Abstract
Vascular endothelial growth factor (VEGF) is produced either as a pro-angiogenic or anti-angiogenic protein depending upon splice site choice in the terminal, eighth exon. Proximal splice site selection (PSS) in exon 8 generates pro-angiogenic isoforms such as VEGF165, and distal splice site selection (DSS) results in anti-angiogenic isoforms such as VEGF165b. Cellular decisions on splice site selection depend upon the activity of RNA-binding splice factors, such as ASF/SF2, which have previously been shown to regulate VEGF splice site choice. To determine the mechanism by which the pro-angiogenic splice site choice is mediated, we investigated the effect of inhibition of ASF/SF2 phosphorylation by SR protein kinases (SRPK1/2) on splice site choice in epithelial cells and in in vivo angiogenesis models. Epithelial cells treated with insulin-like growth factor-1 (IGF-1) increased PSS and produced more VEGF165 and less VEGF165b. This down-regulation of DSS and increased PSS was blocked by protein kinase C inhibition and SRPK1/2 inhibition. IGF-1 treatment resulted in nuclear localization of ASF/SF2, which was blocked by SPRK1/2 inhibition. Pull-down assay and RNA immunoprecipitation using VEGF mRNA sequences identified an 11-nucleotide sequence required for ASF/SF2 binding. Injection of an SRPK1/2 inhibitor reduced angiogenesis in a mouse model of retinal neovascularization, suggesting that regulation of alternative splicing could be a potential therapeutic strategy in angiogenic pathologies.
Keywords: Growth Factors, RNA/Splicing, ASF/SF2, Angiogenesis, SRPK1, VEGF
Introduction
Vascular endothelial growth factor (VEGF-A, hereafter referred to as VEGF)5 is a key regulatory component in physiological and pathological angiogenesis. Inhibition of VEGF has shown to be effective in cancer (1) and ocular angiogenesis (2), and it is up-regulated by a number of growth factors also implicated in these conditions, including insulin-like growth factor-1 (IGF-1) (3). VEGF is generated as multiple isoforms by alternative splicing (4). There are two principal families of VEGF isoforms, the pro-angiogenic VEGFxxx isoforms, generated by proximal splice site selection in the terminal exon, exon 8a (5), and the anti-angiogenic VEGFxxxb isoforms (6), generated by use of a distal splice site 66 bp further into exon 8, generating mRNA isoforms that contain exon 8b. As the stop codon for the protein is encoded in exon 8, these two isoforms contain alternate six amino acids at the C terminus (Fig. 1A). The pro-angiogenic isoforms such as VEGF165 encode a terminal six amino acid sequence of CDKPRR, and the anti-angiogenic isoforms such as VEGF165b encode SLTRKD (7). Many normal tissues, including the eye generate both isoforms (8), and previous studies have shown that the anti-angiogenic isoforms dominate in non-angiogenic tissues such as the normal colon (9) and the vitreous (8). However, there is a splicing switch in angiogenic conditions such as proliferative diabetic retinopathy (8), colon (9), prostate (10), renal (7), and skin cancers (11), and in Denys Drash Syndrome (12). In contrast, in non-angiogenic conditions where VEGF is up-regulated, such as glaucoma and rhegmatogenous retinal detachment associated with proliferative vitreoretinopathy (13) or glaucoma (14), the anti-angiogenic isoforms are up-regulated. We have previously shown that IGF-1 can switch splicing in cultured epithelial cells from anti-angiogenic to pro-angiogenic isoforms (15). As IGF-1 has been implicated in a number of angiogenic conditions including diabetic retinopathy and colon cancer, we hypothesized that the mechanism through which IGF-1 mediates this change in splicing may be a potential therapeutic target to prevent angiogenesis. To this end, we have investigated the signaling pathways, the splicing factors involved, and the possibility of therapeutic intervention in the pathway in an animal model of diabetic retinopathy.
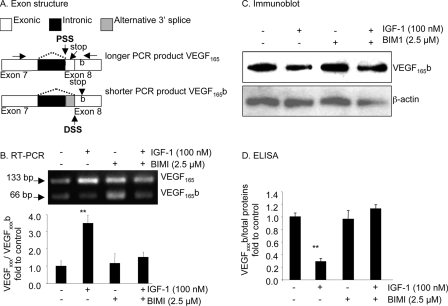
FIGURE 1.
Inhibition of PKC by BIM1 prevents the down-regulation of VEGFxxxb by IGF. A, exon structure of the VEGF pre-mRNA. Alternative splicing of exon 8 to either 8a or 8b results in use of proximal (PSS) or distal splice sites (DSS) resulting in shorter mRNA for distal splicing. Because the last stop codon is missing, the final six amino acid open reading frame is replaced by an identically sized open reading frame encoding six different amino acids. The primer position is shown by horizontal arrows. B–D, podocytes were treated with BIM1 (2.5 μm) alone or in combination with IGF-1 (100 nm). B, RT-PCR showed that BIM1 reduced the VEGFxxx:VEGFxxxb ratio at the RNA level. C, Western blot demonstrating that BIM1 inhibited the IGF-mediated down-regulation of VEGFxxxb expression at the protein level. D, ELISA results confirming that BIM1 specifically attenuated the IGF-1-dependent down-regulation of VEGFxxxb, but does not affect endogenous expression of VEGFxxxb. **, p < 0.01 compared with untreated.
EXPERIMENTAL PROCEDURES
Proliferating Podocytes
PCIPs (courtesy of Moin Saleem, University of Bristol, Bristol, UK) were derived from a cell line conditionally transformed from normal human podocytes with a temperature-sensitive mutant of immortalized SV-40 T-antigen. At the permissive temperature of 33 °C, the SV-40 T-antigen is active and allows the cells to proliferate rapidly (16). PCIPs were cultured in T75 flasks (Greiner) in RPMI 1640 medium (Sigma) with 10% fetal bovine serum, 1% ITS (insulin transferrin selenium) (Sigma), 0.5% penicillin-streptomycin solution (Sigma), and grown to 95% confluency. Then cells were split into 6-well plates (2 × 105 cells per well) and grown until 95% confluency.
Treatments with IGF-1 and Pharmacological Inhibitors
To investigate the inhibitory effect of IGF-1 on VEGFxxxb mRNA and protein synthesis, pharmacological inhibitors and IGF-1 with PKC-BIMI (Calbiochem), and SRPK1/2 (SR protein kinases 1 and 2)-SRPIN340 (SR protein phosphorylation inhibitor 340) (17) were used. 24 h before treatment, cultured medium was replaced with serum-free RPMI 1640 medium (Sigma) containing 1% ITS (Sigma) and 0.5% penicillin-streptomycin (Sigma). Subsequently, the medium was replaced with fresh serum-free RPMI 1640 medium (Sigma) containing 1% ITS, 0.5% penicillin-streptomycin, and either 2.5 μm BIMI (bisindolylmaleimide 1) or 10 μm SRPIN340 for 60 min before treatment with IGF-1. 12 h after stimulation, RNA was extracted, and 48 h after stimulation, proteins were extracted.
