Abstract
Cellular migration is an essential prerequisite for metastatic dissemination of cancer cells. This study demonstrates that the neuron/testis-specific F-actin-targeted inositol 1,4,5-trisphosphate 3-kinase-A (ITPKA) is ectopically expressed in different human tumor cell lines and during tumor progression in the metastatic tumor model Balb-neuT. High expression of ITPKA increases invasive migration in vitro and metastasis in a xenograft SCID mouse model. Mechanistic studies show that ITPKA promotes migration of tumor cells by two different mechanisms as follows: growth factor independently high levels of ITPKA induce the formation of large cellular protrusions by directly modulating the actin cytoskeleton. The F-actin binding activity of ITPKA stabilizes and bundles actin filaments and thus increases the levels of cellular F-actin. In growth factor-stimulated cells, the catalytically active domain enhances basal ITPKA-induced migration by activating store-operated calcium entry through production of inositol 1,3,4,5-tetrakisphosphate and subsequent inhibition of inositol phosphate 5-phosphatase. These two functional activities of ITPKA stimulating tumor cell migration place the enzyme among the potential targets of anti-metastatic therapy.
Keywords: Calcium/Cellular Regulation, Cell/Migration, Cytoskeleton, Diseases/Cancer, RNA/Interference/RNAi, Signal Transduction/Calcium, Tissue/Organ Systems/Thyroid, Actin Cytoskeleton
Introduction
The formation of distant metastases is a complex process involving escape of cancer cells from the primary tumor, dissemination to distant organs, and finally re-colonization and expansion (1). For metastatic dissemination, the cancer cell must acquire the ability to migrate, which is associated with cytoskeletal re-arrangements. Migrating cells extend actin-based filopodia and lamellipodia at the leading edge. To initiate this process, a local formation of F-actin is required, which can be mediated by actin nucleating (e.g. Arp2/3 and formins (2)), F-actin bundling (e.g. fascin (3)), and by F-actin cross-linking proteins (e.g. filamins (4)). The actin-severing proteins ADF/cofilin and gelsolin depolymerize F-actin and thus increase actin turnover (5). The balance between stimulation of actin-polymerizing proteins and actin-depolymerizing proteins is tightly regulated by distinct signaling pathways (e.g. phospholipase C and phosphoinositide 3-kinase (6–8)). As activation of these pathways can also result in induction of proliferation, “fine-tuning” of continuous signal inputs determines the cellular response. This fine-tuning is mediated by various small molecules, including calcium, cyclic AMP, phosphatidylinositol phosphates, and inositol phosphates. Among the inositides, membranous phosphatidylinositol 4,5-bisphosphate plays a central role in the control of migration as it regulates the activity of cofilin, gelsolin, and profilin (9, 10) and serves as a substrate for the production of the calcium-mobilizing second messenger inositol 1,4,5-trisphosphate (Ins(1,4,5)P3).2 Phospholipase C-mediated hydrolysis of phosphatidylinositol 4,5-bisphosphate increases Ins(1,4,5)P3 levels and subsequent calcium release from the endoplasmic reticulum. Calcium plays an important role in cell migration because calcium transients activate gelsolin and indirectly ADF/cofilin (9, 10).
Inositol 1,4,5-trisphosphate 3-kinase isoenzymes (ITPKA, ITPKB, and ITPKC) metabolize Ins(1,4,5)P3 to inositol-1,3,4,5-tetrakisphosphate (Ins(1,3,4,5)P4), and thus regulate Ins(1,4,5)P3-induced calcium signals (11). The ITPK isoenzymes are highly conserved in their catalytically active C-terminal domains but show large differences in their N-terminal regulatory domains mediating mainly cellular targeting. The isoenzymes differ in subcellular localization and tissue expression patterns. ITPKB and ITPKC mRNAs are ubiquitously expressed, whereas mRNA of ITPKA was only identified in neurons and testis (12). In neurons, ITPKA was shown to be targeted to F-actin via an N-terminal actin binding domain (amino acids 1–66 (13)) and was suggested to be relevant for long term potentiation and spatial learning (14, 15). Remarkably, in some malignant transformed cells, ectopic expression of the neuronal/testicular ITPK isoform has been found. ITPKA expression levels are increased in two different MDA mammary carcinoma cell lines overexpressing ErbB2 (16, 17). In rat fibroblasts transformed by p60v-src (18), ITPKA activity is increased 7-fold, and enhanced protein levels of ITPKA were detected in the center of cutaneous malignant melanoma (19). Overexpression of ITPKA in the lung carcinoma cell line H1299 induces the formation of actin-based cell protrusions and increases migration (20), indicating a role of ITPKA in actin remodeling and cell motility. In this study, we examined the mechanistic role of ITPKA in tumor cell migration and metastasis.
EXPERIMENTAL PROCEDURES
Cell Lines
NCI-H1299 (H1299) cells were kindly provided by Cagatay Günes (Hamburg, Germany); A549 and skin fibroblasts cells were a gift from Ulla Kasten-Pisula (Hamburg, Germany), and Mevo and T47D cells were provided by Udo Schumacher (Hamburg, Germany). For characteristics of these cells, see American Type Culture Collection (ATCC, Manassas, VA). The cell lines HepG2, MDA-MB-231 (MDA231), and SKBR-3 were purchased from ATCC, and normal lung (IMR-90) and mammary epithelial cells (HmEpc) were obtained from Cell Culture Service (Hamburg, Germany). The cell lines HepG2, MCF-7, MDA-MB-231, NCI-H1299, A549, and SKBR-3 were cultured in Dulbecco's modified Eagle's medium; Mevo and T47D were grown in RPMI 1640 medium; both media were supplemented with 10% (v/v) fetal calf serum (FCS), 4 mm l-glutamine, 100 μg/ml streptomycin, and 100 units/ml penicillin. IMR-90 cells were cultivated in α-minimal essential medium containing 10% FCS. These media were purchased from Invitrogen. HmEpc were grown in mammary epithelial cell media supplemented with 10% FCS (Cell Applications, San Diego).
Immunohistochemistry
The Balb-neuT tissue samples (21) were fixed in 3.7% formaldehyde solution and embedded in paraffin. Paraffin sections of all samples were deparaffinized in xylene and rehydrated in a graded series of ethanol. Microwave antigen retrieval was performed for 20 min using 20 mm citrate buffer, pH 6.0. Consecutive incubation steps with peroxidase-blocking solution (Dako, Glostrup, Denmark) and 2.5% normal horse serum were performed to block endogenous peroxidase activity and nonspecific antibody binding, respectively. The primary antibody, affinity-purified goat anti-ITPKA (Santa Cruz Biotechnology, Santa Cruz, CA), was used at a concentration of 1.3 μg/ml and incubated with the tissue section overnight at 4 °C. As a negative control, duplicate sections were incubated with normal goat IgG (Santa Cruz Biotechnology) at the same dilution. The secondary antibody polymer (Vector Laboratories, Burlingame, CA) and the diaminobenzidine chromogen system (Dako, Glostrup, Denmark) were used according to the manufacturer's instructions. Finally, the slides were counterstained with hematoxylin, dehydrated, and coverslipped with permanent mounting medium. To evaluate the specificity of the anti-ITPKA antibody, paraffin-embedded samples of H1299 control, H1299 kd cells, and of H299 cells overexpressing ITPKA were used for comparative stainings.
Western Blot and Immunocytology
For the preparation of lysates, cells were washed with phosphate-buffered saline (PBS) and harvested in MPER buffer (Promega, Mannheim, Germany) supplemented with protease inhibitor mixture (Roche Applied Science). The resulting suspension was frozen in liquid nitrogen, thawed, and then centrifuged (15 min, 15,000 × g, 4 °C). Protein concentrations of the supernatants were determined using the Bradford assay, and the remaining supernatant was immediately added to Laemmli sample buffer (20 mm Tris-HCl, 2% SDS, 25% glycerol, 0.3% mercaptoethanol, 0.032% bromphenol blue). 20 μg of protein were analyzed by Western blot using a standard protocol. For detection of expression of ITPKA, the same antibody as used in the immunohistochemistry experiments was employed in a dilution of 1:2000. The secondary anti-goat antibody (Dako, Carpinteria, CA) was diluted 1:5000. Specific immune complexes were visualized by employing the ECLTM Advance Western blotting detection kit (Amersham Biosciences). Signal intensities were analyzed by a luminescent image analyzer (model LAS-3000 Plus, Fuji Film).
For immunocytology, paraformaldehyde-fixed cells were treated with 0.3% Triton X-100 for 3 min and blocked with 5% bovine serum albumin for 20 min at room temperature. Between each step, the cells were washed three times with PBS. The cells were then treated with the anti-ITPKA antibody (see above) diluted 1:200 in PBS overnight at 4 °C. After washing three times with PBS, the cells were incubated with a fluorescein isothiocyanate-labeled anti-goat antibody (Molecular Probes, Eugene, OR) in a dilution of 1:2000 for 1 h at room temperature and were finally washed three times with PBS.
Transient Transfections
Cells were transfected with LipofectamineTM LTX and Plus Reagent (Invitrogen) according to the manufacturer's instructions.
Immunoprecipitation
This experiment was performed as described previously (20).
Genetic Modification of High and Low ITPKA-expressing Cell Lines for Functional Studies
For establishment of A549 or H1299 cells overexpressing ITPKA, the cDNA fragment encoding for ITPKA was cloned into the retroviral vector MigRI. For virus production, this construct (or the original vector as control) was co-transfected with SVgp (M57) and pHCMV-VSV-G (M75) into HEK293 phoenix ampho cells. Vectors were kindly provided by Manfred Jücker (from our laboratory). After 48 h of incubation, the supernatant containing the viruses was filtered and transferred to A549 or H1299 cells, respectively. After 1 week of cultivation, the virus-transduced cells were sorted by fluorescence-activated cell sorter based on their GFP fluorescence.
