Abstract
Yellow proteins form a large family in insects. In Drosophila melanogaster, there are 14 yellow genes in the genome. Previous studies have shown that the yellow gene is necessary for normal pigmentation; however, the roles of other yellow genes in body coloration are not known. Here, we provide the first evidence that yellow-e is required for normal body color pattern in insect larvae. In two mutant strains, bts and its allele bts2, of the silkworm Bombyx mori, the larval head cuticle and anal plates are reddish brown instead of the white color found in the wild type. Positional cloning revealed that deletions in the Bombyx homolog of the Drosophila yellow-e gene (Bmyellow-e) were responsible for the bts/bts2 phenotype. Bmyellow-e mRNA was strongly expressed in the trachea, testis, and integument, and expression markedly increased at the molting stages. This profile is quite similar to that of Bmyellow, a regulator of neonatal body color and body markings in Bombyx. Quantitative reverse transcription-PCR analysis showed that Bmyellow-e mRNA was heavily expressed in the integument of the head and tail in which the bts phenotype is observed. The present results suggest that Yellow-e plays a crucial role in the pigmentation process of lepidopteran larvae.
Keywords: Development Differentiation, Evolution/Protein, Gene/Mapping, Gene/Regulation, Genetics, Organisms/Insect, Insect, Melanine Synthesis, Silkworm
Introduction
Major royal jelly proteins (MRJPs)2 were initially identified as major constituents of royal jelly; they constitute 80–90% of total royal jelly proteins, which play a central role in honeybee development (1). The yellow gene family of Drosophila melanogaster is known to encode the MRJP domain-containing protein (2) and to consist of 14 genes (3). The D. melanogaster yellow gene (Dmyellow) determines the degree and pattern of melanization by cooperating with the ebony gene (4). Dmyellow also controls male courtship behavior by its temporal expression in the brain (5). Furthermore, biochemical experiments revealed that the products of two other yellow genes, Dmyellow-f and Dmyellow-f2, have dopachrome-conversion enzyme activity (6). Despite the widespread use of the yellow gene as a visible genetic marker, there is little information regarding the functions of other members of the MRJP/YELLOW protein family in Drosophila and other organisms.
Larval color variations are often observed in many lepidopteran insects. About 40 body color mutants have been reported in the silkworm, Bombyx mori (7, 8). Recently, two mutants, chocolate (ch) and sooty (so), were characterized molecularly. In the ch mutants, the body color of neonatal larvae and the body markings of older instar larvae are reddish brown instead of normal black. Mutations at the so locus produce smoky larvae and black pupae. Linkage analysis and genomic studies revealed that Bombyx yellow and ebony are the genes responsible for the ch and so mutation, respectively (9). These results suggest that Yellow promotes and Ebony inhibits melanization in Lepidoptera and that melanin-synthesis enzymes play a critical role in the color pattern of lepidopteran larvae.
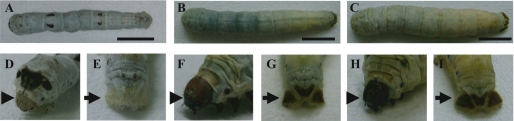
In the bts (brown head and tail spot) body color mutant strain of B. mori, the larval head cuticle and anal plates are reddish brown instead of the white color found in the wild type (Fig. 1). In the bts2 strain, which is an allele of the bts gene, head and tail spots are darker than those of the bts strain (Fig. 1) (7, 8). Both genes were mapped at 30.1 centimorgan units in the Bombyx genetic linkage group 17 (10); these genes are closely linked to the Nid-1 gene (controls nonsusceptibility to infection by BmDNV-1, 31.1 centimorgans) and the ow gene (larval skin moderately translucent, 36.4 centimorgans) (11).
FIGURE 1.
Wild-type and brown head and tail spot (bts and bts2) strains of B. mori larvae. A, wild-type strain (+bts/+bts or +bts2/+bts2) p50T. B, bts strain (bts/bts) w17. C, bts2 strain (bts2/bts2) No. 743. D and E, head and anal plates of the wild-type strain; both plates are white. F and G, head and anal plates of the bts strain; the color is reddish brown. H and I, head and anal plates of the bts2 strain; the color is dark brown. Arrowheads and arrows indicate head and anal plates, respectively. Scale bar, 10 mm.