RT-PCR
1 μg of mRNA was reverse transcribed using MMLV RT, RNase H Minus, point mutant (Promega), and oligo(dT)15 (Promega) as a primer. The reaction was carried out in Bio-Rad cycler for 60 min at 40 °C, and then the enzyme was inactivated at 70 °C for 15 min. Ten percent of the cDNA was then amplified using primers designed to pick up proximal and distal splice forms. 1 μm of each primer (exon 7b 5′-GGCAGCTTGAGTTAAACGAAC-3′, exon 8b 5′-ATGGATCCGTATCAGTCTTTCCTGG-3′) and PCR Master Mix (Promega) were used in reactions cycled 30 times, denaturing at 95 °C for 60 s, annealing at 55 °C for 60 s, and extending at 72 °C for 60 s. PCR products were run on 2.5% agarose gels containing 0.5 μg/ml ethidium bromide and visualized under a UV transilluminator. This reaction usually resulted in one amplicon of 130 bp (VEGFxxx) and one amplicon of 64 bp (VEGFxxxb). For HEK293 and HeLa cells, RT-PCR was performed using primers specific to exon 7a and the 3′-untranslated region of the VEGF mRNA. The primers used were 5′-GTAAGCTTGTACAAGATCCGCAGACG-3′ and 5′-ATGGATCCGTATCAGTCTTTCCTGG-3′. The reaction was set up in a 20-μl reaction using the 2× FastStart Universal SyBR Master Mix (Roche, cat. no: 04913850001) and 1 μm each primer. The reaction was performed on the ABI 7000 cycler for 95 °C for 10 min, followed by 30 cycles of 95 °C for 15 s and 55 °C for 30 s.
Western Blotting
Protein samples were dissolved in Laemmli buffer, boiled for 3–4 min, and centrifuged for 2 min at 20,000 × g to remove insoluble materials. 30 μg of protein per lane were separated by SDS/PAGE (12%) and transferred to a 0.2-μm nitrocellulose membrane. The blocked membranes were probed overnight (4 °C) with antibodies against panVEGF (R&D; MAB 293, 1:500), VEGFxxxb (R&D Systems; MAB3045; 1:250), ASF/SF2 antibody (Santa Cruz Biotechnology; sc-10254; 1:1000), and β-tubulin (Sigma, 1:2000). Western blotting has previously shown that all the proteins recognized by the VEGFxxxb antibody are also recognized by commercial antibodies raised against VEGF165. It binds recombinant VEGF165b, and can be used to demonstrate expression of VEGF165b, VEGF189b, and VEGF121b (collectively termed VEGFxxxb) but does not recognize VEGF165, conclusively demonstrating that this antibody is specific for VEGFxxxb (6). Subsequently, the membranes were incubated with secondary horseradish peroxidase-conjugated antibody, and immunoreactive bands were visualized using ECL reagent (Pierce). Immunoreactive bands corresponding to panVEGF and VEGFxxxb in each treatment were quantified by ImageJ analysis and normalized to those of β-tubulin or β-actin. Blots are representative of at least three experiments. Densitometry was carried out by scanning in gels and using ImageJ to determine gray levels of bands and background.
Construction of Plasmids
The VEGF sequence of interest (from 35-bp upstream of exon 8a to 35-bp downstream of exon 8b) was amplified from a BAC DNA template using 50 ng of BAC DNA, 10 μm of each primers (see Table 1), 10 mm dNTP mix (Promega), and Taq polymerase (Promega). A modified ADML-MS2 plasmid was digested with EcoR1 and BamH1, and PCR products ligated into the vector and subsequently transformed. Colonies were selected, and plasmid extraction (Qiagen) was performed. The identities of the plasmids were confirmed by sequencing.
TABLE 1.
Primer sequences
| Construct A | Forward:5′-GAATTCCTCATCGCCAGGCCTCCTCACTTG-3′ |
| Reverse:5′-GGATTCCCTTCGCCGGAGTCTCGCCCTC-3′ | |
| Construct B | Forward:5′-GAATTCCGCCCTAACCCCAGCCTTTGTTTTCCATTTCCC-3′ |
| Reverse:5′-GGATCCGGACTGTTCTGTCGATGGTG-3′ | |
| Construct C | Forward:5′-GAATTCCGCCCTAACCCCAGCCTTTGTTTTCCATTTCCC-3′ |
| Reverse:5′-GGATCCTGGTTCCCGAAACCCTGAGCG-3′ | |
| Construct D | Forward:5′-GATTCCGAGGAAGGAAGGAGCCTCCCTCAGGG-3′ |
| Reverse:5′-GGATCCGGACTGTTCTGTCGATGGTG-3′ | |
| Construct E | Forward:5′-GAATTCATGTGACAAGCCGAGGCGG-3′ |
| Reverse:5′-GGATCCCTGGTTCCCGAAACCCTGAGCG-3′ | |
| Mouse VEGF-A ex 7 F | Forward 5′ GTTCAGAGCGGAGAAAGCAT-3′ |
| Mouse VEGF-A ex 8a R | Reverse 5′TCACATCTGCAAGTACGTTCG-3′ |
| Mouse VEGF-A ex 2 F | Forward 5′ AAGGAGAGCAGAAGTCCCATGA-3′ |
| Mouse VEGF-A ex 3 R | Reverse 5′ CTCAATCGGACGGCAGTAGCT-3′ |
| β-Actin F | Forward 5′ AGCCATGTACGTAGCCATCC-3′ |
| β-Actin R | Reverse 5′ CTCTCAGCTGTGGTGGTGAA-3′ |
Expression of the MS2-MBP (Maltose-binding Protein) Fusion Protein
MS2-MBP (a gift from Robin Reed, Harvard University) was expressed in Escherichia coli DH5α. The cells were grown to an optical density of ∼0.5 at 600 nm and induced for expression for 3 h with 0.2 mm isopropyl-1-thio-β-d-galactopyranoside. The MS2-MBP protein was purified by amylose beads according to the manufacturer's protocol (NEB, Beverly, MA). The protein was dialyzed with 10 mm sodium phosphate, pH 7, overnight at 4 °C, to remove existing salts that are present and further purified over a Heparin Hi Trap column using a NaCl gradient (GE Healthcare). An immunoblot analysis was performed on the purified fusion proteins using rabbit anti-MS2 antibody (gift from Peter Stockley, Leeds University) to confirm the identity of the protein.
Assembly of the MS2-MBP System
1 μg of the VEGF-MS2 plasmid was linearized with Xba1 and in vitro transcribed with T7 RNA polymerase (NEB) in 0.5 mm rNTP (Ambion), 40 mm Tris-HCl, 6 mm MgCl2, 10 mm dithiothreitol, 2 mm spermidine at 40 °C for 1 h to make VEGF-MS2 RNA. A 100-fold molar excess of MS2-MBP fusion protein and VEGF-MS2 RNA were incubated in a buffer containing 20 mm HEPES, pH 7.9 and 60 mm NaCl on ice for 30 min. 75 mg of HEK293 nuclear extract were added to the MS2-MBP fusion protein/VEGF-RNA mix in 0.5 mm ATP, 6.4 mm MgCl2, 20 mm creatine phosphate for 1 h at 30 °C. Proteins that bound to the MS2-MBP/VEGF-MS2 RNA complex were affinity selected on amylose beads by rotating for 4 h at 4 °C and eluted with 12 mm maltose, 20 mm HEPES pH 7.9, 60 mm NaCl, 10 mm β-mercaptoethanol, and 1 mm phenylmethylsulfonyl fluoride.
