Abstract
A-kinase anchoring proteins (AKAPs) include a family of scaffolding proteins that target protein kinase A (PKA) and other signaling proteins to cellular compartments and thereby confine the activities of the associated proteins to distinct regions within cells. AKAPs bind PKA directly. The interaction is mediated by the dimerization and docking domain of regulatory subunits of PKA and the PKA-binding domain of AKAPs. Analysis of the interactions between the dimerization and docking domain and various PKA-binding domains yielded a generalized motif allowing the identification of AKAPs. Our bioinformatics and peptide array screening approaches based on this signature motif identified GSKIP (glycogen synthase kinase 3β interaction protein) as an AKAP. GSKIP directly interacts with PKA and GSK3β (glycogen synthase kinase 3β). It is widely expressed and facilitates phosphorylation and thus inactivation of GSK3β by PKA. GSKIP contains the evolutionarily conserved domain of unknown function 727. We show here that this domain of GSKIP and its vertebrate orthologues binds both PKA and GSK3β and thereby provides a mechanism for the integration of PKA and GSK3β signaling pathways.
Keywords: Evolution/Protein, Phosphorylation/Kinases/Serine-Threonine, Protein/Protein-Protein Interactions, Signal Transduction/Adapter Proteins, Signal Transduction/Cyclic Nucleotides/Cyclic AMP, Signal Transduction/Protein Kinases, Signal Transduction/Protein Kinases/Cyclic Nucleotide, Signal Transduction/Protein Kinases/Serine/Threonine, Glycogen Synthase Kinase 3, AKAP
Introduction
A-kinase anchoring proteins (AKAPs)3 are a family of scaffolding proteins characterized by the ability to bind cAMP-dependent protein kinase (protein kinase A (PKA)). They tether PKA in the vicinity of its substrates, thereby facilitating their phosphorylation. In addition, AKAPs bind further signaling molecules, including other protein kinases (e.g. protein kinase C and protein kinase D), phosphodiesterases (e.g. PDE4D), and protein phosphatases (e.g. PP1 and PP2B/calcineurin). A few AKAPs possess catalytic activity. For example, AKAP-Lbc is a Rho guanine nucleotide exchange factor (1–3). Thus, AKAPs assemble multiprotein complexes and thereby coordinate cellular signaling. AKAPs are required for many cellular processes, including vasopressin-mediated water reabsorption in renal principal cells and β-adrenoreceptor-dependent increases of cardiac myocyte contractility (2, 4, 5).
The PKA holoenzyme consists of a dimer of regulatory RI or RII subunits and two catalytic subunits, each bound to one R subunit. Upon binding of two molecules of cAMP to each R subunit, the catalytic subunits dissociate and phosphorylate their substrates (6, 7). The interaction of AKAPs with PKA is mediated by the PKA-anchoring domain of AKAPs and the dimerization and docking (DD) domain of R subunit dimers. Because most AKAPs preferentially anchor RII subunits, PKA-anchoring domains are termed RII-binding domains (RIIBD). These domains are structurally conserved amphipathic helices, 14–18 amino acid residues in length (8, 9). Based on recently described determinants of the RIIBD/DD domain interaction (8–10), we developed a bioinformatics and peptide array screening approach to identify new AKAPs. For one of the discovered proteins, GSKIP (GSK3β interaction protein), we show that it functions as an AKAP.
Mammalian cells express two isoforms of glycogen synthase kinase-3 (GSK3), GSK3α and GSK3β, which are constitutively active and phosphorylate a broad range of substrates, thereby participating in the regulation of various processes, including energy metabolism, protein synthesis, sorting and degradation, and transcription (11, 12). The activity of GSK3 isoforms is decreased through phosphorylation by other protein kinases such as Akt/protein kinase B, p70/p85-ribosomal S6 kinase, p90-ribosomal S6 kinase, and PKA (13–15). Phosphorylation of Ser-21 of GSK3α and Ser-9 of GSK3β by any of the abovementioned kinases leads to inactivation. The different kinases appear to regulate different pools of GSK3β. For instance, stimulation of adrenoceptors on skeletal muscle cells leads to PKA-mediated phosphorylation of GSK3β, whereas stimulation of the insulin receptor on these cells leads to protein kinase B-mediated phosphorylation of GSK3β (16). The formation of such pools is likely to require scaffolding proteins. The AKAPs AKAP220 and MAP2 (microtubule-associated protein 2) each recruit PKA and GSK3β and facilitate PKA phosphorylation of GSK3β (17, 18). The phosphorylation by PKA appears to require binding of both PKA and GSK3β to the respective AKAP (13, 19).
Here, we demonstrate that the evolutionarily conserved and widely expressed protein GSKIP (20) functions as an AKAP, recruits PKA and GSK3β, and facilitates PKA phosphorylation of GSK3β. GSKIP and its vertebrate and invertebrate orthologues from fungi to Homo sapiens contain the evolutionarily conserved domain of unknown function 727 (DUF727), which we show here to interact with RII subunits in the vertebrate orthologues, thus conferring an AKAP function. RII binding was not observed for invertebrate orthologues. The interaction with GSK3β, however, appears to be conserved in both invertebrates and vertebrates.
EXPERIMENTAL PROCEDURES
Peptide Arrays, RII Overlay, and Protein Overlay
Peptide spots were generated by automatic SPOT synthesis as described previously (10, 21, 22). Peptides containing “difficult sequences” (23) were excluded from the analysis. For example, cysteines may oxidize to sulfoxides or undergo dimerization. β-Sheet-forming sequences cause collapse of the peptide-resin matrix. Under such conditions, diffusion of reagents into the matrix is limited, and coupling and deprotection reactions may be incomplete (21, 23). The interaction of spot-synthesized peptides with RII subunits of PKA was investigated by RII overlay assay using radioactively labeled RII subunits (10, 24, 25). To determine the binding of GSK3β, peptide spot membranes were blocked for 1 h with blocking buffer (3% bovine serum albumin in Tris-buffered saline + 0.05% Tween 20 (TBS-T)) before incubation with 1 μg/ml glutathione S-transferase (GST) or 1 μg/ml GST-GSK3β (Cell Signaling Technology, Danvers, MA), at 4 °C overnight. Membranes were washed three times with TBS-T and incubated with an anti-GST antibody (Millipore, Billerica, MA) in blocking buffer for 1 h at room temperature. Thereafter, horseradish peroxidase (HRP)-coupled anti-rabbit antibody (Dianova, Hamburg, Germany) was added (1 h, room temperature), and after washing three times with TBS-T, an ECL reaction was carried out using LumiLight substrate (Roche Diagnostics). Signals were visualized with the Lumi-Imager F1 (Roche Diagnostics).
Cloning
Total RNA was isolated from human neuroblastoma SH-SY5Y cells using TRIzol and reverse-transcribed using SuperScript one-step reverse transcription-PCR with Platinum Taq according to the manufacturer's instructions (Invitrogen). The resulting cDNA served as a template to obtain full-length GSKIP (420 bp) by PCR (forward primer 5′-ATGCGCTAGCTATGGAAACAGACTGTAATCCCATG-3′ and reverse primer 5′-GCATACCGGTCCTGACTGTCCATCTCTTTTCAAAG-3′). PCR products were ligated into the vector pECFP-N1 (BD Biosciences) via the restriction sites NheI and AgeI to yield the vector CFP-GSKIP. Valine 41 and leucine 45 were substituted by proline residues (GSKIP-V41P/L45P) by site-directed mutagenesis (QuikChange site-directed mutagenesis kit, Stratagene, La Jolla, CA) to disrupt the helical structure of the RIIBD.
YFP-RIIα used in this work was described previously (24). To generate CFP-AKAP18α, human cDNA (see above) was amplified by PCR (forward primer 5′-CCCCGAATTCCATGGGCC-3′ and reverse primer 5′-CGCGGTCGACCATTTCCTGTTCTCATTGTCA-3′) and inserted into pECFP-N1 via EcoRI and SalI restriction sites. All constructs were sequenced prior to expression.
Generation of Recombinant GSKIP
To obtain His-tagged, full-length GSKIP, a cDNA fragment encoding full-length GSKIP (139 amino acid residues) was generated by PCR using the vector CFP-GSKIP (see above) as template. The forward (5′-CAGCCATATGGAAACAGACT-3′) and reverse primers (5′-TGTGCTCGAGTCAGGACT-3′) contained NdeI and XhoI restriction sites, respectively. The cDNA was cloned into the vector pET28(+) (Merck) via these restriction sites to yield the vector His-GSKIP. His-tagged proteins were expressed in Escherichia coli strain BL21-DE3 and affinity-purified. The His tag was removed by thrombin protease cleavage as recommended by the supplier of the vector pET28(+).
Preparation of Affinity-purified Anti-GSKIP Antibodies
A rabbit polyclonal antiserum was raised against recombinant full-length GSKIP (139 amino acid residues; Biogenes, Berlin, Germany). Specific antibodies were isolated by affinity chromatography of the antisera using GSKIP immobilized on thiopropyl-Sepharose 6B (GE Healthcare) (24).
Cell Culture and Transfection
HEK293 cells (ATCC, Manassas, VA) were grown to 40–60% confluency in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Cells were transfected 48 h after seeding with plasmid DNA (1 μg/60-mm dish) encoding CFP-GSKIP or CFP-GSKIP-V41P/L45P using Transfectin (Bio-Rad) and analyzed 24–48 h post-transfection. SH-SY5Y cells were grown in Dulbecco's modified Eagle's medium/F-12 supplemented with 10% fetal calf serum and 1% nonessential amino acids.