We recently succeeded in positional cloning of the genes responsible for two Bombyx mutants, ow and nsd-2 (controls nonsusceptibility to infection by BmDNV-2), which also map to linkage group 17 (12, 13). In this study, using PCR markers and genomic information obtained in previous studies, we performed positional cloning of bts. We found that it is the Bombyx homolog of the Drosophila yellow-e gene (Bmyellow-e) and that independent deletions were responsible for the bts and bts2 mutations. Reverse transcription (RT)-PCR analysis revealed that Bmyellow-e mRNA was strongly expressed in the trachea, testis, and integument and that its expression markedly increased at the molting and newly ecdysed stages. Also, Bmyellow-e mRNA was detected at a high level in the integument of the head and tail in which the bts phenotype is observed. This is the first report showing that a Yellow homolog other than Yellow is involved in the determination of larval body color patterns in insects.
EXPERIMENTAL PROCEDURES
Silkworm Strains
The bts strains (bts/bts) u49, w17, and w43 were obtained from Kyushu University, and the bts2 strain (bts2/bts2) No. 743 was given to us by National Institute of Agrobiological Sciences. Three strains, p50T, C108T (University of Tokyo), and u42 (Kyushu University), were used as wild-type strains (+bts/+bts or +bts2/+bts2). F1 offspring were produced from a single pair cross between a female p50T and male w17. For linkage and recombination analysis, F2 progeny from the cross (p50T × w17) × (p50T × w17) and BC1 progeny from w17 × (p50T × w17) were used. All silkworm larvae were reared on mulberry leaves at 25 °C.
Positional Cloning
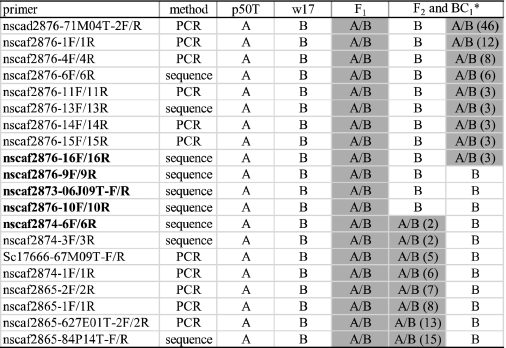
Positional cloning of the bts candidate gene was performed as described previously (12, 13). PCR and single nucleotide polymorphism (SNP) markers that showed polymorphism between the parents was generated at each position in linkage group 17. One hundred nine F2 and 464 BC1 individuals with the bts phenotype were used for mapping studies (Table 1 and supplemental Table 1). Candidate genes in the region narrowed by linkage analysis were annotated using KAIKObase, KAIKO blast, and NCBI blast (blast.ncbi.nlm.nih.gov).
TABLE 1.
Linkage analysis of F2 and BC1 segregants
A indicates p50T homozygous; B indicates w17 homozygous, and A/B indicates heterozygous genotype. Shaded sections indicate heterozygous genotypes. The numbers shown in parentheses indicate the number of heterozygous genotypes (A/B). The bts-linked region is between nscaf2876-16F/16R and nscaf2874-6F/6R.
* In linkage analysis, 109 F2 and 464 BC1 individuals with the bts phenotype were used.
Genomic PCR and RT-PCR
Genomic DNA was isolated from a small portion of the body of 3rd or 4th instar larvae using DNAzol (Invitrogen) according to the manufacturer's protocol. PCR was performed using Ex Taq (Takara Bio, Japan) and the primer sets listed in supplemental Table 1.
Total RNA was prepared using TRIzol (Invitrogen); it was then reverse-transcribed using oligo(dT)12–18 primer (Invitrogen) and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's protocol. RT-PCR was performed using Ex Taq (Takara) as described previously (13).
Cloning and Sequencing of the bts Candidate Genes
To clone the bts candidate genes, we performed RT-PCR using cDNA prepared from the whole body of day 0 3rd instar larvae. Primer sets for PCR of 12 bts candidate genes (Chinese gene models BGIBMGA007217, 007218, 007219, 007220, 007221, 007222, 007236, 007237, 007238, 007253, 007308, and 007309) are listed in supplemental Table 1. PCR products were cloned into a pGEM-T easy vector (Promega, Madison, WI) and sequenced using an ABI3130xl genetic analyzer (Applied Biosystems, Foster City, CA).
Cloning and Sequencing of the Full-length Bmyellow-e cDNA
The full-length Bmyellow-e cDNA of the wild-type strain (p50T) was cloned by 5′- and 3′-rapid amplification of cDNA ends, using the GeneRacer kit (Invitrogen) according to the manufacturer's protocol. Primer sets used in 5′- and 3′-rapid amplification of cDNA ends experiments are listed in supplemental Table 1.
Prediction of BmYellow-e Motifs
Motifs of BmYellow-e were predicted using an NCBI blastp search (blast.ncbi.nlm.nih.gov), a MOTIF search, and an InterProScan sequence search.
