Abstract
A typical plasmid replicon of Escherichia coli, such as ori γ of R6K, contains tandem iterons (iterated initiator protein binding sites), an AT-rich region that melts upon initiator-iteron interaction, two binding sites for the bacterial initiator protein DnaA, and a binding site for the DNA-bending protein IHF. R6K also contains two structurally atypical origins called α and β that are located on either side of γ and contain a single and a half-iteron, respectively. Individually, these sites do not bind to initiator protein π but access it by DNA looping-mediated interaction with the seven π-bound γ iterons. The π protein exists in 2 interconvertible forms: inert dimers and active monomers. Initiator dimers generally function as negative regulators of replication by promoting iteron pairing (“handcuffing”) between pairs of replicons that turn off both origins. Contrary to this existing paradigm, here we show that both the dimeric and the monomeric π are necessary for ori α-driven plasmid maintenance. Furthermore, efficient looping interaction between α and γ or between 2 γ iterons in vitro also required both forms of π. Why does α-γ iteron pairing promote α activation rather than repression? We show that a weak, transitory α-γ interaction at the iteron pairs was essential for α-driven plasmid maintenance. Swapping the α iteron with one of γ without changing the original sequence context that caused enhanced looping in vitro caused a significant inhibition of α-mediated plasmid maintenance. Therefore, the affinity of α iteron for π-bound γ and not the sequence context determined whether the origin was activated or repressed.
Keywords: DNA Helicase, DNA Polymerase, DNA Primase, DNA-protein Interaction, DNA replication, DNA Synthesis, DNA Looping, Initiator Protein, Origin-Origin Interaction, Replication Control
Introduction
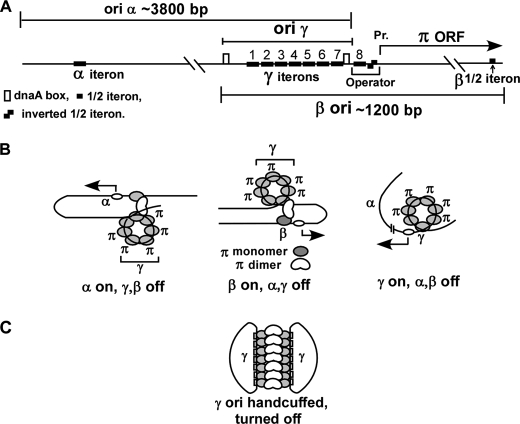
Investigations of bacterial plasmid replicons have been invaluable in revealing the mechanism of replication initiation, termination, and copy number control (1–3). There are generally three types of replication initiation mechanism in plasmids: (i) control by an antisense repressor RNA that hybridizes to the primer RNA and negatively regulates initiation frequency and copy control, as in the case of Col E1(4), (ii) binding of an initiator protein to short repeated DNA sequences called iterons located at an ori (5–7) followed by replisome assembly by protein-protein interaction with the initiator protein, and (iii) a rolling circle mechanism in which the plasmid initiator is not only an ori-specific “nickase” but also functions as a site-specific topoisomerase (8). The plasmid R6K has 3 origins of replication called α, γ, and β, and all three are functionally dependent upon the plasmid-encoded initiator protein called π (Refs. 9–12, see Fig. 1). The π protein binds to γ but not to the α and β iterons; the latter requires the presence of ori γ sequence in cis for initiating replication and enabling plasmid maintenance (13, 14). The replication origins α and β are located ∼3800 and ∼1200 bp away, respectively, from ori γ and at its opposite side (11, 15). We have previously reported that π-mediated DNA looping between the single iteron at α or the half-iteron at β with the γ iteron array appears to activate the two distantly located origins, respectively (9, 13, 16). Models of activation of each of the three origins, that are mutually exclusive, and the postulated looped structures involved in the process are shown in Fig. 1.
FIGURE 1.
Schematic representation of the R6K replicon and models corresponding to origin activation. A, replicon of R6K showing the approximate locations of the origins α, β, and γ; note that ori γ contains 7 iterons, ori β a half-iteron, and ori α a single iteron, respectively. The operator of π ORF contains the 8th iteron and two half-iterons present as inverted repeats. There are 2 dnaA boxes at ori γ. B, models showing looped structures, which are mutually exclusive and correspond to the turning on of α, β, or γ. C, postulated structure of a pair of handcuffed γ origins that leads to turning off of both. In the active origins, the iteron DNA is believed to be wrapped around a core of monomeric π that is postulated to be unraveled in the handcuffed ori γ. C represents negative control of replication in trans, whereas the models in B show origin activation in cis.