For stable knockdown of ITPKA expression in MDA231 and H1299 cells, a lentiviral transduction approach was employed. Five different vectors (pLKO.1 shRNA) from Sigma expressing shRNA against ITPKA were tested. An almost complete down-regulation of ITPKA was achieved with one of them, and this vector was used for all further experiments. As a control, cells were transduced with the control vectors (expressing GFP or scrambled shRNA). For producing lentiviral particles, HEK293T cells were co-transfected with pLKO.1 shRNA plasmid, psPAX2 packaging plasmid, and pMD2.G envelope plasmid (both Addgene, Cambridge, MA). After 24 h of incubation, the supernatant containing the viruses was mixed with 8 μg/ml Polybrene (Sigma) and transferred to MDA231 or H1299 cells, respectively. After 96 h of incubation, the cells were changed to fresh medium containing 1.5 μg/ml puromycin to select for infected cells over a period of 8 days.
Ca2+ Imaging
H1299 control and kd cells were plated at low density on glass bottom culture dishes (35 mm, MatTek, Ashland, MA). After 16 h of incubation, the cells were serum-starved for 24 h. Thee Ca2+ indicator Fura 2/AM (Calbiochem) was then added to the medium at a final concentration of 4 μm. After 30 min (37 °C), the cells were washed twice, and medium was replaced with buffer (140 mm NaCl, 5 mm KCl, 1 mm MgSO4, 1 mm CaCl2, 1 mm Na2HPO4, 5.5 mm glucose, and 20 mm Hepes, pH 7.4). Imaging of cells was performed using a Leica DM-IRBE fluorescence microscope with a ×40 objective (1.3 numerical aperture). Alternating excitation at 340 and 380 nm was achieved using a monochromator system (Polychrome II; TILL Photonics, Graefelfing, Germany). Two images were acquired every 2 s with a grayscale CCD camera (type C4742-95-12NRB; Hamamatsu, Enfield, UK) in 8 bit-mode. Raw data were stored on a hard disk, and ratio images (340:380 nm) were calculated from median filtered (3 × 3 pixels) raw images using Openlab software (version 3.09; Improvision, Tübingen, Germany). For the determination of the duration of the signal, cells were selected as regions of interest, and the mean ratio of the regions of interest over time was used for further analysis.
Culture of Tumor Cells in Gelatin
96-Well plates were coated with 15% gelatin (diluted 1:2 in PBS/Dulbecco's modified Eagle's medium) and incubated for 10 min at room temperature. 1 × 104 cells (in Dulbecco's modified Eagle's medium) were then seeded per well and cultivated for 48 h in 5% CO2 at 37 °C.
Transwell Migration Assay
The transwell assay was performed as described previously (20).
Wound Healing Assay
See supplemental Fig. S1.
Determination of Migratory Potential of Tumor Cell Lines in Mouse Xenografts
H1299 control and H1299 cells overexpressing ITPKA (H1299-ITPKA) cells were injected subcutaneously in BALB/c severe combined immunodeficient SCID/SCID (SCID) mice (n = 9–10) as described previously (22). The mice were sacrificed when solid tumors were about the same size; the tumor weight was about 4 g, and the diameter was about 1 cm. The lungs were dissected and treated as described previously (22). In brief, the lungs were fixed en bloc in formalin and then embedded in agar, which was processed to wax. The blocks were cut in 5-μm sections, and every 10th section was stained with hematoxylin and eosin. To evaluate the presence of spontaneous lung metastases, the stained sections of each lung were examined at a 1 × 200 magnification. Irrespective of the size, all distant lung metastases found in one section were counted, and then the results of all sections were summed to calculate the total number of metastases per lung according to an established formula (22).
Establishment of H1299 Cells Stably Overexpressing GFP-ITPKA Fusion Proteins
The cDNA of GFP-ITPKA cloned into the vector pEGFP-C1 (20) was mutated by QuikChange PCR to generate a silent mutation causing resistance to anti-ITPKA-shRNA. Transfection of this modified vector for stable expression of GFP-ITPKA was performed using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. H1299 kd cells were transfected with GFP-ITPKA, GFP-ITPKA-D416N, or GFP, and lines were then selected using G418. Expression of GFP-ITPKA fusion proteins was examined by Western blot analysis using anti-GFP or anti-ITPKA antibodies.
Bacterial Expression of ITPKA
A cDNA fragment encoding the full-length form of ITPKA was inserted into the vector pGEX-4T-3 encoding an N-terminal glutathione S-transferase (GST) tag. From transformed E. coli cell lysates (BL21 DE3, the fusion protein was purified utilizing a glutathione matrix. The purified GST or GST-ITPKA proteins were dialyzed against polymerization or G-buffer (see below) and centrifuged at 50,000 × g for 30 min, and protein concentrations of supernatants were determined by Coomassie Blue-stained SDS-PAGE using bovine serum albumin as standard.
Actin Polymerization and Depolymerization Assays
Polymerization assays were performed essentially according to the instructions of the manufacturer (Cytoskeleton, Denver, CO) using rabbit muscle actin and pyrene-labeled rabbit muscle actin (Cytoskeleton) in a ratio of 1:10. For depolymerization of F-actin, actin solutions (40 μm actin in G-buffer: 10 mm Tris-HCl, pH 7.5, 0.2 mm CaCl2, 0.2 mm ATP) were incubated on ice for 4 h and centrifuged for 30 min at 100,000 × g in a Beckmann OptimaTM TL ultracentrifuge. The G-actin concentration of the supernatant was determined by Coomassie Blue-stained SDS-PAGE using bovine serum albumin as standard. To measure polymerization, 10 μm G-actin and 1 μm pyrene-labeled G-actin were mixed. The G-buffer and GST or GST-ITPKA proteins (diluted in G-buffer) were then added to a final volume of 180 μl. After incubation for 10 min at room temperature, the basal fluorescence was measured in an F-2000 fluorescence spectrometer (HITACHI; excitation 365 nm, emission 407 nm) for ≥2 min. Polymerization was started by adding 20 μl of ×10 polymerization buffer (end concentration in 200 μl: 1 μm G-actin, 0.1 μm pyrene-labeled G-actin, 10 mm Tris-HCl, pH 7.5, 100 mm KCl, 1 mm MgCl2, 1 mm EGTA, 0.2 mm ATP).
For determination of dilution-induced F-actin depolymerization, 40 μm rabbit muscle actin, 8 μm pyrene-labeled rabbit muscle actin dissolved in G-buffer were diluted in 1× polymerization buffer to 1 μm actin, 0.2 μm pyrene actin. This solution was incubated at room temperature for 4 h in the presence or absence of GST or GST-ITPKA proteins (diluted in polymerization buffer). Spontaneous depolymerization of F-actin was initiated by performing a 10-fold dilution (0.1 μm actin, 0.02 μm pyrene actin) in polymerization buffer, and pyrene-actin fluorescence was continuously measured.
To measure gelsolin induced F-actin depolymerization, 1 μm actin, 0.1 μm pyrene actin diluted in polymerization buffer was incubated for 1 h at room temperature in the presence of GST-ITPKA, GST, or GST-ITPKA L34P. Gelsolin-induced depolymerization was measured after addition of 0.05 μm gelsolin by decrease of actin pyrene fluorescence.
F-actin Bundling Assay
2 μm rabbit muscle actin (Cytoskeleton) diluted in polymerization buffer was incubated with 0.6 μm GST, 0.6 μm GST-ITPKA L34P, or different concentrations of GST-ITPKA (0.0375, 0.075, 0.15, 0.3, and 0.6 μm) for 30 min at room temperature. After centrifugation for 15 min at 7000 × g, the supernatants and the pellets were dissolved in SDS sample buffer and subjected to 12% SDS-PAGE. 2 μg of actin were used as standard. To visualize actin, the gel was stained with Coomassie Blue. To quantify the amount of actin, the signal intensity of the actin bands derived from pellets and supernatants were determined with an image analyzer.
DNase I Inhibition Assay for Determination of Intracellular Actin
The G/total actin ratio in cells was measured according to Katsantonis et al. (23). The principle of this assay is that only G-actin and not F-actin inhibits the activity of DNase I. The cellular G-actin content is directly determined from cell lysates by measuring inhibition of DNase I. For determination of total cellular actin, cell lysates are treated with guanidine HCl to depolymerize F-actin. The F-actin content can be calculated by subtracting the G-actin amount from the total cellular actin (F = T − G). 5 × 106 cells were washed twice with PBS and treated with 100 μl of lysis buffer (10 mm K2HPO4, 100 mm NaF, 50 mm KCl, 2 mm MgCl2, 1 mm EGTA, 0.2 mm dithiothreitol, 0.5% Triton X-100, and 1 m sucrose, pH 7.0). For determination of the G-actin content, the suspension was diluted 1:10 in lysis buffer, and 10 μl were added to 10 μl of DNase I solution (0.1 mg/ml DNase I in 50 mm Tris-HCl, 10 mm phenylmethylsulfonyl fluoride, 0.5 mm MgCl2, pH 7.5). 980 μl of DNA solution (40 μg of DNA in 100 mm Tris-HCl, 4 mm MgCl2, and 1.8 mm CaCl2, pH 7.5) were pipetted into a quartz cuvette (Suprasil), and basal absorption was monitored for 5 min at 30 °C at 260 nm in a Lambda 20 UV-visible spectrometer (PerkinElmer Life Sciences). The DNase I/cell suspension mixture (20 μl) was than added, and DNase I activity was monitored continuously. The concentration of cellular G-actin was determined by reference to a G-actin standard curve for inhibition of DNase I activity. To measure total actin, 10 μl of the 1:10 dilution of the cell suspension was treated with 10 μl of guanidine HCl buffer (1.5 m guanidine HCl, 1 m sodium acetate, 1 mm CaCl2, 1 mm ATP, 20 mm Tris-HCl, pH 7.5) to depolymerize F-actin. After 20 min of incubation at room temperature, 10 μl of DNase I solution was added, and the sample (30 μl) was mixed with 970 μl of DNA solution to measure DNase I inhibition (see above). Finally, the G-actin content was related to total protein concentration, which was determined by using the Bradford assay.