Quantitative RT-PCR (qRT-PCR)
qRT-PCR experiments were performed with 2× Power SYBR® Green PCR Master Mix (Applied Biosystems) using an ABI PRISM 7000 sequence detection system (Applied Biosystems), on the manufacturer's protocol. Primers used in qRT-PCR are listed in supplemental Table 1. Amounts of Bmyellow-e mRNA were normalized with those of 18 S RNA.
Alignment and Phylogenetic Analysis
Amino acid sequences of Yellow proteins were aligned using the ClustalX program (14), and phylogenetic trees were constructed by neighbor-joining methods, using the MEGA4 program (15) as described previously (16). The amino acid sequences used in the phylogenetic studies are listed in Table 2.
TABLE 2.
Abbreviation table of proteins used in phylogenetic trees in Fig. 4
| Abbreviation | Species | Protein | GenBankTM accession no. |
|---|---|---|---|
| BmYellow | B. mori | Yellow | AB438999 |
| BmYellow-b | B. mori | Yellow-b | DQ358083 |
| BmYellow-c | B. mori | Yellow-c | DQ358081 |
| BmYellow-d | B. mori | Yellow-d | DQ358079 |
| BmYellow-e (bts) | B. mori | Yellow-e | AB489224 |
| BmYellow-fa | B. mori | Yellow-fa | DQ358080 |
| BmYellow-fb | B. mori | Yellow-fb | DQ358082 |
| BmYellow-f2 | B. mori | Yellow-f2 | DQ358084 |
| DmYellow | D. melanogaster | Yellow | AAF45497 |
| DmYellow-b | D. melanogaster | Yellow-b | NP_523586 |
| DmYellow-c | D. melanogaster | Yellow-c | AAQ09899 |
| DmYellow-d | D. melanogaster | Yellow-d | NP_523820 |
| DmYellow-d2 | D. melanogaster | Yellow-d2 | NP_611788 |
| DmYellow-e | D. melanogaster | Yellow-e | NP_524344 |
| DmYellow-e2 | D. melanogaster | Yellow-e2 | NP_650289 |
| DmYellow-e3 | D. melanogaster | Yellow-e3 | NP_650288 |
| DmYellow-f | D. melanogaster | Yellow-f | NP_524335 |
| DmYellow-f2 | D. melanogaster | Yellow-f2 | NP_650247 |
| DmYellow-g | D. melanogaster | Yellow-g | NP_523888 |
| DmYellow-g2 | D. melanogaster | Yellow-g2 | NP_647710 |
| DmYellow-h | D. melanogaster | Yellow-h | NP_651912 |
| DmYellow-k | D. melanogaster | Yellow-k | NP_648772 |
| ApYellow-e | Acyrthosiphon pisum | Yellow-e | XP_001948479 |
| AaYellow-e | Aedes aegypti | Yellow-e | XP_001655634 |
| AgYellow-e | Anopheles gambiae str. PEST | Yellow-e | XP_313014 |
| AmYellow-e | Apis mellifera | Yellow-e | XP_001123314 |
| CqYellow-e | Culex quinquefasciatus | Yellow-e | XP_001847272 |
| DaYellow-e | Drosophila ananassae | Yellow-e | XP_001953104 |
| DeYellow-e | Drosophila erecta | Yellow-e | XP_001980269 |
| DgYellow-e | Drosophila grimshawi | Yellow-e | XP_001989896 |
| DmoYellow-e | Drosophila mojavensis | Yellow-e | XP_001999125 |
| DpeYellow-e | Drosophila persimilis | Yellow-e | XP_002013176 |
| DpsYellow-e | Drosophila pseudoobscura pseudoobscura | Yellow-e | XP_001359002 |
| DseYellow-e | Drosophila sechellia | Yellow-e | XP_002031328 |
| DsiYellow-e | Drosophila simulans | Yellow-e | XP_002103565 |
| DvYellow-e | Drosophila virilis | Yellow-e | XP_002056072 |
| DwYellow-e | Drosophila willistoni | Yellow-e | XP_002073083 |
| DyYellow-e | Drosophila yakuba | Yellow-e | XP_002097532 |
| NvYellow-e | Nasenia vitripennis | Yellow-e | XP_001603485 |