Detection of GSKIP in Cells by Laser Scanning Microscopy
Glass coverslips (30 mm) were coated with 100 μg/ml poly-l-lysine in 35-mm cell culture dishes. 24 h prior to transfection, 200,000 HEK293 cells were seeded in Dulbecco's modified Eagle's medium/fetal calf serum. Cells were transfected with 0.5 μg of YFP-RIIα and 1 μg of CFP-GSKIP, CFP-GSKIP-V41P/L45P or CFP-AKAP18α using LipofectamineTM 2000 (Invitrogen). 24 h after transfection, cell were covered with phosphate-buffered saline and analyzed by confocal laser scanning microscopy, using a Zeiss LSM 710 (Carl Zeiss GmbH, Jena, Germany).
Immunoprecipitation and cAMP-Agarose Pulldown, Preparation of Tissue Lysates, and Western Blotting
Untransfected HEK293 cells or HEK293 cells transiently expressing CFP-GSKIP or CFP-GSKIP-V41P/L45P and tissues from wild type Wistar rats were lysed in lysis buffer (10 mm K2HPO4, 150 mm NaCl, 5 mm EDTA, 5 mm EGTA, 1% Triton X-100, pH 7.4) supplemented with protease inhibitors (1 mm benzamidine, 0.5 mm phenylmethanesulfonyl fluoride, 3.2 μg/ml trypsin inhibitor I-S, 1.4 μg/ml aprotinin) and protein phosphatase inhibitors (50 mm NaF and 100 μm Na3VO4) (24). The lysates were cleared by centrifugation (20,000 × g, 4 °C, 10 min). Endogenous GSKIP, CFP-GSKIP, and CFP-GSKIP-V41P/L45P were immunoprecipitated with affinity-purified rabbit anti-GSKIP antibody (see above) and protein A-conjugated agarose (Sigma). For cAMP-agarose pulldown, the lysates were incubated with 8-AHA-cAMP-agarose (4 °C, 3 h; 8-aminohexyl amino adenosine 3′-5′-cyclic monophosphate; Biolog, Bremen, Germany) in the absence or presence of cAMP (50 mm). Agarose-bound proteins were washed four times with lysis buffer and eluted with Laemmli sample buffer (95 °C). The following primary antibodies were used for Western blot detection: rabbit anti-GSKIP (see above); rabbit glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phospho-GSK3α/β (p-GSK3α/β), and total GSK3β antibodies (Cell Signaling Technology, Danvers, MA); mouse anti-β/γ-tubulin (Calbiochem); goat anti-lamin A/C (Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-RIIα (BD Biosciences). CFP was detected with mouse anti-green fluorescent protein (Clontech) (26). Corresponding HRP-coupled secondary antibodies were from Dianova, Hamburg, Germany. Signals were visualized with the Lumi-Imager F1 (Roche Diagnostics). p-GSK3β was quantified by densitometric analysis of Western blots (LumiAnalyst 3.0 software, Roche Diagnostics).
Subcellular Fractionation of SH-SY5Y Cells
SH-SY5Y cells were scraped from 100-mm cell culture dishes in 500 μl of fractionation buffer (250 mm sucrose, 20 mm HEPES, pH 7.4, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol) supplemented with protease and phosphatase inhibitors (see above). Cells were homogenized using a glass-Teflon homogenizer and centrifuged (10 min, 4 °C, 700 × g). The resulting nuclear pellet was washed with fractionation buffer, centrifuged again, and resuspended in RIPA buffer. The supernatant was centrifuged at 100,000 × g for 60 min. The pellet was considered the particulate and the supernatant the cytosolic fraction (24).
Nuclear Magnetic Resonance Measurements
Isotopically enriched GSKIP was expressed in E. coli BL21-DE3 strain using 15N-NH4Cl as nitrogen source in M9 minimal medium. 1H,15N correlation experiments with 15N-labeled GSKIP and unlabeled DD domain were recorded using a standard heteronuclear single quantum coherence pulse program employing WATERGATE for solvent suppression. All NMR experiments were recorded on a Bruker 600 MHz Avance spectrometer equipped with a triple channel cryoprobe. The temperature was set to 30 °C in all experiments. For interaction studies, the typical protein concentration was on the order of 0.1 mm. The acquisition time in the direct dimension was restricted to 150 ms. A total of 256 increments was recorded in the indirect dimension.
Surface Plasmon Resonance Measurements
Interaction studies were performed employing Biacore 2000 or 3000 instruments (Biacore AB, GE Healthcare) (27). Measurements were performed in running buffer containing 20 mm MOPS, 150 mm NaCl, 0.005% Tween 20, pH 7.0, at 25 °C instrument temperature. In brief, 8-AHA-cAMP (BioLog, Bremen, Germany) was covalently coupled to CM5 sensor chips (research grade) using NHS/EDC chemistry. RI or RII subunits, prepared according Bertinetti et al. (28) using cAMP affinity chromatography, were injected in running buffer containing 1 mg/ml bovine serum albumin and reversibly captured on an 8-AHA-cAMP surface (surface concentration of 120–200 resonance units for each subunit). GSKIP was injected at different concentrations (8–256 nm) at a flow rate of 30 μl/min. Both association and dissociation phases were recorded for 300 s. After each GSKIP injection, the entire protein complex (R subunits and GSKIP) was removed by three short injections (30 s each) of 3 m guanidinium hydrochloride. Rate constants (ka and kd) and equilibrium binding constants (KD) were calculated based on nonlinear regression analysis assuming a 1:1 Langmuir binding model using the BIAEvaluation software, version 3.0 (Biacore AB, GE Healthcare). For control, experiments were performed in the presence of 5 μm PKA anchoring disruptor peptides (Ht31 or AKAP18δL314E (10)) using 500 nm GSKIP. As negative controls, double proline substituted peptides were used under the same conditions.
Enzyme-linked Immunosorbent Assay for Monitoring the GSKIP, PKA, and GSK3β Interaction
Enzyme-linked immunosorbent assays were conducted in Corning 384-well clear or white flat bottom polystyrene high bind microplates (Corning B.V., Schiphol-Rijk, the Netherlands). The plates were coated by adding 20 μl/well RIIα (50 nm; prepared as described in Ref. 27) in coating buffer (phosphate-buffered saline containing 1 mm benzamidine, 0.5 mm phenylmethanesulfonyl fluoride, 3.2 μg/ml trypsin inhibitor type I-S, 1.4 μg/ml aprotinin, and 1 mm dithiothreitol) and incubation for 1 h at room temperature. Thereafter, the coating buffer was removed, and blocking buffer (coating buffer containing 0.3% skimmed milk powder and 0.05% Tween 20) was added (100 μl, 1 h, room temperature). After removal, monitoring of interactions of RIIα with GSKIP and/or GSK3β was carried out in newly added blocking buffer (20 μl/well, 1 h, room temperature). Protein concentrations are indicated in the legend to Fig. 6A. Unbound protein was removed by washing the wells three times with 100 μl of washing buffer (phosphate-buffered saline containing 0.05% Tween 20). Bound His-GSKIP was detected with monoclonal anti-polyhistidine HRP-conjugated antibody (Sigma; 1:8000 in blocking buffer, 1 h, room temperature). Bound GSK3β was detected by incubation with rabbit anti-GSK3β antibody (Cell Signaling Technology, Danvers, MA; 1:1500 in blocking buffer) and HRP-conjugated anti-rabbit IgG (1:3000, 1 h, room temperature). Each antibody incubation step was followed by washing. The HRP reaction was initiated by addition of 3,3′,5,5′-tetramethylbenzidine enzyme-linked immunosorbent assay substrate solution (Sigma) and terminated after 30 min by adding HCl (1 m; 20 μl/well). The colored reaction product was quantified by measuring A450 nm in a GeniosPro plate reader (Tecan, Durham, NC) with 10 flashes/well. Curves were fitted based on a one-site-binding model, and KD values were calculated using Prism 4.0 (GraphPad Software, San Diego, CA).
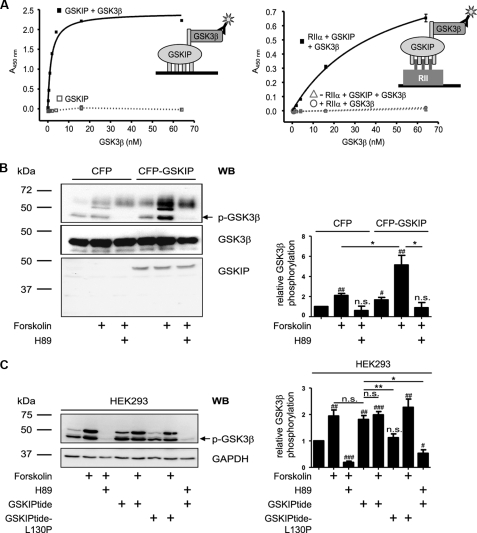
FIGURE 6.