RNA Immunoprecipitation
HEK293 cells were transfected with plasmid containing the last 131 nt of intron 7 and the first 152 nt of exon 8 inserted downstream of the CMV promoter in pTARGET. Two variants of this were generated by site-directed mutagenesis: a deletion of 11 nucleotides upstream of the PSS from −4 to −24 nt, and the second is a mutation of the sequence CTTTGTTTTCCATTTC to GGGGGGGGGGCAGGGG. Cells were cross-linked for 10 min at 4 °C with 1% formaldehyde in phosphate-buffered saline and blocked by the addition of glycine, pH 8, at a final concentration of 250 mm. The cells were washed twice with PBS, and the cell pellet resuspended in 500 μl of radioimmune precipitation assay buffer (50 mm, Tris-Cl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS) and incubated on ice for 20 min. The cells were sonicated three times for 15 s with an XL ultrasonic homogenizer (setting 5) and incubated on ice for 2 min between each sonication. The extract was centrifuged for 10 min at 10,000 rpm and precleared by a 1-h incubation at room temperature with protein A-agarose beads (previously coated with 0.5 mg/ml bovine serum albumin and 0.2 mg/ml herring sperm DNA). The antibodies mouse IgG (vector I-200) or anti-ASF/SF2 (SC96, Santa Cruz Biotechnology) were incubated for 1 h at room temperature with protein A-agarose beads (Sigma, coated as above). ASF/SF2-containing complexes were pulled down after a 2-h incubation of the precleared extract with the antibody/beads and washed six times for 10 min each in 50 mm Tris-Cl, pH 7.5, 1 m NaCl, 1 mm EDTA, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 1.5 m urea. The complexes were eluted and cross-link reversed by treatment for 45 min at 70 °C with 50 mm Tris-Cl, pH 7.5, 5 mm EDTA, 10 mm dithiothreitol, and 1% SDS. RNA was then extracted with Tri-reagent solution (Ambion) according to the manufacturer's protocol, precipitated with 0.8 volumes of isopropyl alcohol in the presence of glycoblue (Ambion), the pellet resuspended in water, and subjected to DNase treatment for 1 h at 37 °C and reverse transcription for 1 h at 42 °C using MMLV (Promega) and oligo(dT)15. PCR was carried out using the Master Mix from Promega and specific primers for the plasmid VEGF sequence. 5′-CTAGCCTCGAGACGCGTGAT-3′ and 5′-GGCAGCGTGGTTTCTGTATC-3′ or GAPDH.
Oxygen-induced Retinopathy (OIR)
The OIR model was performed as previously described (18, 19) with minor modifications. Neonatal C57/Bl6 mice and nursing CD1 dams were exposed to 75% oxygen between P7 and P12. Return to room air induced hypoxia in the ischemic areas. On P13, mice received either HBSS or SRPIN340 (10 pmol) in Hank's-buffered solution in a 1-μl intraocular injections using a Nanofil syringe fitted with a 35-gauge needle (WPI, Sarasota, FL) into the left eye under isoflurane anesthesia. On P17, both eyes were dissected, fixed in 4% paraformaldehyde overnight at 4 °C, and retinas were dissected. Retinas were permeabilized in PBS containing 0.5% Triton X-100 and 1% bovine serum albumin, stained with 20 μg/ml biotinylated isolectin B4 (Sigma Aldrich) in PBS, pH 6.8, 1% Triton X-100, 0.1 mm CaCl2, 0.1 mm MgCl2, followed by 20 μg/ml ALEXA 488-streptavidin (Molecular Probes, Eugene, OR) and flat mounted in Vectashield (Vector Laboratories, Burlingame, CA). Retinas were examined under a Nikon Eclipse 400 epifluorescence microscope and areas of neovascularization identified under a 4× objective. Images were captured and imported into Image J, and neovascular, ischemic, and normal areas were traced and measured. Imaging was done by investigator, blinded to treatment.
Real-time PCR on Mouse OIR Retina
HBSS or 100 pmol of SRPIN340 in 1 μl was injected intraocularly into OIR pups on day 13 (day 7–12 in 75% O2), and after 48 h, the eyes were enucleated and placed in RNAlater (Sigma Aldrich), and the retinae were excised. Total RNA was extracted using RNAeasy (Qiagen) according to the manufacturer's manual, and 0.3 μg of DNase-digested total RNA was reverse transcribed using the oligo(dT15) primer. Real-time PCR was performed on a Cepheid Real time thermocycler using ABsolute QPCR SYBR green mix (Thermo Scientific) and 70 nm primers specific for VEGF165 (exon 7/8a) or total VEGF (exon 2/3) at 95 °C for 15 min, then 95 °C for 15 s, and 60 °C for 30 s × 40 cycles or for the housekeeping gene (β-actin) 95 °C 15 min, at 95 °C for 15 s, 55 °C for 30 s, 72 °C for 30 s × 40 cycles.
Immunocytofluorescence
Cells were washed with PBS, fixed for 5 min with 4% (w/v) PFA, washed with PBS in 0.05% Triton X (PBS-T) blocked in 5% horse serum in PBS-T (1 h), washed three times, and incubated overnight with 2 μg/ml of anti-ASF/SF2 (SC10255) or a nonspecific goat IgG, washed, and incubated with donkey anti-goat Alexa Fluor 594 for visualization and counterstained for the nucleus with Hoechst. Images were taken at 40× magnification with the Nikon Eclipse 400 epifluorescence microscope or 60× on a Perkin Elmer Ultraview-Fret H confocal microscopy system.
Statistical Analysis
Statistical analyses were carried out on raw data using the Friedman test (Dunnet post-test), and a p value of less than 0.05 was considered statistically significant. Values are expressed as means ± S.E. For all data, n represents the number of independent cell populations or derived from different donors.
RESULTS
The IGF-1-dependent Switch between Isoforms Is PKC-dependent
To determine whether the IGF-1-mediated switch in splicing was regulated by PKC inhibition, podocytes were incubated with pharmacological inhibitors of PKC (BIMI). Treatment with 100 nm IGF-1 and 2.5 μm BIMI, the PKC inhibitor, followed by RNA extraction and RT-PCR using primers that detect both proximal (VEGFxxx, 130-bp amplicon) and distal splice isoforms (VEGFxxxb, 64 bp amplicon) was carried out. Treatment with IGF-1 increased the relative intensity of the VEGFxxx (upper) band to the VEGFxxxb band (lower) from 1.39 ± 0.42 to 4.84 ± 0.65 (p < 0.01, Fig. 1B). Treatment with 100 nm IGF-1 and 2.5 μm BIMI, the PKC inhibitor, resulted in a VEGFxxx:VEGFxxxb density of 2.12 ± 0.39, which was lower than treatment with IGF-1 alone (4.84 ± 0.65, p < 0.05) but was not different from treatment with BIMI alone (1.62 ± 0.76, p > 0.05, Fig. 1B).