| PhYellow-e | Pediculus humanus corporis | Yellow-e | EEB16617 |
| TcYellow-e | Tribolium castaneum | Yellow-e | XP_001812634 |
RESULTS
Mapping of the bts Mutation
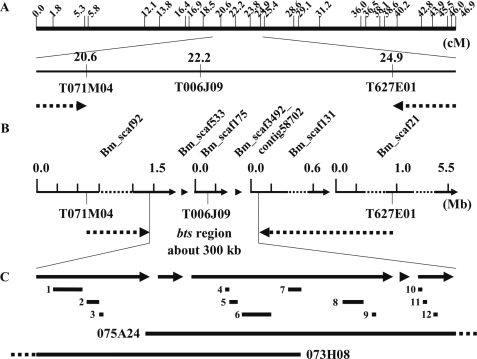
To identify a candidate region for the bts mutation, we performed genetic linkage analysis using primer sets designed for the SNP linkage map (17) and the Bombyx genome sequence (supplemental Table 1) (18). First, we roughly mapped the bts mutation using 109 F2 and 464 BC1 individuals and narrowed the bts-linked region to between the T7 ends of two bacterial artificial chromosome clones, 071M04 and 627E01 (Fig. 2A). This region was estimated to be 4.3 centimorgans long according to the SNP map (17). In this region, we identified six scaffolds, Bm_scaf92, Bm_scaf533, Bm_scaf175, Bm_scaf3492_contig58702, Bm_scaf131, and Bm_scaf21, using KAIKObase (Fig. 2B). By further mapping with primers on these scaffolds, we narrowed the bts-linked region to a position between 1,490,311 bp of Bm_scaf92 and the 28,906 bp of Bm_scaf131 (Fig. 2B and Table 1). This region was covered by two bacterial artificial chromosome clones, 073H08 and 075A23 (12), and was estimated to be ∼300 kb in length (Fig. 2C).
FIGURE 2.
Mapping of the bts mutation in linkage group 17. A, SNP markers and genetic analysis. Upper panel indicates the positions of SNP markers from bacterial artificial chromosome end sequences (17). Distances are shown in centimorgans(cM). In the lower panel, the dotted arrows indicate the result of rough mapping to narrow the region linked to the bts mutation. B, genomic scaffolds on the narrowed region and fine structure mapping. The dotted arrows indicate the result of detailed linkage analysis to narrow the region linked to the bts mutation. C, gene annotation in the bts-linked region. Arrows correspond to the scaffolds shown in B. Upper bars indicate 12 predicted genes and lower bars indicate 2 bacterial artificial chromosome clones, 075A24 and 073H08. 1, BGIBMGA007253; 2, 007308; 3, 007309; 4, 007220; 5, 007219; 6, 007221; 7, 007218; 8, 007217; 9, 007222; 10, 007238; 11, 007237; and 12, 007239.
Identification of the bts Candidate Gene
We predicted 12 bts candidate genes, BGIBMGA007217, 007218, 007219, 007220, 007221, 007222, 007236, 007237, 007238, 007253, 007308, and 007309, using a Chinese gene model search within the narrowed region (Fig. 2C). First, we investigated whether these candidate genes were expressed in the whole body of the day 0 3rd instar larvae. RT-PCR analysis showed evidence for expressions of 7 of the 12 candidate genes: BGIBM007218, 007222, 007236, 007237, 007238, 007253, and 007309 (data not shown). Next, we cloned and sequenced these seven genes and compared the sequences between wild-type (p50T, C108T, and u42) and bts strains (u49, w17, and w43). These data revealed that one of the genes, BGIBMGA007253, had a deletion in the open reading frame of all of the bts strains (data not shown). On the other hand, we did not detect any mutations in the coding regions of the other genes. Below, we characterize BGIBMGA007253 in detail as the candidate for bts.
