Abstract
GILZ (glucocorticoid-induced leucine zipper) is an ubiquitous protein whose expression is induced by glucocorticoids in lymphoid cells. We previously showed that GILZ expression is rapidly induced upon interleukin 2 deprivation in T-cells, protecting cells from apoptosis induced by forkhead box subgroup O3 (FOXO3). The aim of this work is to elucidate the molecular mechanism of FOXO factor inhibition by GILZ. We show in the myeloid cell line HL-60 and the lymphoid CTLL-2 T-cell line that GILZ down-regulates the expression of p27KIP1 and Bim, two FOXO targets involved in cell cycle regulation and apoptosis, respectively. GILZ inhibits FOXO1, FOXO3, and FOXO4 transcriptional activities measured with natural or synthetic FOXO-responsive promoters in HL-60 cells. This inhibitory effect is independent of protein kinase B and IκB kinase phosphorylation sites. GILZ does not hinder FOXO3 DNA-binding activity and does not physically interact with FOXO3. However, using fluorescence microscopy, we observe that GILZ expression provokes a Crm-1-dependent nuclear exclusion of FOXO3 leading to its relocalization to the cytoplasm. Moreover, GILZ exclusive cytoplasmic localization is a prerequisite for FOXO3 inhibition and relocalization. We propose that GILZ is a general inhibitor of FOXO factors acting through an original mechanism by preventing them from reaching target genes within the nucleus.
Keywords: Gene/Regulation, Nucleus/Nuclear Export, Protein/Binding/DNA, Transcription/Regulation, Protein-Protein Interactions, Crm1, FOXO, GILZ
Introduction
Forkhead box subgroup O1 (FOXO1 or FKHR), FOXO3a (FKHRL1), FOXO4 (AFX), and FOXO6 constitute the mammalian FOXO family of transcription factors and achieve important functions in the regulation of genes involved in cell cycle regulation, apoptosis, DNA repair, stress response, energy metabolism, and control of lifespan (for review, see Ref. 1). These highly related members are ubiquitously expressed in all mammalian tissues, interact with the same core consensus DNA sequence, and display overlapping patterns of transcriptional activities (2). Interest about FOXO factors in the hematopoietic system is increasing due to their role in regulation of immune responses. In vitro, FOXO3 has been shown to participate in cytokine withdrawal-induced apoptosis of lymphocytes through up-regulation of Bim (3) or Puma (4). Moreover, the Fas Ligand gene has been described as a downstream target of FOXO3 in Jurkat T-lymphocytes (5). Pink1 was recently described as an anti-apoptotic FOXO3 target gene whose induction upon growth factor deprivation paradoxically prolongs lymphocyte survival (6). In vivo, FOXO3 appears to be predominant in peripheral lymphoid organs and to regulate lymphoid and myeloid homeostasis. Indeed, mice bearing a mutated FOXO3 allele presented spontaneous T-cell activation and a multisystemic inflammatory syndrome associated with lymphadenopathy (7). Moreover, adult mice with conditional deletion of FOXO1, FOXO3a, and FOXO4 showed hematopoietic stem cells with increased cell cycling and apoptosis and defective long term repopulating activity in the bone marrow (8). Somatic disruption of the three FOXO genes in mice resulted in thymic lymphomagenesis (9).
Nuclear import of FOXO factors follows stress signals such as oxidative stress or growth factor deprivation, whereas nuclear export results from interaction with the exportin Crm1 (chromosomal region maintenance) and Ran-GTP and from the phosphorylation by the serine/threonine kinase Akt (also called protein kinase B (PKB)),3 generating two binding sites for the 14-3-3 family of proteins. These post-translational modifications also impair DNA binding (5) and promote proteasomal degradation (10). FOXO factors have been recently shown to be regulated by Akt-independent pathways such as phosphorylation, acetylation, or interaction with numerous signaling molecules, suggesting that multiple mechanisms can regulate FOXO transcriptional activity. Indeed, IκB kinase (IKKβ) has been shown to interact with and to phosphorylate FOXO3a at Ser-644, promoting nuclear exclusion and proteasomal degradation independently of PKB phosphorylation (11).
Glucocorticoid-induced leucine zipper (GILZ) is a ubiquitous 17-kDa protein belonging to the TSC-22 family of proteins characterized by the presence of common domains termed the TSC box and leucine zipper. GILZ has been described as a regulator of gene transcription through protein-protein interactions resulting in inhibition of AP-1 (12) and NF-κB (13, 14) transcriptional activities, thereby regulating transduction pathways essential to inflammation and immune response. GILZ was initially identified as a dexamethasone-responsive gene from a thymus subtraction library (15) and was further shown to be regulated by glucocorticoids in immune cells (15–17) and in human airway epithelial cells (18), appearing as an important mediator of glucocorticoid immunomodulatory and anti-inflammatory actions. GILZ expression is also regulated by IL-4 and IL-10 in monocytes, macrophages, and dendritic cells (17). We previously demonstrated that GILZ expression was rapidly induced upon IL-2 deprivation of T-lymphocytes, protecting these cells from the onset of apoptosis (19). This protection was conferred through inhibition of FOXO3 transcriptional activity, resulting in down-regulation of the pro-apoptotic Bim protein (19). The objective of this work was to determine whether GILZ is a general inhibitor of FOXO transcription factors and to investigate the mechanism of FOXO3 inhibition by GILZ in hematopoietic cells.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
The dual luciferase reporter assay system was purchased from Promega (Madison, WI). G418 sulfate was from PAA Laboratories (Pasching, Austria). Leptomycin B was purchased from Sigma.
Cell Culture and Transfection
CTLL-2, an IL-2-dependent murine cytotoxic T-cell line, CTLL-2-Myc clone 1, and CTLL-2-Myc-GILZ clone 6 stably transfected with empty vector or pcDNA3-Myc-GILZ, respectively, were previously described (19). The human HL-60 cell line, established from a patient with acute promyelocytic leukemia, was stably transfected with empty vector or pcDNA3-Myc-GILZ. Selection of stably transfected cells was initiated 48 h after electroporation using 800 μg/ml of G418 for 2 weeks. Cells were then cloned by limiting dilution. The clones were selected based on GILZ expression (HL-60-Myc-GILZ clones 27 and 32). HL-60-Myc clones 3 and 7 were randomly selected. HL-60 cells and HL-60 clones were cultured in RPMI 1640 medium containing 0.1 mg/ml of streptomycin, 100 units/ml of penicillin, 1% sodium pyruvate, 10% fetal calf serum (Fisher Scientific). Transient transfections were performed using electroporation as previously described (20). The total amounts of transfected DNA were kept constant by addition of empty control vector. Cells were then cultured for 24 h before harvesting.
