Abstract
The angiotensin converting enzyme (ACE) catalyzes the extracellular formation of angiotensin II, and degradation of bradykinin, thus regulating blood pressure and renal handling of electrolytes. We have previously shown that exogenously added ACE elicited transcriptional regulation independent of its enzymatic activity. Because transcriptional regulation generates from protein-DNA interactions within the cell nucleus we have investigated the initial cellular response to exogenous ACE and the putative internalization of the enzyme in smooth muscle cells (SMC) and endothelial cells (EC). The following phenomena were observed when ACE was added to cells in culture: 1) it bound to SMC and EC with high affinity (Kd = 361.5 ± 60.5 pm) and with a low binding occupancy (Bmax = 335.0 ± 14.0 molecules/cell); 2) it triggered cellular signaling resulting in late activation of focal adhesion kinase and SHP2; 3) it modulated platelet-derived growth factor receptor-β signaling; 4) it was endocytosed by SMC and EC; and 5) it transited through the early endosome, partially occupied the late endosome and the lysosome, and was localized to the nuclei. The incorporation of ACE or a fragment of it into the nuclei reached saturation at 120 min, and was preceded by a lag time of 40 min. Internalized ACE was partially cleaved into small fragments. These results revealed that extracellular ACE modulated cell signaling properties, and that SMC and EC have a pathway for delivery of extracellular ACE to the nucleus, most likely involving cell surface receptor(s) and requiring transit through late endosome/lysosome compartments.
Keywords: Cell/Endocytosis, Cell/Sorting, Cell/Trafficking, Nucleus, Protein/Intracellular Trafficking, Receptors
Introduction
Two isoforms of the secretory peptidyl dipeptidase angiotensin-converting enzyme (ACE)3 are present in mammals: somatic ACE and testicular ACE. These enzymes are both type I transmembrane proteins consisting of an extended N-terminal extracellular domain and a short C-terminal cytoplasmic domain (1). Both isoforms are derived from a single gene by alternative use of transcription initiation sites (2). The two active sites of somatic ACE are located in the extracellular portion of the molecule (3). Particularly relevant to this study is a well known process whereby the enzymatic cleavage of the entire N-terminal extracellular portion of ACE generates the so called “soluble form” of the enzyme that is released into circulation (1, 4).
Among the various peptide substrates of ACE in the extracellular space are angiotensin I leading to formation of angiotensin II, the effector hormone of the renin-angiotensin system, and bradykinin, which is degraded to inactive metabolites. As both are major contributors to cardiovascular regulation, inhibition of ACE has become the standard therapy for hypertension, ischemic heart disease, and heart failure (6).
The classical peptidyl dipeptidase-dependent role of ACE proteins in the renin-angiotensin system has been recently broadened to areas of cell surface activity and transcriptional regulation. Thus, ACE has been shown to manifest a glycosylphosphatidylinositol (GPI)-anchored protein releasing activity (GPIase activity), which is independent of its peptidase activity and sheds various GPI-anchored proteins from the cell surface by cleavage within the GPI moiety, and is associated with its role in fertilization (7). It is not known if ACE acts directly on GPI-anchored proteins, or indirectly through another protein(s) (8). The formation of a complex between ACE and bradykinin B2 receptors was recently reported (9). Also membrane-bound ACE has been shown to function as a receptor, which upon activation by binding of ACE inhibitors (10, 11), triggers a signal transduction pathway leading to changes in gene expression (12, 13).
We have recently reported that exogenous ACE added to smooth muscle cells (SMC) resulted in transcriptional stimulation of the genes of bradykinin receptors B1 and B2 after 3–4 h (14). We hypothesized that this transcriptional regulation by exogenous ACE may involve its nuclear localization. An early immunohistochemistry study showed that when the ACE cDNA was transfected into intact rat carotid arteries, ACE protein was localized in the SMC as well as in the intima endothelial cells (EC) (15). A fact not mentioned in this report was that ACE appeared to be localized in the nuclei of SMC and EC.4 In another early report an elevated endogenous expression of ACE in the abdominal aorta neointima SMC was observed in response to experimentally induced vascular injury (16). Remarkably, this injury-induced increment in ACE protein and activity was restricted to injured tissue and was not detected in serum or lung ACE levels. Thus, it is possible that SMC and EC can be exposed to locally produced exogenous ACE under certain pathophysiological conditions such as vascular injury. In a more recent report (17) using the 9B9 monoclonal antibody, ACE was detected inside cell nuclei of mesangial cells from spontaneously hypertensive rats where it colocalized with angiotensin II and angiotensin III. However, these previous reports neither enquire on the origin of the nuclear ACE, nor did they focus on the molecular mechanisms involved.
Here we present characterization of the initial, intermediate, and late events of the interaction of exogenous ACE with SMC and human umbilical vein endothelial cells (HUVEC). The binding of ACE to the cell surface was succeeded by its endocytosis to the early and late endosomes, the lysosome, and the nuclei. Nuclear localization of exogenous ACE coincided with changes in cell signaling response manifested as late activation of focal adhesion kinase (FAK) and SHP2, as well as modulation of PDGFR-β signaling.
EXPERIMENTAL PROCEDURES
Materials
125I-Monoiodo Bolton-Hunter reagent was from Amersham Biosciences. ACE was purified from pig kidney cortex as described elsewhere (18). BSA (≥98.0% pure) was purchased from Sigma. Rabbit polyclonal antibodies against phosphoproteins of the PDGFR-β signal transduction pathway are described in the text. The source of other reagents are as indicated.
Cell Culture Preparation
Rat aorta SMC were isolated and expanded as described (14). Cells were seeded at a density of 7.5 × 104 cell/ml in 2 ml of medium per well of a 6-well plate. Freshly isolated cells were cultured in Dulbecco's modifies Eagle's medium (DMEM) (Invitrogen) supplemented with 10% bovine calf serum, 1 mm sodium pyruvate (Invitrogen), 0.1 mm non-essential amino acids (Invitrogen), and 1% penicillin/streptomycin (Invitrogen). The normal rat lung fetal fibroblast cell line RFL-6 (American Type Culture Collection) was cultured and propagated as described above for SMC. Pooled HUVEC (Lonza Walkersville Inc.) were cultured and propagated as described above for SMC in complete endothelial basal medium 2. Cell counting was carried out by hemocytometry in triplicate cultures using the trypan blue exclusion method.
Iodination of ACE and Binding of 125I-ACE to Cell Monolayers
ACE was iodinated using the Bolton-Hunter reagent (19). Briefly, 50 μg of freeze-dried, affinity purified ACE was resuspended in 50 μl of 100 mm sodium borate, pH 8.5, and added to [125I]monoiodo-Bolton-Hunter reagent (1000 μCi), which had been dried under nitrogen and incubated for 15 min at 4 °C. 200 μl of quenching solution (0.1 m sodium borate, pH 8.5, and 0.2 m glycine) was added and the mixture incubated for an additional 10 min at 4 °C followed by addition of 50 μl of 100 mm sodium borate containing 5 mg/ml of BSA. Two 5-μl aliquots of the mixture were used to assess the efficiency of iodination and the remaining 295 μl was loaded onto a 5-ml d-salt polyacrylamide desalting column (6 kDa cut-off, Pierce) that had been equilibrated in phosphate-buffered saline (PBS) containing 1 mg/ml of BSA. Fractions of 500 μl were collected and 125I-ACE was eluted in the first peak of radioactivity (fractions 4th to 6th) with 25,000 cpm/ng of specific radioactivity. 125I-ACE retained around 70% of its enzymatic activity. BSA (50 μg/50 μl of 100 mm borate buffer, pH 8.5) was iodinated under similar conditions, except that BSA was not included in the buffers used after addition of the quenching solution. Binding of 125I-ACE was measured in monolayers of cells grown in 24-well cluster culture plates. Cells were seeded on the plates at a density of 5 × 104 cells/well, grown to near confluence, starved in serum-free medium for 48 h, washed three times with cold binding buffer (25 mm Hepes, pH 7.4, 125 mm NaCl, 5 mm MgSO4, 5 mm KCl, and 1 mm CaCl2, containing 2 mg/ml of BSA), and incubated in 1 ml of binding buffer for 30 min at 4 °C. The binding buffer was aspirated and 1 ml of fresh binding buffer containing the indicated concentrations of 125I-ACE at a specific radioactivity of 88,000 cpm/ng was added and further incubated at 4 °C for 4 h. After washing four times with 1 ml of binding buffer at 4 °C, cells were solubilized with 1 ml of 1% Triton X-100, 10% glycerol, 25 mm Hepes, pH 7.5, 1 mg/ml of BSA. Radioactivity was measured in a γ-counter. Nonspecific binding was determined in the presence of unlabeled ACE at 500 ng/ml. Specific binding was obtained by subtracting nonspecific binding from total binding. The dissociation constant (Kd) and maximum binding capacity (Bmax) for 125I-ACE were determined by least squares non-linear regression fitting of the hyperbolic function for ligand binding to one-site saturation. Cell numbers were determined in triplicate parallel cultures by trypsinization and counting the detached cells in a hemocytometer.