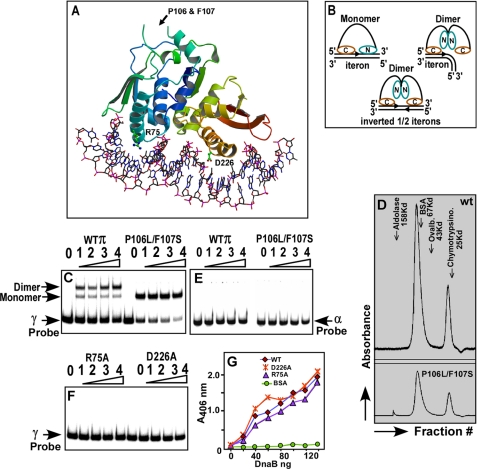
More recently, the replication initiation from ori γ has been reconstituted in vitro using 22 purified proteins and that has illuminated part of the initiation mechanism at ori γ (17). Further progress has been made by solving the crystal structure of the monomeric γ iteron complex (18). The π protein has a winged helix structure (see Fig. 2, A and B) with N-terminal and C-terminal DNA recognition helices. In the monomeric form of the protein called π cop (for high copy number), the C-terminal recognition helix contacts base pairs (bps) located in the 5′-half of the iteron DNA, in the major groove, whereas the N-terminal recognition helix contacts the 3′-half of the iteron (see Fig. 2B and Ref. 18). Chemical and enzymatic footprinting data are consistent with the structure (19, 20). In contrast, the dimeric form of the protein contacts the bp located only in the 5′-half of the iteron probably through the C-terminal recognition helix; no contacts are seen with those located in the 3′-half (19). The dimers also fully contact the operator sequence that consists of two inverted half-repeats corresponding to the 5′-halves of the iteron (Fig. 2B) (19). Results suggest that dimerization of π causes significant alterations in its structure that probably prevent the N-terminal recognition helix from contacting the iteron DNA.
FIGURE 2.
Structure of the monomeric π iteron complex and the postulated configuration of the dimeric and monomeric π protein bound to iteron DNA. A, crystal structure of monomeric π-iteron complex showing the two DNA recognition α helices; the N terminus is blue-green and the C terminus is red-brown. The locations of the various mutations used in this work are shown, and the structure is a winged helix. B, models based on footprinting data and crystal structure showing the postulated binding of monomeric and dimeric π to the iterons and the binding of dimeric π to the inverted half-iterons present at the π operator sequence shown in Fig. 1A; note that the dimer is postulated to undergo a structural alteration that prevents the N-terminal recognition helix from binding to the 3′-half of a full iteron sequence. C, gel mobility shift experiments showing that the dimer binds to a labeled γ iteron, giving a major shift corresponding to the dimer and a minor one corresponding to the monomer. The monomeric πP106L F107S (called π cop) binds to the γ iteron probe and exclusively generates a band corresponding to a monomeric π iteron complex; note that π cop binds with higher affinity to the DNA than the wt π as revealed by the intensity of the unbound probe. D, gel filtration profiles of the dimeric wt π (upper panel) and π cop (lower panel)) with marker proteins, showing a majority of dimeric protein (with a lesser peak corresponding to the monomer). E, neither the wt π nor the π cop binds to the α iteron probe under conditions identical to that shown in C. F, gel mobility shift experiments showing that the R75A and D226A mutants fail to bind to DNA. In all gel mobility shift experiments, 20 fmol of labeled DNA were incubated with 0, 8, 16, 32, and 64 pmol of π (wt, cop, R75A, and D226A forms). G, ELISA showing the binding of DnaB to various forms of purified π; note that although the R75A and D226A mutant forms are completely defective in iteron binding, these are almost as effective as wt π in binding to DnaB.
The dimeric form of wt π is inert in replication initiation but gets converted to active monomers by the action of the DnaJ-DnaK-GrpE chaperone system, in an ATP hydrolysis-dependent reaction (21). Iteron binding, as shown in this work, also induces some of the mutant forms such as π cop to monomerize almost completely.
Replication initiation at ori γ is greatly enhanced by the binding of the DNA-bending protein, the integration host factor (IHF),2 to its cognate site that apparently promotes protein-protein interactions between DnaA and π by bending the intervening DNA between a dnaA box and the iterons (22, 23). Ori γ can replicate feebly in an ihfΔ host, and the resultant plasmid copy number in this situation is drastically reduced so that the host cells, harboring the plasmid marked by an ampicillin resistance (ampR) marker and containing the open reading frame (ORF) of wt π, do not form visible colonies on plates containing 40 μg/ml of ampicillin. However, inclusions of an α or a β ori sequence in cis in the proper sequence context with γ rescues the growth on antibiotic plates. This observation has been used as a tool to distinguish between initiation and plasmid maintenance by ori γ from that by ori α or β (e.g. see Refs. 9, 24).
