Abstract
E3 ubiquitin ligases catalyze the final step of ubiquitin conjugation and regulate numerous cellular processes. The HECT class of E3 ubiquitin (Ub) ligases directly transfers Ub from bound E2 enzyme to a myriad of substrates. The catalytic domain of HECT Ub ligases has a bilobal architecture that separates the E2 binding region and catalytic site. An important question regarding HECT domain function is the control of ligase activity and specificity. Here we present a functional analysis of the HECT domain of the E3 ligase HUWE1 based on crystal structures and show that a single N-terminal helix significantly stabilizes the HECT domain. We observe that this element modulates HECT domain activity, as measured by self-ubiquitination induced in the absence of this helix, as distinct from its effects on Ub conjugation of substrate Mcl-1. Such subtle changes to the protein may be at the heart of the vast spectrum of substrate specificities displayed by HECT domain E3 ligases.
Keywords: Enzymes, Enzymes/Structure, Protein/Structure, E3 Ubiquitin Ligase, Ubiquitination
Introduction
Ubiquitin (Ub)6 conjugation regulates many cellular processes, including protein stability, cell cycle control, DNA repair, transcription, signal transduction, and protein trafficking (1–3). An enzymatic cascade consisting of Ub-activating enzyme (E1), Ub-conjugating enzyme (E2), and Ub ligase (E3) is responsible for catalyzing Ub conjugation to target proteins. The E1 enzyme activates Ub for transfer by adenylating its C terminus in an ATP-dependent step. Ub is then transferred to the E1 active site cysteine, forming a thioester bond between E1 and Ub. The activated Ub is transferred to E2 enzyme in a transthiolation reaction and is conjugated to target proteins by the action of E3 Ub ligases (3). The E2 and E3 enzymes provide substrate specificity to the Ub conjugation reaction (4). Approximately 600 E3s exist in the human proteome, 28 of which belong to the HECT (homologous to E6AP C terminus) domain family (5). HECT domain E3s possess a ∼350-amino acid C-terminal HECT domain containing a conserved catalytic cysteine that participates in the transfer of Ub to substrate. Mutation, abnormal expression, or misregulation of these enzymes predisposes to the development of cancer and disease (1, 6).
Structural studies describe the canonical HECT domain architecture and provide insight into its mechanism of catalysis (7–9). HECT domains are composed of two subdomains connected by a flexible peptide linker. The N-terminal (N) lobe contains the E2 binding region, and the C-terminal (C) lobe contains the catalytic cysteine. In the structure of the E6AP-UbcH7 complex (Protein Data Bank codes 1C4Z and 1D5F), the catalytic cysteines of E2 and E3 are separated by 41 Å, suggesting that a substantial conformational rearrangement is required to achieve Ub transfer (7). Analysis of the structure of the WWP1 HECT domain (PDB code 1ND7) partially addresses how Ub is transferred from the E2 to the E3 catalytic cysteine by illustrating conformational flexibility of the HECT domain (9). In the WWP1 structure modeled in a complex with UbcH5, the C-lobe is rotated about the hinge region, placing it in closer proximity to the E2 cysteine and closing the distance between active site cysteines to 16 Å. Mutations in this hinge loop that restrict HECT domain rotation decrease activity (9). Additional structural elements within the HECT domain that modulate conformation or activity remain unknown.
HUWE1 (also called ARF-BP1, Mule, Lasu1, Ureb1, E3 histone, and HectH9) is a 482-kDa HECT domain E3 Ub ligase implicated in the regulation of cell proliferation, apoptosis, DNA damage response, and base excision repair (10–16). We recovered this enzyme in immunoprecipitations using Ub C-terminal electrophilic probes (17). After an initial biochemical characterization (17), we completed a structural and biophysical analysis of the HECT domain to understand modulation of its robust in vitro activity. Here we present crystal structures of the HUWE1 HECT domain and characterize a structural element that both stabilizes this domain and modulates its activity. This structural element, the α1 helix, is an important component of the HECT domain that largely restricts its autoubiquitination activity while only nominally affecting Mcl-1 ubiquitination activity.
EXPERIMENTAL PROCEDURES
Plasmids
HECT domain constructs of HUWE1 (amino acids 3993–4374 or amino acids 4012–4374) were cloned into a modified pET-28a plasmid (Novagen) containing a human rhinovirus 3C (HRV3C) protease site to generate an N-terminal His6 fusion protein for use in biochemical assays. The mutants C4341A and C4099A/C4184A/C4367A were generated using site-directed mutagenesis (Stratagene). For biochemical assays with radiolabeled substrate, FLAG-Mcl-1 (amino acids 1–327) and Ub were both cloned with an N-terminal protein kinase A site for 32P labeling into pET-16b and pET-28a with the HRV3C site, respectively, as previously reported (17). UBE2L3 was cloned into the pET-28a plasmid (Novagen).
Bacterial Protein Expression and Purification
All versions of the HUWE1 HECT domain were expressed and purified as previously reported (17). 32-Labeled proteins were purified and labeled as previously described (17). Native UBE2L3 was expressed in Rosetta (DE3) cells (Novagen). UBE2L3 was precipitated from bacterial lysate by the addition of saturated ammonium sulfate to 90%. The precipitated protein was resuspended in 50 mm HEPES, pH 7.4, 200 mm NaCl, and purified by gel filtration (Superdex 75 PC 3.2/30, GE Healthcare).