GSKIP forms a ternary complex with regulatory RIIα subunits of PKA and GSK3β and facilitates PKA phosphorylation of GSK3β. A, left panel, wells of microtiter plates were coated with His-GSKIP (500 nm) and incubated with GSK3β (0.25–64 nm) in blocking buffer or with blocking buffer without GSK3β. GSK3β was detected with anti-GSK3β and secondary HRP-conjugated antibodies and an HRP-catalyzed reaction with a chromogenic substrate solution. Right panel, wells of microtiter plates were coated with RIIα (50 nm) in blocking buffer or with blocking buffer in the absence of RIIα. GSKIP (250 nm) in combination with increasing amounts of GSK3β (0.25–64 nm) was added. To quantify unspecific signals, the GSKIP-GSK3β-combination was incubated in wells blocked in the absence of RIIα (▵). Direct binding of GSK3β to RIIα was assessed by incubation of RIIα-coated wells with GSK3β (0.25–64 nm; ○). Detection was carried out as above. B, left panel, CFP or CFP-GSKIP was transiently expressed in HEK293 cells. The cells were left untreated, incubated with the direct activator of adenylyl cyclases, forskolin (20 μm, 30 min), or pretreated with the PKA inhibitor H89 (30 μm, 30 min prior to FSK treatment). Cells were lysed, and CFP-GSKIP, phosphorylated (p-GSK3β), and total GSK3β were detected with specific antibodies by Western blotting (WB). Right panel, signals obtained for p-GSK3β and GSK3β by Western blotting were analyzed densitometrically. The ratio of p-GSK3β to GSK3β was calculated (n = >5 independent experiments for each experimental condition, mean ± S.E.). C, left panel, HEK293 cells were treated with FSK or a combination of FSK and H89 as in B or with stearate-coupled GSKIPtide, which displaces GSK3β from GSKIP (20), or the inactive control peptide GSKIPtide-L130P (100 μm, 30 min). Cells were lysed and phosphorylated GSK3β (p-GSK3β) and GAPDH (loading control) were detected by Western blotting. Right panel, signals obtained for p-GSK3β and GAPDH by Western blotting were analyzed densitometrically. The ratio of p-GSK3β to GAPDH was calculated (n = 7 independent experiments for each experimental condition, mean ± S.E.). B and C, significantly different from untreated control: #, p < 0.05; ##, p < 0.01; ###, p < 0.001. Significant differences between groups connected by lines: *, p < 0.05; **, p < 0.01). n.s., not significant.
Statistical Analysis
GraphPad Prism 5.01 for Windows (GraphPad Software, San Diego) was used to perform two-tailed t tests. One sample t tests were applied to determine significance in comparison with the basal value.
RESULTS
Bioinformatics-based Approach to Identify New AKAPs
Hydrophobic amino acid residues in conserved positions of RIIBDs of AKAPs are important contacts for RII subunit binding (Fig. 1A) (8–10). A consensus pattern summarizing these properties is suitable for the identification of RIIBDs within a given AKAP (29). However, searching a protein data base with such a general pattern for new AKAPs identified too many false positives (data not shown). Our recent observation that H-bonds and salt bridges influence AKAP-RII binding (10) provided the possibility to refine the search pattern. Charged amino acid residues, putative salt bridge, or H-bond partners influence the isoelectric point (pI) of a peptide. Thus, although the number and positions of amino acids that function as acceptors or donors of H-bonds or salt bridges vary among AKAPs, the calculated pI for RIIBDs should be limited to a certain range. Indeed, the calculated pI for peptides derived from several RIIBDs of AKAPs range from 3.43 to 6.23 (Fig. 1A). Taking into account the range of pI values, the conserved positioning of hydrophobic amino acid residues, and that the DD-domain of the RII subunit dimer forms a nearly symmetric groove (30), and thus assuming a 2-fold mirror symmetry for the RIIBDs led to the following search pattern: (AVLISE)XX(AVLIF)(AVLI)XX(AVLI)(AVLIF)XX(AVLISE), where amino acid residues in parentheses denote alternatives of the amino acid residues in conserved positions of RIIBDs, and X stands for any amino acid (Fig. 1, A and B). Utilizing this pattern and limiting the range of pI values to 3.0–6.4, the Swiss-Prot Database was searched for potential RII-binding proteins with the ScanSite program. The search retrieved ∼11,700 hits from Dictyostelium discoideum, Saccharomyces cerevisiae, Drosophila melanogaster, Danio rerio, Mus musculus, Rattus norvegicus, and Homo sapiens. The cognate sequences of the human proteins were retrieved utilizing the bioperl package and the corresponding identification numbers (31). Amino acid sequences were extracted from the cognate proteins and extended by 9 amino acid residues at the N terminus and 4 amino acid residues at the C terminus of the hit sequence to obtain 25-mer amino acid sequences. In total, 4519 peptides were derived from 2352 human proteins. These peptides were further filtered to exclude duplicates and so-called difficult sequences not accessible to peptide spot synthesis (see “Experimental Procedures”) (21, 23). For example, the RIIBD of mAKAP (also termed AKAP6) was excluded as it contains a cysteine residue. The filtering reduced the number of peptides to 2572. They were spot-synthesized and subjected to RII overlay assay to test their ability to bind RII subunits (supplemental Fig. 1 and supplemental Table 1). Peptide spots were overlaid with 32P-labeled RII subunits of PKA, and binding was detected by autoradiography. This approach is summarized in Fig. 1B.
FIGURE 1.
Bioinformatics approach to identify new AKAPs. A, multiple sequence alignment of RIIBDs of several AKAPs. Amino acid residues in conserved positions are shaded in gray. The isoelectric points (pI) of the peptides are indicated. All sequences except murine (Mm) AKAP-Lbc, rat (Rn) AKAP18δ, and D. melanogaster (Dm) DAKAP550 are derived from human AKAPs. AKAPIS is a peptide designed in silico (10, 52). B, flow chart for the identification of RII-binding domains. Depicted is the AKAP search pattern for screening of the Swiss-Prot Database for potential RII-binding proteins. For the data base search, the pI value range was limited to 3.0–6.4. Duplicates and peptides not accessible to synthesis were removed (Filter). The remaining peptides (2572) were spot-synthesized and subjected to RII overlay assay (supplemental Fig. 1). 829 peptides bound RII subunits. C, 829 peptides were synthesized on three membranes and subjected to RII overlay (Blocks 1–3; for sequences see supplemental Table 1). Signals were detected by autoradiography. Row A on each block contains the indicated control peptides, which are established RII-binding peptides or inactive versions thereof. Dashed boxes, RIIBDs from previously identified AKAPs. Solid box, peptide derived from the protein GSKIP. D, of the 829 RII-binding peptides, 27 representing putatively new AKAPs (and additionally 9 derived from previously described AKAPs) were spot-synthesized and subjected to RII overlay in the absence (control) or presence of the PKA anchoring disruptor peptide AKAP18δ-L314E or the inactive control peptide AKAP18δ-PP (10 μm each). Row A of each membrane contains the indicated control peptides; row B contains peptides derived from previously identified AKAPs; and rows C–E contain the putative RII-binding domains of the indicated proteins (given as abbreviations; for sequences see Table 1). Box, RII-binding domain of GSKIP. Displayed are representative membranes from at least three independent experiments.
Approximately one-third (829) of the 2572 peptides bound RII subunits (Fig. 1C, Blocks 1–3). The peptide sequences are indicated in supplemental Table 1. To select peptides binding to RII subunits in a typical AKAP manner at the DD domain, all RII-binding peptides were spot-synthesized again and probed for RII binding with human RII subunits preincubated with the peptide AKAP18δ-L314E. The peptide was derived from the RIIBD of AKAP18δ. It binds RII subunits with subnanomolar affinity (KD = 0.7 ± 0.5 nm for binding to RIIα) at the DD domain and thereby prevents the interaction of RII with AKAPs (10). As a control, RII subunits were incubated with the peptide AKAP18δ-PP, which contains two prolines and does not bind RII subunits (10). Among the 829 peptides, 27 (and additionally 9 derived from previously described AKAPs) reproducibly interacted with RII subunits in the absence of peptide (control) and in the presence of peptide AKAP18δ-PP (Fig. 1D; for peptide sequences see Table 1). Preincubation of RII subunits with peptide AKAP18δ-L314E reduced or abolished RII binding to the 27 RII-binding peptides. Thus, these peptides represent putative RIIBDs of their cognate proteins (Fig. 1D). Secondary structure predictions (32) of the candidate peptides revealed high probabilities to form α-helices (data not shown).
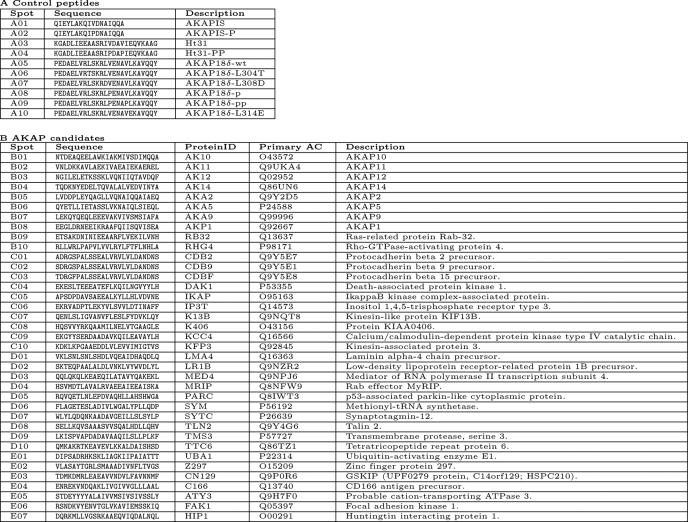
TABLE 1.
Sequences of peptides from Fig. 1D
Spot numbering corresponds to positions of peptide spots on the membrane in Fig. 1D. Part A, control peptides. Part B, peptides derived from AKAP candidates. Protein identification (ID) and primary accession numbers (AC) relate to entries in the Swiss-Prot Database. Abbreviations for proteins as shown in Fig. 1D, and the corresponding full names of the cognate proteins are indicated.
GSKIP Binds PKA with High Affinity in Vitro
Among the cognate proteins containing a putative RIIBD, GSKIP was characterized with regard to its AKAP function. The sequence of the putative RIIBD (TDMKDMRLEAEAVVNDVLFAVNNMF) is located between amino acid residues 28–52 of full-length GSKIP (Fig. 1D and Fig. 2A; Table 1). GSKIP is also named C14orf129 after its chromosomal localization, chromosome 14, open reading frame 129 (Swiss-Prot ID, GSKIP_human).