Bombyx Yellow-e (Bmyellow-e) Is Responsible for the bts Phenotype
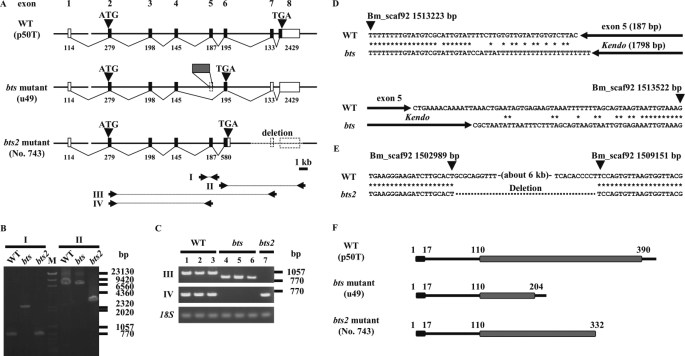
We first attempted to find expressed sequence tag clones corresponding to the bts candidate gene; however, we did not find the transcripts in any cDNA library. Therefore, we cloned and determined complete cDNA sequences of the bts genes from the wild-type (p50T), bts (u49), and bts2 (No. 743) strains. The wild-type candidate gene was 3691 bp long, potentially encoding a protein of 410 amino acids, and consisted of 8 exons and 7 introns (Fig. 3A). The open reading frame of the bts strain was spliced from exon 4 to exon 6 and lacked exon 5 (Fig. 3A). This deletion resulted in a frameshift mutation and generated a premature stop codon (Fig. 3A). DNA sequencing of the corresponding genomic region showed that deletion of exon 5 was due to insertion of the retrotransposon, non-long terminal repeat Kendo (Fig. 3, A and D) (19). We observed that the open reading frame from the bts2 strain had an abnormal exon 6, which was 385 bp longer than that from the wild-type strain, and also caused a premature stop codon (Fig. 3A). Genomic sequencing revealed that the bts2 strain had a deletion of ∼6 kb, thus resulting in a loss of exons 7 and 8 (Fig. 3, A–C, and E).
FIGURE 3.
Schematic structures of the bts candidate gene from the wild-type (WT), bts, and bts2 strains. A, genomic structure of the bts candidate gene from the WT strain p50T (upper), the bts strain u49 (middle), and the bts2 strain No. 743 (lower). Bars and boxes indicate genomic DNA sequences and exons (black box, coding regions; white box, noncoding regions), respectively. Exon 1 is located on scaffold Bm_scaf175, whereas exons 2–8 are Bm_scaf92. Numbers show the lengths of exons. Arrowheads show the start and stop codons. The gray box of u49 indicates insertion of the retrotransposon, non-LTR Kendo. The dotted line in the structure for the bts2 strain indicates the observed genomic deletion. Primer sets used for genomic PCR and reverse transcription PCR are also shown (I–IV). B, genomic PCR. M, λ, and ϕ×174 DNAs were digested separately with HindIII and HincII, and mixed as molecular markers. I and II indicate the primer set of A. C, RT-PCR analysis in seven strains. Lane 1, p50T; lane 2, C108T; lane 3, u42; lane 4, u49; lane 5, w17; lane 6, w43; lane 7, No. 743. Lanes 1–3, WT strains; lanes 4–6, bts strains; lane 7, bts2 strain. III and IV indicate the primer set of A. D, genomic position and flanking sequences of the transposable insertion detected in the bts strain. Asterisks indicate identical nucleotides. E, genomic position of a deletion found in the bts2 strain. F, protein structure of WT, bts mutant, and bts2 mutant strains. Black and gray boxes indicate the signal-peptide and major royal jelly protein motif (MRJP), respectively.
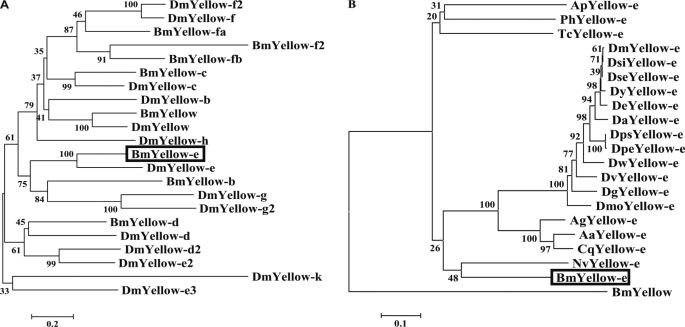
MOTIF and InterProScan searches suggested that this protein had an MRJP motif and a putative signal sequence (Fig. 3F). A portion of the MRJP motif was lacking in both of the bts and bts2 mutants (Fig. 3F), suggesting that it is a critical part of this protein responsible for the normal skin color phenotype in Bombyx. Together with these results, we concluded that this candidate is the gene responsible for the bts phenotype. A BLAST search revealed that the deduced amino acid sequence of the candidate gene showed high homology with the product of the Drosophila yellow-e gene. Phylogenetic studies using 22 Yellow proteins from D. melanogaster and B. mori indicated that this gene is a Bombyx homolog of Drosophila yellow-e (Fig. 4A), which we termed Bmyellow-e (B. mori yellow-e). BmYellow-e clustered with Yellow-e related proteins from dipteran insects such as fruit fly, mosquito, and wasp and was most closely related to that of the parasitic wasp, Nasonia vitripennis (Fig. 4B). Although yellow-e-related genes from some insects have been deposited in public data bases, their functions are still unknown.
FIGURE 4.