Gel Filtration Chromatography of ACE and 125I-ACE
To assess if iodination of ACE induced polymerization (aggregation), the native molecule (0.5 μg) and its iodinated form (125I-ACE containing 105 cpm) were subjected to gel filtration chromatography in a HiPrep Sephacryl S-300 using aprotinin (6.5 kDa), cytochrome c (12.4 kDa), carbonic anhydrase (28 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), and apoferritin (481 kDa) as molecular mass markers. The column was equilibrated and resolved in PBS, pH 7.4. The elution patterns of native ACE and 125I-ACE were monitored by determining ACE activity and radioactivity, respectively. ACE activity was assayed in medium (50 μl) containing 100 mm Tris-HCl, pH 7.0, 50 mm NaCl, 10 μm ZnCl2, and the quenched fluorogenic substrate Abz-XXK(Dnp)P (10 μm). The activity was monitored continuously in a fluorescence microplate reader at 320/420 nm.
ACE Signaling
The activation of SHP2, AKT, FAK, PDGFR-β, and ERK1/ERK2 (p44/42 MAPK) by exogenously added ACE to cell monolayers was assessed by monitoring their phosphorylated state using specific anti-phosphoprotein. Cells grown to confluence on 6-well plates were incubated for 24 h in serum-free medium and preincubated without or with 10 nm ACE for 120 min. Then cells were stimulated with: 10 nm ACE, 2 nm PDGF-BB, or 2 nm PDGF-BB in the presence of 10 nm ACE, for the indicated times at 37 °C in a CO2 incubator. The reaction was stopped by aspirating the medium, and cells were immediately lysed with 200 μl/well of extraction buffer containing 20 mm Tris, pH 7.5, 5 mm EGTA, 150 mm NaCl, 1% Nonidet P-40, 0.1 mm Na3VO4, 1 mm sodium fluoride, 10 mm sodium β-glycerophosphate, 0.5 mm phenylmethylsulfonyl fluoride, 1 μl/ml of protease inhibitor mixture (Sigma). Cell extracts were sonicated to break down genomic DNA, clarified by centrifugation at 10,000 × g for 10 min at 4 °C, and the supernatants were saved. Protein concentrations were determined by the bicinchoninic acid method (Pierce) using BSA as the standard. Total protein extracts (30 μg/lane) were resolved by 4.0–20.0% gradient SDS-polyacrylamide gel electrophoresis. Proteins were transferred to polyvinylidene difluoride membranes in 25 mm Tris, 192 mm glycine, 20% methanol using a Mini Trans-Blot blotter (Bio-Rad) at 30 V overnight and under continuous cooling at 4 °C. Blots were blocked in PBS buffer containing 0.1% Tween 20 and 5% (w/v) nonfat dry milk for 1 h and then incubated overnight with gentle agitation at 4 °C with a mixture of the anti-phosphoprotein antibodies, anti-P-SHP2, anti-P-AKT, anti-P-ERK1/ERK2 (p44/42 MAPK), anti-P-FAK, and anti-P-PDGFR-β, each diluted 1:1000 in PBS buffer containing 0.1% Tween 20 (PBS/T) and 5% (w/v) BSA. Membranes were washed three times for 5 min each with PBS/T buffer and incubated for 1 h with horseradish peroxidase-conjugated, goat anti-rabbit IgG antibody (1:2000) in PBS/T, 5% (w/v) BSA. After washing four times for 10 min each with PBS/T, membranes were developed with a chemiluminescence substrate for detection of horseradish peroxidase (Pierce) and images were captured in a Chemi Doc XRS imager (Bio-Rad) using the Quantity One version 4.6 imaging software.
Preparation of TRITC-ACE and TRITC-BSA
ACE and BSA (90 μg/200 μl) were diluted in 200 mm sodium bicarbonate, pH 9.0, and reacted with 0.5 mg of TRITC for 1 h at room temperature with gentle shaking. The reaction was terminated by the addition of 100 μl of 1.5 m hydroxylamine, with further incubation for 1 h at room temperature. Finally, TRITC-ACE and TRITC-BSA were separated from unreacted TRITC by two passages through a 5-ml d-salt polyacrylamide desalting column as described above. The resulting TRITCT-ACE retained around 80% of its enzymatic activity.
Fluorescent Microscopy
Cells were grown in the wells of the Nunc Lab-Tek II CC2 Chamber Slide System to ∼80% confluence under the conditions as described herein, including starvation in serum-free medium and washing steps. Cells were fixed in PBS containing 3% formaldehyde for 20 min at room temperature, washed three times with PBS, incubated for 10 min at room temperature in PBS containing 10 mm ammonium chloride to quench the excess of formaldehyde, and then washed twice with PBS. For detection of late endosome and lysosome, fixed cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min at room temperature, blocked with 10% fetal bovine serum in PBS, and incubated overnight at 4 °C with 1:200 dilution of the commercially available primary antibodies indicated in the corresponding figures. Then slides were incubated with 1:1000 dilution of FITC-labeled commercially available secondary antibodies for 1 h at room temperature and rinsed three times with PBS solution. Slides were mounted on Vectashield (Vector Laboratories) mounting solution containing DAPI and fluorescent images were captured using a ×60 NA 0.5–1.25 oil objective on an inverted Nikon TE-2000 microscope with a Photometrics Cool SNAP HQ camera. Images were captured using NIS-Elements software (Nikon) and processed with a three-dimensional deconvolution plug-in (Media Cybernetics) in ImageJ. Time lapse live cell imaging was performed using the Live Cell device (Pathology Devices) adapted to the Nikon TE-2000 microscope for feedback control of temperature, CO2, and humidity. Images presented in the figures are representative of 15 fields observed by scanning the entire slide area. Imaging was done in the BUMC Cellular Imaging Core Facility.
125I-ACE in Subcellular Fractions and Nuclei Purified by Isopicnic Sedimentation
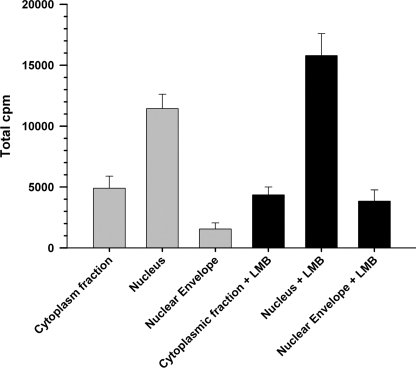
SMC were grown to 80% confluence, serum starved for 24 h in serum-free DMEM, and incubated in the same medium with 100 μCi of [3H]thymidine at 37 °C or with 10 nm (100 μCi) 125I-ACE for 120 min at 37 and 4 °C. In some experiments cells were preincubated for 30 min with inhibitors of endocytosis before incubation with 125I-ACE. The inhibitors used were: 3 μm phenylarsine oxide, 100 μg/ml of concanavalin A, 10 mm methyl-β-cyclodextrin, 200 μm monodansyl cadaverin, 10.0 μm amiloride, 10.0 μg/ml of dextran sulfate, 2.0 μg/ml of polyinosine, and 20.0 μg/ml of fucoidan. Cells were washed three times with Hanks' balanced salt solution (HBSS) and disrupted mechanically by homogenization in a Dounce homogenizer (20 strokes on ice) in an isotonic grinding medium (0.5 ml/60-mm plate) containing 250 mm sucrose, 25 mm KCl, 5 mm MgCl2, 20 mm Hepes, pH 7.9, Sigma protease inhibitor mixture (50 μl/20 ml), 200 μg/ml of phenylmethylsulfonyl fluoride in dimethyl sulfoxide (50 μl/20 ml), and 1% Nonidet P-40 detergent. Nuclei and cell debris were recovered by centrifugation for 10 min at 1,000 × g. The supernatant, referred to as the cytoplasmic fraction, was saved at 4 °C. The resulting pellet was gently re-suspended in 500 μl of isotonic grinding medium and loaded on top of a discontinuous Opti-Prep gradient by diluting the original 60% solution in medium containing 150 mm KCl, 30 mm MgCl2, and 120 mm Tricine, pH 7.8. The gradient was composed of three layers of 15 (2 ml), 20 (5 ml), and 35% (2 ml) loaded on a 14-ml Sorvall SH80 rotor tube. Samples were spun down for 60 min at 10,000 × g and purified nuclei were recovered from the 20/35% interface. Finally, purified nuclei were diluted 10 times with grinding buffer without detergent and recovered by centrifugation at 5,000 × g for 15 min. The resulting pellet was suspended in a minimal volume of grinding medium without detergent and the number of nuclei was determined in 10 μl of this suspension after adding 10 μm DAPI. Nuclei were counted in a hemocytometer chamber using a fluorescent Nikon Eclipse E400 microscope. Alternatively protein content of the purified nuclei was determined using the bicinchoninic acid (BCA) method (Pierce). This method was also applied to the study of subcellular distribution of internalized 125I-ACE and the effect of 3 nm leptomycin B (LMB) on nuclear localization of 125I-ACE presented in Fig. 8. In those studies cells were incubated with 10 nm ACE containing 6.0 × 105 cpm.