Measurement of ITPK Activity
The metabolism of Ins(1,4,5)P3 by total cytosolic Ins(1,4,5)P3-3-kinase (ITPK) and Ins(1,4,5)P3-5-phosphatase (5-PPT) activity was analyzed by using an ammonium sulfate-precipitated and thoroughly dialyzed cell extract. For that, four culture dishes with 2 × 107 cells each were washed three times with PBS at room temperature in their culture dishes, and after complete removal of PBS, the cells were lysed by adding 3 ml of ice-cold lysis buffer per dish (10 mm Tris-HCl, pH 7.5, 150 mm KCl, 100 mm NaF, 12 mm mercaptoethanol, 0.5% Nonidet P-40, phosphatase-stop and proteinase inhibitor mixture, Roche Diagnostics) and freezing in liquid nitrogen and re-thawing three times. After 20 min of centrifugation at 13,000 × g and at 4 °C, the supernatant was diluted to 17 ml with ice-cold buffer A containing 20 mm Tris-HCl, 150 mm NaCl, 1 mm dithiothreitol, pH 7.5, and extracted protein was precipitated by adding ammonium sulfate to 50% saturation for 1 h at 4 °C. After centrifugation for 15 min at 20,000 × g and at 4 °C, the pellet was suspended in 1 ml of buffer A and dialyzed against this buffer for 4 h at 4 °C, whereby the dialysis buffer was changed every 30 min. After dialysis, the solution was centrifuged for 5 min at 13,000 × g and 4 °C, and protein concentration of the supernatant was determined by using the Bradford assay. To analyze Ins(1,4,5)P3 metabolism, about 420 μg of protein extract was assayed in a total volume of 800 μl. 160 μl of dialyzed protein was diluted into 600 μl of assay buffer composed of (final concentrations) 20 mm Hepes, pH 7.5, 7 mm MgCl2, 30 mm KCl, 1 mm dithiothreitol, 2 mm ATP, 3 mm phosphoenolpyruvate, 10 μg/ml pyruvate kinase and preincubated to 30 °C, and the assay was started with 40 μl of Ins(1,4,5)P3 (Alexis, San Diego) giving an initial concentration of 10.6 μm. Five aliquots of 150 μl were withdrawn between 0 and 40 min of incubation at 30 °C. For activation of ITPK by Ca2+/calmodulin (CaM), 2.15 μg/ml calmodulin and 10–20 μm free CaCl2 were in the assay mixture. Samples were treated for HPLC analysis as described previously (20), and Ins(1,4,5)P3 and its metabolites Ins(1,3,4,5)P4, Ins(1,3,4)P3, and Ins(1,3,4,6)P4 were analyzed by Micro MDD-HPLC as described previously (24). Total ITPK activity was determined for individual intervals as the sum of the rates of formation of Ins(1,3,4,5)P4, Ins(1,3,4)P3, and Ins(1,3,4,6)P4. 5-PPT activity over time of incubation was defined as the rate of decrease of Ins(1,4,5)P3 minus the actual rate of ITPK. Ins(1,3,4,5)P4 was averaged for each interval. Specific activities were determined by referring to cellular extract protein concentration.
To measure 5-PPT activity under kinase-inactive conditions, ATP, phosphoenolpyruvate, and protein kinase were omitted from the assay buffer, and total MgCl2 was reduced to 4 mm. Due to rapid further dephosphorylation, only the Ins(1,4,5)P3 decrease was used to determine specific activity.
Total cellular activity of the ITPKA isoforms was determined after immunoprecipitation of the enzyme from cell lysates prepared as described previously (20). ITPKA assays in the absence and presence of Ca2+/CaM were performed as described above. In contrast to the assays with total extracted protein, an Ins(1,4,5)P3 decrease precisely corresponding to the Ins(1,3,4,5)P4 formation was found in assays, indicating that no 5-PPT activity was co-immunoprecipitated. The activity found was referred to cellular extract protein concentration. Kinetic parameters were obtained by fitting corresponding kinetic functions to the data sets with GraphPad PRISM.
Statistical Analyses
If not stated explicitly, statistical comparisons of normalized values were made using Student's t test for unpaired samples. p values ≤0.05 were taken to indicate significant differences between groups.
RESULTS
Ectopic Expression of Neuron-specific ITPKA Modulates Tumor Cell Shape and Increases Migration
ITPKA (GenBankTM accession number X54938, Swiss-Prot entry P23677) was reported to be up-regulated in four malignant cell types (16–19). Here, we re-examined ITPKA protein levels in a larger set of cell lines. Transformed and nontransformed cell lines were tested for ITPKA expression (Fig. 1A). Western blot analyses revealed that nontransformed cells expressed low (IMR-90, HSF-14) to nearly undetectable (HmEpc, leukocytes) levels of ITPKA. The cancer cell lines showed large differences in their ITPKA protein levels. Whereas SKBR-3 and T47D cells expressed very low ITPKA levels, very high expression levels were found in the highly metastatic cell lines H1299, MDA231, and HepG2. The data reveal that most of the transformed cell lines (H1299, MDA231, Mevo, Jurkat T-cells, HEK293, and HepG2) examined expressed increased levels of ITPKA compared with the low levels in nontransformed cells (IMR-90, HmEpc, HSF-14, and leukocytes).
FIGURE 1.
ITPKA controls cell motility. A, whole cell lysates from cells cultivated in complete medium were analyzed for ITPKA expression by Western blot. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression served as loading control. IMR-90, normal lung epithelial cells; A549 and H1299, non-small lung cancer cells; HmEpc, normal breast epithelial cells; T47D, SKBR-3, and MDA231, breast cancer cells; HSF14, normal skin fibroblasts; Mevo, melanoma cells; human leukocytes; Jurkats, leukemic T-cells; HEK293, transformed embryonal kidney cells; HepG2, liver cancer cells. B, ITPKA expression was stably up-regulated in A549 cells and stably down-regulated in MDA231 and H1299 cells. C, cell lines were seeded in wells covered with 15% gelatin solution and monitored by light microscopy after 48 h. One out of three representative experiments is shown for each cell line. D, migration of cells cultivated in complete medium was examined by the transwell assay. Migration (“trans-migrated cells”) was expressed as relative amounts with respect to control cells (considered as 100%). Data represent mean values ± S.D. of four independent experiments. ***, p < 0.0001.
Subsequently, we analyzed the phenotypic consequences of ITPKA expression in tumor cells. Therefore, ITPKA was stably knocked down (MDA231 kd and H1299 kd, Fig. 1B) in two cell lines with high ITPKA expression levels (MDA231 and H1299) and was stably overexpressed (A549-ITPKA, Fig. 1B) in low ITPKA-expressing A549 cells. To examine cell shape and motility, modified and control cell lines were grown in a highly viscous gelatin matrix. MDA231 kd, H1299 kd, and A549-ITPKA cells showed dramatically changed phenotypes compared with their respective control cells (Fig. 1C). Whereas highly metastatic MDA231 control cells migrated completely through the matrix and grew in an evenly scattered pattern, ITPKA-knockdown variants grew as cell clusters inside the gelatin layer or adhered to the bottom of the culture dish in tight aggregates. Also, H1299 control cells completely migrated through the gelatin. H1299 kd cells by contrast remained inside the gelatin matrix, forming tight aggregates of round cells. In cells with up-regulated ITPKA expression, we found the opposite behavior as observed in cells with low ITPKA expression. Most of the A549-ITPKA cells migrated through the gelatin layer and adhered to the bottom of the culture dish. In contrast, fewer A549 control cells migrated through the gelatin. Nonmigrating cells remained in the matrix, and cell clusters had a rounded morphology. To analyze the influence of ITPKA on invasive migration, we performed transwell migration assays, an in vitro model for invasive migration (Fig. 1D). Transwell migration of MDA231 kd cells was reduced by 73 ± 9%, mean ± S.D. (p < 0.0001), and that of H1299 kd cells by 58 ± 11%, mean ± S.D. (p < 0.0001), as compared with the respective control cells exhibiting high levels of ITPKA. Approximately twice as many A549-ITPKA cells penetrated as A549 control cells (101 ± 15%, mean ± S.D. increase, p < 0.0001). In addition, migration of modified A549 and H1299 cells was determined by a standard wound healing assay. As shown in supplemental Fig. S1, the results obtained with the gelatin and the transwell assays were confirmed by determination of wound healing migration. A549-ITPKA cells showed 2.5-fold higher migration compared with control A549 cells, whereas migration of H1299 kd cells was reduced 3.4-fold relative to H1299 control cells. In summary, the data reveal that expression of high levels of ITPKA controls shape and motility of tumor cells.
ITPKA Controls Migration by Stabilizing and Bundling Actin Filaments
The results above demonstrate that ITPKA increases motility of tumor cells without addition of growth factors (Fig. 1C and supplemental Fig. S1), indicating that ITPKA induced cell motility is not dependent on growth factor simulation. To confirm this observation, transwell migration of high ITPKA-expressing H1299 control cells and of H1299 ITPKA kd cells was determined for serum-starved cells (supplemental Fig. S2) in comparison with cells cultivated in complete medium. We found that the number of migrating control cells was reduced by about 30% by serum starvation, whereas that of H1299 kd cells was reduced by about 84%. The lower degree of serum dependence of migration in ITPKA-expressing control cells as compared with ITPKA kd cells supports the hypothesis that ITPKA promotes migration independent of growth factor stimulation.