Phylogenetic analysis of BmYellow-e. A, phylogenetic tree constructed from 22 Yellow proteins of D. melanogaster (Dm) and B. mori (Bm). B, phylogenetic analysis of BmYellow-e and Yellow-e proteins from other insects. Amino acid sequences were aligned using the ClustalX program, and the phylogenetic tree was constructed by the neighbor-joining method. Bootstrap values after 1000 replications are shown. GenBankTM accession numbers and abbreviation of each protein are given in Table 2. Boxes indicate the bts gene (BmYellow-e).
Expression Profiles of Bmyellow-e
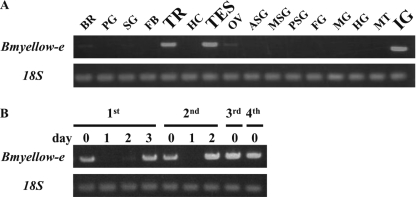
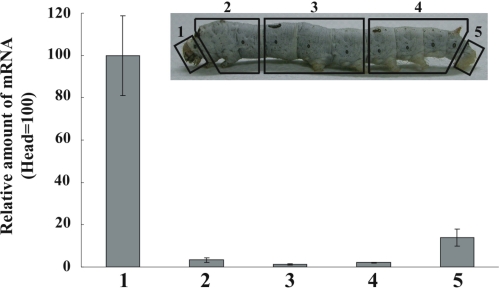
RT-PCR analysis of expression profiles of Bmyellow-e showed that it was strongly expressed in the trachea, testis, and integument in 5th instar larvae (Fig. 5A); furthermore, its expression increased markedly at the molting and newly ecdysed stages (Fig. 5B). These expression profiles are quite similar to those of Bmyellow, a Bombyx homolog of Drosophila yellow (9), suggesting that these two genes coordinately play crucial roles in determining body color pattern in Bombyx. To examine spatial expression of Bmyellow-e in larval integument, we performed qRT-PCR analysis using cDNA prepared from the integument of five compartments (Fig. 6A), and we found that it was heavily expressed in head and anal plates, where reddish coloration occurs in the bts strain (Fig. 6B). Expression of Bmyellow has been shown to correlate with the presumptive black markings but not with the white striped region (9). These results suggest that, although the temporal expression patterns of these two yellow genes are similar, their spatial regulation is different.
FIGURE 5.
Expression profiles of Bmyellow-e. A, RT-PCR analysis using cDNAs from 16 tissues of the WT strain p50T. 1st lane, brain (BR); 2nd lane, prothoracic gland (PG); 3rd lane, salivary gland (SG); 4th lane, fat body (FB); 5th lane, trachea (TR); 6th lane, hemocyte (HC); 7th lane, testis (TES); 8th lane, ovary (OV); 9th lane, anterior silk gland (ASG); 10th lane, middle silk gland (MSG); 11th lane, posterior silk gland (PSG); 12th lane, foregut (FG); 13th lane, midgut (MG); 14th lane, hindgut (HG); 15th lane, Malpighian tubule (MT); and 16th lane, integument (IG). B, RT-PCR analysis using cDNAs from whole body of different larval stages of the WT strain p50T. Day 0 from 1st, 2nd, 3rd, and 4th is the newly ecdysed stages and day 3 from 1st and day 2 from 2nd are molting stages.
FIGURE 6.
Quantitative RT-PCR analysis of Bmyellow-e mRNA in the integument of five compartments. Upper figure shows the positions of the five compartments (1–5). Numbers correspond to those in the lower figure. Lower figure indicates the expression profiles of Bmyellow-e in the larval integument. Expression levels are shown relative to that detected in the head region (1). 18 S RNA was used as an internal control. Data are shown as means ± S.E. (n = 3).
DISCUSSION
To identify the mechanisms determining body color pattern in insects, we used a Bombyx color mutant, bts, in which the larval head cuticle and anal plates are reddish brown instead of the white color found in normal strains. Positional cloning revealed mutations in the candidate gene of bts and bts2 strains, both of which resulted in the generation of a truncated gene product. Homology searches and phylogenetic analysis revealed that this gene is Bmyellow-e, a Bombyx homolog of the Drosophila yellow-e gene. Together with mRNA expression profiles of Bmyellow-e, we concluded that BmYellow-e is required for the normal pattern of larval body color in Bombyx. To our knowledge, this is the first study showing a role for yellow-e.