Plasmid Constructs
Expression vectors pcDNA3-Myc- GILZ, pcDNA3-Myc-FOXO3-WT (wild type) (19), and pcDNA3-Myc-FOXO3-TM (T32A,S253A,S315A) were previously described (19). pcDNA3-Flag-FKHR-WT (FOXO1-WT) and pcDNA3-Flag-FKHR-AAA (FOXO1-TM) were a kind gift from Dr. Tang (University of Michigan Medical School, Ann Arbor, MI). pMT2-AFX-WT (FOXO4-WT) was a kind gift from Dr. Burgering (University Medical Center, Utrecht, The Netherlands). pMT2-FOXO4-TM was obtained by site-directed mutagenesis of pMT2-FOXO4-WT (T28A,S193A,S258A). pcDNA3-Myc-NES-GILZ and pcDNA3-Myc-NLS- GILZ plasmids were obtained by insertion of double-stranded oligonucleotides from the NES sequence of MAPK/ERK kinase (MEK) (5′-GGATGAACCTGGTGGACCTCCAAAAGAAGCTGGAGGAGCTGGAGCTGGACGAGCAGCAGG-3′) or the NLS sequence of the simian virus 40 large-T antigen (5′- GGATCGATCCAAAAAAGAAGAGAAAGGTAGATCCAAAAAAGAAGAGAAAGGTAGATCCAAAAAAGAAGAGAAAGGTAG-3′) in SacII/BamHI-digested pcDNA3-Myc- GILZ. pEGFP-FOXO3-WT was a kind gift from Dr. Hung (11) (University of Texas, Houston, TX). pEGFP-FOXO3-TM was generated by site-directed mutagenesis of pEGFP-FOXO3-WT (Thr-32, Ser-253, and Ser-315 were replaced by alanine). The EcoRI-blunt GILZ fragment was subcloned into the pDsRed-N1 vector (Clontech Laboratories) to obtain pDsRed-GILZ. pcDNA3-Myc-FOXO3-TM-NESm (L390A,L391A,I394A) was obtained by site-directed mutagenesis of pcDNA3-Myc-FOXO3-TM. The sequences of the primers were as follows: I394 forward (5′-GGATAACGCCACGCTCCCGCCATC-3′), I394 reverse (5′-GGATGGCGGGAGCGTGGCGTTATC-3′), L390 forward (5′-CCTCATGGACGACGCGCTGATGGATAAC-3′), L390 reverse (5′-GTTATCCAGCGCGTCGTCCATGAGG-3′), L391 forward (5′-ATGGACGACGCGGCGGATAACG-3′), and L391 reverse (5′-CGTTATCCGCCGCGTCGTCCAT-5′).
Reporter Plasmids
pBim-Luc (3.6 kb) was a kind gift from Dr. Bouillet (WEHI, Melbourne, Australia), p27-Luc was a kind gift from Dr. Coffer (University Medical Center, Utrecht, The Netherlands), and pFasL-Luc was a kind gift from Dr. Green (La Jolla, CA). p3xIRS-MLP-Luc contains three copies of the insulin response sequence (IRS) upstream of the adenovirus major late promoter (MLP) and was a kind gift from Dr. Fukamizu (University of Tsukuba, Japan).
Dual Luciferase Reporter Assay
Reporter assays were performed as previously described (16). Normalized relative luciferase units were calculated as follows: firefly luciferase units/Renilla luciferase units. Results were expressed as percentages relative to the FOXO3-normalized relative luciferase units (100%). Data represent the mean ± S.E.M. of three independent experiments, each performed in duplicate or triplicate.
Antibodies, Western Blot
Cells were harvested 24 h after transfection and Western blotting was performed as described previously (16) using the following antibodies: polyclonal anti-GILZ (19), monoclonal anti-β-tubulin (T4026, Sigma), polyclonal anti-FOXO3 (07-702, Millipore, Billerica, MA), polyclonal anti-FOXO4 and anti-FOXO1 (9472 and 9462 respectively, Cell Signaling Technology, Danvers, MA). Anti-Bim (sc-11425), anti-14-3-3 (sc1657) and anti-p27KIP1 (sc-528) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Monoclonal anti-Myc 9E10 antibody was produced in the laboratory. Densitometric analysis of the blots was performed using the ImageQuant® software (GE Healthcare, Saclay, France). Nuclear extracts were performed using a Kontes all-glass Dounce homogenizer (kimble/kontes, Vineland, NJ) as described previously (16).
Immunofluorescence
HL-60-Myc and HL-60-Myc-GILZ cells were transfected with 10 μg of pEGFP-FOXO3-WT or pEGFP-FOXO3-TM and fixed in buffer containing 2% paraformaldehyde and 1.5% sucrose for 15 min. Cells were then quenched with 50 mm NH4Cl for 10 min. Permeabilization was performed using 0.05% Triton in phosphate-buffered saline medium for 4 min followed by two washes with 1× phosphate-buffered saline. Cells were then blocked with 5% bovine serum albumin for 1 h, stained with the anti-Myc antibody for 90 min at room temperature, and washed 3 times with buffer before incubation with a secondary anti-mouse IgG antibody conjugated with Alexa 546 (Molecular Probes) in darkness for 90 min at room temperature. Cells were stained with 4′,6-diamidino-2-phenylindole (bibenzimide H 33258, Sigma) for nucleus labeling. Dako mounting medium was used (Glostrup, Denmark). The immunolabeled cells were examined with a Zeiss Imager Z1, camera Axio Cam R3. The fluorescence intensity of the nuclear and cytoplasmic compartments was quantified using ImageJ software, and at least 200 cells were counted to calculate the nuclear/cytoplasmic ratio (N/C) of either EGFP-FOXO3-WT or EGFP-FOXO3-TM. The fluorescence of the EGFP-FOXO3 protein was higher in the nucleus than in the cytoplasm (N/C > 1) and the fluorescence was higher in the cytoplasm than in the nucleus (N/C < 1), but fluorescence was similar in the two compartments (N/C = 1). Results were expressed as percentages.
DNA Affinity Precipitation of FOXO3 Proteins
HL-60 cells transfected with 5 μg of pcDNA3-FOXO3-TM and with or without 10 μg of pcDNA3-Myc-GILZ were harvested 24 h after transfection. Double-stranded 5′-biotinylated IRS oligonucleotides of the IGFBP-1 promoter were coupled to streptavidine-agarose beads (Sigma) and nuclear and cytoplasmic extracts were incubated with the precoated beads. Beads were then washed and boiled in a reducing sample buffer containing 40% glycerol, 125 mm Tris (pH 6.8), 4% SDS, 5% β-mercaptoethanol, and 0.025% bromophenol blue to elute bound proteins. Western blots were performed using the anti-FOXO3 antibody.
Co-immunoprecipitation Assay
Cells were transfected with 2.5 μg of pcDNA3-Myc-GILZ and/or 5 μg of pcDNA3- FOXO3-WT plasmids and then cultured overnight before harvesting. Cells were lysed in a lysis buffer containing 50 mm Hepes (pH 7.3), 150 mm sodium chloride, 1 mm EDTA, 1.5 mm magnesium chloride, 100 mm sodium fluoride, 10 mm sodium pyrophosphate, 200 μm sodium orthovanadate, 10% glycerol, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml of aprotinin, and 1 μg/ml of leupeptin. A total of 600 μg of total protein extract was precleared by incubation with 30 μl of Protein G-Sepharose 4 fast flow beads (GE Healthcare) at 4 °C for 45 min with preimmune serum. Precleared samples were then incubated for 3 h with 30 μl of protein G-Sepharose beads and 5 μg of anti-FOXO3 or anti-GILZ antibodies at 4 °C. The beads were washed 5 times with lysis buffer containing 0.1% Triton X-100 supplemented with proteases and phosphatases inhibitors and boiled in reducing sample buffer before performing Western blotting.
Statistical Analysis
Experiments were performed at least three times and presented as mean ± S.E.M. p values were determined using the Student's test. Results were considered significant for p < 0.05. For microscopy statistical analysis, data were analyzed with one-way analysis for variance and regression analysis for correlations using GraphPad InStat 3 (San Diego, CA). Statistical significance was set at p < 0.05.