Although the π protein makes a positive contribution to initiation of replication by assembling a replisome at the ori γ by protein-protein interactions with host-encoded replisomal proteins (25, 26), it also negatively controls replication in two ways: (i) it binds to an inverted half-iteron sequence at its operator (19) (Fig. 1A) and represses its own synthesis at the transcriptional level (28, 29), (ii) it promotes pairing between the two sets of 7 iterons of γ present in 2 daughter plasmids and shuts down their replication. This pairing mechanism has been called handcuffing (5, 6) (see Fig. 1C). Using the F plasmid as a model system, we have shown in vitro that dimeric initiators cause origin shutdown by promoting handcuffing in cis between the origin iterons and a second set of iterons located at incC (incompatibility), which is a negative regulatory element of plasmid maintenance. The dimeric initiator protein, RepE-mediated iteron pairing blocks the melting of the AT-rich region at the F plasmid origin (30). In summary, the existing paradigm of replication control, based on the data from several different plasmid systems, is that dimeric initiators are negative regulators of copy control and of initiation of replication where as the monomeric form catalyzes replication in association with other proteins and that the monomer-dimer equilibrium in the host cell contributes to plasmid copy number control.
The present work was designed to address two mechanistic questions about DNA looping-dependent replication control in R6K: (i) do DNA looping in vitro and replication activation at α and β in vivo both require the dimeric and the monomeric forms of π? (ii) why does iteron-iteron interaction, which generally suppresses replication initiation at iteron-regulated plasmid origins (6, 30), induce replication at ori α rather than its repression? Using an in vitro DNA-looping assay (30) and in vivo replication/plasmid maintenance measurements, we show that both intrachromosomal, π-mediated iteron-iteron pairing, and looping out of the intervening DNA and plasmid maintenance in vivo by α require not only the monomeric but also the dimeric form of π. Using an iteron swap experiment in vivo, we show further that origin activation requires very transient, weak interactions between the α and γ iterons. Enhancement of the looping interaction by replacement of the weak α iteron with a consensus, stronger γ iteron without changing the original sequence context, did not cause a corresponding enhancement of ori α activity. Rather, the stronger looping reaction between the 2 iterons generated by the iteron swap greatly diminished plasmid maintenance in vivo. The mutant forms of the dimeric π that did not bind to DNA could neither collaborate with the monomeric π to promote DNA looping in vitro nor induce replication initiation from ori α in vivo, thereby supporting the interpretation that both monomer-dimer protein-protein interaction and binding of each form of the protein with DNA, are essential for productive initiation at α.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Oligonucleotides
Escherichia coli strain DH5α was used for all cloning experiments. HN356 and HN840 cells were used for in vivo replication assays. BL 21(DE3) pLysS, codon plus cells were used for the preparation of the proteins. The list of oligonucleotides used is given in Table 1. All strains were grown at 37 °C (unless otherwise mentioned). Mini alpha (pAM 738α) plasmid has been described (9). All chemicals were purchased from Sigma. The custom oligonucleotides were purchased from Integrated DNA Technologies.
TABLE 1.