FIGURE 8.
Effect of leptomycin B on nuclear localization of ACE. Nuclei, cytoplasmic fraction, and nuclear envelopes from SMC incubated with 125I-ACE were prepared as described under “Experimental Procedures.” Subcellular fractions were obtained from untreated cells (gray bars) and cells treated with 3 nm LMB (black bars). Error bars are the S.E. ± mean values of three determinations (n = 3).
Effect of Endocytosis Inhibitors on 125I-BSA Internalization
SMC were incubated with 10 nm (100 μCi) 125I-BSA for 120 min at 37 °C, in the absence (control) or presence of the indicated inhibitors, as described above for internalization of 125I-ACE. Non-internalized 125I-BSA was eliminated by three washes with HBSS medium at 4 °C. Cell were solubilized with 1 ml of 1% Triton X-100, 10% glycerol, 25 mm Hepes, pH 7.5, 1 mg/ml of BSA, and radioactivity was measured in a γ-counter.
Preparation of Nuclear Envelope
Nuclei from SMC cells were prepared as described above. The nuclei (500 μg of protein) were suspended in 1.0 ml of ice-cold buffer containing 10 mm Tris-HCl, pH 7.4, 10 mm NaH2PO4/Na2HPO4, pH 7.4, Sigma protease inhibitor mixture (50 μl/20 ml), 200 μg/ml of phenylmethylsulfonyl fluoride in dimethyl sulfoxide (50 μl/20 ml), 250 μg/ml of heparin, 1 mm Na3VO4, 10 mm sodium fluoride, and 400 units of Benzon Nuclease (Merck). After mild stirring for 90 min at 4 °C, nuclear envelopes were sedimented by centrifugation (10,000 × g) for 30 min at 4 °C and resuspended in a buffer containing 0.25 buffer (20 mm Tris-HCl, pH 7.4, 0.25 m sucrose, 5 mm MgSO4, 1 mm Na3VO4, and the protease inhibitors indicated above). Protein concentrations were determined by the BCA method (Pierce).
Kinetics of 125I-ACE Nuclear Localization
SMC were grown in 60-mm plates to ∼80% confluence, rinsed twice with HBSS, and starved for 24 h in serum-free DMEM. After addition of fresh serum-free DMEM, ACE was added at a final concentration of 10 nm containing 2.0 × 106 cpm of 125I-ACE and incubated for the indicated times (see Fig. 11) at 37 °C in a CO2 incubator. Nuclei were isolated and counted as described above. The radioactivity in samples containing 105 nuclei was measured in a γ-counter.
FIGURE 11.
Transactivation domains in the ACE sequence. The primary sequence of somatic pig porcine kidney ACE (GenBank accession number EF121312.1) used in these studies was analyzed for 9-amino acid TAD consensus sequence and cathepsin D cleavage sites using the ExPASy Proteomic tools. Cathepsin D cleavage sites (in italics and highlighted in gray), 6-amino acid TAD (highlighted in black), and ACE active site (bold and underlined in the 10.0-kDa peptide) are indicated. A leucine-rich nuclear export signal predicted in the 8.7-kDa cathepsin fragment (DINYLLKMALE) is underlined.
Analysis of ACE Degradation
For in vitro analysis of ACE degradation by cathepsin D, 125I-ACE (10 ng, containing 25,000 cpm/ng) was incubated with 0.1 units of cathepsin D (R&D Systems) in 25 μl of 100 mm glycine HCl, pH 4.5, at 37 °C for the indicated time intervals (see Fig. 12). The reaction was terminated by the addition of 2 μl of 100 mm phenylmethylsulfonyl fluoride in dimethyl sulfoxide and 10 μl of SDS-PAGE sample buffer containing 50 mm dithiothreitol. Samples (35 μl) were resolved in a 4.0–20.0% gradient SDS-polyacrylamide electrophoresis gel. The analysis of intracellular degradation of ACE was carried out as follows: SMC were grown to 80% confluence in 10-mm plates, serum-starved for 24 h, and rinsed three times with HBSS buffer and serum starved as indicated above. Cells were preincubated with pepstatin A (10 μg/ml) in serum-free DMEM or DMEM alone (control) for 2 h at 37 °C in a CO2 incubator. Then 125I-ACE (400 ng/ml, containing 2,000 cpm/ng) was added to the cells and further incubated for an additional 120 h. Cell plates were placed at 4 °C and rinsed four times with cold HBSS medium to eliminate the bulk of the extracellular 125I-ACE. Finally, cell monolayers were dispersed and their total protein content determined as indicated above. Samples (30 μg) in SDS-polyacrylamide gel electrophoresis sample buffer containing 50 mm dithiothreitol were resolved in 4.0–20.0% gradient SDS-polyacrylamide electrophoresis gels. Gels were dried and analyzed by autoradiography at −80 °C using a Bio-Max TranScreen HE (Kodak).
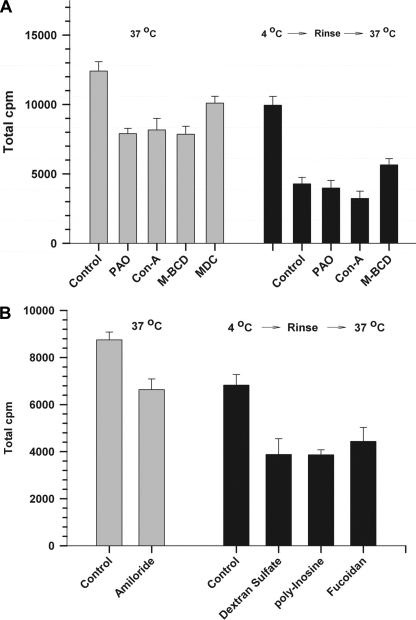
FIGURE 12.
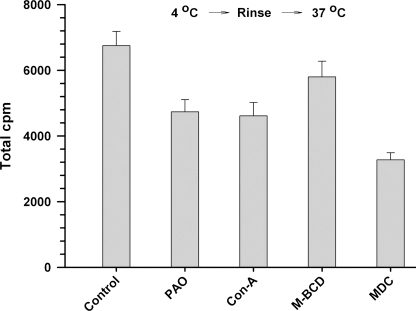
Nuclear localization of 125I-ACE is impaired by selective inhibitors of endocytosis. A, content of 125I-ACE in nuclei (106) that were isolated from cells incubated with 125I-ACE in the absence (control) or presence of endocytosis inhibitors of phenylarsine oxide (PAO), concanavalin A (Con-A), methyl-β-cyclodextrin (M-BCD), and momodansyl cadaverin (MDC), as described under “Experimental Procedures.” 125I-ACE in the nuclei of control cells and cells were incubated with the inhibitors at 37 °C (gray bars). 25I-ACE in the nuclei of control cells and cells were incubated with the inhibitors at 4 °C and then transferred to 37 °C after elimination of unbound 25I-ACE (black bars). B, as in A except that the inhibitors tested were amiloride, dextran sulfate, polyinosine, and fucoidan. Error bars are the S.E. ± mean of three determinations (n = 3).