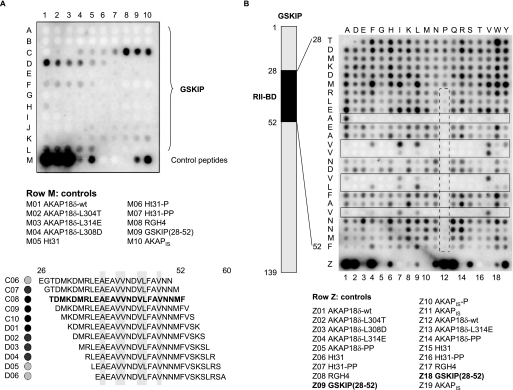
FIGURE 2.
RII-binding domain of GSKIP. A, mapping the RII-binding domain of GSKIP. The entire sequence of GSKIP (139 amino acid residues) was spot-synthesized as overlapping peptides (25-mers, 24 amino acid residue overlap) and probed for RII binding by RII overlay assay. Signals were detected by autoradiography. Peptide sequences binding RII (position C06–D06) are displayed in the lower panel. Hydrophobic amino acid residues in conserved positions of RII-binding domains are shaded in gray (compare Fig. 1A). As controls, PKA anchoring disruptor peptides (row M) were synthesized. B, influences of single amino acid residues within the GSKIP RII-binding domain on the interaction with RII subunits. The scheme (left) depicts the location of the RII-binding domain (RII-BD) within GSKIP. Peptides derived from the RII-binding domain (positions 28–52 within GSKIP; vertical) and the indicated control peptides in row Z were spot-synthesized. The amino acid residues of the RII-binding domain were substituted with the residues indicated at the top of the membrane. Interaction of the peptides with RII subunits was revealed by RII overlay assay and detection by autoradiography. Solid boxes, peptides substituted at amino acid residues in conserved positions of the RII-binding domain. Dashed box, introduction of prolines into the RII-binding domain reduces or abolishes RII binding. Amino acid residues are indicated by single letter code. A and B, displayed are representative membranes from at least three independent experiments.
Initially, we aimed to validate the localization of the RIIBD within GSKIP. For this purpose, an array of overlapping peptides covering the entire sequence of GSKIP (139 amino acids) was spot-synthesized (25-mers, shift 1) and subjected to RII overlay assay. The cysteines in positions 5, 59, and 77 were substituted with serines as cysteine-containing peptides are not accessible to spot-synthesis (see “Experimental Procedures”). The RII subunits bound to a series of overlapping peptides (Fig. 2). A, C4–D6) such as is expected for a continuous binding domain, i.e. decreasing affinity upon increasing distance from the region of maximal binding. The RII subunits bound the previously identified peptide TDMKDMRLEAEAVVNDVLFAVNNMF with the highest apparent binding affinity (Fig. 2A, peptide C08). The lower panel of Fig. 2A shows the sequences of the binding peptides with hydrophobic amino acid residues highlighted. They are in conserved positions of the amphipathic helix binding the DD domain (for comparison see Fig. 1, A and B). The RII subunits bound additional peptides (F1–5 and J5–L5) with low apparent binding affinities suggesting that GSKIP contains a second RII-binding site. However, further experiments indicated that sites other than the RIIBD in positions 28–52 of GSKIP are not involved in RII binding in cells (data not shown).
The influence of individual amino acid residues within the RIIBD of GSKIP on the interaction with RII subunits was determined by probing a peptide substitution array of the GSKIP RIIBD for RII binding (Fig. 2B). In this array every amino acid residue of the RIIBD (vertical sequence) was substituted with the amino acid residues indicated above the array (Fig. 2B). When hydrophobic amino acid residues in conserved positions of the RIIBD were substituted by polar or charged amino acid residues, RII bound with apparently lower affinity, or binding was abolished. The nature (e.g. the size) of the hydrophobic residues at particular positions appears to be important for the interaction with RII subunits because substitution of a hydrophobic amino acid residue with another hydrophobic residue frequently decreased the interaction with RII. For example, the Val-Val sequence at positions 40 and 41 within GSKIP shows variable tolerance for isoleucine, leucine, tyrosine, and tryptophan (Fig. 2B, solid boxes). As expected from a typical AKAP, introduction of proline into the RIIBD reduces or abrogates the interaction with RII subunits as the α-helical structure of the core RIIBD is lost (Fig. 2B, dashed box).
Next, it was investigated whether full-length GSKIP functions as an AKAP. cDNA encoding full-length GSKIP (amino acid residues 1–139) was obtained from human neuroblastoma cells (SH-SY5Y) and utilized for the generation of recombinant GSKIP fused with a His tag (His-GSKIP). The tag was removed by thrombin protease cleavage, and the interaction of GSKIP with the DD domain of RIIα was analyzed by NMR experiments (Fig. 3A). The spectra confirmed that the above described RIIBD is located at positions 28–52 of the protein and binds the DD domain. Resonance attenuation in the presence of the DD domain could be assigned to residues within the RIIBD (Leu-35, Val-41, Phe-46, and Ala-47) but also within the β-sheet region of GSKIP and in the C-terminal helix in proximity to the RIIBD (Fig. 3A, D112, L114, F122, and G123).
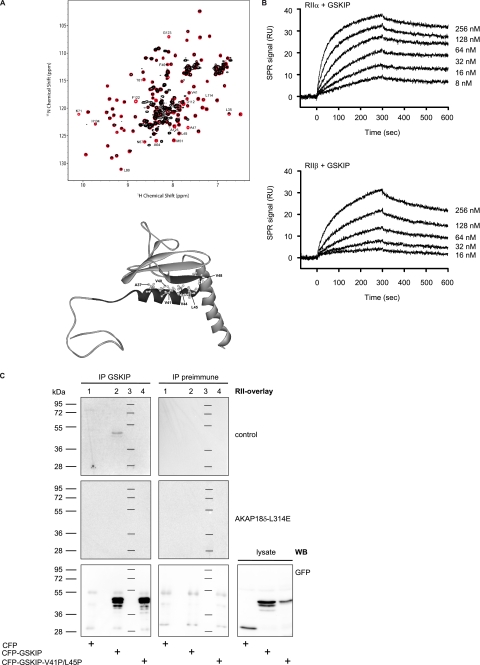
FIGURE 3.
GSKIP functions as an AKAP in vitro. A, GSKIP and the DD domain of human regulatory RIIα subunits of PKA were generated and NMR measurements performed. 1H-15N heteronuclear single quantum coherence spectra of the GSKIP domain in the absence (red) or presence of DD (black) at 1:0.5 ratio are shown. Disappearance of the GSKIP resonances is caused because of chemical exchange broadening and/or increased transverse relaxation rate due to high molecular weight of the complex. Lower panel, Protein Data Bank structure of GSKIP (Protein Data Bank code 1sgo) The RII-binding domain (black) encompasses the amphipathic α1-helix of GSKIP. The conserved hydrophobic amino acid residues Ala-37, Val-40, Val-41, Val-44, Leu-45, and Val-48 (see also Fig. 1A) are depicted as ball-and-stick models. The contact interface for RII binding is hidden by a β-sheet. B, surface plasmon resonance measurements to determine the association and dissociation rate constants (see Table 2) for the binding of GSKIP to PKA regulatory RII subunits. Human RIIα and RIIβ subunits (upper and lower panel, respectively) were captured on 8-AHA-cAMP sensor chips, and His-GSKIP was injected into the Biacore instrument in the indicated concentrations. The plots show representative experiments. Each experiment was repeated at least three times using different protein preparations and different immobilization rates of the regulatory subunits. RU, resonance units. C, immunoprecipitated (IP) GSKIP binds RII subunits. Cyan fluorescent protein (CFP; lane 1) and the indicated fusions of human full-length GSKIP with CFP (lane 2) and GSKIP-V41P/L45P with CFP (lane 4) were transiently expressed in HEK293 cells. CFP-GSKIP-V41P/L45P contains prolines in the RII-binding domain disrupting the α-helical structure and thus preventing the interaction with RII. The cells were lysed, and GSKIP was immunoprecipitated with anti-GSKIP antibodies, or the precipitation was carried out with the corresponding preimmune serum. Precipitated proteins were probed for RII binding by RII overlay assay in the absence (control) or in the presence of the PKA anchoring disruptor peptide, AKAP18δ-L314E (10 μm). Signals were detected by autoradiography. Lane 3, molecular weight standard. Lower panel, Western blot (WB) with anti-green fluorescent protein antibody (also detects CFP (26)) for the detection of CFP, CFP-GSKIP, or CFP-GSKIP-V41P/L45P in HEK293 cells transiently expressing the proteins. Proteins were detected in precipitates obtained with rabbit anti-GSKIP antibody (IP, left panel) or obtained with preimmune serum (middle panel) or in cell lysates.
To quantify the interaction of GSKIP with recombinant human RIIα- and RIIβ-subunits, Biacore technology was employed. R subunits were captured on 8-AHA-cAMP sensor surfaces to yield an average surface concentration of 120–200 resonance units (see “Experimental Procedures” for details). His-tagged GSKIP was injected as an analyte at different concentrations (8–256 nm). Association and dissociation rate constants were determined, and equilibrium binding constants of 5 and 43 nm were calculated for RIIα and RIIβ, respectively (Fig. 3B and Table 2). The interactions were reduced by 80% if the experiments were carried out in the presence of the peptide AKAP18δ-L314E (data not shown). GSKIP did not bind to RI subunits (data not shown).
TABLE 2.