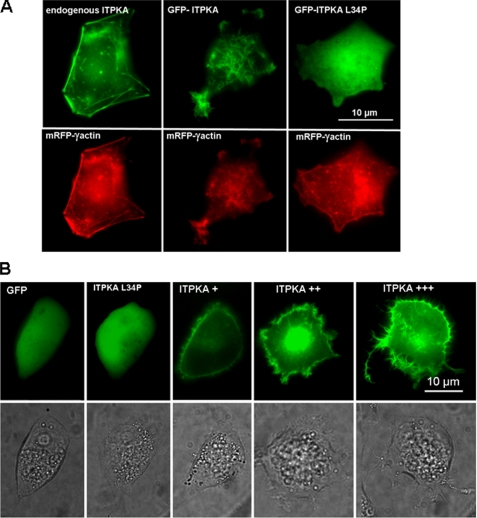
In the next step, we analyzed the mechanism underlying ITPKA-induced cell motility in growth factor-poor medium in more detail. Migration requires formation of cellular protrusions, and we demonstrated that high levels of ITPKA induce the formation of lamellipodia-like and neurite-like cell processes (20). As ITPKA has been shown to bind to F-actin in neurons (13), and we found that overexpression of ITPKA alters the actin cytoskeleton (20), we examined whether ITPKA-induced formation of cell processes may result from direct interaction between ITPKA and F-actin. For this purpose, we first confirmed that in tumor cells ITPKA also binds to F-actin. Therefore, mRFP-γ-actin (the vector was kindly provided by Dr. Michael Schell) was overexpressed in H1299 cells (Fig. 2A, left, lower panel), and endogenous ITPKA was stained with an anti-ITPKA antibody (Fig. 2A, left, upper panel). In addition, a GFP-ITPKA fusion protein was co-transfected with mRFP-γ-actin (Fig. 2A, middle panel). Both endogenous ITPKA and the fusion protein showed co-localization with F-actin. Furthermore, many large protrusions were visible in cells expressing GFP-ITPKA, confirming our results published in Ref. 20. To show whether formation of these protrusions may be dependent on binding of ITPKA to actin, a point mutant (ITPKA L34P (25)) that blocks actin binding was overexpressed in H1299 cells. As shown in Fig. 2A (right panel), the morphology of cells expressing GFP-ITPKA L34P and those of cells expressing GFP did not show visible differences. Thus, binding of ITPKA to actin seems to be required to alter the morphology of cellular protrusions. To show whether ITPKA-induced formation of large cell protrusions may be dependent on the expression level of ITPKA, processes of cells expressing high levels of GFP or GFP-ITPKA L34P and low (+), medium (++), or high (+++) levels of GFP-ITPKA were quantified. For this approach, the same cell was visualized by transmitted light and by fluorescence images, and protrusions were quantified in cells visualized by transmitted light. We found that control cells (H1299-GFP, H1299-GFP-ITPKA L34P) extended multiple short (<1 μm) thin filopodia-like protrusions (see also Ref. 20) and observed that cells with intermediate and high expression of ITPKA extended many large protrusions, whereby the number of these cell processes increased with increasing concentrations of GFP-ITPKA (Fig. 2B and Table 1). On the other hand, cells with low expression of GFP-ITPKA did not show significant differences relative to control cells. From these data, we conclude that the effect of ITPKA on protrusion morphology is concentration-dependent and that a minimum ITPKA level is required to alter the structure of cell protrusions.
FIGURE 2.
Effects of ITPKA on formation of F-actin-based cell protrusions. A, to analyze co-localization between endogenous ITPKA and F-actin, H1299 cells were transfected with mRFP-γ actin (red, left panel) and stained with an anti-ITPKA antibody (green, left panel). In addition, H1299 cells were co-transfected with GFP-ITPKA (green, middle panel) and mRFP-γ-actin (red, middle panel) fusion proteins. Right panel, actin binding of ITPKA was blocked by L34P (25), and cells were co-transfected with GFP-ITPKA L34P (green) and mRFP-γ-actin (red). Fluorescence of the fusion proteins was monitored by confocal fluorescence microscopy. B, cells were transfected with GFP, GFP-ITPKA L34P, and with GST-ITPKA. After 16 h of incubation, the cells were washed and covered with PBS. Viable cells were monitored by light and by fluorescence microscopy. The protrusions were determined from cells expressing high levels of GFP and GFP-ITPKA L34P and low (+), intermediate (++), or high (+++) levels of GFP-ITPKA (see Table 1).
TABLE 1.
Effect of ITPKA on protrusion size of H299 cells
H1299 cells were transfected with GFP (H1299-GFP), GFP-ITPKA-L34P (H1299-L34P), or with GFP-ITPKA (H1299-ITPKA), and cellular protrusions were counted from at least 100 cells strongly expressing GFP and GFP-ITPKA-L34P. In addition, cell processes were determined from at least 100 H1299 cells expressing low (+), intermediate (++), or high (+++) levels of GFP-ITPKA. The strength of the expression level was determined by the green fluorescence of GFP and GFP-ITPKA fusion proteins. To quantify the protrusions, the same cell was monitored by light and by fluorescence microscopy, and protrusions were only counted from cells monitored by light microscopy.
| % of cells extending at least one protrusion; length >1 μm, width >0.6 μm | No. of protrusions per cell | % of cells extending at least one protrusion; length >4 μm, width >0.6 μm | No. of protrusions per cell | |
|---|---|---|---|---|
| H1299-GFP | 44 | 1.8 | 2 | 1 |
| H1299-L34P | 51 | 1.9 | 0 | 0 |
| H1299-ITPKA + | 36 | 2.2 | 1 | 1 |
| H1299-ITPKA ++ | 88a | 4.5a | 14a | 1.7a |
| H1299-ITPKA +++ | 95a | 6.9a | 52a | 2.0a |
a p < 0.0001 and statistically significant differences in protrusion number relative to controls (H1299-GFP) are shown.
As our data show that ITPKA-mediated alterations in protrusion morphology result from direct interaction of ITPKA with actin, we examined potential ITPKA-induced alterations of G-actin and F-actin contents in H1299 cells. For that, G- and F-actin contents were investigated in cells stably overexpressing GFP, GFP-ITPKA L34P, or GFP-ITPKA by using the DNase I inhibition assay. As shown in Table 2, expression of GFP-ITPKA increased the level of F-actin by 13% as compared with cells overexpressing GFP, whereas the F-actin concentration of cells expressing GFP-ITPKA L34P was not significantly different from that of H1299-GFP cells. Thus, we conclude that the high expression of ITPKA significantly increases the total cellular level of F-actin in H1299 cells.
TABLE 2.
Effect of ITPKA on relative content of G-actin and F-actin in extracts from H1299 cells
Extracts of H1299 cells stably expressing GFP, GFP-ITPKA, or GFP-ITPKA L34P were examined for G- and F-actin contents by using the DNase I inhibition assay. Values for G-actin and F-actin are % of total actin measured in cell lysates. G-actin was directly measured, and after depolymerizing F-actin with 1.5 m guanidinium chloride, a second DNase I assay was carried out to measure total actin. Data shown were calculated as F-actin = total actin − G-actin. The results are mean values ± S.D. of four experiments.
| G-actin | F-actin | |
|---|---|---|
| % | % | |
| H1299-GFP | 53.65 ± 1.79 | 46.35 ± 1.86 |
| H1299-L34P | 53.03 ± 3.55 | 47.00 ± 3.81 |
| H1299-ITPKA | 40.97 ± 0.05 | 59.03 ± 0.04a |
a p < 0.01 and statistically significant differences in F-actin content as compared with controls (H1299-GFP) are shown.
To analyze how ITPKA promotes formation of F-actin, the effect of a purified GST-ITPKA fusion protein on formation of F-actin was assessed by a pyrene-actin-based polymerization/depolymerization assay in vitro. Prior to this analysis, we examined binding of the GST-ITPKA fusion protein to actin in H1299 cells. For this approach, the N-terminal GFP tag of the GFP-ITPKA cDNA was replaced by an N-terminal GST tag. As we found that GST-ITPKA bound to actin and induced the same morphological effects as observed after overexpression of the GFP fusion protein (data not shown), we considered that the GST-ITPKA protein is a suitable tool for in vitro assays.
