuji S., Ikeda O., Sato N., Aoki N., Aoyama K., Sugiyama K., Matsuda T. (2006) Oncogene 25, 5801–5806 [DOI] [PubMed] [Google Scholar]
- 11.Larsen M., Tremblay M. L., Yamada K. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 700–711 [DOI] [PubMed] [Google Scholar]
- 12.Parsons J. T. (2003) J. Cell Sci. 116, 1409–1416 [DOI] [PubMed] [Google Scholar]
- 13.Schaller M. D., Hildebrand J. D., Shannon J. D., Fox J. W., Vines R. R., Parsons J. T. (1994) Mol. Cell. Biol. 14, 1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calalb M. B., Polte T. R., Hanks S. K. (1995) Mol. Cell. Biol. 15, 954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitra S. K., Hanson D. A., Schlaepfer D. D. (2005) Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 16.Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. (2004) Nat. Cell Biol. 6, 154–161 [DOI] [PubMed] [Google Scholar]
- 17.Hunter T. (1995) Cell 80, 225–236 [DOI] [PubMed] [Google Scholar]
- 18.Hood K. L., Tobin J. F., Yoon C. (2002) Biochem. Biophys. Res. Commun. 298, 545–551 [DOI] [PubMed] [Google Scholar]
- 19.Niwa R., Nagata-Ohashi K., Takeichi M., Mizuno K., Uemura T. (2002) Cell 108, 233–246 [DOI] [PubMed] [Google Scholar]
- 20.Wang J. Y., Lin C. H., Yang C. H., Tan T. H., Chen Y. R. (2006) J. Neurochem. 98, 89–101 [DOI] [PubMed] [Google Scholar]
- 21.Wang J. Y., Yang C. H., Yeh C. L., Lin C. H., Chen Y. R. (2008) J. Neurochem. 107, 1544–1555 [DOI] [PubMed] [Google Scholar]
- 22.Ridley A. J. (2006) Trends Cell Biol. 16, 522–529 [DOI] [PubMed] [Google Scholar]
- 23.Kanner S. B., Reynolds A. B., Vines R. R., Parsons J. T. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 3328–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maa M. C., Leu T. H. (1998) Biochem. Biophys. Res. Commun. 251, 344–349 [DOI] [PubMed] [Google Scholar]
- 25.Arregui C. O., Balsamo J., Lilien J. (1998) J. Cell Biol. 143, 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlucci A., Gedressi C., Lignitto L., Nezi L., Villa-Moruzzi E., Avvedimento E. V., Gottesman M., Garbi C., Feliciello A. (2008) J. Biol. Chem. 283, 10919–10929 [DOI] [PubMed] [Google Scholar]
- 27.Liang F., Lee S. Y., Liang J., Lawrence D. S., Zhang Z. Y. (2005) J. Biol. Chem. 280, 24857–24863 [DOI] [PubMed] [Google Scholar]
- 28.Yu D. H., Qu C. K., Henegariu O., Lu X., Feng G. S. (1998) J. Biol. Chem. 273, 21125–21131 [DOI] [PubMed] [Google Scholar]
- 29.Angers-Loustau A., Côté J. F., Charest A., Dowbenko D., Spencer S., Lasky L. A., Tremblay M. L. (1999) J. Cell Biol. 144, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoyt R., Zhu W., Cerignoli F., Alonso A., Mustelin T., David M. (2007) J. Immunol. 179, 3402–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuvaniyama J., Denu J. M., Dixon J. E., Saper M. A. (1996) Science 272, 1328–1331 [DOI] [PubMed] [Google Scholar]
- 32.Yoon T. S., Jeong D. G., Kim J. H., Cho Y. H., Son J. H., Lee J. W., Ryu S. E., Kim S. J. (2005) Proteins 61, 694–697 [DOI] [PubMed] [Google Scholar]
- 33.Yokota T., Nara Y., Kashima A., Matsubara K., Misawa S., Kato R., Sugio S. (2007) Proteins 66, 272–278 [DOI] [PubMed] [Google Scholar]