Apparent rate and equilibrium dissociation constants of GSKIP binding to RIIα and RIIβ
Surface plasmon resonance measurements using Biacore technology were performed to determine rate constants (ka and kd) and equilibrium binding constants (KD) for the interaction of His-tagged GSKIP with human R subunits. RIIα and RIIβ were reversibly captured via 8-AHA-cAMP, which was covalently coupled on a CM5 sensor chip (not shown in the plot). His-GSKIP was injected over RIIα and RIIβ surfaces (Fig. 3B). Association and dissociation phases were monitored for 5 min, respectively. No binding was detected when RIα and RIβ were captured (data not shown).
| ka | kd | KD | |
|---|---|---|---|
| m−1s−1 | s−1 | nm | |
| RIIα | 1.3 × 105 | 6.6 × 10−4 | 5 |
| RIIβ | 2.8 × 104 | 1.2 × 10−3 | 43 |
In addition, we tested whether immunoprecipitated full-length GSKIP binds RII subunits. Human full-length GSKIP was transiently expressed in HEK293 cells as a fusion with cyan fluorescent protein (Fig. 3C, CFP-GSKIP). A fusion protein containing two prolines in the RIIBD of GSKIP (CFP-GSKIP-V41P/L45P) was generated as a negative control. The fusions were immunoprecipitated with rabbit anti-GSKIP-antibody and tested for RII binding in an RII overlay (Fig. 3C, upper panel). RII subunits bound to immunoprecipitated proteins corresponding in size to CFP-GSKIP (48 kDa). No signal was detected when the precipitation was carried out with the corresponding preimmune serum. In addition, the precipitated proteins bound RII subunits only in the absence of the peptide AKAP18δ-L314E (see above). As expected, the introduction of two prolines into positions 41 and 45 abolished binding to RII subunits. Taken together, the data show that full-length GSKIP binds PKA at its RIIBD between amino acid residues 28 and 52 and thus functions as an AKAP. Similar amounts of the fusions of CFP with wild type GSKIP and CFP with GSKIP-V41P/L45P were precipitated (Fig. 3C, lower panel). This was revealed when the amounts of precipitated proteins were compared by Western blotting with anti-green fluorescent protein antibody. The experiment also confirmed the specificity of the anti-GSKIP antibody. This antibody precipitated the fusions containing the GSKIP versions and CFP but not CFP alone, or other proteins corresponding in size to CFP (28 kDa) were specifically detected (Fig. 3C, lower panel).
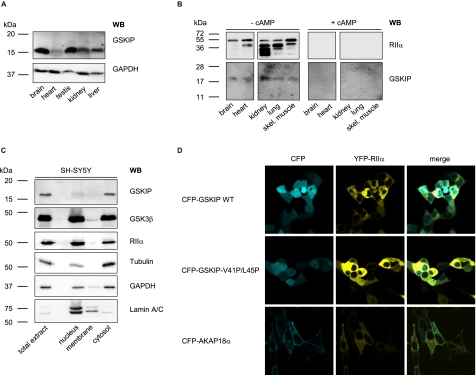
GSKIP Is Widely Expressed and Functions as an AKAP in Vivo
Homogenates from various rat organs were subjected to SDS-PAGE and Western blotting with our rabbit anti-GSKIP antibody (Fig. 4A). GSKIP was detected in all tested organs. The highest GSKIP protein levels were found in testis and brain (Fig. 4A). These results are in accordance with those of Chou et al. (20) who demonstrated ubiquitous expression of GSKIP mRNA and with data base entries showing organism-wide GSKIP expression, e.g. Serial Analysis of Gene Expression (SAGE) studies as summarized under the gene card homepage of the Weizmann Institute. To determine whether endogenous GSKIP binds regulatory subunits of PKA and thus functions as an AKAP in vivo, cAMP-agarose pulldown assays and Western blotting were carried out using lysates derived from various rat tissues (Fig. 4B). Cyclic AMP-agarose precipitates regulatory subunits of PKA, including their interacting partners. Co-precipitated GSKIP was detected by immunoblotting with rabbit anti-GSKIP antibody. Immunoblotting revealed a band of the expected molecular weight (Fig. 4B, 17 kDa, −cAMP), which was not detected if the cAMP-agarose pulldown was carried out in the presence of excess cAMP (+cAMP). As a control, RIIα subunits were detected by Western blot. In addition to full-length RIIα, several other bands were detected. These may be degradation products mainly observed in kidney and lung, which are protease-rich tissues (33). The data show that GSKIP functions as an AKAP in vivo (Fig. 4B).
FIGURE 4.
GSKIP is widely expressed and functions as an AKAP in vivo. A, lysates from the indicated rat organs were subjected to Western blot (WB) analysis with an anti-GSKIP antibody and, as a loading control, anti-GAPDH antibody (167 μg of total protein per lane). B, cAMP-agarose pulldowns were obtained from the indicated rat tissue lysates (3 mg of total protein in each sample) in the absence (− cAMP) or presence (+ cAMP; 50 μm) of cAMP. RIIα subunits of PKA and GSKIP were detected with specific antibodies by Western blotting. skel., skeletal. C, indicated subcellular fractions were obtained from SH-SY5Y cells. 10 μg of total protein from each fraction was analyzed for the presence of the indicated proteins by Western blotting. D, GSKIP and RIIα co-localize in HEK293 cells. HEK293 cells were transiently transfected to express YFP-RIIα and fusions of CFP with wild type GSKIP (CFP-GSKIP WT), GSKIP-V41P/L45P, or as a plasma membrane marker AKAP18α.
To investigate the cellular localization of GSKIP, we prepared subcellular fractions of SH-SY5Y cells expressing GSKIP endogenously. The enrichment of nuclei and cytosol was monitored by detecting lamin A/C (nucleus) and GAPDH and tubulin (both cytosol). Apparently, GSKIP is localized exclusively in the cytosol (Fig. 4C). This result is in line with a prediction using the subcellular localization prediction program pTarget (34), which calculated a cytoplasmic localization of GSKIP with a high confidence of 87.6%. The soluble fraction also contains high amounts of GSK3β and PKA RIIα subunits. These two proteins were also present in the nuclear and membrane fractions. This has been shown previously for both proteins (35). Laser scanning microscopic analysis was carried out to examine whether GSKIP and RIIα co-localize in the cytosol. For this purpose, the two proteins were expressed in HEK293 cells as fusions with CFP and yellow fluorescent protein, respectively. CFP-GSKIP is distributed throughout the cells, including the nucleus (Fig. 4D). The RII-binding deficient mutant CFP-GSKIP-V41P/L45P displays a similar distribution. The nuclear localization of the GSKIP versions is most likely due to the CFP moiety, which is known to target various proteins to the nucleus (36). Similarly to the GSKIP versions, YFP-RIIα was distributed throughout the cytoplasm but was not found in the nucleus, probably due to the higher molecular weight of this fusion protein in comparison with CFP-GSKIP. Together with our molecular interaction and pulldown studies (Figs. 3 and 4), our data indicate that GSKIP and RIIα co-localize and form complexes in the cytosol of cells. We compared the localization of GSKIP to another AKAP, AKAP18α, which is lipid-anchored and thus located in the plasma membrane.
RII-binding Domain of GSKIP Is Located within the Domain of Unknown Function 727
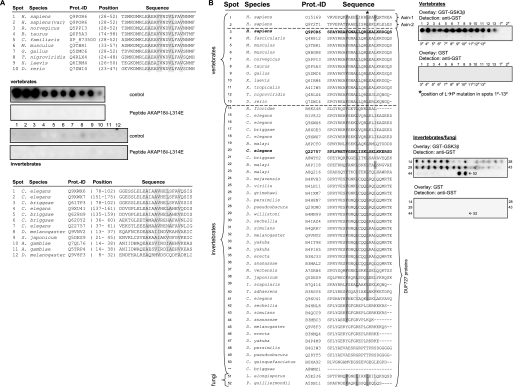
Screening the InterPro data base (37) revealed that GSKIP orthologues contain the “domain of unknown function 727” (DUF727; InterPro data base accession number IPR007967) based on their protein sequence similarity. DUF727 spans amino acid residues 32–139 of human GSKIP. This domain, covering almost the full-length of GSKIP, including the RIIBD, is conserved within metazoans. For example, the domain shows a 68% amino acid identity between zebrafish and human proteins. To test whether the members of the DUF727 family bind RII subunits, peptides homologous to the RIIBD of GSKIP were identified in various vertebrate and invertebrate family members, spot-synthesized, and subjected to RII overlay assay with human RIIα subunits (Fig. 5A). All peptides derived from the indicated vertebrate proteins bind to human RIIα subunits. Binding was abolished if the RII subunits were preincubated with the peptide AKAP18δ-L314E, suggesting that the cognate proteins function as AKAPs. The high degree of conservation implies that the AKAP function of GSKIP and its orthologues is involved in the control of mechanisms common to all vertebrates. The sequence identity of the putative RIIBDs of the vertebrate and invertebrate species is low, e.g. between the human GSKIP (Q6POR6) and the C. elegans orthologue (Q9XWX6) <40% (Fig. 5A). In addition, there are fewer hydrophobic amino acid residues in the invertebrate sequences compared with those in conserved positions of the vertebrate sequences. These differences most likely account for the observation that binding of human RIIα subunits to the peptides derived from the indicated invertebrate proteins was apparently weak or undetectable (Fig. 5A). Thus, it appears that the invertebrate proteins may either be low affinity AKAPs or do not possess an AKAP function. We have confirmed that our approach can identify both vertebrate and invertebrate AKAPs. D. melanogaster DAKAP550 (also termed neurobeachin (38) or rugose) contains two RII-binding sites (39). Both sites were identified by our AKAP candidate search, and binding was confirmed by RII overlay with human RIIα (data not shown).
FIGURE 5.