To determine whether IGF-1-mediated regulation of splicing was apparent at the protein level, podocytes were incubated with the pharmacological inhibitors of PKC (BIMI), and ELISA carried out on the protein extracted from the cells. Treatment with 100 nm IGF-1 and 2.5 μm BIMI, the PKC inhibitor, resulted in cells producing 0.47 ± 0.03 pg/μg of VEGFxxxb, which was significantly greater than cells treated with IGF-1 alone (0.12 ± 0.02 pg/μg, p < 0.001) but was not different from treatment with BIMI alone (0.40 ± 0.06 pg/μg, p > 0.05, Fig. 1D). This was confirmed by Western blot (Fig. 1C).
PKC Activation Induces Proximal Splice Site Selection
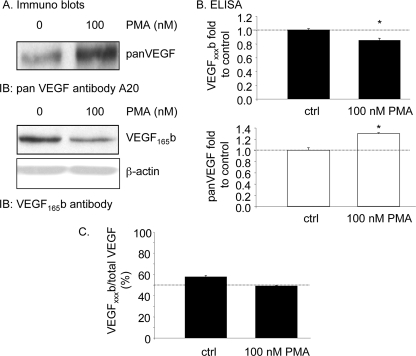
To determine whether the PKC activation was sufficient to cause proximal splice site selection, cells were treated with 100 nm phorbol myristate acetate (PMA), which is known to induce PKC activation. PMA treatment resulted in a significant increase in VEGF expression as determined by Western blot, but a decrease in VEGF165b expression (Fig. 2A). To confirm this quantitatively, ELISA was performed on protein extracted from these cells, and a significant reduction in VEGF165b but increase in total VEGF was seen (Fig. 2B). This results in a decrease in the relative VEGFxxxb levels (Fig. 2C).
FIGURE 2.
Proximal splicing is activated by protein kinase C. A–C, treatment of podocytes with the PKC activator PMA reduced VEGF165b expression, but increased expression of total VEGF as measured by Western blot (A) and ELISA (B). This results in a change of relative expression from 60% (anti-angiogenic) to just under 50% (angiogenic) (C).
SRPK1/2 Inhibition Prevents the Down-regulation of VEGFxxxb by IGF-1
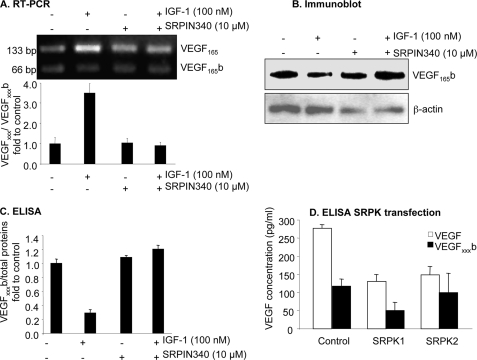
There are a number of splicing factor kinases that are activated by PKC, including the SR protein kinases SRPK1 and SRPK2 (20, 21). To test the effect of SRPIN340, an inhibitor of SRPK1/2, on IGF-1-mediated down-regulation of VEGFxxxb at the protein and mRNA level, cells were treated with SRPIN340 (10 μm) alone and then in combination with IGF-1 (100 nm). Amplification of cDNA from podocytes showed that IGF-1 treatment with 10 μm SRPIN340, the SRPK1/2 inhibitor resulted in a relative VEGFxxx:VEGFxxxb density of 1.26 ± 0.22, which was lower than treatment with IGF-1 alone (4.84 ± 0.65, p < 0.01) but was not different from treatment with SRPIN340 alone (1.45 ± 0.30, p > 0.05, Fig. 3A). At the protein level, SRPIN340 inhibited IGF-1-dependent down-regulation of VEGFxxxb from 0.12 ± 0.02 pg/μg to 0.50 ± 0.05 pg/μg (p < 0.001), but not when SRPIN340 was used alone (0.45 ± 0.01 pg/μg), indicating that SRPK inhibition did not affect endogenous expression of VEGFxxxb (0.42 ± 0.02 pg/μg, p > 0.05) (Fig. 3C). This was again confirmed by Western blot (Fig. 3B). To confirm the involvement of SRPK1 or SRPK2 in the terminal splice site choice, epithelial cells were transfected with expression vectors to overexpress SRPK1 and SRPK2. Fig. 3D shows that overexpression of SRPK1, but not SRPK2, resulted in reduced distal splice site selection and hence reduced overall VEGF levels. By itself, it was not sufficient to simply switch the splicing but resulted in inhibition of VEGF165b without increased VEGF165. Interestingly, SRPK2 overexpression did not affect VEGF165b production, although it did reduce total VEGF expression.
FIGURE 3.
Inhibition of SPRK1/2 by SRPIN340 prevents the down-regulation of VEGFxxxb by IGF. A–D, cells were treated with SRPIN340 (10 μm) alone or in combination with IGF-1 (100 nm). A, RT-PCR showed that SRPIN340 reduced the VEGF165:VEGF165b ratio at the RNA level. B, Western blot demonstrating that SRPIN340 inhibited the IGF-mediated down-regulation of VEGFxxxb expression at the protein level. C, ELISA results confirming that SPRIN340 specifically attenuates the IGF-1-dependent down-regulation of VEGFxxxb, but did not affect endogenous expression of VEGFxxxb. D, ELISA of the protein extract shows that SRPK1 transfection reduces VEGFxxxb expression, and total VEGF expression. SRPK2 reduces total expression, but did not affect VEGFxxxb expression.
IGF1 Treatment Resulted in Nuclear Localization of ASF/SF2, which Was Blocked by SPRK1/2 Inhibition
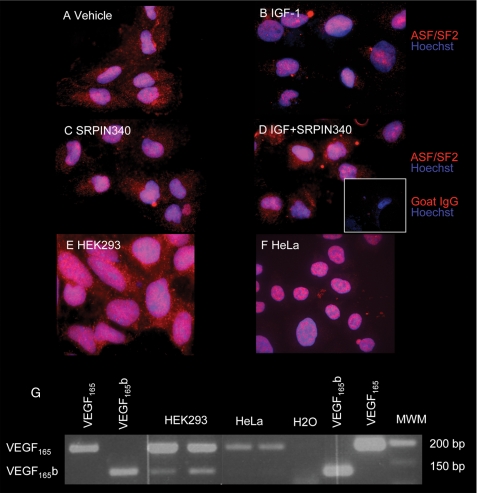
SRPK1 has been shown to phosphorylate ASF/SF2, which we have previously shown to favor proximal splice site selection. To determine whether IGF-1 altered ASF/SF2 localization, podocytes were treated with 100 nm IGF and stained for ASF/SF2. Untreated cells (Fig. 4A) contained both nuclear and cytoplasmic ASF/SF2, whereas after treatment with IGF-1, ASF/SF2 localized specifically to the nucleus (Fig. 4B). This localization was inhibited by SRPIN340 (Fig. 4, B versus D), indicating that IGF1-mediated activation of SRPK1/2 was responsible for the nuclear localization of ASF/SF2. It has previously been demonstrated that ASF/SF2 can be shuttled from the nucleus to the cytoplasm in HeLa cells, but is predominantly nuclear. We therefore investigated ASF/SF2 localization in these and another cell type, HEK293 cells. Fig. 4, E and F shows that whereas in HEK293 cells there is a strong cytoplasmic localization for the ASF/SF2, in HeLa cells expression is predominantly nuclear, as previously described. This subcellular localization of ASF/SF2 was confirmed by the use of a second ASF/SF2 antibody. Furthermore, high resolution gel electrophoresis showed that whereas podocyte cytoplasmic protein contains a single molecular weight ASF/SF2, an additional higher molecular weight band is seen in podocyte nuclei (data not shown), confirming that in podocytes ASF/SF2 is both cytoplasmic and nuclear. We also investigated VEGF165b mRNA expression in these two additional cell types. Fig. 4G shows that HEK cells (with cytoplasmic ASF/SF2) express VEGF165b, whereas HeLa cells (nuclear ASF/SF2) only express VEGF165. This is consistent with the cytoplasmic location of ASF/SF2 being associated with VEGF165b expression.