RESULTS
GILZ Overexpression Down-regulates p27KIP1 and Bim Protein Expression in IL-2-deprived CTLL-2 Cells and HL-60 Cells Transiently Expressing FOXO3
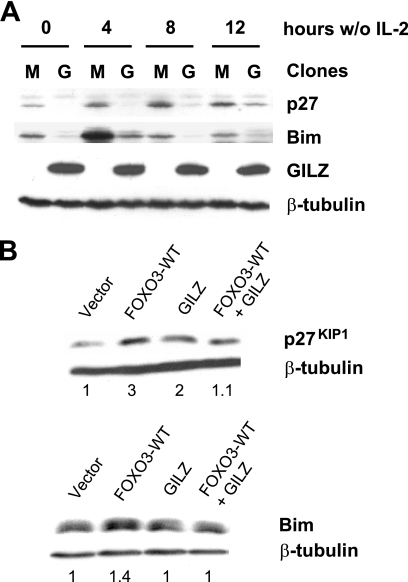
Withdrawal of IL-2 in CTLL-2 cells results in up-regulation of Bim expression. We previously described that in GILZ-overexpressing CTLL-2 cells, Bim expression was delayed (19), protecting these cells from IL-2 deprivation-induced apoptosis. This effect was mediated through inhibition of FOXO3 transcriptional activity, explaining the inhibition of Bim expression. p27KIP1 is a cyclin-dependent kinase inhibitor known to be regulated by FOXO3 upon growth factor deprivation and involved in cell cycle regulation (21). To determine whether GILZ could affect the levels of p27KIP1 in CTLL-2 cells, control or GILZ-overexpressing clones were deprived of IL-2 for 4, 8, and 12 h. p27KIP1 expression was induced in a time-dependent manner between 4 and 12 h after IL-2 deprivation in the control clone (Fig. 1A). In GILZ-overexpressing cells, p27KIP1 expression was down-regulated compared with control clones. These results were observed in all clones tested. Comparable results were obtained for Bim as previously described (Fig. 1A) (19).
FIGURE 1.
GILZ expression down-regulates p27KIP1 and Bim protein in IL-2-deprived CTLL-2 cells and in HL-60 cells. A, CTLL-2-Myc 1 or CTLL-2-Myc-GILZ 6 clones were deprived of IL-2 and lysed for the indicated periods of time. Western blot was performed using anti-p27KIP1 and anti-Bim antibodies. After stripping, membranes were reblotted with an anti-β-tubulin antibody as a loading control. A representative experiment of three is shown. M, CTLL-2-Myc 1; G, CTLL-2-Myc-GILZ 6. B, HL-60 cells were transiently transfected with 10 μg of pcDNA3-FOXO3-WT, pcDNA3-Myc-GILZ, or pcDNA3-Myc (empty vector) and lysed after 24 h of incubation. Western blot was performed using anti-p27KIP1 and anti-Bim antibodies. After stripping, membranes were reblotted with an anti-β-tubulin antibody as a loading control. A representative experiment of three is shown.
To evaluate whether these results were specifically related to FOXO3 inhibition, we used the HL-60 cell line, a hematopoietic cell line independent of cytokines for proliferation and survival. FOXO proteins were undetectable in this cell line under normal growing conditions (first lanes of Figs. 2E, 3E, and 4B). HL-60 cells were cotransfected with the expression vectors for FOXO3-WT and/or GILZ (Fig. 1B). FOXO3 and GILZ expressions are shown on Fig. 2E. Western blot analysis showed that GILZ inhibited FOXO3-induced Bim and p27KIP1 expression in HL-60 cells. These observations suggested that the inhibitory effect of GILZ was not restricted to endogenous FOXO factors activated by IL-2 deprivation in the CTLL-2 cell line but could also be observed using exogenous FOXO3 in another hematopoietic cell line. The HL-60 model will then be used to study the mechanisms of FOXO inhibition by GILZ.
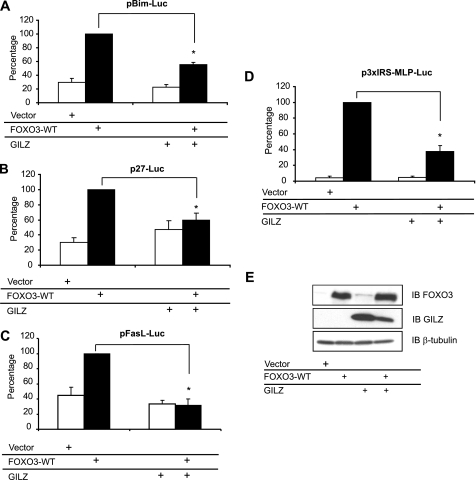
FIGURE 2.
GILZ inhibits FOXO3-WT transcriptional activity in HL-60 cells. HL-60 cells were transiently transfected with 10 μg of pcDNA3-FOXO3-WT and/or 10 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc and with 10 μg of the reporter plasmid pBim-Luc (A), 10 μg of p27-Luc (B), or pFasL-Luc (C). The transcriptional activity was analyzed after 24 h of culture. Results are expressed as the percentage of the reporter activity, with 100% representing the activity of the construct in the presence of FOXO3-WT. Data represent the mean ± S.E.M. of three independent experiments performed in triplicate. *, p < 0.05. D, HL-60 cells were transiently transfected with 5 μg of the reporter plasmid p3xIRS-MLP-Luc and with either 1 μg of pcDNA3-FOXO3-WT and/or 5 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc. Results are expressed as a percentage of p3xIRS-MLP-Luc activity, with 100% representing the activity of the construct in the presence of FOXO3-WT. Data represent the mean ± S.E.M. of three independent experiments performed in triplicate. E, HL-60 cells were transiently transfected with 10 μg of pcDNA3-FOXO3-WT and/or 10 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc and cells were harvested after 24 h of culture. Western blot (IB) was performed using anti-FOXO3 and anti-GILZ antibodies. β-Tubulin was used as an internal control for protein levels. Error bars, S.E.
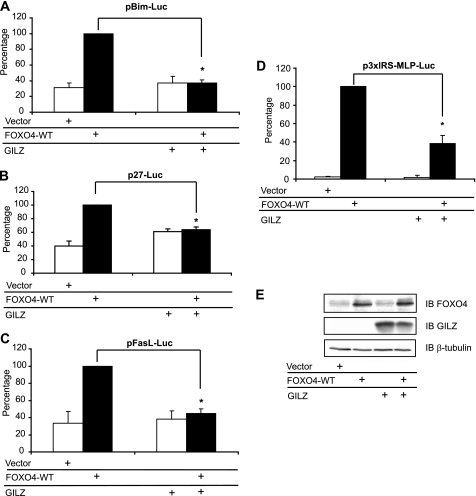
FIGURE 3.
GILZ inhibits FOXO4-WT transcriptional activity in HL-60 cells. HL-60 cells were transiently transfected with 10 μg of pMT2-FOXO4-WT and/or 10 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc and with either 10 μg of the reporter plasmid pBim-Luc (A) or 10 μg of p27-Luc (B) or pFasL-Luc (C). The transcriptional activity was analyzed after 24 h of culture. Results are expressed as the percentage of reporter activity, with 100% representing the activity of the construct in the presence of FOXO4-WT. Data represent the mean ± S.E.M. of 3 independent experiments performed in triplicate. *, p < 0.05. D, HL-60 cells were transiently transfected with the reporter plasmid p3xIRS-MLP-Luc and either 1 μg of pMT2-FOXO4-WT and/or 5 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc. Results are expressed as the percentage of p3xIRS-MLP-Luc activity, with 100% representing the activity of the construct in the presence of FOXO4-WT. Data represent the mean ± S.E.M. of three independent experiments performed in triplicate. *, p < 0.05. E, HL-60 cells were transiently transfected with 10 μg of pcDNA3-FOXO4-WT and/or 10 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc, and cells were harvested after 24 h of culture. Western blot was performed using anti-FOXO4 and anti-GILZ antibodies. β-Tubulin was used as an internal control for protein levels. IB, immunoblot. Error bars, S.E.