Circular Dichroism
HUWE1 Δ α1 and HUWE1 + α1 HECT domains were dialyzed into 5 mm HEPES, pH 7.5, 100 mm NaCl immediately before the scanning and melting CD experiments using an AVIV model 202 CD spectrometer. HUWE1 Δ α1 HECT domain at 2.4 μm and HUWE1 + α1 HECT domain at 2.8 μm were used for scanning experiments between 195 and 280 nm at 25 °C. CD signal at 222 nm of 4.8 μm HUWE1 Δ α1 and 5.6 μm HUWE1 + α1 was recorded every 2 °C degrees over a 20–94 °C temperature ramp with 2 min of equilibration time at every step.
Biochemical Assays
Reaction mixtures (10 μl) for HUWE1 autoubiquitination assay contained 100 nm human E1 (Ube1, Boston Biochem), 5.6 μm E2 (UBE2L3), HECT domain, and 60 μm 32P-labeled Ub with an ATP-regenerating system (50 mm Tris, pH 7.6, 5 mm MgCl2, 5 mm ATP, 10 mm creatine phosphate, 3.5 units/ml creatine kinase). Reactions were incubated at room temperature, and aliquots were removed after the indicated amount of time and terminated in reducing SDS-PAGE sample buffer. Samples were boiled for 10 min and separated on 8% Tris-Tricine SDS-PAGE. Dried gels were exposed to a phosphor screen. 32P-Labeled Ub bands were visualized by autoradiography, and quantitation of data was performed using phosphorimaging. Background correction for each sample was performed by subtracting the counts from an equivalent area of the gel from a lane containing all reaction components except E3 enzyme (lane marked N in Figs. 3 (a and b) and 5 (a and b) and supplemental Fig. 2 (a–d)). The 32P-labeled Ub signal from this lane was used to convert the observed sample counts to a concentration value. The concentrations of HUWE1-32P- labeled Ub were measured, and rates of product formation were determined by fitting the initial linear data points to a least squares regression line.
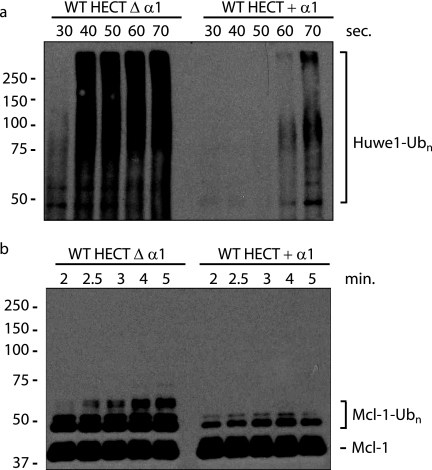
FIGURE 3.
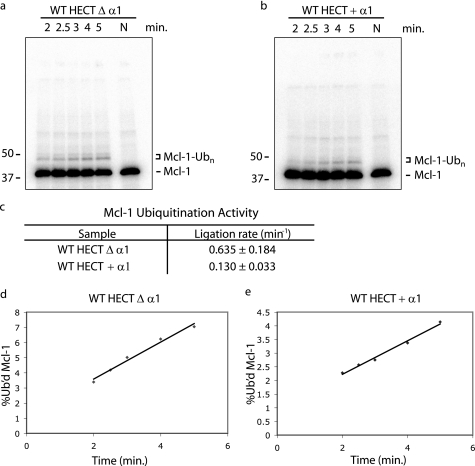
E3 ubiquitin ligase activity of HUWE1 HECT domain. a and b, the autoubiquitination activity of HUWE1 HECT domain was tested using 60 μm 32P-labeled Ub as substrate and 2 μm WT Δ α1 (a) or 2.9 μm WT + α1 (b) HECT domains incubated with UBE1, UBE2L3, and an ATP regenerating system (note the different time scale for the two variants of the HECT domain). The HECT domain is omitted in the lane marked N. The asterisk denotes a likely ubiquitin polymer; the double asterisk denotes likely mono-ubiquitinated UBE2L3. Concentrations of HECT domain were chosen to obtain initial rate conditions. c, ligation activity of the WT Δ α1 and WT + α1 HECT domains in the autoubiquitination assay is shown. Activity is given as the ratio between initial velocity (pmol of total 32P-labeled Ub product/min) and total enzyme concentration E (pmol). Errors are the S.D. calculated from three independent experiments. d, shown is a single turnover assay monitoring transfer of Ub from the UBE2L3∼Ub thioester to a lysine in the WT Δ α1 and WT + α1 HECT domains. The UBE2L3∼Ub thioester is generated in a pulse reaction containing E1, UBE2L3, ATP regenerating system, and a Ub mutant in which all lysines are mutated to arginine (K0 Ub). Ub is chased from the E2 enzyme to the HECT domain added to the reaction. Ub-conjugated HECT domain is visualized by anti-Ub immunoblot. Samples were terminated in reducing or non-reducing sample buffer as indicated. The panel marked N is a chase reaction performed in the absence of HECT domain and terminated in non-reducing sample buffer.
FIGURE 5.
Substrate ubiquitination activity of the HUWE1 HECT domains. a and b, the Mcl-1 ubiquitination activity of HUWE1 HECT domain was tested using 5 μm 32P-labeled Mcl-1 as substrate and 100 nm WT Δ α1 (a) or 300 nm WT + α1 (b) HECT domains incubated with UBE1, UBE2L3, Ub, and an ATP regenerating system. HECT domain was omitted from the lane marked N. Concentrations of HECT domain were chosen to obtain initial rate conditions. c, ligation activity of the HECT domains in the Mcl-1 ubiquitination assay is shown. Activity is given as the ratio between initial velocity (pmol of total 32P-labeled Ub product/min) and total enzyme concentration E (pmol). Errors are the S.D. calculated from three independent experiments. d and e, shown is a graph of the percent ubiquitinated Mcl-1 as a function of time in the reactions shown in panels a and b catalyzed by HUWE1 Δ α1 (d) or HUWE1 + α1 (e) HECT domains.