GSKIP contains the evolutionarily conserved DUF727, which binds PKA and GSK3β. A, RII-binding domain of GSKIP is located within DUF727. Shown are sequence alignments of the RII-binding domain of GSKIP with orthologues from the indicated vertebrate and invertebrate species (upper and lower, panels, respectively). Swiss-Prot protein identification codes (Prot.-ID) and positions of the RII-binding domains within the cognate proteins are indicated (for Canis familiaris, the NCBI protein accession is given as no Swiss-Prot entry was available). Amino acid residues in conserved positions of RII-binding domains are shaded in gray (see also Fig. 1A). Peptides depicted in the alignments were spot-synthesized and assayed for RII binding in the absence (control) or the presence of the PKA anchoring disruptor peptide AKAP18δ-L314E (10 μm). Signals were detected by autoradiography. Numbers of the spots correspond to the respective numbers in the alignments. Shown are representative membranes from three independent experiments. B, GSK3β binding is conserved in GSKIP orthologues. Left panel, alignment of all 51 DUF727 proteins with the C terminus of human GSKIP (amino acids 115–139) and human axin-1 and -2. Hydrophobic residues required for GSK3β interaction, which are identical (dark gray) or similar (bright gray) to human axin-1, are highlighted. Right panel, peptide spots of corresponding sequences in the alignment were spot-synthesized and incubated with GST-GSK3β or GST as a negative control. Peptides marked with a P contain an Leu → Pro (L→P) mutation in the position corresponding to amino acid 130 in human GSKIP (asterisk in left panel). Protein binding was detected with an anti-GST antibody. Both axin peptides and 40 of 50 DUF727 peptides bound GST-GSK3β. This result is representative for three independent experiments.
An interaction of GSKIP with GSK3β has been demonstrated for the human proteins (20, 40) but also for the C. elegans orthologues of GSKIP (Q22757) and GSK3β (41). Axin interacts with GSK3β. The GSK3β-binding sites of GSKIP and of axin are homologous (20). The axin/GSK3β interaction is brought about by four hydrophobic amino acids of an axin α-helix binding to a hydrophobic groove in GSK3β (42). Sequence comparison revealed that three amino acids of axin, essential for the binding to GSK3β, are conserved in 41 of 51 members of the DUF727 family, i.e. the GSKIP orthologues (Phe-122, Leu-126, and Leu-130 in human GSKIP, see Fig. 5B). These 41 proteins also have a hydrophobic amino acid in the position corresponding to Leu-133 in human GSKIP. In this position, various hydrophobic amino acids are permissive for GSK3β binding as seen in axin-1 and axin-2, which contain valine and leucine residues. To test whether the putative GSK3β interaction domains of the DUF727 proteins bind GSK3β, peptide spots of the corresponding sequences were incubated with recombinant GST-GSK3β (serine was used for peptide synthesis instead of cysteine, see “Experimental Procedures”). Binding was detected with an anti-GST antibody. As negative controls, the peptide spots were incubated with GST, and in addition, the leucine (indicated in Fig. 5B by an asterisk) was substituted by proline in the peptides derived from vertebrate sequences. 40 out of 41 putative DUF727 GSK3β interaction peptides bound GST-GSK3β (Fig. 5B). No binding of GST to any of the peptides or of GST-GSK3β to Leu → Pro mutant peptides was observed. In summary, the data indicate that GSK3β binding is conserved in almost all GSKIP orthologues/DUF727 proteins from fungi to mammals and that all vertebrate DUF727 proteins possess an AKAP function.
GSKIP Forms a Ternary Complex with PKA and GSK3β and Facilitates PKA Phosphorylation of GSK3β
We aimed to elucidate the function of the GSKIP/PKA interaction. GSKIP interacts with GSK3β through amino acids 115–139 of GSKIP (20). We therefore hypothesized that GSKIP facilitates PKA phosphorylation of GSK3β. Initially, we investigated whether GSKIP forms a ternary complex with GSK3β and PKA in an enzyme-linked immunosorbent assay-based approach (Fig. 6A). If wells of microtiter plates were coated with GSKIP, GSK3β bound as detected with anti-GSK3β and secondary HRP-coupled antibodies. If GSKIP was omitted, i.e. wells were blocked with blocking solution without GSKIP, signals were hardly detectable (Fig. 6A, left panel). If increasing concentrations of GSK3β in combination with constant amounts of GSKIP were added to RIIα-coated wells, a complex consisting of the three proteins formed. Complexes accumulated with increasing concentrations of GSK3β, as detected with anti-GSK3β and secondary HRP-coupled antibodies. As negative controls, wells were blocked and incubated with GSKIP and GSK3β or coated with RIIα and incubated with GSK3β in the absence of GSKIP. In neither negative control was significant binding of GSK3β observed. This indicates that neither GSKIP nor GSK3β bound to blocked wells and that GSK3β interacts indirectly with RIIα via GSKIP. Thus, GSKIP is a scaffolding protein recruiting GSK3β and RII subunits and positions the two proteins in close proximity.
Next, we examined whether GSKIP facilitates PKA phosphorylation of GSK3β. HEK293 cells expressing GSK3β endogenously (Fig. 6B) were transiently transfected to express CFP or a fusion of CFP with GSKIP and left untreated or were treated with forskolin (20 μm, 30 min). A basal level of phosphorylated GSK3β (p-GSK3β) was detectable in both CFP and CFP-GSKIP-expressing cells. The level, however, was higher in cells expressing CFP-GSKIP. Forskolin is a direct activator of adenylyl cyclases and induces a rise in cAMP activating PKA. In CFP-GSKIP-expressing cells, forskolin treatment induced a 2.5-fold higher increase of GSK3β phosphorylation than in cells expressing CFP alone. In addition, preincubation of the cells with the PKA inhibitor H89 prevented the forskolin-induced and GSKIP-dependent increase in p-GSK3β. The data indicate that GSKIP facilitates PKA-mediated phosphorylation of endogenous GSK3β. To determine whether the AKAP function of GSKIP is required for this, we incubated untransfected HEK293 cells with a peptide derived from the C terminus of GSKIP, GSKIPtide (20), which represents the GSK3β interaction domain of GSKIP (Fig. 6C). GSKIPtide displaces GSK3β from GSKIP and thus from its associated pool of PKA. GSKIPtide caused a 2-fold increase in p-GSK3β (Fig. 6C), indicating that the GSK3β interaction domain of GSKIP is sufficient to increase GSK3β phosphorylation. The observation that GSKIPtide-induced phosphorylation is blocked by H89 shows that this reaction is PKA-dependent. The extent of GSK3β phosphorylation in GSKIPtide-treated cells was comparable with FSK-treated cells. The inactive peptide GSKIPtide-L130P, which does not bind to GSK3β, had no effect on GSK3β phosphorylation. FSK did not increase GSK3β phosphorylation in GSKIPtide-treated cells (Fig. 6, B and C). Altogether, it appears that the interaction of the anchoring protein with the substrate by itself renders the substrate protein a better target for PKA whether or not PKA is anchored. Additionally, the AKAP function of GSKIP enhances PKA-dependent phosphorylation of GSK3β in the ternary complex consisting of GSKIP, PKA, and GSK3β.
DISCUSSION
Here, we describe a bioinformatics and peptide array screening approach to identify new AKAPs. We defined a search pattern for data base screening, which is based on an AKAP signature motif combined with the range of pI values calculated from the peptide sequences of RIIBDs. The search pattern (AVLISE)XX(AVLIF)(AVLI)XX(AVLI)(AVLIF)XX(AVLISE) assigns hydrophobic amino acid residues to conserved positions (with the exception of permitted serine and glutamic acid in the N- and C-terminally conserved position) and allows, within the pI range limitation, charged and polar amino acid residues in the remaining positions between the hydrophobic residues (X in the search pattern). The search pattern reflects an amphipathic helix structure typical for RIIBDs of AKAPs. Thus, a data base search is likely to retrieve RII-binding peptides that possess a high probability to form amphipathic α-helices that dock into the DD domain in a classical AKAP manner. Our procedure identified 36 peptides binding RII subunits in the same way known AKAPs do. Nine of the peptides were derived from previously described AKAPs, the RIIBDs of which are identical with the ones identified in this work, underlining the validity of our approach. The hydrophobic faces of such helices may also bind to hydrophobic regions on RII subunits outside the hydrophobic groove of the DD domain. This may lead to the identification of proteins binding to regulatory subunits of PKA in a noncanonical AKAP manner as was recently observed for α4-integrins, p90-ribosomal S6 kinase, and actin (43–46).
Our approach identified an RIIBD in GSKIP. We show here that full-length GSKIP functions as an AKAP and binds RII subunits in a typical AKAP manner, i.e. through an amphipathic helix-forming RIIBD. GSKIP binds RII subunits with nanomolar affinities in Biacore measurements (RIIα with KD = 5 nm and RIIβ with KD = 43 nm). It is apparently an RII-specific AKAP because binding of RI subunits was not detectable in Biacore experiments (data not shown).
The structure of GSKIP had been previously determined by NMR (Protein Data Bank code 1sgo). Projection of the identified RIIBD onto the Protein Data Bank structure and our own NMR measurements revealed that the RIIBD of GSKIP (amino acid residues 28–52) is located within an α-helix formed by amino acid residues 33–48 that turns into a short β-strand (amino acid residues 49–54) (Fig. 3A, lower panel). The α-helix displays an amphipathic character as the charged and polar amino acid residues (Asp-32, Arg-34, Glu-36, Glu-38, and Asn-42) are located on one side of the helix, and the hydrophobic amino acid residues (Ala-37, Val-40, Val-41, Val-44, Leu-45, and Val-48) are located on the opposite side. The latter, forming the contact surface to the RII subunit dimer (Fig. 2B and Fig. 3A), are masked by the anti-parallel β-sheet. Binding of GSKIP to RII subunits is therefore likely to be associated with major conformational changes of GSKIP. Our NMR experiments confirmed that the RIIBD interacts with the DD domain of RIIα (Fig. 3A, upper panel). Furthermore, shifts were observed in the β-sheet region and in residues of the C-terminal helix, which are in close proximity to the α1-helix. These shifts probably represent altered intramolecular interactions due to DD domain binding.