RESULTS
ACE Binds to Vascular SMC with High Affinity
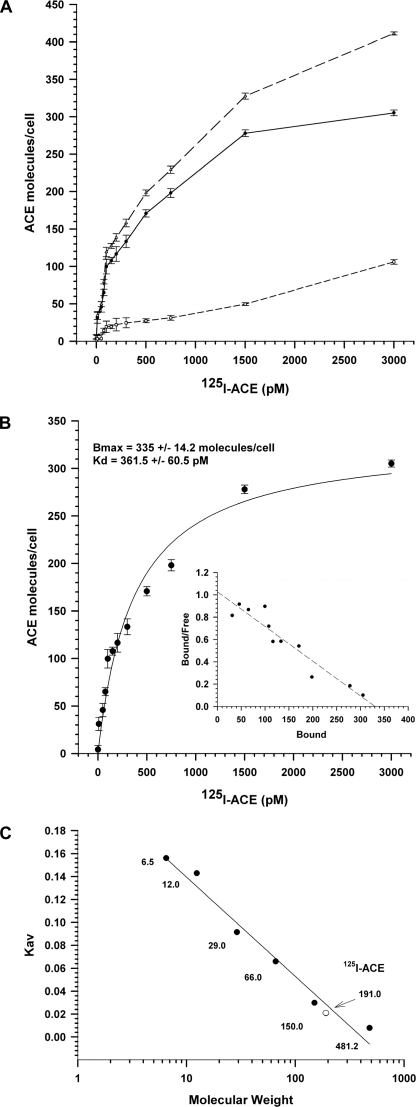
The binding of 125I-ACE to SMC monolayers was studied in the absence and presence of excess non-labeled ACE, to determine the characteristics of total binding and nonspecific binding, respectively. The nonspecific binding of 125I-ACE displayed a linear increment, reaching around 25% of the total binding at 3000 pm 125I-ACE (Fig. 1A). The specific binding, obtained by subtraction of nonspecific binding from total binding, displayed the characteristics of a saturable, single binding site process, with affinity (Kd) of 361.5 ± 60.5 pm in the occupancy (Bmax) of 335.0 ± 14.2 molecules/cell (Fig. 1B). These binding parameters are characteristics of a high affinity (picomolar range), low-abundance receptor involved in binding. Scatchard plot analysis revealed a linear correlation (Fig. 1B, inset), suggesting the lack of cooperativity for binding of ACE to the cells. Iodinated ACE was analyzed by gel filtration chromatography to assess a putative acquisition of a quaternary structure induced by iodination, a phenomenon that would have a significant effect on the Kd value. Both ACE and 125I-ACE eluted as a single peak of 191.0 kDa, close to the predicted value of 180 kDa (Fig. 1C). This result suggests that iodination of ACE does not alter its monomeric structure in solution.
FIGURE 1.
ACE binding to SMC. Binding of ACE to SMC monolayers was done using 125I-ACE as described under “Experimental Procedures.” The binding of ACE was measured over a wide concentration range of the ligand (10–3500 pm). A, total binding (upper plot), nonspecific binding (lower plot), and specific binding (middle plot). B, non-linear regression analysis of the specific binding plot from panel A. Bmax and Kd values are shown on the top. The inset is a Scatchard plot of the specific binding. C, gel filtration chromatography of ACE and 125I-ACE using aprotinin (6.5 kDa), cytochrome c (12.4 kDa), carbonic anhydrase (28 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), and apoferritin (481 kDa) as molecular mass markers. Error bars are the S.D ± mean of eight determinations (n = 8).
ACE Signaling Enhances FAK and SHP2 Activation in Vascular SMC
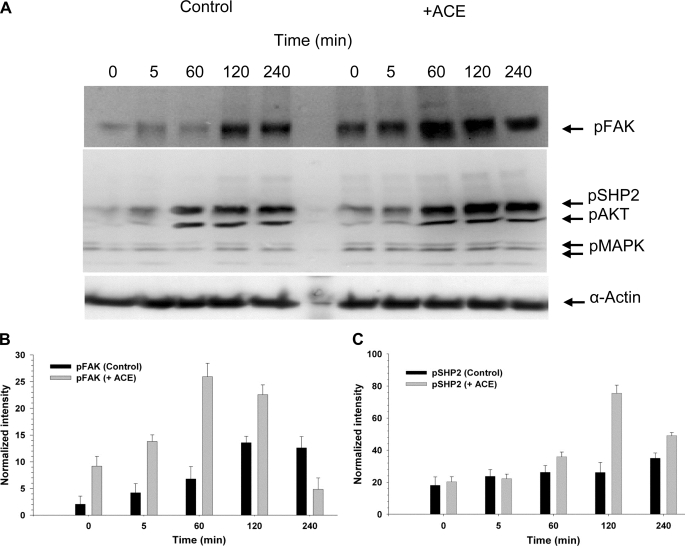
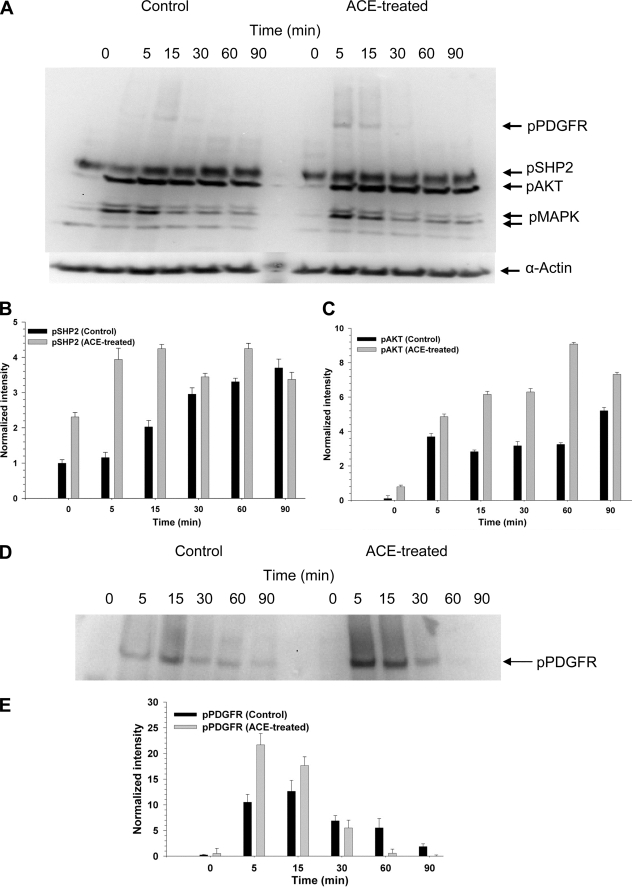
We next studied the possibility that ACE may trigger a signal within the cell by monitoring the putative activation of ubiquitous early responders to various growth factors, hormones, or cytokines. The experiments were designed to examine the effect of exogenous ACE on cells that were preincubated in the presence or absence of ACE for 120 min. We specifically focused on monitoring the putative activation of ubiquitous early responders to various growth factors, hormones, or cytokines. Signaling enzymes SHP2 and FAK were selected. SHP2 is a ubiquitous protein-tyrosine phosphatase containing Src homology 2 (SH2). It is a unique protein-tyrosine phosphatase that promotes activation, rather than down-regulation, of intracellular signaling pathways. SHP2 is considered a key signaling relay of a variety of receptor tyrosine kinases, including the PDGF-R, fibroblast growth factor receptor, and epidermal growth factor receptor (20). FAK is activated by a variety of cell surface receptors and participates in growth factor receptor-mediated signaling pathways, playing key roles in cell survival, proliferation, migration, and invasion (21). SMC that were not preincubated with ACE were then either treated with medium without ACE (control) or with exogenous ACE (+ACE) for the indicated time intervals. The time intervals were chosen to monitor early (5–60 min) and late (120–240 min) stages of activation. In general, background activation of FAK and SHP2 in the absence of added inducer is variable, depending on the cells and treatment preceding the induction assay. We have consistently observed that aorta smooth muscle cells are resilient to reach quiescence under serum deprivation. This reflects on the background level of activation of certain signaling molecules including SHP2 and FAK (Fig. 2A) and excluding others such as AKT and MAPK (see Fig. 3A). Nevertheless, even with a detectable background level of activation, SHP2 and FAK were further activated by exogenous ACE at longer incubation periods (60–240 min) (Fig. 2A). Thus, at 60 min of incubation p-FAK was stimulated around 3.7-fold by ACE (Fig. 2B). pSHP2 was stimulated around 3.2-fold at 120 min (Fig. 2C). Similar levels of stimulation of pFAK and pSHP2 have been reported by others in different cell lines. On the other hand, spontaneous activation of AKT observed at late stages in control SMC was not significantly changed by exogenously added ACE. Cells that were preincubated with ACE for 120 min displayed a similar pattern on FAK and SHP2 activation to the one described above (Fig. 2, +ACE), but in the absence of exogenously added ACE (not shown).
FIGURE 2.
Exogenous ACE triggered a late activation of FAK and SHP2. Analysis of the activation of FAK (125.0 kDa), SHP2 (80.0 kDa), AKT (70.0 kDa), and MAPK (44/42 kDa) was done by monitoring their phosphorylated state as described under “Experimental Procedures.” A, Western blot of total SMC extract, at different times after the addition of serum-free medium (Control) or serum-free medium containing 10 nm ACE, probed with the anti-phosphoprotein antibodies against the proteins indicated on the right-hand side column. The original membrane was cut into three sections, probed with antibodies against the indicated phosphoproteins, recomposed, and developed. Wells between 5 and 60 min, which were left empty, were deleted from the final image. B and C, graphs showing the densitometry plots of the Western blot bands corresponding to the activation of FAK and SHP2, respectively, in the absence and presence of ACE. Error bars are the S.D. ± mean values of eight determinations (n = 3).
FIGURE 3.