Mcl-1 Ubiquitination Assay
Reaction mixtures (10 μl) for the Mcl-1 ubiquitination assay were set up as above except with the addition of 5 μm [32P]-labeled FLAG-Mcl-1 and 100 μm Ub (Sigma). Reactions were quenched with reducing sample buffer and separated on 10% SDS-PAGE. Bands from dried gels were analyzed as above.
Thioester Assay
Reaction mixtures for the thioester assay (10 μl) contained 100 nm human E1 (Ube1, Boston Biochem), 5.6 μm E2 (UBE2L3), 2 μm HUWE1 Δ4 HECT domain, and 60 μm Ub (Sigma) with an ATP regenerating system described above. Reactions were incubated at room temperature, and aliquots were removed after the indicated amount of time, terminated in 4 m urea, and incubated for 15 min at 30 °C or terminated in reducing SDS-PAGE sample buffer. Samples were boiled for 10 min, separated on 10% Tris-glycine SDS-PAGE, and analyzed by immunoblot using anti-Ub antibody (Sigma).
Single-turnover Assay
For the single-turnover assay, the E2 ∼ Ub thioester was generated in a 20-μl reaction containing 200 nm E1 (Boston Biochem), 8 μm E2, the ATP regenerating system described above, 60 μm mutant Ub in which all lysines were mutated to arginine (K0 Ub) (Boston Biochem), and 1 μg/μl bovine serum albumin incubated for 25 min at room temperature. Formation of the E2∼Ub thioester was quenched with 50 mm EDTA on ice for 5 min. The E2∼Ub thioester was diluted into a chase mixture containing 2 μm HECT domain, 100 mm NaCl, 50 mm EDTA, and 1 μg/μl bovine serum albumin or the same reaction components lacking the HECT domain (labeled N in Fig. 3d). Reactions were incubated at room temperature, and aliquots were removed after the indicated amount of time and terminated in either 4 m urea and incubated for 15 min at 30 °C or in reducing SDS-PAGE sample buffer. Samples were boiled for 10 min, separated on 10% Tris-glycine SDS-PAGE, and analyzed by immunoblot using anti-Ub antibody (Sigma).
Crystallization of HUWE1 HECT Domain
Crystallization experiments with purified HUWE1 HECT domain, including the N-terminal His6 tag, HRV3C protease site, and with C4099A, C4184A, and C4341A mutations, were set up in 96-well sitting drop trays using commercially available sparse-matrix screens (Hampton Research, Qiagen). The initial crystals were improved in hanging-drop vapor diffusion setups. The HUWE1 + α1 HECT domain crystallized by mixing 1 μl of protein sample concentrated to 17 mg/ml with a 1-μl solution containing 0.1 m citric acid, pH 5.2, and 1.8 m (NH4)2SO4. Birefringent crystals in the shape of thick rods with dimensions of ∼80 × 40 × 40 μm grew within 2 days of incubation at 18 °C. The HUWE1 Δ α1 HECT domain crystallized by mixing a 1-μl solution containing (Na/K)2PO4 (pH 6.5) and 1.4 m (Na/K)2PO4. Thin rod-shaped crystals grew within 10 days at 23 °C.
Data Collection and Processing
For native x-ray diffraction studies, crystals were cryoprotected by soaking in 0.1 m citric acid, pH 5.2, 1.8 m (NH4)2SO4, 12% glycerol for 30 s before vitrifying in liquid nitrogen. X-ray diffraction data were collected on a single cryogenized crystal at beamline 24ID-E, Advanced Photon Source (Argonne, IL), summarized in Table 1. Data were processed using DENZO and SCALEPACK (18). The crystals belong to the monoclinic space group C2 and diffracted to 1.9 Å. Initial phases were obtained by molecular replacement using PHASER from the CCP4 crystallographic program suite (19, 20), with the coordinates of the E3 ligase WWP1 (PDB accession code 1ND7) as search model. The final model was refined at resolution of 1.9 Å using PHENIX with four TLS groups (9). Details of refinement are given in Table 1. The figures were made using PyMol (21).
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell.
| Data set | HUWE1 HECT + α1 |
|---|---|
| Data collection | |
| Wavelength | 0.9793 |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 119.6, 56.6, 69.6 |
| β (°) | 122.5 |
| Unique reflections | 30,847 |
| Resolution (Å) | 30-1.9 (1.93-1.9) |
| Rsyma | 0.069 (0.392) |
| Rr.i.m.b | 0.084 (0.493) |
| Rp.i.m.c | 0.048 (0.295) |
| Completeness | 98.3 (96.3) |
| Redundancy | 3.2 (2.5) |
| I/σ | 17.2 (2.1) |
| Wilson B factor (Å2) | 25 |
| Refinement | |
| Resolution (Å) | 30-1.9 |
| Nonhydrogen atoms | 3190 |
| Water molecules | 357 |
| Rworkd | 16.6 |
| Rfreee | 22.9 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.20 |
| B factors (Å2) | |
| Protein | 30.0 |
| Water | 39.4 |
| Coordinate error (Å) | 0.68 |
| Ramachandran plotf | |
| Most favored | 376 |
| Allowed | 5 |
| Outliers | 0 |
a Rsym = Σ|Ii − 〈Ii〉|/ΣIi, where Ii is the intensity of the ith observation, and 〈Ii〉 is the mean intensity of the reflection.
b Rr.i.m. = Σhkl[N/(N − 1)]1/2?i|Ii(hkl) − 〈I(hkl) 〉|/ΣhklΣi Ii(hkl), where Ii(hkl) is the observed intensity, and 〈I(hkl) 〉 is the average intensity of multiple observations of symmetry-related reflections.
c Rp.i.m. = Σhkl[1/(N − 1)]1/2?i|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the observed intensity, and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections.
d Rwork = Σ(‖Fobs| − |Fcalc‖/Σ|Fobs|).
e Rfree = R value for a randomly selected subset (5%) of the data that were not used for minimization of the crystallographic residual.
f Number of residues calculated with the program MolProbity (33).