Yellow is an essential protein for normal adult pigmentation patterns in Drosophila (4). Futahashi et al. (9) recently reported that Bmyellow is the gene responsible for the Bombyx ch mutant, thus suggesting that Yellow plays a crucial role in lepidopteran larval color patterns and that the function of Yellow in promoting black coloration is conserved in insects. Although 13 and 6 additional yellow genes have been found in the genomes of Drosophila (3) and Bombyx (20), respectively, there are no reports elucidating the roles of other Yellow proteins in the formation of body color. In genetic studies using Drosophila mutants, Yellow-g and Yellow-g2 were shown to play catalytic roles in cross-linking of chorion and/or underlying vitelline membrane proteins; however, no phenotypic abnormalities in body color were reported in these mutants (21). In contrast, biochemical studies using recombinant proteins provided evidence that the products of Drosophila yellow-f and yellow-f2 have dopachrome-conversion enzyme activity (6), but their functions have not been examined genetically using loss-of-function mutants. Therefore, Yellow-e is a second Yellow protein that is involved in body color patterns in insects.
Expression of Bmyellow-e was markedly increased at the molting stages (Fig. 5B), suggesting that this gene may be regulated by the molting hormone ecdysone. Interestingly, mRNA profiles of Bmyellow-e were very similar to those of Bmyellow (9), indicating that these two genes work concomitantly during the molting process and coordinately control the larval body color in Bombyx. Furthermore, qRT-PCR analysis revealed that Bmyellow-e was heavily expressed in the head and anal plates where reddish coloration occurs in the bts strain, thus indicating that BmYellow-e functions in the integument of the larval head and tail. Sequencing of Bmyellow-e from the bts and bts2 strains showed that the MRJP domain of the bts strain was about 130 amino acids shorter than that of the bts2 strain (Fig. 3F). Moreover, the color of head and tail spots of the bts2 strain is darker than that of the bts strain (Fig. 1) (8). Together, these results suggest that the degree of pigmentation in the head and tail spot likely results from the length or activity of BmYellow-e protein. Although we have no idea how the wild-type BmYellow-e induces white coloration, biochemical studies using recombinant protein will clarify its enzymatic function in larval pigmentation.
There are about 40 spontaneous body color mutants in Bombyx (7, 8). Combined with current post-genomic tools such as the draft sequence information (18), EST Databases (22), microarrays (23–25), and transgenic techniques (26), Bombyx could be a suitable model for clarifying the genetic basis of body color formation in other insects. Using the newly assembled genome sequence, we have already cloned seven genes required for normal color pattern in Bombyx larvae (9, 13, 27–30). Further studies on Bombyx color mutants will identify novel molecules involved in insect body color patterns.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (17018007), MAFF-NIAS (Agrigenome Research Program), National BioResource Project, and Japan Science and Technology Agency (Professional Program for Agricultural Bioinformatics), Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- MRJP
- major royal jelly protein
- RT
- reverse transcription
- qRT
- quantitative RT
- WT
- wild type
- SNP
- single nucleotide polymorphism.
REFERENCES
- 1.Schmitzová J., Klaudiny J., Albert S., Schröder W., Schreckengost W., Hanes J., Júdová J., Simúth J. (1998) Cell. Mol. Life Sci. 54, 1020–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maleszka R., Kucharski R. (2000) Biochem. Biophys. Res. Commun. 270, 773–776 [DOI] [PubMed] [Google Scholar]
- 3.Drapeau M. D. (2001) Biochem. Biophys. Res. Commun. 281, 611–613 [DOI] [PubMed] [Google Scholar]
- 4.Wittkopp P. J., True J. R., Carroll S. B. (2002) Development 129, 1849–1858 [DOI] [PubMed] [Google Scholar]
- 5.Drapeau M. D., Radovic A., Wittkopp P. J., Long A. D. (2003) J. Neurobiol. 55, 53–72 [DOI] [PubMed] [Google Scholar]