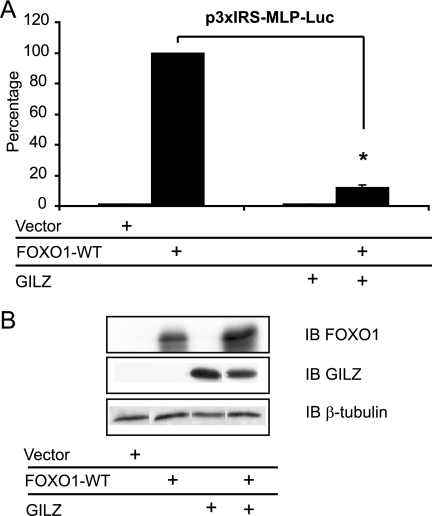
FIGURE 4.
GILZ inhibits FOXO1-WT transcriptional activity in HL-60 cells. A, HL-60 cells were transiently transfected with the reporter plasmid p3xIRS-MLP-Luc and either 1 μg of pcDNA3-FOXO1-WT and/or 5 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc, and transcriptional activity was analyzed after 24 h of culture. Results are expressed as the percentage of p3xIRS-MLP-Luc activity, with 100% representing the activity of the construct in the presence of FOXO1-WT. Data represent the mean ± S.E.M. of three independent experiments performed in triplicate. *, p < 0.05. B, HL-60 cells were transiently transfected with 10 μg of pcDNA3-FOXO1-WT and/or 10 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc and cells were harvested after 24 h of culture. Western blot was performed using anti-FOXO1 and anti-GILZ antibodies. β-Tubulin was used as an internal control of protein levels. IB, immunoblot. Error bars, S.E.M.
GILZ Inhibits FOXO3, FOXO4, and FOXO1 Transcriptional Activities in HL-60 Cells
HL-60 cells were cotransfected with expression vectors for FOXO3-WT and/or GILZ, along with reporter plasmids measuring BIM, p27KIP1, and FASL promoter activities. Results showed that GILZ significantly inhibited BIM promoter transactivation in FOXO3-WT-expressing HL-60 cells (Fig. 2A), confirming results previously described in the CTLL-2 cell line. GILZ also inhibited FOXO3-induced p27KIP1 (Fig. 2B) and FASL (Fig. 2C) promoter activities. Besides acting directly on FOXO3, GILZ could alter recruitment of coactivators on these natural composite promoters. To address this hypothesis and specifically evaluate the effect of GILZ on FOXO3 transcriptional activity, we performed experiments using the p3xIRS-MLP-Luc reporter construct composed of only 3 canonical IRS elements. Results showed that this promoter displayed a very low basal activity and that GILZ strongly inhibited FOXO3-WT-induced transactivation of this synthetic promoter (Fig. 2D). Western blotting experiments revealed that coexpression of GILZ and FOXO3 did not result in a decreased amount of FOXO3 protein (Fig. 2E) suggesting that the inhibitory effect of GILZ was not due to FOXO3 degradation. Surprisingly, GILZ expression was slightly decreased when coexpressed with FOXO3. However, mRNA expression of GILZ measured by semi-quantitative reverse transcriptase-PCR was not modified, showing that this effect was not due to transcriptional interference between the two expression vectors (data not shown).
FOXO factors are known to interact with the same core consensus DNA sequence to modulate common target gene expression (2). Nevertheless, functional as well as structural differences exist between these family members. We then asked whether the inhibitory effect of GILZ was conserved among the FOXO family of transcription factors. HL-60 cells were cotransfected with expression vectors for FOXO4 and/or GILZ, along with reporter plasmids (pBim-Luc, p27-Luc, pFasL-Luc, and p3xIRS-MLP-Luc). As shown in Fig. 3, GILZ significantly reduced BIM (Fig. 3A), p27KIP1 (Fig. 3B), FASL (Fig. 3C), and p3xIRS (Fig. 3D) promoter transactivation in FOXO4-WT-expressing HL-60 cells without affecting FOXO4 protein levels (Fig. 3E). We were not able to measure the transactivation of FOXO1-induced BIM and FASL in HL-60 cells, despite obvious expression of the FOXO1 protein in this cell line upon transfection (Fig. 4B), suggesting that FOXO1 alone cannot transactivate these natural promoters in HL-60 cells. However, FOXO1 expression significantly increased p3xIRS-MLP-Luc activity (Fig. 4A) and GILZ expression strongly inhibited FOXO1-WT transcriptional activity (Fig. 4A) without affecting FOXO1 protein levels (Fig. 4B). Altogether, these results suggest a common mechanism for inhibition of FOXO transcription factors by GILZ.
GILZ Inhibition of FOXO-mediated Transcription Is Not Dependent on PKB and IKK Phosphorylation Sites
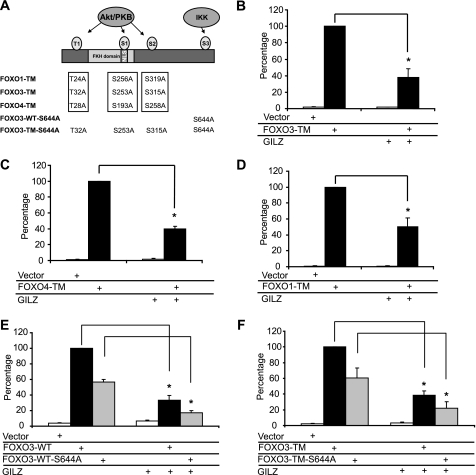
FOXO proteins are regulated by growth factor signaling through the phosphatidylinositol 3-kinase-PKB pathway. PKB phosphorylation of the two N-terminal residues (T1 and S1, Fig. 5A) generates binding sites for 14-3-3 proteins, resulting in nuclear exclusion of the FOXO·14-3-3 complex. This raised the possibility that the inhibitory effect of GILZ might be related to PKB-mediated phosphorylation of FOXO, resulting in inactivation of FOXO activity by translocation from the nucleus to the cytoplasm. To test this hypothesis, we used FOXO-TM mutants in which the three PKB phosphorylation sites were mutated to alanine (Fig. 5A). Due to these substitutions, these constitutively activated mutants are known to be predominantly located in the nucleus (22). HL-60 cells were cotransfected with pcDNA3- FOXO3-TM, pMT2-FOXO4-TM, or pcDNA3-FOXO1-TM and/or pcDNA3-Myc-GILZ, along with the p3xIRS-MLP-Luc plasmid, and transcriptional activity was analyzed after a 24-h culture. Results showed that GILZ significantly inhibits FOXO3-TM- (Fig. 5B), FOXO4-TM- (Fig. 5C), and FOXO1-TM- (Fig. 5D) induced transactivation of p3xIRS-MLP-Luc, suggesting that PKB phosphorylation sites were not involved in FOXO inhibition by GILZ. IKKβ has also been shown to interact with and phosphorylate FOXO3a in vivo and in vitro at Ser-644, promoting nuclear exclusion and proteasomal degradation independently of PKB phosphorylation (11). We performed site-directed mutagenesis experiments to generate FOXO3-WT-S644A and FOXO3-TM-S644A plasmids. We observed that these mutants displayed a lower transcriptional activity than FOXO3-WT and FOXO3-TM, respectively. However, they were still significantly inhibited by GILZ (Fig. 5, E and F) suggesting that Ser-644 phosphorylation did not play a role in the mechanism of FOXO3 inhibition by GILZ.
FIGURE 5.
GILZ inhibition of FOXO-mediated transcription is not dependent on PKB and IKK phosphorylation sites. A, schematic representation of FOXO1-TM, FOXO3-TM, and FOXO4-TM. HL-60 cells were transiently transfected with reporter plasmid p3xIRS-MLP-Luc, with 5 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc, and/or 0.5 μg of pcDNA3-FOXO3-TM (B), 1 μg of pMT2-FOXO4-TM (C), 0.5 μg of pcDNA3-FOXO1-TM (D), 1 μg of pcDNA3-FOXO3-WT-S644A (E), 0.5 μg of pcDNA3-FOXO3-TM-S644A (F), and transcriptional activity was analyzed after 24 h of culture. Results are expressed as the percentage of p3xIRS-MLP-Luc activity, with 100% representing the activity of the construct in the presence of FOXO-TM. Data represent the mean ± S.E. of three independent experiments performed in triplicate. *, p < 0.05. Error bars, S.E.M.