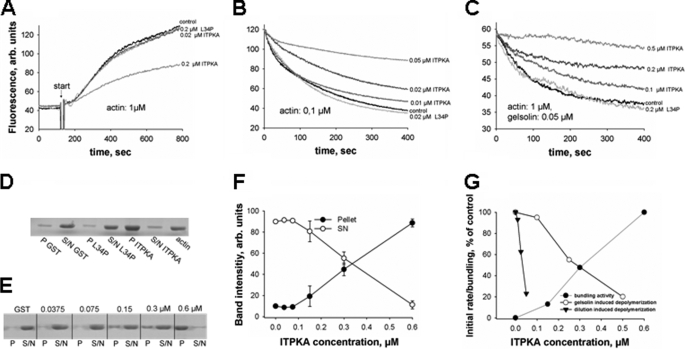
First, we analyzed the effect of GST-ITPKA on actin polymerization, using ITPKA to actin ratios of 1:5 and 1:50. Purified GST and GST-ITPKA L34P served as controls. As shown in Fig. 3A, addition of low concentrations of GST-ITPKA (0.02 μm, 1:50) did not alter actin polymerization, whereas high concentrations (0.2 μm, 1:5) inhibit actin polymerization by about 44% as compared with controls (GST, GST-ITPKA-L34P). This result clearly shows that ITPKA does not increase the cellular level of F-actin by promoting actin polymerization. Therefore, we next analyzed whether ITPKA may stabilize actin filaments and examined the effect of GST-ITPKA on spontaneous, dilution-induced F-actin depolymerization, using GST and GST-ITPKA L34P as controls (Fig. 3B). 1 μm F-actin was diluted to 0.1 μm. We found that GST-ITPKA decreased F-actin depolymerization in a concentration-dependent manner in substoichiometric concentrations (0.01 μm ITPKA, 1:10 ITPKA/actin; 0.02 μm ITPKA, 1:5 ITPKA/actin; 0.05 μm ITPKA, 1:2 ITPKA/actin), whereas GST-ITPKA L34P had no effect on F-actin depolymerization. In addition, we assayed the influence of ITPKA on active, gelsolin-mediated depolymerization of F-actin. In this experiment, ITPKA was used in molecular excess to gelsolin (0.05 μm) and in substoichiometric concentrations (0.1, 0.2, and 0.5 μm) to actin. To minimize spontaneous depolymerization of F-actin, F-actin was used in a concentration of 1 μm (Fig. 3C). We found that ITPKA inhibited gelsolin-mediated degradation of F-actin in similar actin to ITPKA ratios as it inhibits spontaneous actin depolymerization. Based on these results, we assume that ITPKA inhibits actin dynamics (polymerization and depolymerization) by binding to actin filaments, resulting in reduced annealing of new actin molecules and in inhibition of dissociation of actin monomers. However, this result alone does not explain the increased levels of F-actin found in cells overexpressing ITPKA. As Johnson and Schell (25) revealed that the actin binding domain (ABD) of ITPKA bundles F-actin, we analyzed whether this is also true for the full-length protein. To show this, the GST-ITPKA fusion protein was incubated with actin in an ITPKA/actin ratio of 1:3, and bundling of F-actin was examined by pelleting F-actin bundles. Fig. 3D shows that only actin incubated with GST-ITPKA was pelleted, and Fig. 3, E and F, reveals that this effect is dependent on the ITPKA/actin ratio or ITPKA concentration, respectively. We found that GST-ITPKA in an ITPKA/actin ratio of 1:3 nearly completely bundled actin, whereas dilution of GST-ITPKA to an ITPKA/actin ratio of 1:6 decreased the actin bundling activity of GST-ITPKA to about half of that observed at a ratio of 1:3. Dilutions lower than 1:12 did not affect F-actin bundling. Thus, the full-length form of ITPKA bundles F-actin in a concentration-dependent manner. In Fig. 3G, all three effects observed with GST-ITPKA were re-plotted relative to total ITPKA concentration during the assays. The data reveal that inhibition of the initial rate of gelsolin-induced F-actin severing and the degree of bundling were half-maximal at a similar concentration of about 0.3 μm, whereas inhibition of the rate of actin depolymerization after dilution to 0.1 μm actin was half-maximal at about 0.03 μm. The much lower EC50 for the latter effect may reflect a much higher affinity of full-length ITPKA for binding to F-actin as found for the isolated ABD by Johnson and Schell (25). In contrast, the 10-fold higher and similar EC50 values both for actin bundling and inhibition of gelsolin-induced severing may reflect a lower affinity for dimerization of ITPKA, which is a likely prerequisite for bundling. Also, the initial increase of both effects in a quadratic type concentration dependence is indicative for such dimerization, which has also been shown recently by Johnson and Schell (25) for the N-terminal domain of ITPKA.
FIGURE 3.
Effect of ITPKA on actin dynamics in vitro. A, effect of GST-ITPKA on actin/pyrene-actin polymerization was analyzed by measuring increased pyrene actin fluorescence. 0.2 μm GST and 0.2 μm GST-ITPKA L34P were used as controls. Arb, arbitrary units. B, dilution-induced F-actin depolymerization was analyzed by measuring decreased pyrene actin fluorescence in the presence of different concentrations of GST-ITPKA. 0.2 μm GST and 0.2 μm GST-ITPKA L34P served as controls. C, gelsolin-induced F-actin depolymerization was carried out with constant gelsolin and different GST-ITPKA concentrations. The control proteins were used in concentrations of 0.2 μm. Each experiment was performed at least three times, and one representative experiment is shown. D, bundling of F-actin was examined by incubating actin in polymerization buffer together with 0.6 μm GST, GST-ITPKA L34P, or GST-ITPKA, respectively. After 30 min of incubation the solution was centrifuged, and F-actin bundling was examined by analyzing the pellet (P) and the supernatant (S/N) by Coomassie-stained SDS-PAGE. E, different concentrations of ITPKA and 0.6 μm GST as control were examined for its effect on F-actin bundling. F, Coomassie-stained bands derived from this bundling analysis were determined with an image analyzer. The sum of pellet and supernatant were calculated and set to 100%, and then the percentage of signal intensity of pellet and supernatant relative to sum was plotted for each sample. Data represent mean values of three independent experiments. G,“initial” rates from the data obtained from F-actin depolymerization assays (B and C) were determined by calculating the rate of fluorescence decrease (B and C) between 30 and 90 s, and the rates measured without addition of ITPKA were set to 100%. Data were plotted against the GST-ITPKA concentration in the assays. Data obtained from the bundling experiment (F) were also plotted against GST-ITPKA concentration, but in this case bundling activity in samples without addition of ITPKA were set to 0%.
Enzyme Activity of ITPKA Is Required to Completely Restore Migration in H1299 kd Cells
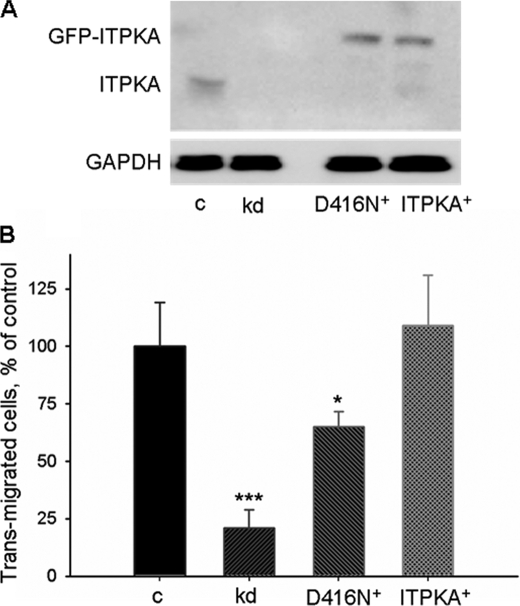
Our data shown in Fig. 1 and supplemental Fig. S1 indicate that under nonstimulating conditions, where Ins(1,4,5)P3 levels are low and ITPKA is not significantly Ca2+/CaM-stimulated, ITPKA promotes migration independent of its enzymatic activity. To examine whether its enzyme activity is also involved in ITPKA-promoted migration, a kinase-dead mutant of GFP-ITPKA (ITPKA-D416N+) or the active form of GFP-ITPKA (ITPKA+) was stably re-expressed in H1299 kd cells. Cells expressing GFP alone were used as control (H1299 kd). Expression levels of these proteins are depicted in Fig. 4A. Migration of the modified H1299 cells (H1299 kd, H1299-ITPKA-D416N+, and H1299-ITPKA+) and that of H1299 control cells expressing only endogenous ITPKA was measured by the transwell assay after addition of 100 nm epidermal growth factor (EGF) (Fig. 4B). We found that re-expression of the kinase-dead mutant in H1299 kd cells incompletely restored migration (65 ± 7%, mean ± S.D.) as compared with H1299 control cells. This result confirms our observation that the kinase-dead form of ITPKA has the potential to promote cell motility. Our finding that only the enzymatically active form of ITPKA re-expressed in H1299 kd cells completely rescued migration of H1299 kd cells (109 ± 22%, mean ± S.D. of the value of H1299 control cells) shows, however, that a second mechanism dependent on the catalytic activity of ITPKA is required for the full migratory potential of ITPKA-expressing cells.
FIGURE 4.
Enzyme activity of ITPKA is required for EGF-mediated migration. A, whole cell extracts of H1299 kd cells stably re-expressing a kinase-dead form of GFP-ITPKA (D416N+), a GFP-ITPKA fusion protein (ITPKA+), or solely GFP (kd) were probed with an anti-ITPKA antibody, and expression of ITPKA in these cells was compared with endogenous expression of ITPKA in H1299 control cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, H1299 control, kd, D416N+, and ITPKA+ cells were serum-starved for 24 h and seeded in Boyden chambers containing Dulbecco's modified Eagle's medium, 100 nm EGF. After 16 h of incubation, migration was measured by the transwell assay. Migration was expressed as relative amounts with respect to H1299 control cells (c) (considered as 100%). Data represent mean values ± S.D. of four independent experiments. *, p < 0.05; ***, p < 0.0001.
ITPKA Modulates Ins(1,4,5)P3-induced Calcium Signaling
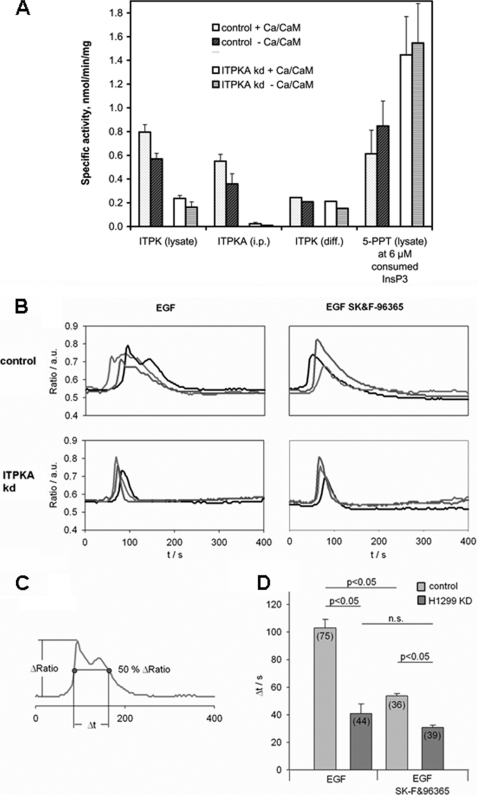
To examine the role of ITPKA activity in EGF-mediated cell migration, the consequences of reduced ITPKA enzyme activity in H1299 kd cells were analyzed. As many cell types express more than one ITPK isoform and also inositol polyphosphate multikinase (IPMK) activity is able to phosphorylate Ins(1,4,5)P3 at the 3-position, we first controlled a possible compensatory expression of ITPKB, ITPKC, and IPMK in H1299 kd cells. Small amounts of mRNA were detectable in H1299 control and H1299 kd cells for both ITPKB and ITPKC (data not shown), and Western blot analysis revealed a low ITPKC signal in H1299 kd cells (supplemental Fig. S3). IPMK was not detectable by Western blot (data not shown). To rule out that the expression of ITPKC or very low expression levels of IPMK might compensate for the loss of ITPKA activity, we prepared substrate and product-free protein extracts from H1299 control and H1299 kd cells and analyzed total ITPK activity in the presence and absence of Ca2+/CaM. In addition, the specific activity of ITPKA was analyzed in H1299 control and H1299 kd cells by measuring Ins(1,4,5)P3 phosphorylation catalyzed by immunoprecipitated ITPKA. We found that the activity of ITPKA was reduced by 94–95% in H1299 kd cells as compared with controls (Fig. 4A), showing that knockdown of ITPKA expression was very efficient. In protein extracts from H1299 kd cells, the total ITPK activity was reduced by 62–66% as compared with H1299 control cells (Fig. 4A) in the presence and absence of Ca2+/CaM, respectively. We also assayed IPMK activity in the protein extracts by employing Ins(1,3,4,6)P4 as an IPMK-specific substrate, but we did not detect any activity (data not shown). The data indicate that the total Ins(1,4,5)P3 3-kinase activity is drastically reduced in H1299 kd cells and reveal that ITPKA is the prominent Ins(1,4,5)P3-metabolizing enzyme in H1299 cells. By subtracting the ITPKA activity from total ITPK activity, both determined per mg of extracted cell protein, we got an indication of remaining cellular ITPK activity not due to ITPKA. This residual ITPK activity is almost equal in control and H1299 kd cells and is only weakly Ca2+/CaM-activated. This indicates further weakly CaM-stimulated isoform(s) of ITPK, either ITPKC alone (see blotting data, above) or N-terminally truncated forms of both ITPKC and ITPKB, which also show this behavior. Because all isoform-specific antibodies recognize epitopes in the N-terminal domains, Western blots would not reveal positive signals for such truncated isoforms. However, the remaining ITPK activity in H1299 kd cells is so low that the rate of formation of Ins(1,3,4,5)P4 is about 3-fold lowered.