ACE enhances the activation of PDGFR-β signaling pathway. Analysis of the activation of signaling enzyme was done as described in the legend to Fig. 2. A, Western blot of total SMC extract, at different times after the addition of PDGF to cells that were not preincubated (Control) or were preincubated with 10 nm ACE for 120 min (ACE-treated). Graphs show the densitometry plots of the Western blot bands corresponding to activation of SHP2 (B) or AKT (C) by PDGF in control and ACE-treated cells. D, a longer exposure of the band corresponding to pPDGFR-β (185 kDa) shown in A. E, densitometry plot of the Western blot bands corresponding to activation of PDGFR-β.
ACE Selectively Enhances the Activation of Early Responders in PDGFR-β Signaling Pathway
We next designed an experiment in which SMC were treated without (control) or with ACE for 120 min and then exposed to PDGF-BB (PDGF henceforth). It is worth recalling that PDGFR-β is activated upon binding of PDGF to its extracellular domain. Following this induction by PDGF several signal transducing enzymes are activated. Some of the early responders in the PDGFR-β signaling pathway are the PDGFR-β, SHP2, and AKT. Activation of PDGFR-β, SHP2, and AKT was enhanced and hastened in cells preincubated with ACE (Fig. 3A). Interestingly, activation of MAPK (ERK1/2), further downstream in the PDGFR-β signaling, was not significantly changed by ACE pretreatment. Fig. 3D shows a longer exposure of the effect of ACE on PDGFR-β activation. Thus, after prolonged incubation (120 min) exogenous ACE helps to propagate PDGFR-β signaling by enhancing and accelerating activation of the early responders, PDGFR-β, SHP2, and AKT.
ACE Localizes to the Nuclei of SMC and HUVEC
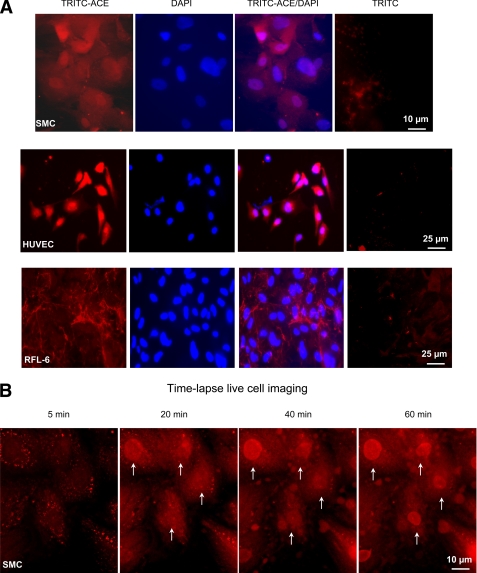
Incubation of SMC with TRITC-ACE for 120 min revealed that TRITC-ACE had reached the nucleus (Fig. 4A). Although some nuclei accumulated ACE, others did not. Some cells also showed a punctuate pattern of fluorescence throughout the cytoplasm, probably corresponding to TRITC-ACE at endosomal compartments. A visual inspection of fluorescence intensity indicates that the steady-state level of ACE accumulated in the nuclei is variable among cells in the field. By scoring 190 nuclei from 15 different fields we estimated that 80 nuclei were not labeled, 40 nuclei displayed a intense signal, and 70 nuclei displayed a normal signal. Thus, the pathway of ACE accumulation in the cell nucleus seems to involve its transit through an endosomal compartment. TRITC-ACE was localized in the nuclei of HUVEC. It seems that the nuclei of HUVEC cells accumulated more TRITC-ACE than the nuclei of SMC. At this point is not clear what controls the level of nuclear ACE in both cells. TRITC-ACE added to the normal rat lung fetal fibroblast cell line RFL-6 was bound to the fibrous material of the extracellular matrix but did not accumulate in the nuclei. Thus, selective nuclear localization of ACE in SMC and HUVEC may require the expression of specific protein receptors in these cells. The incorporation of TRITC-ACE into SMC was also visualized by live fluorescence microscopy (Fig. 4B). Ten minutes after addition of TRITC-ACE, the typical punctuated pattern of the endosomal compartment was observed. At 20 min, a clear increment of TRITC-ACE was observed in the cytoplasm and in one nucleus (Fig. 4, upper left corner, 20 min frame). TRITC-ACE entered most of the nuclei at 60 min. This result indicates that the rate of accumulation of ACE in the nucleus is heterogeneous within the cell population.
FIGURE 4.
ACE is localized to the nuclei of SMC and HUVEC. A, indirect immunofluorescence images were obtained from SMC, HUVEC, and RFL-6 monolayers incubated with 10 nm TRITC-ACE (red) for 120 min as described under “Experimental Procedures.” TRITC-ACE/DAPI is the merging of TRITC-ACE and DAPI images. Scale bars are at the lower right-hand corners. B, time lapse live cell confocal imaging of TRITC-ACE uptake by SMC. Arrows indicate nuclei that are progressively incorporating TRITC-ACE.
Kinetic Analysis of the Nuclear localization of ACE
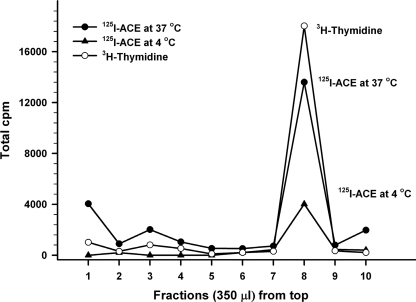
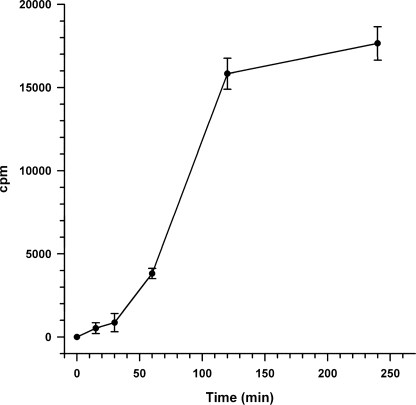
Nuclear localization of ACE was alternatively monitored by quantitation of 125I-ACE recovered in nuclei purified by isopicnic sedimentation in iodixanol (Opti-Prep) gradients from SMC that had been incubated with 125I-ACE. Fig. 5 shows that the accumulation of 125I-ACE in the nucleus (filled circles) was largely prevented when SMC were incubated with 125I-ACE at 4 °C (filled triangles). The nuclear fraction recovered in the 20–35% Opti-Prep interface was ascertained by resolving through the gradient nuclei from SMC whose DNA was labeled with [3H]thymidine during growth. In these studies we have applied the method presented in Fig. 5 to perform a kinetic analysis of the nuclear localization of 125I-ACE. The analysis revealed that incorporation of the enzyme into the nuclei is preceded by a lag time of around 30 min (Fig. 6). This lag time is indicative of a priming event, such a processing of ACE, at some stage during its traffic to the nuclei.
FIGURE 5.
125I-ACE in nuclei purified by isopicnic sedimentation. Nuclei sedimented in the 20–35% Opti-Prep interface (open circles) corresponding to fraction number 8. 125I-ACE nuclear localization at 37 °C (closed circles) was inhibited at 4 °C (closed triangles).
FIGURE 6.
Kinetic of the nuclear localization of ACE. Incubation of SMC with 125I-ACE, isolation of nuclei, and measurement of radioactivity was done as described under “Experimental Procedures.” Error bars are the S.D. ± mean values of eight determinations (n = 6).
Nuclear Co-localization of ACE
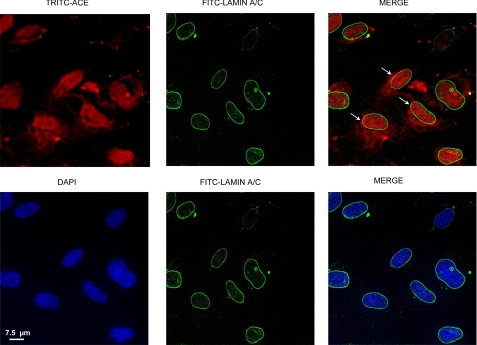
We next studied the possibility that TRITC-ACE could be localized, at least in part, in the nuclear envelope. SMC that were grown in microscope slides were incubated for 120 min with TRITC-ACE and then reacted with anti-lamin A/C antibody for detection of the nuclear envelope by immunofluorescence as described under “Experimental Procedures.” Confocal images of the nuclear envelope protein lamin A/C displayed a typical ring-like, peripheral labeling of the nucleus (Fig. 7). TRITC-ACE showed a limited co-localization with restricted areas of the nuclear envelope (Fig. 7, arrows). As expected, lamin A/C staining did co-localize with DAPI staining (blue) of nuclear DNA.
FIGURE 7.
Localization in the nucleus and nuclear envelope. SMC were grown in slide chambers, incubated with TRITC-ACE, and processed for immunofluorescence detection as described under “Experimental Procedures.” Arrows indicate the areas in which lamin A/C (green) co-localize with TRITC-ACE (red). DAPI-stained nuclear DNA (blue) and lamin A/C do not colocalize.