RESULTS
Structure of the HUWE1 HECT Domain
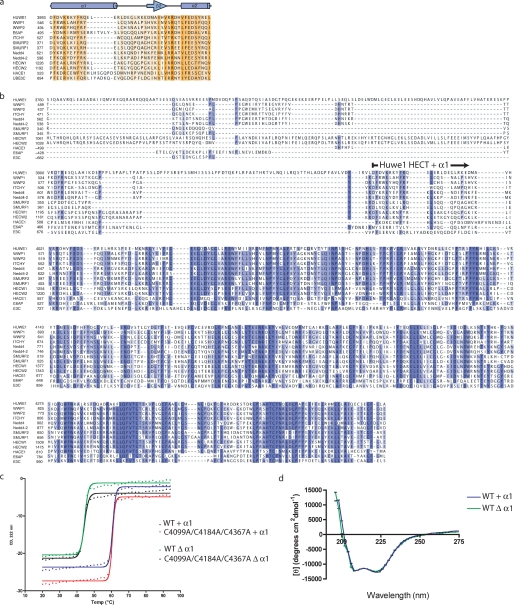
We first attempted to crystallize the HUWE1 HECT domain using a fragment defined by the founding member of the HECT domain E3 ligase family, E6AP (PDB codes 1C4Z and 1D5F) (7). The crystals diffracted to only 3.5 Å (data not shown), with fairly high temperature factors indicating vibrational disorder within the protein crystals. By sequence comparison, we noted the significance of a conserved N-terminal helix that seals the hydrophobic core of the N-lobe in the structures of WWP1, Smurf2, and Nedd4-like (residues 546–560 in WWP1 and 371–387 in Smurf2) (8, 9). The presence of this helix is conserved in over 13 HECT domain E3 ligases based on sequence comparison (Fig. 1a) and Verdecia et al. (9), highlighting its structural importance. The α1 helix has been previously described as a critical element for structural stability yet an element dispensable for HECT domain function (7, 9). Initial model-building into the 3.5-Å electron density showed a noticeable hydrophobic groove on the surface of helices 5, 11, 12, and 13, possibly indicating an additional helix being bound here. As sequence conservation drops off N-terminal of this α1 helix (Fig. 1b), we hypothesized that this element is an important part of the HECT domain. We note that expression of the HECT domain of Nedd4 yielded soluble folded product when the homologous helical segment was included in the expression construct but was not successful in its absence.7 We, therefore, asked whether the addition of helix α1 would not only assist in our crystallographic efforts but also affect the catalytic activity of the HECT domain.
FIGURE 1.
The α1 helix stabilizes the HUWE1 HECT domain. a, multiple sequence alignment of helix α1 with a diverse set of human HECT E3 ligases is shown. Residue conservation is indicated by degree of shading ranging from orange (most conserved) to light yellow (least conserved). Secondary structure is illustrated with α-helices as cylinders and β-sheets as arrows. b, shown is multiple sequence alignment with a diverse set of human HECT E3 ligases indicating that sequence conservation drops off N-terminal to the α1 helix. The N terminus of the HECT domain + α1 helix is indicated. c, thermostability of the HUWE1 HECT domain was measured in a CD melting experiment. HUWE1 HECT domain, +/Δ α1 helix, WT, or with cysteine mutations, was heated in a circular dichroism cuvette, and unfolding was measured at 222 nm as a loss of helical content. Deletion of helix α1 results in a drastic reduction of thermostability. d, a CD scan experiment demonstrates the structural similarities of the +/Δ α1 helix versions of the HECT domain.