To examine whether Ins(1,4,5)P3-induced calcium release playing an essential role in growth factor-induced cell migration (26) might be affected by down-regulation of ITPKA expression, Ins(1,4,5)P3-induced calcium signaling was analyzed in EGF-simulated H1299 cells. In H1299 kd cells, treatment with 100 nm EGF resulted in a rapid transient calcium (Ca2+) peak, whereas in most of the H1299 control cells the initial peak was followed by a second much broader phase of increased Ca2+ (Fig. 5B). To get a quantitative measure for the duration of this second signal, we determined the time that the signal stays above 50% of the maximum amplitude as indicated in Fig. 5C. This analysis showed that the duration of the signal in cells expressing ITPKA was significantly longer than in the H1299 kd cells (103 ± 6 s in H1299 control versus 41 ± 7 s in H1299 kd cells, mean ± S.E., p < 0.05; Fig. 5D). To investigate a possible role of Ca2+ entry in the second phase of the Ca2+ signal we used SK&F-96365, an inhibitor of store-operated Ca2+ entry (SOCE). Inhibiting the Ca2+ entry turned the prolonged signal observed in H1299 control cells into a single transient peak, largely reducing the duration of the signal (103 ± 6 to 54 ± 2 s with SK&F-96365, mean ± S.E., p < 0.05; Fig. 5D). The signal returned slightly slower to its basal value than in the H1299 kd cells indicating a longer lasting Ca2+ release from intracellular stores (Fig. 5, B and D). In contrast to the pronounced effect on the H1299 control cells, the signal in H1299 kd cells was not significantly affected by inhibition of Ca2+ entry. In summary, the results indicate that cells expressing ITPKA show a longer lasting Ca2+ release from intracellular stores and thereby activate Ca2+ entry more efficiently.
FIGURE 5.
Effect of ITPKA on EGF-stimulated calcium entry in H1299 cells. A, total cellular ITPK activity, cellular ITPKA-specific activity, calculated remaining cellular ITPK activity, and cellular Ins(1,4,5)P3-5-PPT activity, the latter assayed under “active ITPK” conditions (i.e. under Ins(1,3,4,5)P4 inhibition) was measured with and without Ca2+/CaM activation from dialyzed cellular protein extracts from control and ITPKA kd cells. Total protein extracts from H1299 control and H1299 kd cells or ITPKA specifically immunoprecipitated from total protein extracts were incubated with 10.75 μm Ins(1,4,5)P3 for 0, 5, 10, 20, and 40 min, and phosphorylation and dephosphorylation of Ins(1,4,5)P3 was analyzed by metal dye detection-HPLC. Specific activity (all referred to 1 mg of total cellular protein extract) was determined as described under “Experimental Procedures.” a.u., arbitrary units. B, Ca2+-imaging experiments on Fura 2-loaded H1299 control and H1299 kd cells in the presence or absence of the SOCE inhibitor SK&F-96365 (30 μm). Overlay of ratio tracings from individual representative cells stimulated with 100 nm EGF (at 20 s). C, duration of the signals was determined as the time the signal stays above 50% of the maximal amplitude. D, quantitative analysis of signal duration (data represented as mean ± S.E., two-way analysis of variance with Holm-Sidak post test was used to determine the difference between cell types and inhibitor treatment. Differences were considered significant at p < 0.05). n.s., not significant.
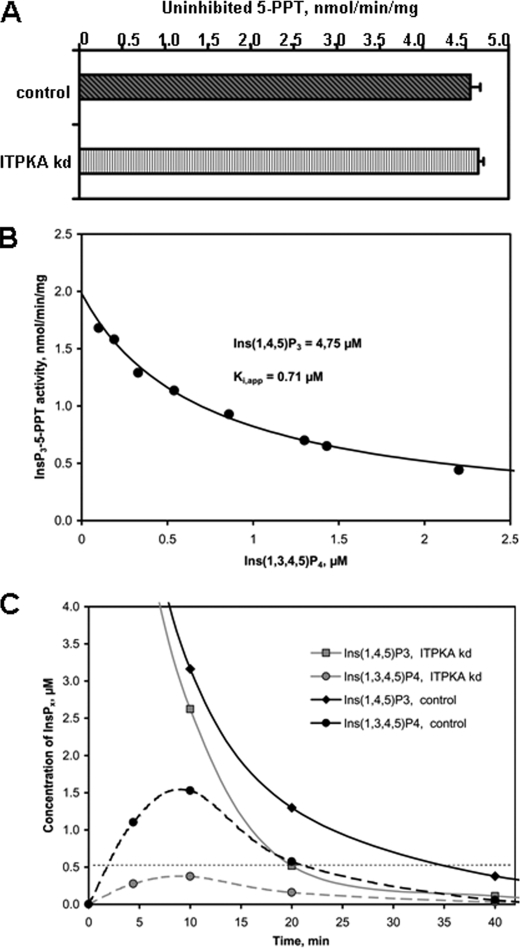
To understand the correlation between ITPKA activity and activation of SOCE, one has to keep in mind that the role of ITPKA action on Ca2+ signaling can be dual, i.e. high ITPKA activity can significantly reduce the concentration of the Ins(1,4,5)P3 signal by rapidly removing the Ca2+-releasing second messenger. On the other hand, the product of ITPK, Ins(1,3,4,5)P4 has been shown to be a very potent “alternative substrate inhibitor” of 5-PPT (27). Based on these considerations, we analyzed the activity of 5-PPT in H1299 control and ITPKA kd protein extracts under “inactive” and “active kinase” conditions, only in the latter case allowing formation of Ins(1,3,4,5)P4. We found that the maximal activity of 5-PPT, determined in protein extracts of both cell lines under “inactive kinase” conditions, was 1 order of magnitude higher than total ITPK activity (about 5 nmol/min/mg of cell extract protein) and was not influenced by knocking down ITPKA (Fig. 5A). Also under active kinase conditions, where the presence of phosphoenolpyruvate and ATP increased the apparent Km value of the 5-PPT for Ins(1,4,5)P3 to 8.9 μm (supplemental Fig. S5), the maximal activity at >30 μm Ins(1,4,5)P3 was unchanged. In these assays, the higher level of Ins(1,3,4,5)P4 generated by the high ITPK activity in H1299 control as compared with ITPKA kd cell extracts (see Fig. 5A) resulted in reduction of apparent 5-PPT activity and thus led to protection of Ins(1,4,5)P3 against rapid dephosphorylation by 5-PPT. To allow a comparison of the Ins(1,3,4,5)P4 inhibited 5-PPT activities, we compared these activities after an identical decrease of Ins(1,4,5)P3 from the initial 10.75 to 4.75 μm. Resulting 5-PPT activities are shown by the outer right set of bars in Fig. 4A. Clearly, the inverse relationship between cellular ITPK activity and 5-PPT is evident. Therefore, in H1299 cells 5-PPT activity is likely to be the main terminating activity for Ins(1,4,5)P3-induced Ca2+ release.
We determined in detail the dependence of 5-PPT on the concentration of both Ins(1,4,5)P3 and Ins(1,3,4,5)P4 simultaneously present at a certain time after addition of 10.75 μm InsP3 (supplemental Fig. S4). The initial 5-PPT activity was not significantly different between H1299 control and H1299 kd cells, but the further decrease of Ins(1,4,5)P3 over time in fact was significantly delayed in assays made with extracts from control cells relative to those from H1299 cells. Ca2+/CaM activation in both cases exaggerated this delay. Inhibition can be nicely explained by increased production of Ins(1,3,4,5)P4 (see horizontal bars in supplemental Fig. S4). A more detailed analysis of reduction of 5-PPT activity by increasing Ins(1,3,4,5)P4 concentrations, all derived at 4.75 μm Ins(1,4,5)P3 from eight different transients, confirmed this hypothesis by revealing a remarkably high degree of competitive-like “weak alternative substrate inhibition” by Ins(1,3,4,5)P4 with an apparent Ki of 0.71 μm (Fig. 6B). An apparent Km value for Ins(1,3,4,5)P4-5-PPT of 0.79 μm, determined in presence of between about 1 and 3 μm Ins(1,4,5)P3 from six different transients, indicates that both values are close to the real Kd of 5-PPT for Ins(1,3,4,5)P4 which is ≥12-fold lower than the Km for Ins(1,4,5)P3. Because this alternative substrate has a 14-fold lower Vmax than Ins(1,4,5)P3 (see legend to supplemental Fig. S5) and thus is only slowly dephosphorylated (Fig. 5C), its inhibitory effect is long lasting. From our data showing simple enzyme kinetics for 5-PPT, it is likely that only one major isoform of 5-PPT is dominating Ins(1,4,5)P3 dephosphorylation, but we have not further investigated the 5-PPT enzymatic isomerism, e.g. by Western blotting due to a lack of isoform-specific antibodies.