Effect of Leptomycin B on Nuclear localization of ACE
SMC were incubated with 125I-ACE for 120 min in the presence and absence of LMB, an irreversible inhibitor of crn-1-mediated nuclear export, and nuclei and nuclear envelopes were isolated as described under “Experimental Procedures.” In cells that were not treated with LMB, distribution of 125I-ACE in the cytoplasmic fraction, nucleus, and nuclear envelope was 27, 65, and 8%, respectively (Fig. 8). The small amount of 125I-ACE localized in the nuclear envelope is consistent with the immunofluorescence detection shown in Fig. 7. Pretreatment of the cells with LMB resulted in a 28% stimulation of 125I-ACE accumulation in the nucleus. Interestingly, inhibition of nuclear export by LMB resulted in a 2.5-fold increment of 125I-ACE associated with the nuclear envelope. Comparing the amount of radioactive ACE (6.0 × 105 cpm) initially added to the cells with the amount recovered in the nuclei (12,000 cpm) we can estimate that only around 2% of the external ACE is localized in the nuclei. Thus, the small fraction of nuclear-localized ACE probably reflects the involvement of a low abundance receptor for endocytosis, as predicted by binding studies (Fig. 1B) and partial recycling back to the cell surface (see Fig. 9A). It is also possible that the internalized ACE is a fraction of the total ACE that acquired specific covalent modifications, such as the non-enzymatic advanced glycation end products (22), transglutamination (23), enzymatic oxidation (24), or limited proteolysis, leading to its recognition for internalization.
FIGURE 9.
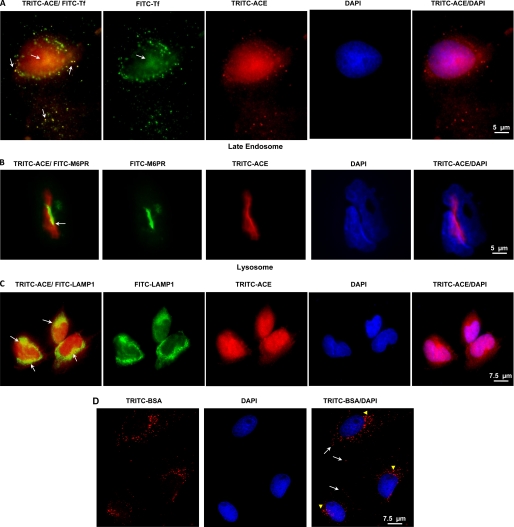
ACE in endosomal compartments and lysosome of SMC. A, SMC monolayers incubated with 10 nm TRITC-ACE (red) and 10 nm FITC-transferrin (FITC-Tf) (green) for 120 min. Cells were also incubated for 120 with 10 nm TRITC-ACE and then with anti-mannose 6-phosphate receptor (M6PR) (B) or anti-LAMP1 (C). Arrows point to ACE localization in the indicated compartment. D, cells were incubated with 10 nm TRITC-BSA as described above for TRITC-ACE. Internalized TRITC-BSA occupied the early endosome (arrows) and late endosome/lysosome (arrowheads) but not the nuclei. Indirect fluorescent microscopy analysis was done as described under “Experimental Procedures.” Scale bar is at the lower right hand corner.
ACE in Endosomal Compartments of SMC
The subcellular localization of exogenous TRITC-ACE in SMC was further assessed by comparison with transferrin, mannose 6-phosphate receptor, and LAMP-1, markers of the recycling endosome, late endosome, and lysosome, respectively. In SMC, TRITC-ACE was partially localized to the recycling endosome (Fig. 9A, TRITC-ACE/FITC-Tf panel) and nuclei (Fig. 9A, TRITC-ACE/DAPI) after 120 min. Under the same conditions, TRITC-ACE was detected in the late endosome (Fig. 9B, TRITC-ACE/FITC-M6PR panel) and lysosome (Fig. 9C, TRITC-ACE/FITC-LAMP1). A similar pattern of subcellular occupancy by TRITC-ACE was observed in HUVEC (data not shown). To learn if nuclear localization of ACE is a specific or at least a selective phenomenon, monomeric cellular fibronectin (cFN), a glycoprotein localized in the extracellular milieu, was labeled with TRITC and added to vascular smooth muscle monolayers. TRITC-cFN (TRITC-cFN) displayed the predicted pattern of cFN interacting with the cell surface and the fibrotic structure of the extracellular matrix both at 37 and 4 °C (not shown). Thus, although cFN binds to integrin receptors at the cell surface it is not internalized. SCM internalize serum albumin by a saturable endocytic pathway (25). TRITC-labeled albumin was predictably internalized, populated the endocytic compartment, but did not localize to the nuclei of SMC (Fig. 9D).
Intracellular Processing of ACE
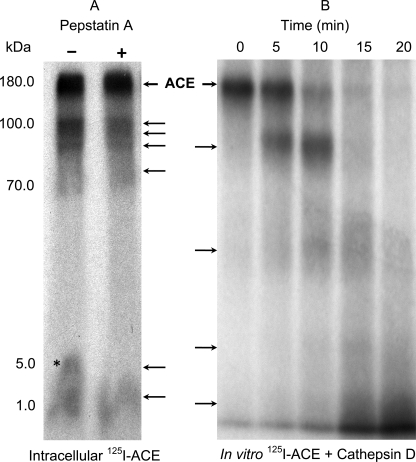
Maximal nuclear localization of ACE, taking place after 120 min of incubation, coincided with partial intracellular generation of large (∼80 kDa) and small (5.0–1.0 kDa) fragments of ACE (Fig. 10, panel A). This intracellular cleavage of ACE was partially prevented by pepstatin A, a cell-permeating, potent inhibitor of lysosomal/endosomal aspartyl proteases (26). This result pointed to the possible contribution of cathepsin D, a major representative of pepstatin-sensitive aspartyl proteases in the lysosomal/endosomal compartments. Is ACE a substrate of cathepsin-D in vitro? Incubation of ACE with cathepsin D, under optimal conditions for cathepsin D activity, resulted in progressive degradation of ACE, characterized by the initial generation of a major fragment of around 80 kDa succeeded by generation of smaller fragments, some of which are in the size range observed for intracellular degradation of 125I-ACE (Fig. 10, panel B). Thus, the potential for in vivo processing of ACE by cathepsin D exists provided both molecules share the same acidic compartment within the endocytic pathway. Several predicted cathepsin D cleavage sites exist in the primary sequence of ACE (Fig. 11). Unexpectedly, primary sequence analysis of ACE also revealed the presence of three predicted, perfect matches for the 9-amino acid long transactivation domain (TAD), a recently defined motif common to transactivation domains of many transcription factors ranging from Gal4 to p53 to NF-κB (27). Interestingly, the 8.7-kDa fragment also contains a predicted leucine-rich nuclear export signal overlapping the 9-amino acid long TAD predicted domain (Fig. 11, underlined). The observation that these 9-amino acid long TADs are flanked by cathepsin D cleavage sites brings about the possibility that fragments of ACE containing the 9-amino acid long TAD could be generated in vivo within the lysosomal/endosomal compartments. Because cleavage of ACE in vivo was not fully protected by pepstatin A (Fig. 10, panel A) it is possible that proteases, such as the pepstatin A-insensitive aspartyl protease, cathepsin E, predominant in aortic SMC (28), or cysteinyl or seryl proteases, could be involved in the processes.
FIGURE 10.
Intracellular and in vitro degradation of ACE. A, SMC preincubated with 125I-ACE displayed the pattern of large (100 to 70 kDa) and small (5.0 to 1.0 kDa) fragments. Pretreatment of the cells with pepstatin A partially prevented generation of small fragments. The asterisk indicates a 5.0-kDa fragment that is absent when cells are pretreated with pepstatin A. Arrows indicate the observed fragments. B, a time course incubation of 125I-ACE with cathepsin D also revealed the production of several fragments.