FIGURE 4.
Nuclear localization of ASF/SF2 is increased by IGF-1. A–F, cells were treated with vehicle or IGF in the presence or absence of SRPIN340 and stained for ASF/SF2 and counterstained with Hoechst. A, podocytes show expression of ASF/SF2 in the nucleus and in the cytoplasm. B, IGF induces nuclear localization of cytoplasmic ASF/SF2. C, SRPIN340 by itself does not affect localization of ASF/SF2. D, SRPIN340 inhibited this IGF-mediated localization. E, HEK cells also show cytoplasmic localization of ASF/SF2. F, in contrast, HeLa cells have nuclear ASF/SF2 localization. G, RT-PCR of mRNA from HEK cells shows VEGF165b expression, but not in HeLa cells. MWM, molecular weight marker.
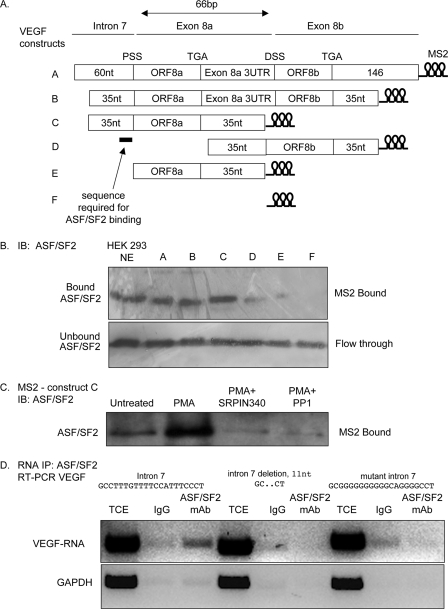
ASF/SF2 Requires a 35-nt Region around the Proximal Splice Site of Exon 8 to Bind to VEGF mRNA
To determine whether ASF/SF2 could bind directly to the proximal splice site RNA, we used the MS2-MBP system to pull-down proteins that could interact with the RNA. Fig. 5A shows that ASF/SF2 was present both in crude nuclear extract and in the pull-down of nuclear extract incubated with an RNA containing the MS2 binding domain RNA fused to an 88-nt fragment containing the initial 35 nucleotides upstream of exon 8a, the coding sequence of exon 8a, and 35 nucleotides of 3′-UTR. In contrast, less ASF/SF2 was seen in the pull-down of nuclear extract incubated with the mRNA that did not have the 35 nucleotides upstream of exon 8a and did not bind the MS2 binding domain by itself, indicating that ASF/SF2 required a 35-nt fragment of exon 8a upstream of the proximal splice site to most efficiently bind the VEGF pre-mRNA. To determine whether binding of ASF/SF2 was PKC-dependent and whether this was SRPK1-dependent, HEK293 cells were treated with the PKC activator PMA in the presence or absence of the SRPK inhibitor SRPIN340, and then the protein run on an MS2-MBP column with the VEGF exon 8 construct. Fig. 5B shows that in cells treated with PMA, more ASF/SF2 binds to the VEGF construct than in untreated cells. This increase was inhibited by treatment with 10 μm SRPIN340 or by treatment with the phosphatase PP1. The sequence 35 nucleotides upstream of the PSS contains ASF/SF2 and U2AF65 consensus binding sequences. To determine more precisely the sequence required for ASF/SF2 binding to the RNA, a short sequence upstream of the proximal splice site was mutated (14 nt) or deleted (11 nt) in a plasmid containing just the terminal part of intron 7 and the proximal part of exon 8. Fig. 5D shows that the three constructs express the recombinant VEGF RNA (total cell extract, TCE). However, only the wild-type RNA was pulled-down with the ASF/SF2 protein, suggesting that the region we have mutated or deleted is required for ASF/SF2 binding to the VEGF pre-mRNA.
FIGURE 5.
ASF/SF2-1 binds a 35-nt region of VEGF pre-mRNA upstream of the proximal splice site of exon 8. A, constructs were generated containing fragments of the exon 7/exon 8 boundary, fused to a sequence encoding the stem loop structures recognized by the MBP-MS2-binding protein, which can bind maltose. These were transcribed in vitro. B, Western blot of HEK cell crude nuclear extract (NE) or NE incubated with mRNA constructs as above and run over a maltose column to isolate proteins that bind to the RNA constructs and probed with an ASF/SF2 antibody. Whereas mRNA containing the 5′ regions of the intron 7/exon 8 boundary contained ASF/SF2 immunoreactivity, RNA encoding the exon 8 region did not, identifying the binding site for ASF/SF2 in the intron 7/exon 8a boundary. C, immunoblot of HEK cell NE of cells treated as shown incubated with the RNA construct C and run over the MS2-MBP column. PMA activation increased binding, and this was blocked by SRPIN340 and phosphatase treatment. D, RNA immunoprecipitation of ASF/SF2 in cells expressing constructs with a mutated or deleted intron 7 sequence. The top shows RT-PCR of total cell extract (TCE) or immunoprecipitated RNA using a nonspecific mouse IgG (IgG), or using a mouse monoclonal antibody to ASF/SF2 using primers to detect the VEGF sequence. The bottom blot shows the same treatments subjected to GAPDH amplification. The wild-type sequence showed a stronger band in the ASF/SF2 IP, whereas the mutants showed no difference between mouse IgG and ASF/SF2.