FOXO3 Is Mainly Cytoplasmic in the Presence of GILZ
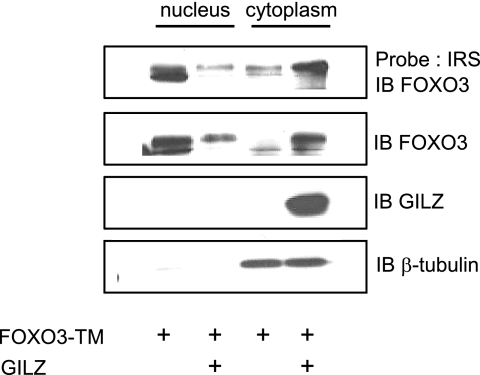
We then tested the hypothesis that GILZ could reduce the amount of FOXO3 in the nucleus. HL-60 cells were transiently transfected with pcDNA3-Myc-GILZ and/or pcDNA3-FOXO3-TM. Western blotting and DNA affinity precipitation experiments were performed with nuclear and cytoplasmic extracts. In the absence of GILZ, FOXO3-TM was predominantly located in the nucleus and bound to the IRS probe as described in the literature (Fig. 6). In the presence of GILZ, FOXO3-TM expression was decreased in the nucleus concomitantly with an increase in the cytoplasm, suggesting a cytoplasmic relocalization of FOXO3-TM (Fig. 6) that has never been described before. DNA binding experiments confirmed the Western blot with a decrease of FOXO3 DNA binding in nuclear extracts in the presence of GILZ, probably related to relocalization of FOXO3-TM in the cytoplasm. Altogether, these results suggested that GILZ did not hinder FOXO3 from binding in vitro to the IRS probe. It is also interesting to note that GILZ was localized in the cytoplasm independently of FOXO3-TM expression. The β-tubulin revelation was used to exclude a possible nuclei contamination with cytoplasmic material.
FIGURE 6.
FOXO3 is mainly cytoplasmic in the presence of GILZ. HL-60 cells transfected with 5 μg of pcDNA3-FOXO3-TM, with or without 10 μg of pcDNA3-Myc-GILZ were harvested 24 h after transfection, and nuclear and cytoplasmic extracts were performed. The 5′-biotinylated IRS probe coupled to streptavidine-agarose beads was incubated with cell extracts. Bound proteins were identified by Western blotting using the specific anti-FOXO3 antibody. A representative experiment of three is shown. IB, immunoblot.
GILZ Expression Regulates FOXO3-WT and FOXO3-TM Nucleocytoplasmic Trafficking
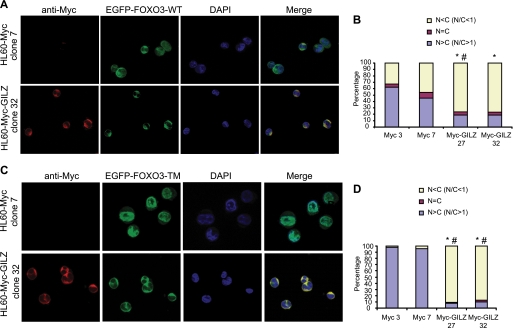
Results presented above suggested an effect of GILZ on FOXO subcellular localization. To address this hypothesis, we performed immunofluorescence experiments in GILZ-expressing HL-60 clones and in randomly selected control clones transiently transfected with pEGFP-FOXO3-WT (Fig. 7, A and B) or pEGFP-FOXO3-TM (Fig. 7, C and D). In all clones tested, GILZ was localized in the cytoplasm (Fig. 7, A and C). As expected, FOXO3-WT was distributed throughout the cell (Fig. 7A) and FOXO3-TM was predominantly localized in the nucleus in HL-60-Myc clones (Fig. 7C). However, in the presence of GILZ, FOXO3-WT was clearly relocalized to the cytoplasm (Fig. 7A). Quantification is shown on Fig. 7B, and statistical differences are found between control and GILZ clones. GILZ also had a profound and statistically significant effect (Fig. 7D) on subcellular distribution of EGFP-FOXO3-TM (Fig. 7C), causing the majority of fluorescence to be retained in the cytoplasm and excluded from the nucleus. These experiments confirmed the results obtained with nuclear and cytoplasmic extracts (Fig. 6) and showed that GILZ-induced FOXO3 cytoplasmic relocalization was independent of PKB phosphorylation.
FIGURE 7.
GILZ expression provokes FOXO3-WT and FOXO3-TM accumulation within the cytoplasm. HL-60-Myc and HL-60-Myc-GILZ clones were transiently transfected with 10 μg of pEGFP-FOXO3-WT plasmid (A) or 10 μg of pEGFP-FOXO3-TM plasmid (C). After overnight expression of exogenous proteins, cells were fixed in paraformaldehyde and stained with anti-Myc antibody and 4′,6-diamidino-2-phenylindole (DAPI) for nuclei detection. Cells were analyzed using fluorescence microscopy. 200 cells were scored according to the nuclear/cytoplasmic ratio of EGFP-FOXO3-WT (B) or EGFP-FOXO3-TM (D) fluorescence as described under “Experimental Procedures.” Percentages of HL-60-Myc-GILZ clones 27 or 32 with a N/C < 1 were compared with percentages of HL-60-Myc control clones 3 or 7 with a N/C < 1. *, p < 0.05 compared with HL-60-Myc clone 3. #, p < 0.05 compared with HL-60-Myc clone 7.
Cytoplasmic Localization of GILZ Is Required for Efficient Inhibition of FOXO-mediated Transcription
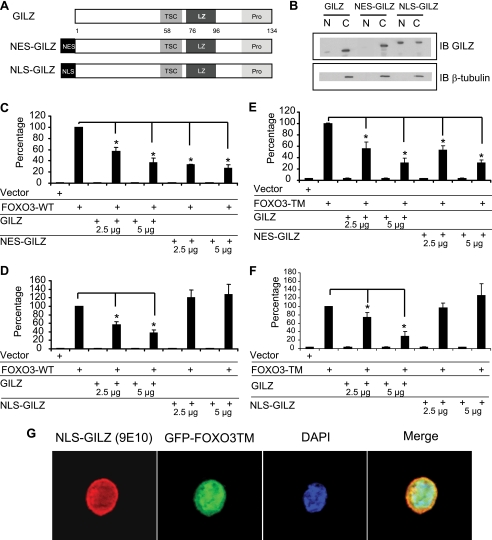
We then wondered whether the cytoplasmic localization of GILZ was necessary for FOXO3 inhibition. We constructed a NES-GILZ mutant (Fig. 8A), where the potent NES of MEK was fused to the GILZ protein, resulting in an exclusive cytoplasmic distribution of the fusion protein as assessed by Western blot on nuclear and cytoplasmic extracts (Fig. 8B). A NLS-GILZ mutant was also constructed, where the NLS of the SV40 T antigen was fused to GILZ, targeting GILZ in the nucleus (Fig. 8B). Results showed that NES-GILZ mainly behaves like wild-type GILZ with the inhibitory effect of this mutant comparable with GILZ-WT (Fig. 8, C and E). However, the NLS-GILZ protein failed to inhibit FOXO3-WT (Fig. 8D) and FOXO3-TM (Fig. 8F) transcriptional activities, suggesting that cytoplasmic localization of GILZ was mandatory for inhibition of FOXO3 transcriptional activity. Interestingly, despite the strong NLS sequence used, a minor fraction of NLS-GILZ was still cytoplasmic, but the level was probably insufficient for FOXO3 inhibition. Levels of FOXO3-WT, FOXO3-TM, GILZ, NLS-GILZ, and NES-GILZ were assessed by Western blot (supplemental Fig. S1). Results showed that NES-GILZ and NLS-GILZ did not affect FOXO3-WT or FOXO3-TM expression. Interestingly, NLS-GILZ, which no longer inhibits FOXO3 transcriptional activity (Fig. 8F), only caused a discrete cytoplasmic relocalization of GFP-FOXO3-TM as assessed by fluorescence microscopy (Fig. 8G) after transient transfection of pcDNA3- NLS-GILZ and pEGFP-FOXO3-TM. The majority of GFP-FOXO3-TM remained in the nucleus in the presence of NLS-GILZ and these results were confirmed by Western blot after nuclear and cytoplasmic fractionation (data not shown).