Mechanism of ACE Endocytosis
The mechanisms involving receptor-mediated endocytosis can be broadly distinguished by their sensitivity to specific inhibitors (29). Fig. 12A (left panel) shows that nuclear localization of 125I-ACE was inhibited around 40% by inhibitors of receptor-mediated endocytosis, phenylarsine oxide and concanavalin A, and by the inhibitor of raft-dependent endocytosis methyl-β-cyclodextrin. On the other hand, a specific inhibitor of clathrin-dependent endocytosis, monodansyl cadaverine, afforded a mild inhibitory effect of around 20%. This partial inhibition of 125I-ACE endocytosis suggested that a fraction of the uptake was receptor mediated. To evaluate this possibility SMC were subjected to the following treatments: 1) incubation with 125I-ACE at 4 °C for 60 min to allow for cell surface binding; 2) elimination of the excess of 125I-ACE; and 3) transfer to 37 °C to initiate the internalization on cell surface-bound 125I-ACE. This procedure resulted in a higher level of inhibition by the inhibitors tested (Fig. 12, right panel), reaching a value of around 70% inhibition for methyl-β-cyclodextrin. Interestingly, at least 20% 125I-ACE uptake seems to be mediated by a receptor-independent mechanism, probably fluid phase endocyotosis. Indeed, nuclear localization of 125I-ACE was inhibited around 28% by amiloride (Fig. 12B), a specific inhibitor of fluid phase endocytosis (30). These results indicate that the majority of ACE is internalized by a receptor-mediated endocytosis, through a mechanism involving primarily, but not exclusively, lipid rafts. In addition, a small fraction of extracellular ACE is internalized by fluid phase endocytosis. The partial inhibition of ACE nuclear localization afforded by polyanions dextran sulfate, polyinosine, and fucoidan (Fig. 12B) suggests the involvement of cationic domain(s) in the ACE receptor(s) essential, for interaction with ACE. Such domains are found in the general scavenger receptors, some of which are present or inducible in smooth muscle and endothelial cells (31).
Like ACE, BSA is internalized by receptor-mediated endocytosis and fluid phase endocytosis. However, only ACE reaches the cell nucleus (Fig. 4). In addition, whereas methyl-β-cyclodextrin inhibited by 70% the internalization of ACE, it only inhibited the internalization of BSA by around 15% (Fig. 13).
FIGURE 13.
Effect of endocytosis inhibitors on 125I-BSA internalization. Content of 125I-BSA in SMC (106) that were incubated with 125I-BSA for 120 min in the absence (control) or presence of the endocytosis inhibitors described in the legend of Fig. 11A. The incubation was carried out at 37 (gray bars) or at 4 °C and then switched to 37 °C after elimination of unbound 25I-ACE (black bars).
DISCUSSION
We have previously reported that exogenous ACE increased the level of mRNA expression of BK1 and BK2 receptors in cultured SMC after a 3–4 h incubation (14). This phenomenon, whereby exogenous ACE modulates the transcriptional response of SMC, prompted us to gain further insight into the molecular events involved. We have based our research strategy on the assumption that ACE may be taken up by the cell and localized to the nucleus; i.e. despite being an extracellular protein, it may be taken up by the cell and function intracellularly as well as a transcriptional factor in the nucleus. Consequently we have formulated our experiments on the premise that the most likely steps involved in nuclear localization of an extracellular protein are: 1) specific binding to cell surface receptor(s); 2) endocytosis; 3) endocytic transport; 4) exit from the endosomal lumen to the cytoplasm; 5) possible interaction with cytoplasmic factors; and 6) transport through the nuclear pore for a final nuclear delivery. These steps are followed during intracellular trafficking of the adenovirus subgroup B, such as Ad7 (32), as well as in the nonenveloped canine parvovirus (33). The recent discovery of “endosomal escape peptides” has expanded the field of genes (34) and active macromolecule delivery (35) for therapeutical purposes.
The initial interaction of ACE with the cell surface displayed characteristics of a binding to a low abundance receptor (36, 37). In addition, exogenous ACE promoted late activation of FAK and SHP2 and modulation of PDGFR-β response in the ACE nuclear localization time line. Considering the central role of PDGRFR-β signaling in sustaining the SMC synthetic/proliferative phenotype and due to the critical link of this phenotype to known pathological conditions (38), this modulation of PDGRFR-β signaling uncovers a new, unexplored domain of biological functions for ACE. In addition, the modulation of SHP2, FAK, and AKT signaling by ACE may impact on the activation of several receptor tyrosine kinases (24), on cell proliferation, migration, and invasion (38), or on cell survival as well as normal and malignant tissue growth (39).
Shortly after interacting with the plasma membrane, exogenous ACE populated the endosomal compartment, partially sharing the early and recycling endosome with the transferring receptor, a marker of those compartments (33). Subsequently, ACE was observed in the nuclei of some, but not all SMC (Fig. 4). This suggests that the steady-state level of nuclear ACE may be subjected to a dynamic turnover. Alternatively, nuclear localization of ACE may only take place at specific stages of the cell cycle. ACE was also localized in the HUVEC nuclei. These results imply that SMC and HUVEC possess an active mechanism to internalize exogenous ACE and target it to their nuclei. Nuclear localization of ACE was largely prevented by incubation at 4 °C, a condition that blocks endocytosis, and was partially impaired by specific inhibitors of receptor-mediated endocytosis. The inhibitory effect was enhanced when the unbound excess of 125I-ACE was eliminated before uptake took place (Fig. 12), suggesting the participation of a receptor independent uptake such as fluid phase endocytosis. Nuclear localization of ACE is a relatively slow process consisting of an initial lag time of around 40 min, succeeded by a more rapid phase that levels off at around 120 min (Fig. 6). Remarkably, internalized ACE undergoes partial degradation generating small fragments in the 5.0–1.0 kDa range. The partial protection afforded by pepstatin A pointed to possible involvement of cathepsin D in generation of these fragments. ACE is indeed a substrate of cathepsin D, as confirmed by in vitro analysis. The possibility that ACE and cathepsin D may share the same compartment was affirmed by the observation that internalized ACE reached the late endosome and the lysosome in SMC and EC. Several secretory proteins containing the signal sequence for targeting to the ER but no nuclear localization signal, as in the case of ACE, have been reported to localize in the nucleus. Those proteins include receptors (angiotensin II receptor, endothelin receptor, prostaglandin E2 receptor, etc.) and hormones (growth hormone, insulin, actoferrin, etc.) (40). The prevalent hypothesis explaining the escape of endosomal cargo proteins to the cytoplasm is based on the acid nature of the late endosomal/lysosomal compartment leading to a selective increase of membrane permeability as well as the favorable unfolding of the translocated molecule (41). This mechanism was demonstrated for retention and escape of the adenovirus 7 from the endosomal compartment to the cytosol (32). Surprisingly, analysis of the ACE primary sequence revealed the presence of three predicted, perfect matches for the 9-amino acid long TAD, a recently defined motif common to transactivation domains of many transcription factors ranging from Gal4 to p53 to NF-κB (27). The observation that these 9-amino acid long TADs are flanked by cathepsin D cleavage sites (Fig. 11) suggests that fragments of ACE containing the 9-amino acid TAD could be generated in vivo within the lysosomal/endosomal compartment. Interestingly, whereas no nuclear localization signal is predicted in the entire ACE sequence, the putative 8.7-kDa cathepsin fragment (Fig. 11) contains a predicted nuclear export signal overlapping the 9-amino acid long TAD. Thus, nuclear import of an ACE fragment, such as the 8.7-kDa fragment, could be accomplished by a piggyback mechanism (42) after binding to a NLS-containing piggyback partner. The moderate enhancing effect of LMB on nuclear localization of ACE (Fig. 8) suggests that the predicted nuclear export signal could be involved in exit of ACE, or a fragment of it, from the nucleus.
Because in vivo cleavage of ACE was not fully protected by pepstatin A it is possible that other proteases, such as the pepstatin A-insensitive aspartyl protease, cathepsin E, predominant in aortic SMC (27), or cysteinyl or seryl proteases, may also cleave endocytosed ACE. These results support the notion that a fragment of ACE, not necessarily the whole molecule, may reach the nucleus. The putative transcriptional activity of the ACE fragments depicted in Fig. 11 is now under investigation. The six steps outlined above for internalization and final nuclear localization of ACE are the foundation of our working hypothesis to approach future studies on the molecular events leading to the nuclear localization of exogenous ACE.
Although the extracellular enzymatic activities of ACE are well recognized and have led to therapeutic cardiovascular applications, its intracellular and endonuclear properties are only beginning to be appreciated. The potential implications of these novel properties remain to be seen, but might be relevant to certain aspects of carcinogenesis (5, 43–45) or other hitherto uncovered pathophysiological conditions.
Acknowledgments
We gratefully acknowledge the expert support by Dr. Irene Gavras from the Hypertension and Atherosclerosis Section and Dr. Michael T. Kirber from the Cellular Imaging Core Facility, Boston University, School of Medicine.
This work was supported in part by Alapis Research Laboratories.