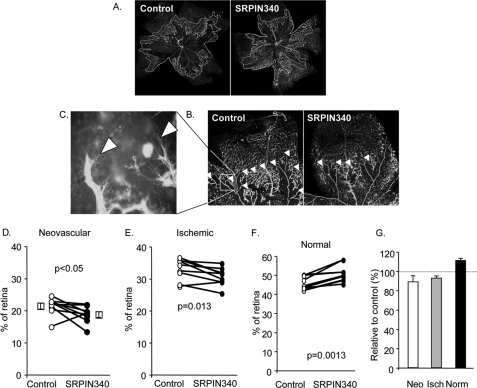
SRPK1/2 Inhibition Inhibits Angiogenesis in a Mouse Model of Retinal Neovascularization
To determine whether the inhibition of proximal splice site selection by blocking SRPK1/2, and hence increased anti-angiogenic VEGF165b production, we used a mouse model of retinal neovascularization where angiogenesis is driven by hypoxia, a process known to favor proximal splice site selection. Injection of SRPIN340 into mouse retina resulted in a significant inhibition of neovascular area of the retina, as well as a significant reduction in ischemic area (Fig. 6, C, D, and F). This resulted in a significant increase in the normally vascularized area, a result that is qualitatively consistent with injection of recombinant VEGF165b into the vitreous in this model (Fig. 6, E and F). To determine whether VEGF levels were altered by SRPIN340 treatment, mRNA was extracted from the retinae and subjected to Q-PCR for all VEGF isoforms using primers in exon 2 and 3. Treatment of eyes with SRPIN340 made no difference in overall VEGF levels (2.3 ± 0.5% of actin compared with 2.2 ± 2.0%). However, exon 8a containing mRNA was altered from 1.1 ± 0.5 to 0.3 ± 0.2% of actin).
FIGURE 6.
Neovascularization induced by hyperoxia is inhibited by a single dose of SRPK inhibitor, SRPIN340. A, low power fluorescence micrograph of FITC-labeled lectin staining of retinal whole mounts with areas of NV (white) and ischemic (orange) outlined. B, higher power view of a single retinal quadrant, with angiogenic areas highlighted by arrowheads. C, high power view of retinal angiogenic area showing sprouting endothelial cells. D, quantification of neovascular areas shows a small but significant inhibition by a single injection of 1 μl of 10 μm SRPIN340 1 day after removal from oxygen. E, ischemic area was also reduced in these mice. F, normal area was consequently increased. G, data shown as relative to control, uninjected contralateral eye. Neo, neovacular; isch, ischemia; Norm, normal.
DISCUSSION
VEGF induction by IGF-1 occurs via different signaling pathways including PKC (22) and PI3-K (23–25). There is increasing evidence that transducing components that link the cell surface with the nuclear splicing machinery implicate signaling pathways such as PKC (26), PI3-K (27, 28), or PKB/Akt (29, 30). IGF-1 modulates splicing of VEGF isoforms by preferential use of the PSS to increase expression of pro-angiogenic isoforms (15). Moreover, previously we have shown that ASF/SF2 overexpression preferentially increases usage of the proximal splice site (15) and gives the same effect as IGF-1. SRPK1 has been shown specifically to phosphorylate 12 serines of the RS domain in ASF/SF2 (31), and SRPK2 has been involved in the localization of ASF/SF2 within the nucleus. Thus, in this report, we have investigated the link between the splicing machinery and IGF-1 signaling.
We have shown that the IGF-1-mediated increase in VEGF isoforms using the proximal splice site is inhibited by blocking PKC and SRPK1/2, and that this can be overcome by the use of a PKC inhibitor or mimicked by a PKC agonist or overexpression of SRPK1. This firmly suggests that this kinase cascade is involved in splice site selection in the VEGF gene. We have used RT-PCR, ELISA, and Western blotting to investigate VEGF splicing. VEGF isoform mRNA expression depends on transcription, splicing, and degradation of mRNA, and protein expression additionally depends upon translational rate and degradation rate. The finding that the mRNA and protein isoforms are both altered in a similar way by each intervention suggests that this is an mRNA switch, at least in part. As the mRNAs are generated by alternative splicing it is unlikely that differential isoform production is due to differential transcription, as both isoforms are transcribed from the same promoter region. However, VEGF has been shown to have two alternate transcription start sites. Although there is no evidence to date to show that these are differentially used for the different exon 8 isoforms, they do confer different exon 7 inclusion (32). If alternate transcription start sites are used, the splicing machinery would still need to be different in order for the transcription complex to recognize the different exon 8 splice sites. It is possible that these two different isoform families are differentially degraded, and that IGF mediates a decrease in degradation of mRNA encoding the proximal splice site. This is a possibility that we have not as yet excluded. However, the finding that ASF/SF2, a known splicing factor, requires the presence of a specific short sequence in the polypyrimidine tract upstream of the proximal splice site and that ASF/SF2 is induced to nuclear localization by SRPK1 activation and IGF-1 activation strongly suggest that this is a splicing mechanism rather than a degradation mechanism. We have shown that ASF/SF2 requires this sequence, which contains both a U2AF65 and consensus ASF/SF2 sequence, for binding to the VEGF pre-mRNA, but does not demonstrate that this is the sequence it binds to. ASF/SF2 binding to a region upstream of a splice site is generally considered a splicing repressor. There is evidence that SR proteins can interact with sequences upstream of the splice site that act as intronic splicing enhancer or silencer regions (reviewed in Ref. 33); for instance in the FGFR2 gene mutations in 50% of the sequential 6 nucleotide sequences in the intronic region upstream of the splice site resulted in altered splicing (34). However, an alternative explanation is that ASF/SF2 requires the U2AF65 consensus sequence adjacent to exon 8a to be present in order for it to bind to consensus sequences downstream and repress distal splice site selection, and hence when mutated or deleted DSS repression is lifted resulting in preferential proximal splice site selection. More research is required to pinpoint the exact mechanism of splicing regulation by ASF/SF2.
It is still not clear if the IGF-1 system plays a direct role in the growth of new blood vessels. However, there are increasing examples that IGF-1 indirectly increases angiogenesis by up-regulation of VEGF (22, 25, 35). Moreover, IGF-1 induces angiogenesis in the rabbit cornea (36) and stimulates migration, proliferation (37), and tube formation of human endothelial cells (38). IGF-1 is also widely implicated in pathological angiogenesis. Expression of IGF-1 was increased in the vitreous of patients with diabetic retinopathy (39). There is a positive correlation between elevated VEGF-A and IGF-1 in different types of cancers such as colorectal cancer (40), breast cancer (41), and head and neck squamous cell carcinomas (25).
Moreover, the SFRS1 gene, encoding ASF/SF2, fulfills the criteria of a proto-oncogene (42). Overexpression of ASF/SF2 in immortalized rodent fibroblasts resulted in formation of high-grade sarcomas after injection into nude mice (43). Knock-down of ASF/SF2 in lung carcinoma, which has high expression of that molecule, inhibited tumor formation in nude mice (43).