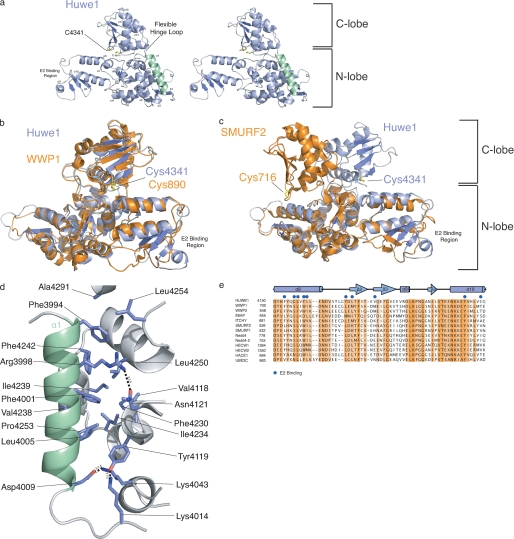
The addition of helix α1 greatly stabilizes the HECT domain, as made evident in thermal denaturation experiments (Fig. 1c), shifting the transition midpoint by 16 °C from 44 to 60 °C. Although thermal stability differs between the two versions of the HECT domain, the level of secondary structure remains the same (Fig. 1d), indicating that the absence of helix α1 does not lead to unfolding but to less rigidity of the domain. We solved the structure of the helix-extended HECT domain by molecular replacement using the E3 ligase WWP1 (9) as a search model. The final model (R/Rfree 16.6/22.9%) was built and refined to 1.9 Å resolution (Table 1). The structure of HUWE1 HECT domain closely resembles that of WWP1, with which it shares 41.3% sequence identity (Fig. 2a). The HUWE1 HECT domain contains two distinct lobes similar to previously determined HECT domain structures (E6AP, Smurf2, WWP1). The larger N lobe (residues 3993–4252) contains the E2 binding region, and the smaller C lobe (residues 4259–4374) contains the conserved catalytic cysteine (C4341). The N lobe is composed of 13 α-helices and 7 β-strands, and the C lobe is composed of 4 α-helices and 4 β-strands. Residues 4253–4258 form the hinge that connects the two lobes. A rotary movement about this linker likely repositions the N and C lobes to bring the catalytic cysteine of the cognate Ub-loaded E2 in proximity to its E3 counterpart (9). Like WWP1, HUWE1 is oriented in an inverted T shape (⊥), in which the C lobe is positioned over the middle of the N lobe, with ∼800 Å2 of contact surface area (Fig. 2b). Hydrogen bonds between Glu-4248 (N lobe) and Ser-4304 (C lobe) as well as Gln-4245 (N lobe) and Gln-4298 (C lobe) and a salt bridge between Glu-4246 and Lys-4295 stabilize the ⊥ conformation. The ⊥ conformation is further stabilized by water-mediated hydrogen bonds between the two lobes, involving residues Arg-4130, Glu-4147, Ser-4148, and Glu-4244 from the N lobe and Gln-4298, Thr-4340, Gly-4302, and Lys-4295 in the C lobe. The orientation of the N and C lobes of the HUWE1 HECT domain differs from the more open conformation observed in the crystal structures of E6AP and Smurf2 (Fig. 2c) (7, 8), although we cannot exclude the possibility that crystal contacts influence the observed orientation of the C lobe. The stabilizing nature of helix α1 is apparent from the extended structure, as it closes the hydrophobic core of the N-lobe (Fig. 2, a and d).
FIGURE 2.
Structure of the HUWE1 HECT domain. a, a stereo view of HUWE1 HECT domain (residues 3993–4374) shows the N and C lobes connected by the hinge loop. Helix α1 is colored green. The N-lobe contains the E2 binding region, and the C-lobe contains the catalytic cysteine (Cys-4341). b, overlay of HUWE1 (blue) and WWP1 (orange; PDB 1ND7) crystal structures is shown. c, overlay of HUWE1 (blue) and Smurf2 (orange; PDB 1ZVD) crystal structures is shown. d, helix α1 plays a significant role in mediating hydrophobic contacts that maintain the core of the HUWE1 HECT domain. Hydrophobic residues in the α1 helix, Phe-3994 and Phe-4001, pack into hydrophobic pockets in the N lobe. Arg-3998 and Asp-4009 form hydrogen bonds stabilizing the N-lobe. Lys-4014 and Tyr-4119, C-terminal to the α1 helix, orient the α1 helix to further stabilize the N-lobe. e, multiple sequence alignment of E2 binding region with a diverse set of human HECT E3 ligases is shown. Residues important for E2 binding are indicated with blue circles.
The most notable difference between HUWE1 HECT domain and previously solved structures concerns the E2 binding region (residues 4150–4200). Most of the hydrophobic residues in WWP1 that mediate contact with the E2 are similar to those in HUWE1, obvious from the alignment between HECT E3 ligases (Fig. 2e). The HUWE1 E2 binding region differs from that of WWP1 in that it contains additional structured elements; that is, mainly ordered β-strands not previously identified. The well ordered β-strands in the E2 binding region of the HUWE1 HECT domain extend farther from the helical core of the protein than seen in the structure of WWP1, and the loop is folded back on itself to complete the β3 strand and form the α8 helix (Fig. 2a). It is possible that HUWE1 uses its unique E2 binding region to interact with a specific set of E2 enzymes in vivo that differ from WWP1.
A comparison of our two HUWE1 HECT domain structures shows that they are nearly identical with respect to the positioning of the N and C lobes (supplemental Fig. 1, a and b). For the structure lacking the α1 helix, the additional β-strands and α-helix seen in the E2 binding region remain unresolved, likely due to the low resolution data and high temperature factors.
Catalytic Activity of the HECT Domain
As the addition of the α1 helix to the HECT domain stabilized the protein, we asked whether the presence of this structural element affected HECT domain catalytic activity. We hypothesized that its addition might confer altered catalytic properties to the HECT domain compared with its helix-lacking counterpart. We, therefore, examined the ability of the HECT domain to catalyze self-ubiquitination in the presence of E1 and E2 (UBE2L3) enzymes, an ATP regenerating system, and 32P-labeled Ub (Fig. 3). The use of 32P-labeled Ub allowed us to quantify the amount of Ub adducts formed and calculate initial rates of product formation in HECT domain-limiting conditions. In this assay the HECT domain catalyzes the formation of a complex mixture of self-ubiquitinated species (Fig. 3, a and b) that are not observed in absence of the HECT domain (lane marked N). Immunoblotting using an anti-His antibody confirmed that these species are ubiquitinated E3 enzyme, as it is the only species in this reaction that contains a polyhistidine tag (data not shown). The pattern of autoubiquitination observed is similar regardless of the presence of the α1 helix; both versions form multi- and polyubiquitinated species (Fig. 3, a and b). Although the pattern of product formation is similar, the presence of the α1 helix suppresses the autoubiquitination activity of the HECT domain by more than 25-fold (Fig. 3c). As autoubiquitination is observed for many Ub ligases and is often used as a criterion of E3 Ub ligase activity, we sought to further characterize the reasons for its modulation.