FIGURE 6.
Effect of ITPK-induced Ins(1,3,4,5)P4 generation on activity of Ins(1,4,5)P3 5-PPT in H1299 cells. A, uninhibited 5-PPT activity in control and ITPKA kd cells. Ins(1,4,5)P3-5-PPT activity was analyzed in cell lysates from H1299 control and H1299 kd cells under inactive kinase conditions where ATP and the ATP reconstitution system were omitted from the assay buffer. Shown are the averages of four determinations. B, inhibitory effect of Ins(1,3,4,5)P4 on Ins(1,4,5)P3-5-PPT activity during Ins(1,4,5)P3 transients generated with total cellular protein extracts. Ins(1,4,5)P3-5-PPT activity was analyzed in cell lysates from control and ITPKA kd cells under active kinase conditions with and without Ca2+/CaM activation. From a total of eight different transients of Ins(1,4,5)P3 and Ins(1,3,4,5)P4 versus time, an inhibition curve of Ins(1,4,5)P3-5-PPT activity by Ins(1,3,4,5)P4 was derived at a constant Ins(1,4,5)P3 of 4.75 μm. C, transients of Ins(1,4,5)P3 are strongly prolonged by high total cellular ITPK activity. Transients of Ins(1,4,5)P3 and Ins(1,3,4,5)P4 obtained with cell extracts from control and ITPKA kd cells under active kinase conditions with and without Ca2+/CaM activation of ITPK are plotted against time. Data show one representative series of experiments out of three performed with different cell extracts.
From the transients of Ins(1,4,5)P3 and Ins(1,3,4,5)P4 depicted in Fig. 6C, we in fact can make the link to the prolonged SOCE observed in control versus kd cells (Fig. 4B). First, we consider that only a level of Ins(1,4,5)P3 ≥0.5 μm would lead to a Ca2+ release. With extracts from H1299 control cells, transiently generating up to 3 μm inhibitory Ins(1,3,4,5)P4, the period where the initially added Ins(1,4,5)P3 remained above 0.5 μm in our assays was almost doubled from about 20 min observed with ITPKA kd cell extracts to about 36 min with control cell extracts (Fig. 6C). Because inside the cell the enzyme concentrations and activities likely are about 10 times those in our assays, duration of transients would correspondingly be 10-fold shorter. The period facilitating calcium release should then last about 100 s in H1299 kd cells and about 180 s in H1299 control cells. This is in good agreement with the duration of SOCE observed by ratiometric calcium measurements in these two cell lines (see above).
Altered Calcium Signaling Directly Influences Migration of H1299 Cells
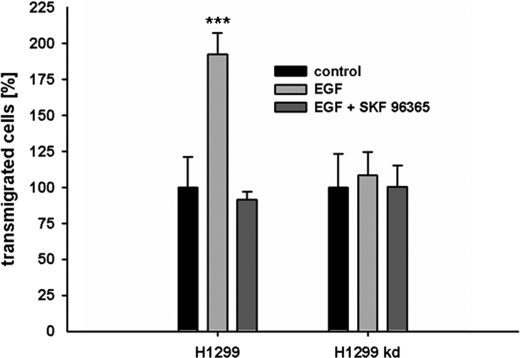
In a next step we analyzed whether ITPKA-mediated increase in Ca2+ entry and cell migration are in fact functionally coupled. For this analysis, we tested the requirement of ITPKA-mediated calcium entry for EGF-induced migration of H1299 cells and performed migration experiments with EGF-stimulated cells in the presence or absence of the calcium entry inhibitor SK&F-96365. Treatment with EGF significantly increased migration of H1299 control cells (by 92 ± 15%, mean ± S.D., p < 0.0001), and incubation with the Ca2+ entry inhibitor SK&F-96365 reduced migration of cells expressing ITPKA to the level of unstimulated cells (Fig. 7). In H1299 kd cells we found neither a significant EGF stimulation of migration nor a reduction of migration by SK&F-96365. These findings clearly show that in EGF-simulated H299 control cells reduced Ca2+ entry and reduced migration are functionally coupled. It is thus likely that ITPKA-mediated activation of Ca2+ entry represents the migration-stimulating potential attributed to the catalytic activity of ITPKA.
FIGURE 7.
ITPKA activity promotes migration by positive regulation of store-operated calcium influx. Serum-starved cells (0.1% FCS) were seeded in Boyden chambers and treated with 100 nm EGF or with 100 nm EGF and 30 μm SK&F-96365 for 16 h. Migration was expressed as relative amounts with respect to cells grown in 0.1% FCS (control). ***, p < 0.0001.
Impact of ITPKA Expression on Invasion and Metastatic Dissemination
Because the tumor cells expressing high and low levels of ITPKA show significant different migratory potentials, we finally were interested whether the expression level of ITPKA has an influence on metastatic dissemination in vivo. First, we examined a xenograft model using SCID mice. Suspensions of cell lines with H1299 control and H1299 cells stably overexpressing ITPKA (H1299-ITPKA) were injected subcutaneously in the neck of the mice (22). Formation of distant metastases in the lung was examined at different time points (in a period of 4 weeks), when the primary tumors formed at the site of inoculation had about the same size. We found that overexpression of ITKPA increased the number of metastases 3.7-fold (p < 0.005) (Table 3). This result shows that the level of ITPKA expression in H1299 cells influences metastasis formation in the SCID mouse model.
TABLE 3.
Influence of ITPKA expression on formation of metastasis
H1299 control and H1299 cells stably overexpressing ITPKA (H1299-ITPKA) were injected subcutaneously in SCID mice. After 6 weeks, the first tumors had formed. When the primary tumors were about the same size, the mice were sacrificed, and the weight of the primary tumor and the number of lung metastases were determined. The mean ± S.E. of tumor weights and the number of metastases detected in lungs from 9 to 10 mice are shown.
| Cell line | Tumour weight, in grams, mean ± S.E | No. of metastases, mean ± S.E. |
|---|---|---|
| H1299 control | 3.93 ± 0.7 | 24 ± 5.9 |
| H1299-ITPKA | 4.33 ± 0.5 | 89 ± 20 |
| p < 0.53 | p < 0.005 |
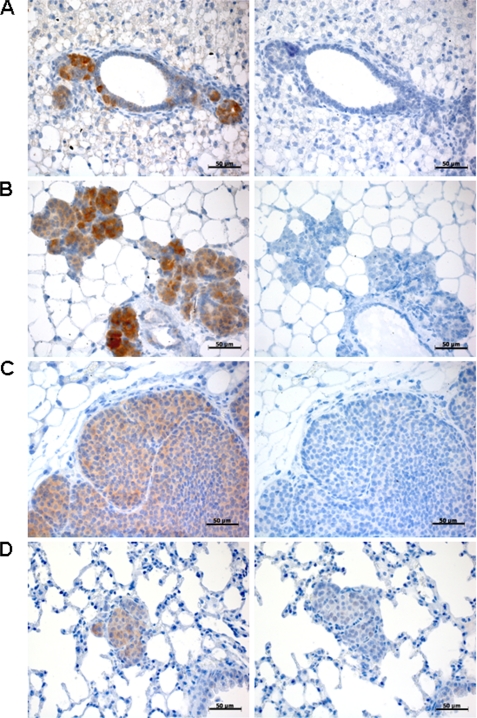
In a second approach, we examined endogenous expression of ITPKA in highly invasive tumors, using the established metastatic tumor model Balb-neuT (21). Females of this mouse strain spontaneously form highly invasive ductal mammary carcinomata that metastasize to the lung due to overexpression of constitutively active ErbB2 in their mammary epithelium. Breast tissues from 5-, 11-, and 20-week-old mice as well lung tissue from 28-week-old mice were paraffin-embedded and examined for expression of ITPKA by immunohistochemistry using an ITPKA-specific antibody. We found high expression of ITPKA during invasion of tumor cells from the primary tumor to the neighboring tissue (Fig. 8, A–C), as well as in distant metastasis (Fig. 8D). Together with the finding that high levels of ITPKA increase invasiveness and motility of tumor cells (Fig. 1 and Table 3), this result indicates that ITPKA plays an important role in invasion and metastatic dissemination.
FIGURE 8.
ITPKA expression in highly invasive mouse tumors. Paraffin sections of Balb-neuT tissue were stained for ITPKA (left panels), whereas normal goat IgG was used as a negative control (right panels). Shown are sections of breast tissues of 5-week-old (A), 11-week-old (B), 20-week-old mice (C), and lung metastasis from 24-week-old mice (D). Scale bars, 50 μm.
DISCUSSION
Cell motility is essential for tumor cell invasion, and it is thought to be a key event in metastasis, the primary cause of death in cancer patients. Our study shows that ectopic expression of ITPKA observed in certain cell types of highly invasive tumor cells (Fig. 1 and Fig. 8) is critical for invasiveness and migration (Fig. 1D and Table 3), and it demonstrates that in these cells ITPKA controls migration by both an enzyme activity-independent and an activity-dependent mechanism. Independent of its InsP3 kinase activity, high levels of ITPKA induce the formation of large cellular protrusions (Fig. 2 and Table 1), which are required for migrating cells to adhere to the substrate and to provide the force to pull the cell body forward (28). Obviously, ITPKA mediates the induction of cell processes by direct interaction with F-actin. The F-actin binding activity of ITPKA bundles and stabilizes actin filaments, resulting in reduced actin dynamics (Fig. 3, A–F) and increased levels of cellular F-actin (Table 2). This observation was confirmed by Johnson and Schell (25) who analyzed the effect of the isolated ABD of ITPKA on actin dynamics. In addition to our results, they reveal that the ABD bundles F-actin by formation of homodimers. For F-actin bundling proteins, it is characteristic that they induce alterations in actin dynamics (29), as their binding to actin filaments decreases annealing of new actin monomers and inhibits dissociation of actin molecules. Based on these conclusions, we assume that the migration-promoting effect of ITPKA in growth factor-poor media results from its interaction with the actin cytoskeleton.