There are increasing examples that manipulation of the splicing machinery can be used as new therapeutic targets (44). Small molecules that can target splicing factors and kinases involved in splicing are promising candidates for drugs (17, 45–47). Alternatively, the use of antisense oligonucleotides such as morpholinos, which can bind to the specific splice sites to modulate aberrant splicing has been investigated (48, 49). SRPK1 seems to be a relatively new target for future cancer therapies but is receiving more attention recently as higher expression of SRPK1 has been observed in breast and colonic tumors, and its increased expression is associated with the grade of a tumor (50). Moreover, it has been shown that known anticancer drugs such as gemcitabine and cisplatin increase cell apoptosis with a much stronger effect when phosphorylation of SR proteins was inhibited by using siRNA against SRPK1 (51). SRPK2 is able to bind and phosphorylate acinus, an SR protein, and moves it from nuclear speckles to the nucleoplasm, resulting in the activation of cyclin A1 (52). Moreover, overexpression of acinus or SRPK2 increased leukemia cell proliferation. SRPK2 and acinus were also overexpressed in human acute myelogenous leukemia patients and correlate with elevated cyclin A1 expression levels (52). These two kinases are able to phosphorylate the splicing factor, ASF/SF2 and all these components are involved in the choice of the PSS in VEGF. The inhibitor of SRPK1/2 kinase, SRPIN340 prevents the down-regulation of the VEGFxxxb isoforms. Moreover, SRPK1 and SRPK2 are known to phosphorylate the ASF/SF2 splicing factor with high specificity (21, 31, 53). These results indicate that an overactivity of SRPK1 can cause phosphorylation of ASF/SF2. However, SRPKs have other targets, including those known to up-regulate VEGFxxxb such as SRp55 (15), and the contribution of SRp55 phosphorylation to IGF-mediated effects (perhaps by inhibiting its binding to the distal splice enhancer region) cannot be ruled out. It thus appears likely that SRPK1-mediated phosphorylation of ASF/SF2 could support an activation of the PSS and increased production of pro-angiogenic isoforms.
The overexpression findings, however, suggest that whereas SRPKs are necessary, they are not sufficient for proximal splice site selection. Thus these findings indicate that SRPK1 inhibitors may be potentially anti-angiogenic, and to that end we set out to investigate this in a model of angiogenesis in the eye. A single dose of SRPIN340 resulted in significant inhibition of angiogenesis and increased normal vascularization. Whereas, we have not measured VEGF165b levels in these eyes, there is a discrepancy between total and exon 8a-containing isoforms that is most likely a result of altered splicing. The angiogenesis is known to be mediated by pro-angiogenic VEGF and can be inhibited by anti-angiogenic VEGF165b (18); thus, allowing us to draw a parallel between anti-angiogenic splice forms and inhibition of splice factors that cause pro-angiogenic splicing in the same retinal angiogenesis model. These data suggest that anti-SRPK1 inhibitors may be useful anti-angiogenic agents, suggesting a use in cancer as well as diabetic retinopathy or age-related macular degeneration.
This work was supported by the British Heart Foundation (FS/04/09 and BS/06/005), the Wellcome Trust (79633), a University of West of England Bristol PhD Studentship, the Richard Bright VEGF Research Trust, Fight for Sight, Cancer Research UK (C11392/A10484), and the Skin Cancer Research Fund.
- VEGF
- vascular endothelial growth factor
- IGF-1
- insulin-like growth factor
- nt
- nucleotide
- PBS
- phosphate-buffered saline
- PKC
- protein kinase C
- ELISA
- enzyme-linked immunosorbent assay
- MBP
- maltose-binding protein
- PSS
- proximal splice site selection
- OIR
- oxygen-induced retinopathy
- UTR
- untranslated region
- ITS
- insulin transferrin selenium
- PMA
- phorbol myristate acetate
- HEK
- human embryonic kidney.
REFERENCES
- 1.Kerbel R. S. (2008) N. Engl. J. Med. 358, 2039–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duh E., Aiello L. P. (1999) Diabetes 48, 1899–1906 [DOI] [PubMed] [Google Scholar]
- 3.Slomiany M. G., Rosenzweig S. A. (2004) Invest. Ophthalmol. Vis. Sci. 45, 2838–2847 [DOI] [PubMed] [Google Scholar]
- 4.Harper S. J., Bates D. O. (2008) Nat. Rev. Cancer 8, 880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houck K. A., Ferrara N., Winer J., Cachianes G., Li B., Leung D. W. (1991) Mol. Endocrinol. 5, 1806–1814 [DOI] [PubMed] [Google Scholar]
- 6.Woolard J., Wang W. Y., Bevan H. S., Qiu Y., Morbidelli L., Pritchard-Jones R. O., Cui T. G., Sugiono M., Waine E., Perrin R., Foster R., Digby-Bell J., Shields J. D., Whittles C. E., Mushens R. E., Gillatt D. A., Ziche M., Harper S. J., Bates D. O. (2004) Cancer Res. 64, 7822–7835 [DOI] [PubMed] [Google Scholar]
- 7.Bates D. O., Cui T. G., Doughty J. M., Winkler M., Sugiono M., Shields J. D., Peat D., Gillatt D., Harper S. J. (2002) Cancer Res. 62, 4123–4131 [PubMed] [Google Scholar]
- 8.Perrin R. M., Konopatskaya O., Qiu Y., Harper S., Bates D. O., Churchill A. J. (2005) Diabetologia 48, 2422–2427 [DOI] [PubMed] [Google Scholar]
- 9.Varey A. H., Rennel E. S., Qiu Y., Bevan H. S., Perrin R. M., Raffy S., Dixon A. R., Paraskeva C., Zaccheo O., Hassan A. B., Harper S. J., Bates D. O. (2008) Br. J. Cancer 98, 1366–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rennel E., Waine E., Guan H., Schüler Y., Leenders W., Woolard J., Sugiono M., Gillatt D., Kleinerman E., Bates D., Harper S. (2008) Br. J. Cancer 98, 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritchard-Jones R. O., Dunn D. B., Qiu Y., Varey A. H., Orlando A., Rigby H., Harper S. J., Bates D. O. (2007) Br. J. Cancer 97, 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher V. A., Jeruschke S., Eitner F., Becker J. U., Pitschke G., Ince Y., Miner J. H., Leuschner I., Engers R., Everding A. S., Bulla M., Royer-Pokora B. (2007) J. Am. Soc. Nephrol. 18, 719–729 [DOI] [PubMed] [Google Scholar]
- 13.Dieudonné S. C., La Heij E. C., Diederen R. M., Kessels A. G., Liem A. T., Kijlstra A., Hendrikse F. (2007) Ophthalmic. Res. 39, 148–154 [DOI] [PubMed] [Google Scholar]