The autoubiquitination reaction described above produces a complex mixture of products. We examined HECT domain activity in a single-turnover reaction to monitor the first round of Ub addition to the HECT domain. This assay encompasses two steps. In the first step, E2 ∼ Ub thioester is generated by incubating E1, E2, an ATP regenerating system, and Ub. After the E2 ∼ Ub thioester has formed, this reaction is quenched by the addition of EDTA to prevent further E1-catalyzed activation of Ub. In the second step, the HUWE1 HECT domain is added, and Ub is chased from the E2∼thioester onto the HECT domain (22). The use of a mutant version of Ub, in which all lysines are mutated to arginine (K0 Ub), prevents polyubiquitin chain formation on the HECT domain. Ub-conjugated HECT domain is visualized using anti-Ub immunoblot (Fig. 3d). We find that the HUWE1 Δ α1 HECT domain shows increased activity under single turnover conditions compared with the HECT domain containing helix α1 (Fig. 3d), confirming the rate differences observed in the autoubiquitination assay.
Thioester Formation in the HECT Domain
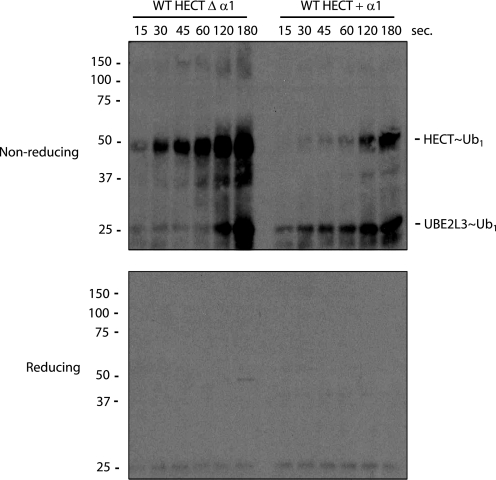
Catalysis by HECT domain E3 enzymes is a multistep process. The E3 enzyme binds Ub-loaded E2 and substrate followed by Ub transfer between the E2 and E3 catalytic cysteines. The E3 then catalyzes isopeptide bond formation between Ub and a lysine residue on the substrate, which may be the E3 itself, Ub, or another protein. We next determined whether the presence of the α1 helix affects this upstream step, in which the catalytic cysteine of the E3 enzyme forms a thioester bond with ubiquitin.
We first attempted to assay thioester formation using the wild-type HECT domain, but the enzyme efficiently catalyzes formation of the isopeptide bond on a time scale too fast to measure (23, 24). Instead, we analyzed thioester formation using a four-amino acid, C-terminal truncation of the HECT domain (25). This truncation removes a crucial determinant for isopeptide bond formation, a conserved phenylalanine located four amino acids from the C terminus of most HECT E3s (25). HUWE1 Δ4 displays a diminished rate of isopeptide bond formation, allowing us to monitor the formation of thioester-linked Ub to the enzyme. After incubation of the HECT domain with E1, the E2 UBE2L3, Ub, and an ATP regenerating system, the reaction was quenched with SDS-PAGE loading buffer with or without β-mercaptoethanol, and after electrophoretic resolution, was analyzed by anti-Ub immunoblot. The presence of the α1 helix greatly reduces the rate of thioester formation (Fig. 4), proportional to its suppression of autoubiquitination activity. The α1 helix, located on the back surface of the N lobe, is clearly not sufficiently close to interact with the E2 binding region of the HECT domain (Fig. 2a), suggesting that the vibrational disorder in this protein contributes to the HECT domain-E2 interaction.
FIGURE 4.
Detection of Ub∼thioesters in the HUWE1 HECT domains. A Ub-thioester assay with the indicated HUWE1 HECT domain proteins is shown. Purified HECT domains containing a C-terminal truncation of the last four amino acids were incubated with E1, UBE2L3, an ATP regenerating system, and Ub for the indicated amounts of time. Reactions were stopped with 4 m urea and non-reducing sample buffer (upper panel) or reducing sample buffer (lower panel), separated by SDS-PAGE, and analyzed by immunoblot with anti-Ub antibody.
Substrate Ubiquitination Catalyzed by the HECT Domain
Having seen that removal of helix α1 destabilizes the HECT domain and increases its autoubiquitination activity, we asked whether this effect is also observed during substrate ubiquitination. The anti-apoptotic Bcl-2 family member Mcl-1 is an in vivo target of HUWE1 (14). HUWE1 recruits Mcl-1 via its BH3-domain, whereas the HECT domain presented here catalyzes Mcl-1 ubiquitination. Although Mcl-1 is a substrate of the full-length HUWE1, we use this assay with the isolated HECT domain here as a measure of non-self-ubiquitination activity with an in vivo verified substrate of HUWE1. We examined initial rates of product formation under HECT domain-limiting conditions to determine whether the intrinsic activity of the HECT domain toward substrate is altered by the destabilizing effect of removing the α1 helix (Fig. 5, a and b). We find that the HECT domain-lacking α1 helix is ∼5-fold more active in catalyzing Mcl-1 ubiquitination than the more stable HECT domain-containing helix α1 (Fig. 5c). These results also suggest that autoubiquitination of the HECT domain does not impair catalytic activity toward substrate. The two versions of the HUWE1 HECT domain, which differ 25-fold in autoubiquitination rates, show only a 5-fold difference in their Mcl-1 ubiquitination rates. A similar observation was made for the heterodimeric complex of the minimal catalytic domains of Ring1a/Bmi1, in which autoubiquitination of the Ring1b protein did not affect E3 ligase activity toward its substrate, histone H2A, in an in vitro reconstituted system (26). We also observe similar differences in autoubiquitination and Mcl-1 ubiquitination activity between the two versions of the HECT domain at 37 °C (Fig. 6).