FIGURE 8.
Localization of GILZ in the cytoplasm is necessary for inhibition of FOXO3-WT and FOXO3-TM transcriptional activities. A, schematic representation of NES-GILZ and NLS-GILZ constructions. B, expression of GILZ, NES-GILZ, and NLS-GILZ in the cytoplasm and nucleus of HL-60 cells. Western blot was performed using anti-GILZ and anti-β-tubulin antibody. C, cytoplasmic extract; N, nuclear extract. A representative experiment of three is shown. HL-60 cells were transiently transfected with the reporter plasmid p3xIRS-MLP-Luc (5 μg) and/or increasing amounts of pcDNA3-Myc-GILZ (2.5 and 5 μg), pcDNA3-Myc-NES-GILZ (2.5 and 5 μg), and/or pcDNA3-FOXO3-WT (5 μg) (C) or pcDNA3-FOXO3-TM (0.5 μg) (E). HL-60 cells were transiently transfected with p3xIRS-MLP-Luc (5 μg) and/or increasing amounts of pcDNA3-Myc-GILZ (2.5 and 5 μg), pcDNA3-Myc-NLS-GILZ (2.5 and 5 μg), and/or pcDNA3-FOXO3-WT (5 μg) (D) or pcDNA3-FOXO3-TM (0.5 μg) (F). Results are expressed as the percentage of p3xIRS-MLP-Luc activity with 100% representing the activity of FOXO3-WT or TM. Data represent the mean ± S.E.M. of three independent experiments performed in duplicate. *, p < 0.05 compared with cells transfected with pcDNA3-FOXO3-TM. G, HL-60 cells were transiently transfected with pEGFP-FOXO3-TM plasmid (10 μg) and pcDNA3-NLS-GILZ (10 μg) or pcDNA3-Myc. After overnight expression of exogenous proteins, cells were fixed in paraformaldehyde and stained with the anti-Myc antibody and 4′,6-diamidino-2-phenylindole (DAPI). Cells were analyzed by fluorescence microscopy. Error bars, S.E.
GILZ Inhibitory Effect Requires a Functional Crm1-dependent Export of FOXO3
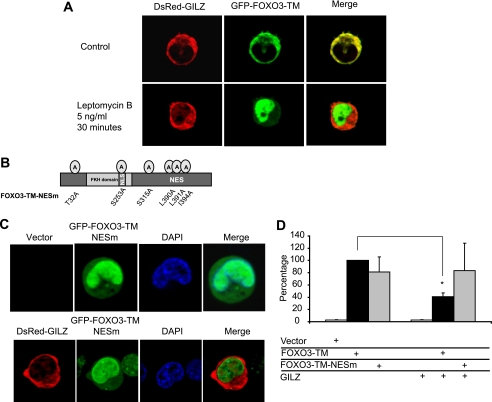
A functional Crm1-dependent hydrophobic leucine-rich nuclear export signal is located C-terminal to the DNA-binding domain of FOXO3. We wondered whether the cytoplasmic localization of FOXO3 in the presence of GILZ could be due to inhibition of nuclear import or an acceleration of nuclear export. We performed fluorescence microscopy experiments with HL-60 cells transiently transfected with DsRed-GILZ and GFP-FOXO3-TM. Within 30 min of treatment with 5 ng/ml of leptomycin B, a specific inhibitor of Crm1-dependent nuclear export, GFP-FOXO3-TM rapidly accumulated in the nucleus, in the presence or absence of GILZ (Fig. 9A). These results suggest that FOXO3-TM nuclear import is not impeded by GILZ, that GILZ does not trap FOXO3-TM in the cytoplasm, and that the effect of GILZ on FOXO3-TM cytoplasmic localization requires a functional Crm1-dependent export of FOXO3-TM that is interestingly not dependent on PKB phosphorylation.
FIGURE 9.
GILZ inhibition of FOXO3 activity requires a FOXO3 functional NES sequence. A, HL-60 cells were transiently transfected with pEGFP-FOXO3-TM plasmid (10 μg) and pDsRed-GILZ (10 μg). After overnight expression, cells were treated with or without 5 ng/ml of leptomycin B, maintained at 37 °C, and analyzed by confocal microscopy for 30 min. The images presented were acquired at 30 min after leptomycin B addition. B, schematic representation of FOXO3-TM-NESm. C, HL-60 cells were transiently transfected with pEGFP-FOXO3-TM-NESm plasmid (10 μg) and pDsRed-GILZ (10 μg) or pcDNA3-Myc. After overnight expression of exogenous proteins, cells were fixed in paraformaldehyde and stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were analyzed by fluorescence microscopy. D, HL-60 cells were transiently transfected with the reporter plasmid p3xIRS-MLP-Luc, with either 0.5 μg of pcDNA3-FOXO3-TM or pcDNA3-FOXO3-TM-NESm, 5 μg of pcDNA3-Myc-GILZ or pcDNA3-Myc, and the transcriptional activity was analyzed after 24 h of culture. Results are expressed as the percentage of p3xIRS-MLP-Luc activity, with 100% representing the activity of the construct in the presence of FOXO3-TM. Data represent the mean ± S.E. of three independent experiments performed in duplicate. *, p < 0.05. Error bars, S.E.
We then wondered whether the NES sequence of FOXO3 was necessary for the inhibitory effect of GILZ to occur. The FOXO3-TM NES sequence was invalidated by site-directed mutagenesis (Fig. 9B). Leucines 390 and 391 and isoleucine 394 of NES were replaced by alanines (FOXO3-TM-NESm). Using fluorescence microscopy, we observed that EGFP-FOXO3-TM-NESm was localized in the nucleus of HL-60 cells, in the absence and also in the presence of DsRed-GILZ (Fig. 9C), as previously demonstrated for leptomycin B treatment. We then compared the effect of GILZ on the transcriptional activity of FOXO3-TM-NESm and FOXO3-TM. HL-60 cells were transiently transfected with pcDNA3-FOXO3- TM, pcDNA3-FOXO3-TM-NESm, and/or pcDNA3-Myc-GILZ, along with the p3xIRS-MLP-Luc reporter plasmid, and the transcriptional activity was analyzed after 24 h culture (Fig. 9D). Interestingly, results showed that GILZ had no significant effect on the transcriptional activity of FOXO3-TM-NESm, suggesting that this functional export sequence played an important role in inhibition of FOXO3 by GILZ.
DISCUSSION
In the present study, we showed that GILZ inhibited expression of FOXO3 targets Bim and p27KIP1 in CTLL-2 cells, as well as in HL-60 cells expressing exogenous FOXO3 and GILZ. We demonstrated that expression of GILZ in HL-60 cells inhibited FOXO3 and FOXO4 transcriptional activities measured with the natural promoters BIM, p27KIP1, and FASL. GILZ also down-regulated FOXO1, FOXO3, and FOXO4 transcriptional activities, as measured with a synthetic promoter composed of 3 IRS sequences. These observations suggest that IRS responsive elements are sufficient for the inhibitory effect of GILZ to occur and that GILZ specifically inhibits FOXO factors independently of the promoter context. These results also rule out a mechanism where GILZ-mediated inhibition would occur through interference with specific factors loading on the BIM, p27KIP1, and FASL natural promoters. Indeed, the BIM promoter is known to be regulated by multiple signaling pathways, including those activating FOXO (5, 23), Myb (24), and AP-1 (25). Specific transcription factors have also been described to control FASL expression: Sp1 (26), NF-AT (27), Egr (early growth response protein) (28), IRF-1 (29), NF-κB (30), and AP-1 (31). Among them, AP-1 and NF-κB were reported to physically interact with GILZ, resulting in modulation of their transcriptional activities.