H. A. Lucero, E. Kintsurashvili, M. E. Marketou, and H. Gavras, unpublished data.
- ACE
- angiotensin converting enzyme
- AKT
- v-akt murine thymoma viral oncogene homolog
- BSA
- bovine serum albumin
- DAPI
- 4′,6-diamidino-2-phenylindole
- EC
- endothelial cells
- ERK
- extracellular signal-regulated kinase
- HUVEC
- human umbilical vascular endothelial cells
- FAK
- focal adhesion kinase
- FITC
- fluorescein isothiocyanate
- MAPK
- mitogen-activated protein kinase
- PBS
- phosphate-buffered saline
- PDGFR-β
- platelet-derived growth factor receptor β
- SHP2
- SRC homology 2 domain-containing protein-tyrosine phosphatase
- SH2
- Src homology 2
- SMC
- rat aorta smooth muscle cells
- TRITC
- tetramethylrhodamine-5-isothiocyanate
- HBSS
- Hanks' balanced salt solution
- LMB
- leptomycin B
- cFN
- cellular fibronectin
- DMEM
- Dulbecco's modified Eagle's medium
- GPI
- glycosylphosphatidylinositol
- TAD
- transactivation domain
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Wei L., Alhenc-Gelas F., Soubrier F., Michaud A., Corvol P., Clauser E. (1991) J. Biol. Chem. 266, 5540–5546 [PubMed] [Google Scholar]
- 2.Howard T. E., Shai S. Y., Langford K. G., Martin B. M., Bernstein K. E. (1990) Mol. Cell. Biol. 10, 4294–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei L., Alhenc-Gelas F., Corvol P., Clauser E. (1991) J. Biol. Chem. 266, 9002–9008 [PubMed] [Google Scholar]
- 4.Beldent V., Michaud A., Bonnefoy C., Chauvet M. T., Corvol P. (1995) J. Biol. Chem. 270, 28962–28969 [DOI] [PubMed] [Google Scholar]
- 5.Rosenthal T., Gavras I. (2009) J. Hum. Hypertens. 23, 623–635 [DOI] [PubMed] [Google Scholar]
- 6.Gavras H. (1994) Hypertension 23, 813–818 [DOI] [PubMed] [Google Scholar]
- 7.Kondoh G., Tojo H., Nakatani Y., Komazawa N., Murata C., Yamagata K., Maeda Y., Kinoshita T., Okabe M., Taguchi R., Takeda J. (2005) Nat. Med. 11, 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leisle L., Parkin E. T., Turner A. J., Hooper N. M. (2005) Nat. Med. 11, 1139–1140 [DOI] [PubMed] [Google Scholar]
- 9.Chen Z., Deddish P. A., Minshall R. D., Becker R. P., Erdös E. G., Tan F. (2006) FASEB J. 20, 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohlstedt K., Brandes R. P., Müller-Esterl W., Busse R., Fleming I. (2004) Circ. Res. 94, 60–67 [DOI] [PubMed] [Google Scholar]
- 11.Kohlstedt K., Gershome C., Friedrich M., Müller-Esterl W., Alhenc-Gelas F., Busse R., Fleming I. (2006) Mol. Pharmacol. 69, 1725–1732 [DOI] [PubMed] [Google Scholar]
- 12.Kohlstedt K., Gershome C., Trouvain C., Hofmann W. K., Fichtlscherer S., Fleming I. (2009) Mol. Pharmacol. 75, 685–692 [DOI] [PubMed] [Google Scholar]
- 13.Kohlstedt K., Busse R., Fleming I. (2005) Hypertension 45, 126–132 [DOI] [PubMed] [Google Scholar]
- 14.Ignjacev-Lazich I., Kintsurashvili E., Johns C., Vitseva O., Duka A., Shenouda S., Gavras I., Gavras H. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H1814–H1820 [DOI] [PubMed] [Google Scholar]
- 15.Morishita R., Gibbons G. H., Ellison K. E., Lee W., Zhang L., Yu H., Kaneda Y., Ogihara T., Dzau V. J. (1994) J. Clin. Invest. 94, 978–984, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rakugi H., Kim D. K., Krieger J. E., Wang D. S., Dzau V. J., Pratt R. E. (1994) J. Clin. Invest. 93, 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camargo de Andrade M. C, Di Marco G. S., de Paulo Castro Teixeira V., Mortara R. A., Sabatini R. A., Pesquero J. B., Boim M. A., Carmona A. K., Schor N., Casarini D. E. (2006) Am. J. Physiol. Renal Physiol. 290, F364–F375 [DOI] [PubMed] [Google Scholar]
- 18.Hooper N. M., Turner A. J. (1987) Biochem. J. 241, 625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolton A. E., Hunter W. M. (1973) Biochem. J. 133, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dance M., Montagner A., Salles J. P., Yart A., Raynal P. (2008) Cell. Signal. 20, 453–459 [DOI] [PubMed] [Google Scholar]
- 21.Schlaepfer D. D., Mitra S. K. (2004) Curr. Opin. Genet. Dev. 14, 92–101 [DOI] [PubMed] [Google Scholar]
- 22.Sparvero L. J., Asafu-Adjei D., Kang R., Tang D., Amin N., Im J., Rutledge R., Lin B., Amoscato A. A., Zeh H. J., Lotze M. T. (2009) J. Translational Med. 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esposito C., Caputo I. (2005) FEBS J. 272, 615–631 [DOI] [PubMed] [Google Scholar]
- 24.Lucero H. A., Ravid K., Grimsby J. L., Rich C. B., DiCamillo S. J., Mäki J. M., Myllyharju J., Kagan H. M. (2008) J. Biol. Chem. 283, 24103–24117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprague E. A., Kelley J. L., Suenram C. A., Valente A. J., Abreu-Macomber M., Schwartz C. J. (1985) Am. J. Pathol. 121, 433–443 [PMC free article] [PubMed] [Google Scholar]
- 26.Fusek M., Větvička V. (2005) Biomed. Papers 149, 43–50 [DOI] [PubMed] [Google Scholar]
- 27.Sandholzer J., Hoeth M., Piskacek M., Mayer H., de Martin R. (2007) Biochem. Biophys. Res. Commun. 360, 370–374 [DOI] [PubMed] [Google Scholar]
- 28.Holycross B. J., Saye J., Harrison J. K., Peach M. J. (1992) Hypertension 19, 697–701 [DOI] [PubMed] [Google Scholar]
- 29.Mayor M., Pagano R. E. (2007) Nat. Rev. Mol. Cell. Biol. 8, 603–612 [DOI] [PubMed] [Google Scholar]
- 30.West M. A., Bretscher M. S., Watts C. (1989) J. Cell Biol. 109, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy J. E., Tedbury P. R., Homer-Vanniasinkam S., Walker J. H., Ponnambalam S. (2005) Atherosclerosis 182, 1–15 [DOI] [PubMed] [Google Scholar]
- 32.Miyazawa N., Crystal R. G., Leopold P. L. (2001) J. Virology 75, 1387–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suikkanen S., Sääjärvi K., Hirsimäki J., Välilehto O., Reunanen H., Vihinen-Ranta M., Vuento M. (2002) J. Virology 76, 4401–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M. E., Rice K. G. (2007) AAPS J. 9, E18–E29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melikov K., Chernomordik L. V. (2005) Cell. Mol. Life. Sci. 62, 2739–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowenthal J. W., Castle B. E., Christiansen J., Schreurs J., Rennick D., Arai N., Hoy P., Takebe Y., Howard M. (1988) J. Immunol. 140, 456–464 [PubMed] [Google Scholar]
- 37.Dower S. K., Kronheim S. R., March C. J., Conlon P. J., Hopp T. P., Gillis S., Urdal D. L. (1985) J. Exp. Med. 162, 501–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawai-Kowase K., Owens G. K. (2007) Am. J. Physiol. Cell Physiol. 292, C59–C69 [DOI] [PubMed] [Google Scholar]
- 39.Testa J. R., Tsichlis P. N. (2005) Oncogene 24, 7391–7393 [DOI] [PubMed] [Google Scholar]
- 40.Arnoys E. J., Wang J. L. (2007) Acta Histochem. 109, 89–110 [DOI] [PubMed] [Google Scholar]
- 41.Rubartelli A., Sitia R. (1995) Trends Cell Biol. 5, 409–412 [DOI] [PubMed] [Google Scholar]
- 42.Steidl S., Tüncher A., Goda H., Guder C., Papadopoulou N., Kobayashi T., Tsukagoshi N., Kato M., Brakhage A. A. (2004) J. Mol. Biol. 342, 515–524 [DOI] [PubMed] [Google Scholar]
- 43.Lever A. F., Hole D. J., Gillis C. R., McCallum I. R., McInnes G. T., MacKinnon P. L., Meredith P. A., Murray L. S., Reid J. L., Robertson J. W. (1998) Lancet 352, 179–184 [DOI] [PubMed] [Google Scholar]
- 44.Lindberg H., Nielsen D., Jensen B. V., Eriksen J., Skovsgaard T. (2004) Acta Oncol. 43, 142–152 [DOI] [PubMed] [Google Scholar]
- 45.Yoshiji H., Kuriyama S., Noguchi R., Fukui H. (2004) Curr. Cancer Drug Targets 4, 555–567 [DOI] [PubMed] [Google Scholar]