FIGURE 6.
Ubiquitination activity of the HUWE1 HECT domains at 37 °C. a, the autoubiquitination activity of HUWE1 HECT domain at 37 °C was tested using 60 μm Ub as substrate and 2 μm WT HECT domains incubated with UBE1, UBE2L3, and an ATP regenerating system (note the different time scale for the two variants of the HECT domain). Reaction mixtures were separated on SDS-PAGE and analyzed by anti-Ub immunoblot. b, the Mcl-1 ubiquitination activity of HUWE1 HECT domain at 37 °C was tested using 5 μm FLAG-Mcl-1 as substrate and 100 nm WT HECT domains incubated with UBE1, UBE2L3, Ub, and an ATP regenerating system. Reaction mixtures were separated on SDS-PAGE and analyzed by anti-FLAG immunoblot.
Catalytic Activity of the C4341A Mutants
Mutation of the conserved catalytic cysteine to alanine (C4341A) abolishes activity of the HECT domain (supplemental Fig. 2). In the case of the helix-lacking HECT domain, we consistently observed that the C4341A mutant is capable of transferring a single ubiquitin to self (supplemental Fig. 2a) or Mcl-1 (supplemental Fig. 2c). These species were not generated when the HECT domain was omitted from the reaction (lane marked N). Although the failure of mutation of the catalytic cysteine to abolish activity has been previously observed (10), quantification of the monoubiquitinated species shows that this activity represents at best a minor fraction of wild-type activity (supplemental Fig. 2, e and f).
DISCUSSION
We present here crystal structures of the HUWE1 HECT domain and identify a conserved structural element, helix α1, that stabilizes the HECT domain and tightly modulates its activity. Helix α1 is present in the structure of the WWP1 HECT domain (referred to in the WWP1 structure as H1′) (9), where the authors note that it plays an obvious role in contributing to HECT domain stability. As the H1′ helix is oriented between the C lobe and domains N-terminal to the HECT domain that presumably mediate protein-protein interactions, the authors suggested that H1′ helix contributes to target protein specificity in reactions catalyzed by the HECT domain. We confirm that the α1 helix is indeed crucial for stability and identify a role for this structural element in modulating HECT domain activity, as judged by autoubiquitination and Mcl-1 ubiquitination assays. Further experiments will determine whether this conserved helix modulates activity of other members of the HECT domain family.
In the absence of the N-terminal α1 helix, the HUWE1 HECT domain gains activity relative to its helix-extended counterpart. What could be the reason for this unexpected behavior? Deletion of helix α1 might expose hydrophobic residues that trigger assembly of HUWE1 HECT domains into oligomers. Such behavior has been observed in the crystals of E6AP (PDB codes 1C4Z and 1D5F) (7). However, HUWE1 HECT Δ α1 behaves as a monomer and is properly folded in solution, as judged by several criteria (Fig. 1d and data not shown).
Our structural data on HUWE1 shows that the HECT domain adopts the same conformation regardless of the presence of helix α1, and both variants contain the same helical content (Fig. 1d). HUWE1 Δ α1, however, is far more active in catalyzing self-ubiquitination and in single-turnover assays. It also accepts Ub from the E2 UBE2L3 more readily than the helix-extended HECT domain. Furthermore, we observed elevated temperature factors, indicative of conformational flexibility, in the crystal structure of HUWE1 HECT Δ α1; a similar observation was made in the crystals of the E3 Ub ligase IpaH from the bacterial pathogen Shigella flexneri (23). We favor the interpretation that removal of helix α1 destabilizes the HECT domain to produce a more relaxed version of the enzyme that exhibits greater intradomain flexibility. This increased flexibility allows the enzyme to sample more conformational states, thereby increasing its level of activity. Some of these conformational states may resemble the extended HECT domain structures observed in the crystal structures of Smurf2 and E6AP, in which the C-lobe has rotated about the flexible linker that connects the two subdomains of the HECT domain. In this scenario, removal of the α1 helix is analogous to the linker-extension mutations made in WWP1 (9). The removal of helix α1 may also shift the conformational equilibrium of the HECT domain into an orientation that facilitates the E2-HECT interaction or product release. This possibility is supported by evidence that enzymes exist in a dynamic range of conformations, and the equilibrium between these different conformers can be shifted by mutation (27).
We did not anticipate that destabilization of the HECT domain would increase enzymatic activity. The Ub transfer reaction involves defined regions including the ordered β-strands that describe the E2 binding region and the catalytic site surrounding residue Cys-4341. However, other steps, such as product release, may contribute to catalytic rate and may be influenced by increased conformational flexibility (28). A correlation between conformational flexibility and promiscuous activity has been observed for several other proteins (28). An example of a flexible enzyme is cytochrome P450, which can adopt a range of different conformations that allow it to act upon a variety of substrates. Among the P450 family of enzymes, the rigid CYP2A6 enzyme exhibits limited substrate specificity, whereas the highly flexible CYP3A4 is far more promiscuous (28). In the case of HUWE1 HECT domain, the α1 helix may serve to impose a constraint on the inherent flexibility of the catalytic domain, thus fine-tuning enzymatic activity.