- 14.Ergorul C., Ray A., Huang W., Darland D., Luo Z. K., Grosskreutz C. L. (2008) Mol. Vis. 14, 1517–1524 [PMC free article] [PubMed] [Google Scholar]
- 15.Nowak D. G., Woolard J., Amin E. M., Konopatskaya O., Saleem M. A., Churchill A. J., Ladomery M. R., Harper S. J., Bates D. O. (2008) J. Cell Sci. 121, 3487–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saleem M. A., O'Hare M. J., Reiser J., Coward R. J., Inward C. D., Farren T., Xing C. Y., Ni L., Mathieson P. W., Mundel P. (2002) J. Am. Soc. Nephrol. 13, 630–638 [DOI] [PubMed] [Google Scholar]
- 17.Fukuhara T., Hosoya T., Shimizu S., Sumi K., Oshiro T., Yoshinaka Y., Suzuki M., Yamamoto N., Herzenberg L. A., Herzenberg L. A., Hagiwara M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11329–11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konopatskaya O., Churchill A. J., Harper S. J., Bates D. O., Gardiner T. A. (2006) Mol. Vis. 12, 626–632 [PubMed] [Google Scholar]
- 19.Smith L. E., Wesolowski E., McLellan A., Kostyk S. K., D'Amato R., Sullivan R., D'Amore P. A. (1994) Invest. Ophthalmol. Vis. Sci. 35, 101–111 [PubMed] [Google Scholar]
- 20.Gui J. F., Lane W. S., Fu X. D. (1994) Nature 369, 678–682 [DOI] [PubMed] [Google Scholar]
- 21.Wang H. Y., Lin W., Dyck J. A., Yeakley J. M., Songyang Z., Cantley L. C., Fu X. D. (1998) J. Cell Biol. 140, 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckert S., Farrahi F., Perveen Ghani Q., Aslam R., Scheuenstuhl H., Coerper S., Königsrainer A., Hunt T. K., Hussain M. Z. (2006) Biochem. Biophys. Res. Commun. 341, 67–72 [DOI] [PubMed] [Google Scholar]
- 23.Miele C., Rochford J. J., Filippa N., Giorgetti-Peraldi S., Van Obberghen E. (2000) J. Biol. Chem. 275, 21695–21702 [DOI] [PubMed] [Google Scholar]
- 24.Poulaki V., Mitsiades C. S., McMullan C., Sykoutri D., Fanourakis G., Kotoula V., Tseleni-Balafouta S., Koutras D. A., Mitsiades N. (2003) J. Clin. Endocrinol. Metab. 88, 5392–5398 [DOI] [PubMed] [Google Scholar]
- 25.Slomiany M. G., Black L. A., Kibbey M. M., Day T. A., Rosenzweig S. A. (2006) Biochem. Biophys. Res. Commun. 342, 851–858 [DOI] [PubMed] [Google Scholar]
- 26.Lynch K. W., Weiss A. (2000) Mol. Cell. Biol. 20, 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel N. A., Chalfant C. E., Watson J. E., Wyatt J. R., Dean N. M., Eichler D. C., Cooper D. R. (2001) J. Biol. Chem. 276, 22648–22654 [DOI] [PubMed] [Google Scholar]
- 28.Blaustein M., Pelisch F., Coso O. A., Bissell M. J., Kornblihtt A. R., Srebrow A. (2004) J. Biol. Chem. 279, 21029–21037 [DOI] [PubMed] [Google Scholar]
- 29.Blaustein M., Pelisch F., Tanos T., Muñoz M. J., Wengier D., Quadrana L., Sanford J. R., Muschietti J. P., Kornblihtt A. R., Cáceres J. F., Coso O. A., Srebrow A. (2005) Nat. Struct. Mol. Biol. 12, 1037–1044 [DOI] [PubMed] [Google Scholar]
- 30.Patel N. A., Kaneko S., Apostolatos H. S., Bae S. S., Watson J. E., Davidowitz K., Chappell D. S., Birnbaum M. J., Cheng J. Q., Cooper D. R. (2005) J. Biol. Chem. 280, 14302–14309 [DOI] [PubMed] [Google Scholar]
- 31.Ma C. T., Velazquez-Dones A., Hagopian J. C., Ghosh G., Fu X. D., Adams J. A. (2008) J. Mol. Biol. 376, 55–68 [DOI] [PubMed] [Google Scholar]
- 32.Bastide A., Karaa Z., Bornes S., Hieblot C., Lacazette E., Prats H., Touriol C. (2008) Nucleic Acids Res. 36, 2434–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voelker R. B., Berglund J. A. (2007) Genome Res. 17, 1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hovhannisyan R. H., Warzecha C. C., Carstens R. P. (2006) Nucleic Acids Res. 34, 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y. Z., Wong Y. C. (1998) Prostate 35, 165–177 [DOI] [PubMed] [Google Scholar]
- 36.Grant M. B., Mames R. N., Fitzgerald C., Ellis E. A., Aboufriekha M., Guy J. (1993) Diabetologia 36, 282–291 [DOI] [PubMed] [Google Scholar]
- 37.Shigematsu S., Yamauchi K., Nakajima K., Iijima S., Aizawa T., Hashizume K. (1999) Endocr. J. 46, S59–S62 [DOI] [PubMed] [Google Scholar]
- 38.Nakao-Hayashi J., Ito H., Kanayasu T., Morita I., Murota S. (1992) Atherosclerosis 92, 141–149 [DOI] [PubMed] [Google Scholar]
- 39.Grant M., Russell B., Fitzgerald C., Merimee T. J. (1986) Diabetes 35, 416–420 [DOI] [PubMed] [Google Scholar]
- 40.Warren R. S., Yuan H., Matli M. R., Ferrara N., Donner D. B. (1996) J. Biol. Chem. 271, 29483–29488 [DOI] [PubMed] [Google Scholar]
- 41.Oh J. S., Kucab J. E., Bushel P. R., Martin K., Bennett L., Collins J., DiAugustine R. P., Barrett J. C., Afshari C. A., Dunn S. E. (2002) Neoplasia 4, 204–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 43.Karni R., de Stanchina E., Lowe S. W., Sinha R., Mu D., Krainer A. R. (2007) Nat. Struct. Mol. Biol. 14, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagiwara M. (2005) Biochim. Biophys. Acta 1754, 324–331 [DOI] [PubMed] [Google Scholar]
- 45.Pilch B., Allemand E., Facompré M., Bailly C., Riou J. F., Soret J., Tazi J. (2001) Cancer Res. 61, 6876–6884 [PubMed] [Google Scholar]
- 46.Muraki M., Ohkawara B., Hosoya T., Onogi H., Koizumi J., Koizumi T., Sumi K., Yomoda J., Murray M. V., Kimura H., Furuichi K., Shibuya H., Krainer A. R., Suzuki M., Hagiwara M. (2004) J. Biol. Chem. 279, 24246–24254 [DOI] [PubMed] [Google Scholar]
- 47.Bakkour N., Lin Y. L., Maire S., Ayadi L., Mahuteau-Betzer F., Nguyen C. H., Mettling C., Portales P., Grierson D., Chabot B., Jeanteur P., Branlant C., Corbeau P., Tazi J. (2007) PLoS Pathog. 3, 1530–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruno I. G., Jin W., Cote G. J. (2004) Hum. Mol. Genet. 13, 2409–2420 [DOI] [PubMed] [Google Scholar]
- 49.Wheeler T. M., Lueck J. D., Swanson M. S., Dirksen R. T., Thornton C. A. (2007) J. Clin. Invest. 117, 3952–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayes G. M., Carrigan P. E., Miller L. J. (2007) Cancer Res. 67, 2072–2080 [DOI] [PubMed] [Google Scholar]
- 51.Hayes G. M., Carrigan P. E., Beck A. M., Miller L. J. (2006) Cancer Res. 66, 3819–3827 [DOI] [PubMed] [Google Scholar]
- 52.Jang S. W., Yang S. J., Ehlén A., Dong S., Khoury H., Chen J., Persson J. L., Ye K. (2008) Cancer Res. 68, 4559–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colwill K., Feng L. L., Yeakley J. M., Gish G. D., Cáceres J. F., Pawson T., Fu X. D. (1996) J. Biol. Chem. 271, 24569–24575 [DOI] [PubMed] [Google Scholar]