- 6.Han Q., Fang J., Ding H., Johnson J. K., Christensen B. M., Li J. (2002) Biochem. J. 368, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii H., Banno Y., Doira H., Kawaguchi Y. (1998) Genetical Stocks and Mutations of Bombyx mori: Important Genetic Resources, 2nd Ed., pp. 1–54, Institute of Genetic Resources, Faculty of Agriculture, Kyusyu University, Fukuoka [Google Scholar]
- 8.Banno Y., Fujii H., Kawaguchi Y., Yamamoto K., Nishikawa K., Nishisaka A., Tamura K., Eguchi S. (2005) A Guide to the Silkworm Mutants: 2005, pp. 1–30, Gene Name and Gene Symbol, Kyusyu University, Fukuoka, Japan [Google Scholar]
- 9.Futahashi R., Sato J., Meng Y., Okamoto S., Daimon T., Yamamoto K., Suetsugu Y., Narukawa J., Takahashi H., Banno Y., Katsuma S., Shimada T., Mita K., Fujiwara H. (2008) Genetics 180, 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chikushi H. (1960) J. Seric. Sci. Jpn. 29, 278 [Google Scholar]
- 11.Eguchi R., Nagayasu K., Ninaki O., Hara W. (2007) Sanshi-Kontyu Biotech. 76, 159–163 [Google Scholar]
- 12.Ito K., Kidokoro K., Sezutsu H., Nohata J., Yamamoto K., Kobayashi I., Uchino K., Kalyebi A., Eguchi R., Hara W., Tamura T., Katsuma S., Shimada T., Mita K., Kadono-Okuda K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7523–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito K., Katsuma S., Yamamoto K., Kadono-Okuda K., Mita K., Shimada T. (2009) Insect Biochem. Mol. Biol. 39, 287–293 [DOI] [PubMed] [Google Scholar]
- 14.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 16.Daimon T., Katsuma S., Iwanaga M., Kang W., Shimada T. (2005) Insect Biochem. Mol. Biol. 35, 1112–1123 [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto K., Nohata J., Kadono-Okuda K., Narukawa J., Sasanuma M., Sasanuma S. I., Minami H., Shimomura M., Suetsugu Y., Banno Y., Osoegawa K., de Jong P. J., Goldsmith M. R., Mita K. (2008) Genome Biol. 9, R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The International Silkworm Genome Consortium (2008) Insect Biochem. Mol. Biol. 38, 1036–1045 [DOI] [PubMed] [Google Scholar]
- 19.Abe H., Seki M., Ohbayashi F., Tanaka N., Yamashita J., Fujii T., Yokoyama T., Takahashi M., Banno Y., Sahara K., Yoshido A., Ihara J., Yasukochi Y., Mita K., Ajimura M., Suzuki M. G., Oshiki T., Shimada T. (2005) Insect Mol. Biol. 14, 339–352 [DOI] [PubMed] [Google Scholar]
- 20.Xia A. H., Zhou Q. X., Yu L. L., Li W. G., Yi Y. Z., Zhang Y. Z., Zhang Z. F. (2006) BMC Genomics 7, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claycomb J. M., Benasutti M., Bosco G., Fenger D. D., Orr-Weaver T. L. (2004) Dev. Cell 6, 145–155 [DOI] [PubMed] [Google Scholar]
- 22.Mita K., Morimyo M., Okano K., Koike Y., Nohata J., Kawasaki H., Kadono-Okuda K., Yamamoto K., Suzuki M. G., Shimada T., Goldsmith M. R., Maeda S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14121–141216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niwa R., Matsuda T., Yoshiyama T., Namiki T., Mita K., Fujimoto Y., Kataoka H. (2004) J. Biol. Chem. 279, 35942–35949 [DOI] [PubMed] [Google Scholar]
- 24.Ote M., Mita K., Kawasaki H., Seki M., Nohata J., Kobayashi M., Shimada T. (2004) Insect Biochem. Mol. Biol. 34, 775–784 [DOI] [PubMed] [Google Scholar]
- 25.Xia Q., Cheng D., Duan J., Wang G., Cheng T., Zha X., Liu C., Zhao P., Dai F., Zhang Z., He N., Zhang L., Xiang Z. (2007) Genome Biol. 8, R162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura T., Thibert C., Royer C., Kanda T., Abraham E., Kamba M., Komoto N., Thomas J. L., Mauchamp B., Chavancy G., Shirk P., Fraser M., Prudhomme J. C., Couble P. (2000) Nat. Biotechnol. 18, 81–84 [DOI] [PubMed] [Google Scholar]
- 27.Fujii T., Abe H., Katsuma S., Mita K., Shimada T. (2008) Insect. Biochem. Mol. Biol. 38, 1072–1079 [DOI] [PubMed] [Google Scholar]
- 28.Fujii T., Ozaki M., Masamoto T., Katsuma S., Abe H., Shimada T. (2009) Genes Genet. Syst. 84, 147–152 [DOI] [PubMed] [Google Scholar]
- 29.Meng Y., Katsuma S., Mita K., Shimada T. (2009) Genes Cells 14, 129–140 [DOI] [PubMed] [Google Scholar]
- 30.Meng Y., Katsuma S., Daimon T., Banno Y., Uchino K., Sezutsu H., Tamura T., Mita K., Shimada T. (2009) J. Biol. Chem. 284, 11698–11705 [DOI] [PMC free article] [PubMed] [Google Scholar]