GSKIP contains the domain of unknown function 727 (DUF727), which was defined on the basis of sequence similarities between GSKIP orthologues. DUF727 covers almost the entire sequence of human GSKIP (amino acids 32–139) and confers the ability to interact with both RII (amino acids 28–52) and GSK3β (amino acids 115–139). We have shown that the AKAP function is conserved among vertebrates, but we have not observed binding of human RIIα to peptide spots of GSKIP invertebrate orthologues (Fig. 5A), although our assay detects invertebrate AKAPs, i.e. DAKAP550 (data not shown). Whereas all 11 vertebrate DUF727 entries contain a putative RIIBD, only 3 of the remaining 40 DUF727 proteins contain this AKAP sequence (Q9XU41, Q628G9, and B0XA92). When we performed RII overlays over peptide spots derived from DUF727 proteins, Q9XU41 from Caenorhabditis elegans and Q628G9 from Caenorhabditis briggsae peptides did not bind human RIIα. Both peptides contain a proline residue 5 amino acids N-terminal of the AKAP pattern which most likely disturbs the helical structure required for RII binding. A proline in this position reduced RII binding of GSKIP (Fig. 2B) and of AKAP18δ (48).
Lancelets (also termed amphioxus or cephalochordata) are the most basal chordate subphylum and are considered predecessors of vertebrates (49). It is noteworthy that the DUF727 protein of the Florida lancelet (Branchiostoma floridae), which has a higher sequence similarity to human GSKIP than lower invertebrates, has only 5 of 6 conserved amino acids of the RIIBD. The absence of a canonical RIIBD in the Florida lancelet and in the invertebrate DUF727 proteins implies that only vertebrate GSKIP orthologues are AKAPs. This reflects the increased regulation and integration of signaling pathways in vertebrates.
An interaction of GSKIP with GSK3β in an axin-like fashion had been reported (20). To investigate whether this GSK3β binding ability is conserved in GSKIP orthologues, we performed a sequence alignment of DUF727 proteins with human axin-1 and -2. It revealed that the key residues of axin essential for an interaction with GSK3β are conserved in 41 out of 51 DUF727 proteins (Fig. 5B). These amino acids are also conserved in the two fungi and the Branchiostoma DUF727 proteins. Employing an overlay approach, we demonstrated that the putative GSK3β interaction peptides interact with GSK3β in vitro (Fig. 5B). This result is in line with the GSKIP/GSK3β interaction previously observed for human and C. elegans proteins (20, 41). GSK3β orthologues are known to play important roles in members of the fungi kingdom (50). However, no axin orthologues are known in fungi, implying that GSKIP is an ancestral and functionally highly conserved GSK3β interaction protein. 9 of the 10 remaining DUF727 proteins without a putative GSK3β-binding motif are from species with two or more DUF727 proteins (all of them invertebrates). In D. melanogaster, for instance, Q9VNV2 contains the putative GSK3β-binding motif, whereas Q9V8F3 does not. This implies that several species have further DUF727 proteins with unknown function in addition to those that can interact with GSK3β.
GSK3β fulfills various functions as it is, for example, involved in energy metabolism, neuronal cell development, and body pattern formation (51). All of these processes are modulated by PKA. Thus, it is conceivable that the interaction of GSKIP with GSK3β, which controls PKA phosphorylation of GSK3β (Fig. 6B), plays a role in elementary cellular processes in vertebrates.
The PKA- and GSK3β-binding sites are located in different regions of GSKIP. Because most AKAPs are scaffolding proteins binding several signaling proteins, we investigated whether GSKIP can interact with PKA and GSK3β simultaneously. We identify here a ternary complex consisting of GSKIP, RIIα subunits of PKA, and GSK3β (Fig. 6A). Complex formation was detected only if GSKIP was present (Fig. 6A). The complex of full-length GSKIP and GSK3β bound RIIα subunits with a KD = 43 ± 5 nm (Fig. 6A).
It has been postulated that PKA-dependent phosphorylation of GSK3β is mediated by binding of PKA to GSK3β via an additional protein (13, 19, 53). Tanji et al. (18) showed a direct interaction of AKAP220 with GSK3β and suggested that the complex contains AKAP220, PKA, PP1, and GSK3β. In this complex, PKA inhibits GSK3β by phosphorylation of Ser-9. MAP2D also interacts with PKA and GSK3β (17). Flynn et al. (17) postulated a complex consisting of MAP2D, PKA, PP2A, and GSK3β in preovulatory granulosa cells that controls PKA phosphorylation and thus inactivation of GSK3β. GSKIP controls PKA-dependent phosphorylation of GSK3β at Ser-9 and thus GSK3β inactivation (Fig. 6B). It appears that GSKIP controls the PKA-dependent phosphorylation of GSK3β by a dual mechanism. 1) The binding of GSKIP itself (or of GSKIPtide) to GSK3β leads to PKA-dependent phosphorylation of GSK3β (Fig. 6, B and C). Thus, it is likely that GSKIP (or GSKIPtide) binding alters GSK3β conformation, causing increased PKA-dependent phosphorylation. This has been shown for other GSK3β inhibitors and interacting peptides (54). For example, the peptide axin GID, which displaces axin from GSK3β, also increases PKA-dependent phosphorylation of GSK3β (54). 2) Forskolin stimulation only enhances GSK3β phosphorylation in cells expressing full-length GSKIP but not in cells treated with GSKIPtide, implying that the anchoring of PKA to GSKIP contributes to PKA-dependent phosphorylation of GSK3β. Surprisingly, the peptides Ht31 and AKAP18δ-L314E, which globally uncouple PKA from AKAPs, did not affect GSKIP-mediated GSK3β phosphorylation (data not shown). However, global displacement of PKA from AKAPs leads to an increase of PKA in the cytosol (55), which in turn might phosphorylate GSKIP-bound GSK3β. This most likely explains that the peptides apparently did not interfere with the forskolin-induced phosphorylation of GSK3β. Experiments using these peptides have also not been shown for the AKAP220- or MAP2D-PKA complexes, which mediate PKA-dependent phosphorylation of GSK3β (17, 18).
We show here by Western blot analysis that GSKIP protein levels are most abundant in brain and testis (Fig. 4A). Also MAP2 and AKAP220, the other AKAPs regulating GSK3β, are predominantly expressed in the brain and reproductive tissues. Moreover, GSK3β protein levels are highest in the brain (56). Although AKAP220 is localized peroxisomally and MAP2 is bound to microtubules, GSKIP seems to be distributed throughout the cytosol (Fig. 4D). Given the high GSK3β levels in the brain, and the complex function and architecture of neurons, it is conceivable that several AKAPs are required to integrate PKA and GSK3β signaling in different brain regions and/or cellular compartments.
Acknowledgments
We are grateful to Angelika Ehrlich, Andrea Geelhaar, Beate Eisermann, Michael Gomoll, and Jürgen Malkewitz for excellent technical assistance. We thank Dr. Katja Faelber and Verena Ezerski for helpful comments on the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Forschergruppe 806, Project KL1415/4-1, the European Union Project Thera cAMP, Proposal 037189, and the GoBio Program of the German Ministry of Education and Science Grants FKZ 0315097 and 0315516.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
- AKAP
- A-kinase anchoring protein
- DUF
- domain of unknown function
- GST
- glutathione S-transferase
- GSK3β
- glycogen synthase kinase 3β
- GSKIP
- GSK3β interaction protein
- HRP
- horseradish peroxidase
- RII
- type II regulatory subunit of PKA
- RIIBD
- RII-binding domain
- 8-AHA-cAMP
- 8-aminohexyl amino adenosine 3′-5′-cyclic monophosphate
- CFP
- cyan fluorescent protein
- PKA
- protein kinase A
- DD
- dimerization and docking
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- FSK
- forskolin
- MOPS
- 4-morpholinepropanesulfonic acid.