The ubiquitin-proteasome system has been shown to be critical for regulation of the levels of FOXO factors in cells. Indeed, PKB phosphorylation and polyubiquitylation of FOXO1 and FOXO3 lead to their degradation by the proteasome (10). Our results exclude an effect of GILZ on FOXOs degradation resulting from enhanced proteasomal degradation.
To analyze the mechanisms underlying the inhibition of FOXO factors by GILZ, we adopted a strategy to focus on FOXO3 and used the HL-60 hematopoietic cell line. In this model, GILZ inhibited the activity of natural or synthetic FOXO-responsive promoters in reporter tests systems, as well as the endogenous expression of Bim or p27KIP1. In addition, FOXO3 was not detectable at the protein level in this cell line.
Results obtained with NES- and NLS-GILZ mutants showed that the GILZ inhibitory effect on FOXO3 required cytoplasmic localization of GILZ. This observation rules out a mechanism where GILZ would behave as a transcriptional repressor in the nucleus by binding to a tandem repeat of CCAAT/enhancer-binding protein (C/EBP) binding sites as previously described for the PPARγ2 promoter (32). We also found that in CTLL-2 and HL-60 cells GILZ was exclusively cytoplasmic, as evaluated either by immunofluorescence or Western blot detection after nuclear and cytoplasmic fractionation. The localization of GILZ is still a matter of debate. In the murine T-cell hybridoma 3DO transfected with a GILZ expression vector, GILZ was detected in the nucleus (15). In adipocytes, GILZ was described as a nuclear protein binding specifically to DNA (32). However, in COS-7 cells transfected with Myc-GILZ and analyzed by fluorescence microscopy, GILZ was found in the cytoplasm (33).
Our observations clearly showed that GILZ inhibited FOXO factors, independently of PKB and IKK phosphorylation sites. This is a surprising result because inactivation of FOXO factors by PKB is the major mechanism described in the literature. FOXO3-WT-S644A and TM-S644A mutants displayed a lower transcriptional activity, which could be attributed to localization of the Ser-644 in the acidic transactivation domain in close proximity to the core region. This was also observed with a FOXO3 ΔCter mutant in which a STOP codon was inserted after amino acid 604, deleting most of the transactivation domain (including Ser-644). Despite a weak transcriptional activity, this mutant was still inhibited by GILZ4 confirming that IKK was not involved in the mechanism of FOXO3 inhibition by GILZ. Nevertheless, FOXO isoforms are also subject to numerous other post-translational modifications that could be affected by GILZ. Recently, phosphorylation of FOXO3 on different regulatory sites by the AMP-activated protein kinase was shown to activate FOXO3 transcriptional activity without affecting its subcellular localization (34). MST1 has been described to interact with FOXO3 and phosphorylate Ser-207, promoting nuclear translocation (35). FOXO1 phosphorylation on Ser-249 by cyclin-dependent kinase 2 has also been shown to result in cytoplasmic localization and inhibition of FOXO1 transcriptional activity (36). Interestingly, in the experiments described in Fig. 6, FOXO3 was mainly cytoplasmic in the presence of GILZ and the cytoplasmic FOXO3 band was shifted, suggesting the existence of a phosphorylated form of FOXO3. Our hypothesis is that inhibition of FOXO3 by GILZ is due to relocalization of FOXO3 to the cytoplasm. GILZ may promote phosphorylation of FOXO factors on regulatory sites affecting their nucleocytoplasmic localization.
GILZ expression in HL-60 cells transfected with EGFP-FOXO3-TM caused the cytoplasmic localization of FOXO3-TM, thus independently of PKB phosphorylation on Thr-32, Ser-253, and Ser-315. Cytoplasmic localization of transcription factors in the presence of GILZ was also described for NF-κB (13), but as the consequence of altered nuclear translocation resulting from formation of GILZ-NF-κB complexes. The proline-rich as well as the leucine zipper regions of GILZ were shown to be crucial for this protein-protein interaction (14). GILZ can also form a ternary complex with activated Ras and Raf, resulting in inhibition of ERK and AP-1 signaling pathways (37). In summary, GILZ has been described to possess various dimerization partners resulting in modulation of transcription factor activity and signaling pathways (13, 37, 38). However, in HL-60 cells, FOXO3 and GILZ, although clearly colocalized in the cytoplasm, did not physically interact as assessed by coimmunoprecipitation experiments (supplemental Fig. S2) or by fishing with a GST-GILZ fusion protein (data not shown) despite high levels of GILZ and FOXO3 expression. As a positive control, we were able to detect in the same experimental conditions the previously described interaction of FOXO3 with 14-3-3. Altogether, our results suggest that GILZ would not directly interact with FOXO3, but could rather interact and/or modulate the activity of another partner involved in regulation of FOXO3 localization.
The mechanism of inhibition could rely on a cytoplasmic retention, a reduced import, or an accelerated export of FOXO3 in the presence of GILZ. Our results obtained with the FOXO3-TM-NESm mutant favor a mechanism where GILZ would promote and/or accelerate the export of FOXO3 from the nucleus to the cytoplasm. When sequestered in the nucleus by invalidation of its export sequence, FOXO3 transcriptional activity was preserved despite the presence of GILZ. Upon inhibition of FOXO3 nuclear export by leptomycin B, import of FOXO3 was rapidly completed, ruling out a role for GILZ on FOXO3 nuclear import and consistent with the absence of physical interaction between GILZ and FOXO3. GILZ could modulate Crm1-NES interactions either through promotion of conformational changes in the NES region or by enhancing the affinity of Crm1 for FOXO3. This could occur through post-translational modifications such as phosphorylations. For example, Engel et al. (39) have observed that the MAPK AP kinase 2 is phosphorylated in the nucleus and is rapidly translocated to the cytoplasm due to unmasking of its NES upon conditions of stress such as ultraviolet light or H2O2. It is interesting to note that our results were obtained with FOXO3-TM, suggesting that these post-translational modifications would occur on residues distinct from the classical PKB phosphorylation sites. In support for this hypothesis, Clavel et al. (40) recently described that activation of the c-Jun NH2-terminal kinase (JNK) signaling pathway in C2C12 muscle cells could induce nuclear export of FOXO3 through Crm1 and independently of PKB.
Our results suggest that GILZ is a natural inhibitor of FOXO factor transcriptional activities in hematopoietic cells with an original mechanism of action. Our hypothesis is an inhibitory effect of GILZ requiring its cytoplasmic localization and promoting nuclear exclusion of FOXO3 in a Crm1-dependent manner. The regulation of nucleocytoplasmic distribution of transcription factors is emerging as one of the most efficient mechanisms to adjust gene expression to the cell environment. This mechanism allows to finely control access of the transcriptional regulators to their target genes as shown here with FOXO factors. More importantly, these findings strongly suggest that expression of GILZ could lead to the general inhibition of FOXO signaling pathways affecting functions such as cell cycle regulation or cell death.