Because in growth factor-simulated H299 cells the maximal migration-stimulating effect of ITPKA was only observed in cells expressing the catalytically active enzyme form, a second mechanism of ITPKA-induced cell migration being dependent on the formation of Ins(1,3,4,5)P4 and/or removal of Ins(1,4,5)P3 by ITPKA was hypothesized and indeed found in our own experiments. In comparison with ITPKA knockdown cells, cells with ITPKA expression responded to EGF-mediated phospholipase Cγ stimulation with a longer lasting Ins(1,4,5)P3-triggered Ca2+ release followed by a second prolonged phase of Ca2+ increase due to SOCE (Fig. 5B). Obviously, this prolonged Ca2+ signal was required for EGF-induced migration as we found that only migration of cells expressing ITPKA was stimulated by EGF, and we demonstrated that this EGF-induced migration was inhibited by the Ca2+ entry blocker SK&F-96365 (Fig. 7). Accordingly, a recent report also showed that SOCE is required for serum-induced migration of MDA231 cells (26). ITPKA-mediated activation of SOCE seems to be functionally coupled to the product of ITPKA, Ins(1,3,4,5)P4, that is discussed to activate SOCE. Ins(1,3,4,5)P4 has a more than 10-fold higher affinity for 5-PPT than Ins(1,4,5)P3 and thus protects Ins(1,4,5)P3 against dephosphorylation. This protection prolongs the effect of Ins(1,4,5)P3 on Ca2+ release from intracellular stores and is sufficient to keep Ca2+ stores depleted, thus inducing activation of SOCE (27). In agreement with this model, we observed prolonged Ins(1,4,5)P3-induced Ca2+ release in H1299 control cells as compared with H1299 kd cells (Fig. 5B), and we determined that increased Ins(1,3,4,5)P4 levels found in H1299 control cells (Fig. 5A) inhibit 5-phosphatase activity with an apparent Ki of 0.71 μm (Fig. 6B). Based on these data, we suggest that in EGF-stimulated tumor cells expressing ITPKA the catalytic activity of ITPKA promotes migration by activation of SOCE and thus factor-dependently enhances the factor-independent effect induced by the N-terminal actin binding domain of ITPKA.
We define here for the first time two functional activities of ITPKA increasing invasive migration of tumor cells, one mediated by the N-terminal actin binding domain via stabilizing and bundling of F-actin and the other via Ins(1,3,4,5)P4 generation mediated by the catalytic domain. ITPKA thus becomes a novel promising target for a therapy directed against invasion and metastasis formation of primary tumors. Lead structures for inhibitors of the catalytic activity of ITPKA have already been identified by our group (30). A complete and isoform-specific down-regulation of ITPKA might be feasible by adenovirus-mediated knockdown approaches (31).
Acknowledgments
We are grateful to Albrecht Wegner for help with measurement of actin polymerization, and we thank Susanne Giehler for excellent technical assistance. We also thank the bachelor course of Life Science, Modul 18, 2009, for help by reproducing the results obtained with actin-bundling assays.
This work was supported by the Roggenbuck-Stiftung Hamburg.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- Ins(1,4,5)P3
- inositol 1,4,5-trisphosphate
- ITPKA
- inositol 1,4,5-trisphosphate 3-kinase-A
- HPLC
- high pressure liquid chromatography
- FCS
- fetal calf serum
- PBS
- phosphate-buffered saline
- EGF
- epidermal growth factor
- SOCE
- store-operated Ca2+ entry
- CaM
- calmodulin
- GFP
- green fluorescent protein
- ITPK
- 1,4,5-trisphosphate 3-kinase
- GST
- glutathione S-transferase
- 5-PPT
- inositol phosphate 5-phosphatase
- IPMK
- inositol polyphosphate multikinase
- ABD
- actin binding domain
- Ins(1,3,4,5)P4
- inositol-1,3,4,5-tetrakisphosphate
- shRNA
- short hairpin RNA
- kd
- knockdown.
REFERENCES
- 1.Comoglio P. M., Trusolino L. (2002) J. Clin. Invest. 109, 857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard T. D. (2007) Annu. Rev Biophys. Biomol. Struct. 36, 451–477 [DOI] [PubMed] [Google Scholar]
- 3.Yamashiro S., Yamakita Y., Ono S., Matsumura F. (1998) Mol. Biol. Cell 5, 993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popowicz G. M., Schleicher M., Noegel A. A., Holak T. A. (2006) Trends Biochem. Sci. 7, 411–419 [DOI] [PubMed] [Google Scholar]
- 5.Stossel T. P., Chaponnier C., Ezzell R. M., Hartwig J. H., Janmey P. A., Kwiatkowski D. J., Lind S. E., Smith D. B., Southwick F. S., Yin H. L. (1985) Annu. Rev. Cell Biol. 1, 353–402 [DOI] [PubMed] [Google Scholar]
- 6.Feldner J. C., Brandt B. H. (2002) Exp. Cell Res. 272, 93–108 [DOI] [PubMed] [Google Scholar]
- 7.Welch H. C., Coadwell W. J., Stephens L. R., Hawkins P. T. (2003) FEBS Lett. 546, 93–97 [DOI] [PubMed] [Google Scholar]
- 8.Hall A. (2005) Biochem. Soc. Trans. 33, 891–895 [DOI] [PubMed] [Google Scholar]
- 9.DesMarais V., Ghosh M., Eddy R., Condeelis J. (2005) J. Cell Sci. 118, 19–26 [DOI] [PubMed] [Google Scholar]
- 10.Gremm D., Wegner A. (2000) Eur. J. Biochem. 267, 4339–4345 [DOI] [PubMed] [Google Scholar]
- 11.Shears S. B. (2004) Biochem. J. 377, 265–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanweyenberg V., Communi D., D'Santos C. S., Erneux C. (1995) Biochem. J. 306, 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schell M. J., Erneux C., Irvine R. F. (2001) J. Biol. Chem. 276, 37537–37546 [DOI] [PubMed] [Google Scholar]
- 14.Jun K., Choi G., Yang S. G., Choi K. Y., Kim H., Chan G. C., Storm D. R., Albert C., Mayr G. W., Lee C. J., Shin H. S. (1998) Learn Mem. 5, 317–330 [PMC free article] [PubMed] [Google Scholar]
- 15.Kim I. H., Park S. K., Sun W., Kang Y., Kim H. T., Kim H. (2004) Brain Res. Mol. Brain. Res. 124, 12–19 [DOI] [PubMed] [Google Scholar]
- 16.Benz C. C., Scott G. K., Sarup J. C., Johnson R. M., Tripathy D., Coronado E., Shepard H. M., Osborne C. K. (1992) Breast Cancer Res. Treat. 24, 85–95 [DOI] [PubMed] [Google Scholar]
- 17.Brandt B., Heyder C., Gloria-Maercker E., Hatzmann W., Rötger A., Kemming D., Zänker K. S., Entschladen F., Dittmar T. (2005) Semin. Cancer Biol. 15, 387–395 [DOI] [PubMed] [Google Scholar]
- 18.Woodring P. J., Garrison J. C. (1996) Biochem. J. 319, 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roesch A., Vogt T., Stolz W., Dugas M., Landthaler M., Becker B. (2003) Melanoma Res. 13, 503–509 [DOI] [PubMed] [Google Scholar]
- 20.Windhorst S., Blechner C., Lin H. Y., Elling C., Nalaskowski M., Kirchberger T., Guse A. H., Mayr G. W. (2008) Biochem. J. 414, 407–417 [DOI] [PubMed] [Google Scholar]
- 21.Muller W. J., Sinn E., Pattengale P. K., Wallace R., Leder P. (1988) Cell 54, 105–115 [DOI] [PubMed] [Google Scholar]
- 22.Jojovic M., Schumacher U. (2000) Cancer Lett. 152, 151–156 [DOI] [PubMed] [Google Scholar]
- 23.Katsantonis J., Tosca A., Koukouritaki S. B., Theodoropoulos P. A., Gravanis A., Stournaras C. (1994) Cell Biochem. Funct. 12, 267–274 [DOI] [PubMed] [Google Scholar]
- 24.Sauer K., Huang Y. H., Lin H., Sandberg M., Mayr G. W. (2009) Current Protocols in Immunology, pp. 11.1.1–11.1.46, Wiley Interscience, New York: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson H. W., Schell M. J. (2009) Mol. Biol. Cell 20, 5166–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S., Zhang J. J., Huang X. Y. (2009) Cancer Cell 15, 124–134 [DOI] [PubMed] [Google Scholar]
- 27.Hermosura M. C., Takeuchi H., Fleig A., Riley A. M., Potter B. V., Hirata M., Penner R. (2000) Nature 408, 735–740 [DOI] [PubMed] [Google Scholar]
- 28.Le Clainche C., Carlier M. F. (2008) Physiol. Rev. 88, 489–513 [DOI] [PubMed] [Google Scholar]
- 29.Murray J. W., Edmonds B. T., Liu G., Condeelis J. (1996) J. Cell Biol. 135, 1309–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayr G. W., Windhorst S., Hillemeier K. (2005) J. Biol. Chem. 280, 13229–13240 [DOI] [PubMed] [Google Scholar]
- 31.Krom Y. D., Fallaux F. J., Que I., Lowik C., van Dijk K. W. (2006) BMC Biotechnol. 28, 6–11 [DOI] [PMC free article] [PubMed] [Google Scholar]