REFERENCES
- 1.Dodge-Kafka K. L., Bauman A., Kapiloff M. S. (2008) Handb. Exp. Pharmacol. 186, 3–14 [DOI] [PubMed] [Google Scholar]
- 2.Hundsrucker C., Klussmann E. (2008) Handb. Exp. Pharmacol. 186, 483–503 [DOI] [PubMed] [Google Scholar]
- 3.McCahill A. C., Huston E., Li X., Houslay M. D. (2008) Handb. Exp. Pharmacol. 186, 125–166 [DOI] [PubMed] [Google Scholar]
- 4.Carnegie G. K., Means C. K., Scott J. D. (2009) IUBMB Life 61, 394–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mauban J. R., O'Donnell M., Warrier S., Manni S., Bond M. (2009) Physiology 24, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. (2007) Cell 130, 1032–1043 [DOI] [PubMed] [Google Scholar]
- 7.Wu J., Brown S. H., von Daake S., Taylor S. S. (2007) Science 318, 274–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold M. G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C. R., Scott J. D., Barford D. (2006) Mol. Cell 24, 383–395 [DOI] [PubMed] [Google Scholar]
- 9.Kinderman F. S., Kim C., von Daake S., Ma Y., Pham B. Q., Spraggon G., Xuong N. H., Jennings P. A., Taylor S. S. (2006) Mol. Cell 24, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hundsrucker C., Krause G., Beyermann M., Prinz A., Zimmermann B., Diekmann O., Lorenz D., Stefan E., Nedvetsky P., Dathe M., Christian F., McSorley T., Krause E., McConnachie G., Herberg F. W., Scott J. D., Rosenthal W., Klussmann E. (2006) Biochem. J. 396, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billadeau D. D. (2007) Pancreatology 7, 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kockeritz L., Doble B., Patel S., Woodgett J. R. (2006) Curr. Drug Targets 7, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 13.Li M., Wang X., Meintzer M. K., Laessig T., Birnbaum M. J., Heidenreich K. A. (2000) Mol. Cell. Biol. 20, 9356–9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugden P. H., Fuller S. J., Weiss S. C., Clerk A. (2008) Br. J. Pharmacol. 153, S137–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taurin S., Sandbo N., Qin Y., Browning D., Dulin N. O. (2006) J. Biol. Chem. 281, 9971–9976 [DOI] [PubMed] [Google Scholar]
- 16.Jensen J., Brennesvik E. O., Lai Y. C., Shepherd P. R. (2007) Cell. Signal. 19, 204–210 [DOI] [PubMed] [Google Scholar]
- 17.Flynn M. P., Maizels E. T., Karlsson A. B., McAvoy T., Ahn J. H., Nairn A. C., Hunzicker-Dunn M. (2008) Mol. Endocrinol. 22, 1695–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanji C., Yamamoto H., Yorioka N., Kohno N., Kikuchi K., Kikuchi A. (2002) J. Biol. Chem. 277, 36955–36961 [DOI] [PubMed] [Google Scholar]
- 19.Fang X., Yu S. X., Lu Y., Bast R. C., Jr., Woodgett J. R., Mills G. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11960–11965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou H. Y., Howng S. L., Cheng T. S., Hsiao Y. L., Lieu A. S., Loh J. K., Hwang S. L., Lin C. C., Hsu C. M., Wang C., Lee C. I., Lu P. J., Chou C. K., Huang C. Y., Hong Y. R. (2006) Biochemistry 45, 11379–11389 [DOI] [PubMed] [Google Scholar]
- 21.Coin I., Beyermann M., Bienert M. (2007) Nat. Protoc. 2, 3247–3256 [DOI] [PubMed] [Google Scholar]
- 22.Stefan E., Wiesner B., Baillie G. S., Mollajew R., Henn V., Lorenz D., Furkert J., Santamaria K., Nedvetsky P., Hundsrucker C., Beyermann M., Krause E., Pohl P., Gall I., MacIntyre A. N., Bachmann S., Houslay M. D., Rosenthal W., Klussmann E. (2007) J. Am. Soc. Nephrol. 18, 199–212 [DOI] [PubMed] [Google Scholar]
- 23.Hyde C., Johnson T., Owen D., Quibell M., Sheppard R. C. (1994) Int. J. Pept. Protein Res. 43, 431–440 [PubMed] [Google Scholar]
- 24.Henn V., Edemir B., Stefan E., Wiesner B., Lorenz D., Theilig F., Schmitt R., Vossebein L., Tamma G., Beyermann M., Krause E., Herberg F. W., Valenti G., Bachmann S., Rosenthal W., Klussmann E. (2004) J. Biol. Chem. 279, 26654–26665 [DOI] [PubMed] [Google Scholar]
- 25.Klussmann E., Maric K., Wiesner B., Beyermann M., Rosenthal W. (1999) J. Biol. Chem. 274, 4934–4938 [DOI] [PubMed] [Google Scholar]
- 26.Schmidt A., Wiesner B., Weisshart K., Schulz K., Furkert J., Lamprecht B., Rosenthal W., Schülein R. (2009) Traffic 10, 2–15 [DOI] [PubMed] [Google Scholar]
- 27.Herberg F. W., Maleszka A., Eide T., Vossebein L., Tasken K. (2000) J. Mol. Biol. 298, 329–339 [DOI] [PubMed] [Google Scholar]
- 28.Bertinetti D., Schweinsberg S., Hanke S. E., Schwede F., Bertinetti O., Drewianka S., Genieser H. G., Herberg F. W. (2009) BMC Chem. Biol. 9, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vijayaraghavan S., Liberty G. A., Mohan J., Winfrey V. P., Olson G. E., Carr D. W. (1999) Mol. Endocrinol. 13, 705–717 [DOI] [PubMed] [Google Scholar]
- 30.Newlon M. G., Roy M., Morikis D., Carr D. W., Westphal R., Scott J. D., Jennings P. A. (2001) EMBO J. 20, 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stajich J. E., Block D., Boulez K., Brenner S. E., Chervitz S. A., Dagdigian C., Fuellen G., Gilbert J. G., Korf I., Lapp H., Lehväslaiho H., Matsalla C., Mungall C. J., Osborne B. I., Pocock M. R., Schattner P., Senger M., Stein L. D., Stupka E., Wilkinson M. D., Birney E. (2002) Genome Res. 12, 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuffin L. J., Bryson K., Jones D. T. (2000) Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 33.Tian J. M., Schibler U. (1991) Genes Dev. 5, 2225–2234 [DOI] [PubMed] [Google Scholar]
- 34.Guda C., Subramaniam S. (2005) Bioinformatics 21, 3963–3969 [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama N., Yin D., Malbon C. C. (2007) J. Mol. Signal. 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seibel N. M., Eljouni J., Nalaskowski M. M., Hampe W. (2007) Anal. Biochem. 368, 95–99 [DOI] [PubMed] [Google Scholar]
- 37.Hunter S., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., Finn R. D., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Laugraud A., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., Mistry J., Mitchell A., Mulder N., Natale D., Orengo C., Quinn A. F., Selengut J. D., Sigrist C. J., Thimma M., Thomas P. D., Valentin F., Wilson D., Wu C. H., Yeats C. (2009) Nucleic Acids Res. 37, D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Herberg F. W., Laue M. M., Wullner C., Hu B., Petrasch-Parwez E., Kilimann M. W. (2000) J. Neurosci. 20, 8551–8565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han J. D., Baker N. E., Rubin C. S. (1997) J. Biol. Chem. 272, 26611–26619 [DOI] [PubMed] [Google Scholar]
- 40.Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., Goldberg D. S., Li N., Martinez M., Rual J. F., Lamesch P., Xu L., Tewari M., Wong S. L., Zhang L. V., Berriz G. F., Jacotot L., Vaglio P., Reboul J., Hirozane-Kishikawa T., Li Q., Gabel H. W., Elewa A., Baumgartner B., Rose D. J., Yu H., Bosak S., Sequerra R., Fraser A., Mango S. E., Saxton W. M., Strome S., Van Den Heuvel S., Piano F., Vandenhaute J., Sardet C., Gerstein M., Doucette-Stamm L., Gunsalus K. C., Harper J. W., Cusick M. E., Roth F. P., Hill D. E., Vidal M. (2004) Science 303, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dajani R., Fraser E., Roe S. M., Yeo M., Good V. M., Thompson V., Dale T. C., Pearl L. H. (2003) EMBO J. 22, 494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houslay M. D. (2006) Sci. STKE 2006, E32. [DOI] [PubMed] [Google Scholar]
- 44.Lim C. J., Han J., Yousefi N., Ma Y., Amieux P. S., McKnight G. S., Taylor S. S., Ginsberg M. H. (2007) Nat. Cell Biol. 9, 415–421 [DOI] [PubMed] [Google Scholar]
- 45.Chaturvedi D., Poppleton H. M., Stringfield T., Barbier A., Patel T. B. (2006) Mol. Cell. Biol. 26, 4586–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rivard R. L., Birger M., Gaston K. J., Howe A. K. (2009) Cell Motil. Cytoskeleton 66, 693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deleted in proof
- 48.Hundsrucker C. (2007) Development of Peptidic Anchoring Disruptors and a Bioinformatics Approach to Identify New Anchoring Proteins of Protein Kinase A. Ph.D. thesis, Free University of Berlin, Germany [Google Scholar]
- 49.Putnam N. H., Butts T., Ferrier D. E., Furlong R. F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J. K., Benito-Gutiérrez E. L., Dubchak I., Garcia-Fernàndez J., Gibson-Brown J. J., Grigoriev I. V., Horton A. C., de Jong P. J., Jurka J., Kapitonov V. V., Kohara Y., Kuroki Y., Lindquist E., Lucas S., Osoegawa K., Pennacchio L. A., Salamov A. A., Satou Y., Sauka-Spengler T., Schmutz J., Shin-I. T., Toyoda A., Bronner-Fraser M., Fujiyama A., Holland L. Z., Holland P. W., Satoh N., Rokhsar D. S. (2008) Nature 453, 1064–1071 [DOI] [PubMed] [Google Scholar]
- 50.Kassir Y., Rubin-Bejerano I., Mandel-Gutfreund Y. (2006) Curr. Drug Targets 7, 1455–1465 [DOI] [PubMed] [Google Scholar]
- 51.Plyte S. E., Hughes K., Nikolakaki E., Pulverer B. J., Woodgett J. R. (1992) Biochim. Biophys. Acta 1114, 147–162 [DOI] [PubMed] [Google Scholar]
- 52.Alto N. M., Soderling S. H., Hoshi N., Langeberg L. K., Fayos R., Jennings P. A., Scott J. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4445–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torii K., Nishizawa K., Kawasaki A., Yamashita Y., Katada M., Ito M., Nishimoto I., Terashita K., Aiso S., Matsuoka M. (2008) Cell. Signal. 20, 1256–1266 [DOI] [PubMed] [Google Scholar]
- 54.Zhang F., Phiel C. J., Spece L., Gurvich N., Klein P. S. (2003) J. Biol. Chem. 278, 33067–33077 [DOI] [PubMed] [Google Scholar]
- 55.Wojtal K. A., de Vries E., Hoekstra D., van Ijzendoorn S. C. (2006) Mol. Biol. Cell 17, 3638–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao H. B., Shaw P. C., Wong C. C., Wan D. C. (2002) J. Chem. Neuroanat. 23, 291–297 [DOI] [PubMed] [Google Scholar]