Acknowledgments
We thank Marine Petitpretz, Angélique Lebouvier, and Stefanie Burleson for help with GILZ and FOXO3 mutant testing. A special thanks to Christophe Klein for assistance in fluorescence images analyses.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
P. Latre de Late, unpublished data.
- PKB
- protein kinase B
- GILZ
- glucocorticoid-induced leucine zipper
- IL
- interleukin
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- EGFP
- enhanced green fluorescent protein
- IRS
- insulin response sequence
- MLP
- major late promoter
- N/C
- nuclear/cytoplasmic ratio
- NES
- nuclear export signal.
REFERENCES
- 1.Huang H., Tindall D. J. (2007) J. Cell Sci. 120, 2479–2487 [DOI] [PubMed] [Google Scholar]
- 2.Furuyama T., Nakazawa T., Nakano I., Mori N. (2000) Biochem. J. 349, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl M., Dijkers P. F., Kops G. J., Lens S. M., Coffer P. J., Burgering B. M., Medema R. H. (2002) J. Immunol. 168, 5024–5031 [DOI] [PubMed] [Google Scholar]
- 4.You H., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T. W. (2006) J. Exp. Med. 203, 1657–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 6.Mei Y., Zhang Y., Yamamoto K., Xie W., Mak T. W., You H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5153–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin L., Hron J. D., Peng S. L. (2004) Immunity 21, 203–213 [DOI] [PubMed] [Google Scholar]
- 8.Tothova Z., Kollipara R., Huntly B. J., Lee B. H., Castrillon D. H., Cullen D. E., McDowell E. P., Lazo-Kallanian S., Williams I. R., Sears C., Armstrong S. A., Passegué E., DePinho R. A., Gilliland D. G. (2007) Cell 128, 325–339 [DOI] [PubMed] [Google Scholar]
- 9.Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J. W., Carrasco D. R., Jiang S., Gilliland D. G., Chin L., Wong W. H., Castrillon D. H., DePinho R. A. (2007) Cell 128, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plas D. R., Thompson C. B. (2003) J. Biol. Chem. 278, 12361–12366 [DOI] [PubMed] [Google Scholar]
- 11.Hu M. C., Lee D. F., Xia W., Golfman L. S., Ou-Yang F., Yang J. Y., Zou Y., Bao S., Hanada N., Saso H., Kobayashi R., Hung M. C. (2004) Cell 117, 225–237 [DOI] [PubMed] [Google Scholar]
- 12.Mittelstadt P. R., Ashwell J. D. (2001) J. Biol. Chem. 276, 29603–29610 [DOI] [PubMed] [Google Scholar]
- 13.Ayroldi E., Migliorati G., Bruscoli S., Marchetti C., Zollo O., Cannarile L., D'Adamio F., Riccardi C. (2001) Blood 98, 743–753 [DOI] [PubMed] [Google Scholar]
- 14.Di Marco B., Massetti M., Bruscoli S., Macchiarulo A., Di Virgilio R., Velardi E., Donato V., Migliorati G., Riccardi C. (2007) Nucleic Acids Res. 35, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Adamio F., Zollo O., Moraca R., Ayroldi E., Bruscoli S., Bartoli A., Cannarile L., Migliorati G., Riccardi C. (1997) Immunity 7, 803–812 [DOI] [PubMed] [Google Scholar]
- 16.Asselin-Labat M. L., Biola-Vidamment A., Kerbrat S., Lombès M., Bertoglio J., Pallardy M. (2005) Mol. Endocrinol. 19, 1752–1764 [DOI] [PubMed] [Google Scholar]
- 17.Berrebi D., Bruscoli S., Cohen N., Foussat A., Migliorati G., Bouchet-Delbos L., Maillot M. C., Portier A., Couderc J., Galanaud P., Peuchmaur M., Riccardi C., Emilie D. (2003) Blood 101, 729–738 [DOI] [PubMed] [Google Scholar]
- 18.Eddleston J., Herschbach J., Wagelie-Steffen A. L., Christiansen S. C., Zuraw B. L. (2007) J. Allergy Clin. Immunol. 119, 115–122 [DOI] [PubMed] [Google Scholar]
- 19.Asselin-Labat M. L., David M., Biola-Vidamment A., Lecoeuche D., Zennaro M. C., Bertoglio J., Pallardy M. (2004) Blood 104, 215–223 [DOI] [PubMed] [Google Scholar]
- 20.Biola A., Lefebvre P., Perrin-Wolff M., Sturm M., Bertoglio J., Pallardy M. (2001) Mol. Endocrinol. 15, 1062–1076 [DOI] [PubMed] [Google Scholar]
- 21.Dijkers P. F., Medema R. H., Pals C., Banerji L., Thomas N. S., Lam E. W., Burgering B. M., Raaijmakers J. A., Lammers J. W., Koenderman L., Coffer P. J. (2000) Mol. Cell. Biol. 20, 9138–9148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin I., Bakin A. V., Rodeck U., Brunet A., Arteaga C. L. (2001) Mol. Biol. Cell 12, 3328–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilley J., Coffer P. J., Ham J. (2003) J. Cell Biol. 162, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas S. C., Liu D. X., Greene L. A. (2005) J. Neurosci. 25, 8349–8358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas S. C., Shi Y., Sproul A., Greene L. A. (2007) J. Biol. Chem. 282, 29368–29374 [DOI] [PubMed] [Google Scholar]
- 26.Xiao S., Marshak-Rothstein A., Ju S. T. (2001) Eur. J. Immunol. 31, 3339–3348 [DOI] [PubMed] [Google Scholar]
- 27.Kasibhatla S., Genestier L., Green D. R. (1999) J. Biol. Chem. 274, 987–992 [DOI] [PubMed] [Google Scholar]
- 28.Mittelstadt P. R., Ashwell J. D. (1998) Mol. Cell. Biol. 18, 3744–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow W. A., Fang J. J., Yee J. K. (2000) J. Immunol. 164, 3512–3518 [DOI] [PubMed] [Google Scholar]
- 30.Savickiene J., Treigyte G., Pivoriunas A., Navakauskiene R., Magnusson K. E. (2004) Ann. N. Y. Acad. Sci. 1030, 569–577 [DOI] [PubMed] [Google Scholar]
- 31.Baumann S., Hess J., Eichhorst S. T., Krueger A., Angel P., Krammer P. H., Kirchhoff S. (2003) Oncogene 22, 1333–1339 [DOI] [PubMed] [Google Scholar]
- 32.Shi X., Shi W., Li Q., Song B., Wan M., Bai S., Cao X. (2003) EMBO Rep. 4, 374–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayroldi E., Zollo O., Macchiarulo A., Di Marco B., Marchetti C., Riccardi C. (2002) Mol. Cell. Biol. 22, 7929–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greer E. L., Oskoui P. R., Banko M. R., Maniar J. M., Gygi M. P., Gygi S. P., Brunet A. (2007) J. Biol. Chem. 282, 30107–30119 [DOI] [PubMed] [Google Scholar]
- 35.Lehtinen M. K., Yuan Z., Boag P. R., Yang Y., Villén J., Becker E. B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T. K., Bonni A. (2006) Cell 125, 987–1001 [DOI] [PubMed] [Google Scholar]
- 36.Huang H., Regan K. M., Lou Z., Chen J., Tindall D. J. (2006) Science 314, 294–297 [DOI] [PubMed] [Google Scholar]
- 37.Ayroldi E., Zollo O., Bastianelli A., Marchetti C., Agostini M., Di Virgilio R., Riccardi C. (2007) J. Clin. Invest. 117, 1605–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redjimi N., Gaudin F., Touboul C., Emilie D., Pallardy M., Biola-Vidamment A., Fernandez H., Prévot S., Balabanian K., Machelon V. (2009) Mol. Cancer 8, 83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel K., Kotlyarov A., Gaestel M. (1998) EMBO J. 17, 3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clavel S., Siffroi-Fernandez S., Coldefy A. S., Boulukos K., Pisani D. F., Dérijard B. (2010) Mol. Cell. Biol. 30, 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]