Autoubiquitination is often used as a criterion of E3 Ub ligase activity and, for some ligases, has been proposed as a mechanism of self-regulation of stability and downstream signaling functions (29, 30). Our data show that this type of activity can be largely suppressed by minor extensions of what has been considered the core catalytic domain. We note, however, that our study focuses on the HECT domain of a multidomain protein, and there may exist other structural elements in the 482-kDa Huwe1 protein that affect its activity. This has been observed for the E3 Ub ligases Smurf2 (30), IpaH9.8 (23), and SspH2 (31), in which domains N-terminal to the catalytic domain suppress autoubiquitination activity. Although autoubiquitination is clear evidence of catalytic activity of Ub ligases, the functional relevance of this reaction remains to be established for many E3s, including HUWE1. The increase in activity seen in the absence of helix α1 appears to stem from increased conformational flexibility in the enzyme. It is difficult to rationalize this behavior from static crystal structures, yet increased thermal motion observed in the Δ helix α1 structure is at least an indirect indicator. The significance of thermal motion within a protein with respect to reaction parameters is an emerging theme (32). HECT domains may have diverged to arrive at their extant spectrum of substrates by modulating flexibility (28) in addition to more tractable changes of surface properties. We conclude that the activity of the HUWE1 HECT domain is tightly modulated through restriction of conformational space rather than steric considerations by the presence of a 19-residue helix α1.
Acknowledgments
We thank Christian Schlieker for helpful discussions and critical reading of the manuscript and Fenghe Du and Xiaodong Wang at UT Southwestern Medical Center for providing cDNA plasmids for HUWE1 (Mule) and Mcl-1.
This work was supported, in whole or in part, by National Institutes of Health Grant Postdoctoral Fellowship F32 AI63854 (to K. R. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (code 3H1D) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
E. Maspero and S. Polo, personal communication.
- Ub
- ubiquitin
- HECT
- homologous to E6AP C terminus
- HUWE1
- HECT, UBA, and WWE domain containing 1
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- WT
- wild type.
REFERENCES
- 1.Bernassola F., Karin M., Ciechanover A., Melino G. (2008) Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 2.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O., Felberbaum R., Hochstrasser M. (2006) Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 4.Christensen D. E., Brzovic P. S., Klevit R. E. (2007) Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 5.Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008) PLoS ONE 3, e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheffner M., Staub O. (2007) BMC Biochem. 8, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. (1999) Science 286, 1321–1326 [DOI] [PubMed] [Google Scholar]
- 8.Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005) Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 9.Verdecia M. A., Joazeiro C. A., Wells N. J., Ferrer J. L., Bowman M. E., Hunter T., Noel J. P. (2003) Mol. Cell 11, 249–259 [DOI] [PubMed] [Google Scholar]
- 10.Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Kogel U., Scheffner M., Helin K., Eilers M. (2005) Cell 123, 409–421 [DOI] [PubMed] [Google Scholar]
- 11.Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. (2005) Cell 121, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 12.Hall J. R., Kow E., Nevis K. R., Lu C. K., Luce K. S., Zhong Q., Cook J. G. (2007) Mol. Biol. Cell 18, 3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X., Heng J. I., Guardavaccaro D., Jiang R., Pagano M., Guillemot F., Iavarone A., Lasorella A. (2008) Nat. Cell Biol. 10, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong Q., Gao W., Du F., Wang X. (2005) Cell 121, 1085–1095 [DOI] [PubMed] [Google Scholar]
- 15.Herold S., Hock A., Herkert B., Berns K., Mullenders J., Beijersbergen R., Bernards R., Eilers M. (2008) EMBO J. 27, 2851–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons J. L., Tait P. S., Finch D., Dianova, Edelmann M. J., Khoronenkova S. V., Kessler B. M., Sharma R. A., McKenna W. G., Dianov G. L. (2009) EMBO J. 28, 3207–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love K. R., Pandya R. K., Spooner E., Ploegh H. L. (2009) ACS Chem. Biol. 4, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z., Minor W. (1997) Macromol. Crystallogr. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19.Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 20.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 22.Eletr Z. M., Huang D. T., Duda D. M., Schulman B. A., Kuhlman B. (2005) Nat. Struct. Mol. Biol. 12, 933–934 [DOI] [PubMed] [Google Scholar]
- 23.Singer A. U., Rohde J. R., Lam R., Skarina T., Kagan O., Dileo R., Chirgadze N. Y., Cuff M. E., Joachimiak A., Tyers M., Sansonetti P. J., Parsot C., Savchenko A. (2008) Nat. Struct. Mol. Biol. 15, 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M., Pickart C. M. (2005) EMBO J. 24, 4324–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salvat C., Wang G., Dastur A., Lyon N., Huibregtse J. M. (2004) J. Biol. Chem. 279, 18935–18943 [DOI] [PubMed] [Google Scholar]
- 26.Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006) EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisenmesser E. Z., Millet O., Labeikovsky W., Korzhnev D. M., Wolf-Watz M., Bosco D. A., Skalicky J. J., Kay L. E., Kern D. (2005) Nature 438, 117–121 [DOI] [PubMed] [Google Scholar]
- 28.Tokuriki N., Tawfik D. S. (2009) Science 324, 203–207 [DOI] [PubMed] [Google Scholar]
- 29.Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 30.Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., Forman-Kay J. D. (2007) Cell 130, 651–662 [DOI] [PubMed] [Google Scholar]
- 31.Quezada C. M., Hicks S. W., Galán J. E., Stebbins C. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4864–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lange O. F., Lakomek N. A., Farès C., Schröder G. F., Walter K. F., Becker S., Meiler J., Grubmüller H., Griesinger C., de Groot B. L. (2008) Science 320, 1471–1475 [DOI] [PubMed] [Google Scholar]
- 33.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]