Oligonucleotides used for mutagenesis and plasmid constructions
| Name | Sequence | Purpose/use |
|---|---|---|
| Nde Pi F | GGGAATTCCATATGAGACTCAAGGTCATGATGGAGCTG | |
| Bam Pi R | GGCGGGATCCTTACTCGAGCCCCTTAGCTTTTTTGGGAGG | Amplification of Pi ORF |
| P106LF106SF | GCTTTAGCTAAAGAGCTTAACCTGCTCTCTACTGCTAAAAACTCC | Generation of P10LF107S mutation in Pi ORF |
| P106LF106SR | CTCTTCAGGGGAGTTTTTAGCAGTAGAGAGCAGGTTAAGCTCTTT | |
| R75A Pi F | CCCCATCGCTTGCTTATGCACAATTAAAAGAGGGTGGTAAATTAC | |
| R75A Pi R | GTAATTTACCACCCTCTTTTAATTGTGCATAAGCAAGCGATGGGG | Generation of R75A mutation in Pi ORF |
| D226A Pi F | CCCTGACTTTCCTATTTTTAAAAGGGCTGTGTTAAATAAAGCCATTGCTG | |
| D226A Pi R | CAGCAATGGCTTTATTTAACACAGCCCTTTTAAAAATAGGAAAGTCAGGG | Generation of D226A mutation in Pi ORF |
| Alpha1 | GATCTGATATCAAAATCTGAGGGATTTCGTAGCTGACTGCA | α iteron cloning in looping vector at BglII-Pst site |
| Alpha2 | GTCAGCTACGAAATCCCTCAGATTTTGATATCA | |
| Gamma 1 | CTAGTGGGCCCAAACATGAGAGCTTAGTACGTAT | γ iteron cloning in looping vector at Spe-XbaI site |
| Gamma2 | CTAGATACGTACTAAGCTCTCATGTTTGGGCCCA | |
| BglII Gamma F | GATCTCCATGGAAACATGAGAGCTTAGTACGTACTGCA | γ iteron cloning in looping vector at BglII-Pst site |
| Pst Gamma R | GTACGTACTAAGCTCTCATGTTTCCAGGTA | |
| Complete Gamma F | GGACTAGTCAAAAATTGCTTTGAGAGGCTCTAAGG | Complete γ cloning in looping vector Spe-XbaI site |
| Xba1 gamma R | GCTCTAGAGATCTGAAGATCGGCAGTTCAACCTG | |
| Alpha MF | GTGCTTCAGTATGTACAGAAATGCAAAACATGAGAGCTTAGTACGCTGAAAGATCGCCAGTCTTCGACCG | Replacement of α iteron by γ iteron |
| Alpha MR | CCTTACGGTCGAAGACTGGCGATCTTTCAGCGTACTAAGCTCTCATGTTTTGCATTTCTGTACATACTGAAG | |
| NotPiF | GAGCGGCCGCACGTTAAAACATGAGATAAAAATTG | Amplification of Pi ORF with its own promoter |
| PstPiR | AACTGCAGTCAAGCTTCATCATCTTTATCGCCAG | |
| KanF | GCGGGAATTCGGGTCTCCGCAAGTGGCACTTTTCGGGGAAATG | Cloning of Kanamycin gene in pUC Kn vector |
| KanR | GCGGGAATTCAGCGTCGATTTTTGTGATGCTCGTCAGGGG | |
| Alpha350 | GCCACAACTGAAATTGTGCTTCAG | Used to detect α iteron in Chip assay |
| Alpha475 | CGAACGCCCCGCACAGGTCTTACAG | |
| Gamma373 | CCTTAGAGGCTATTTAAGTTGCTG | Used to detect γ iteron in Chip assay |
| Gamma R | AGTACGTACTAAGCTCTCATGTTTCAC |
Plasmid Constructions
The plasmid used for the cloning of α and γ iteron had two clusters of multiple cloning sites separated by a 200-bp region in the middle of which there was a single EcoRI site. The oligos used for cloning of single γ and α iterons are given in Table 1. For the cloning of iteron, each pair of oligo was annealed overnight in a buffer containing 10 mm Tris, 1 mm EDTA, pH 7.6, and 600 mm NaCl. The annealed DNA was passed through Sepharose G25 and phosphorylated with ATP and T4 polynucleotide kinase. The inserts were cloned in the plasmid at the compatible restriction sites. The complete γ sequence was cloned after PCR amplification using the primers shown in Table 1, and the amplified product was digested and cloned at the respective sites in the plasmid vector. The plasmid donor (high copy, P106L and F107S) was prepared by cloning of the π ORF with its own promoter and containing the desired mutation in a pUC18-based plasmid that has kanamycin (KmR) instead of ampicillin resistance (ampR) as the selection marker. Mutated π ORF with the R75A mutation was cloned between NdeI-XhoI sites of the “RSF-Duet” expression vector (Novagen).
Replication of α-γ and γ Replicons in Vivo
Isogenic HN356 (IHF+) and HN840 (ihfΔ) cells carrying the π donor plasmid(s) were transformed with either γ or α-γ replicon, and the cells were plated on LB medium containing amp (40 μg/ml) and Km (25 μg/ml) and incubated at 37 °C for 16–18 h. In those cases where π ORF and the origins were located in the same plasmid, transformants of IHF+ and ihfΔ were selected on Luria-Bertani (LB) plates containing ampicillin (40 μg/ml).
Swapping of the α Iteron with a Single γ Iteron
pAM 738α plasmid was used as the template for carrying out site-directed mutagenesis, using the mutagenic primers shown in Table 1. Native Pfu DNA polymerase (Stratagene) was used to catalyze PCR reactions following the manufacturer's instruction. After repeated chain extension by PCR, the reaction products were treated with DpnI for 4 h to eliminate the original template DNA, the products purified and transformed into E. coli DH5α cells. DNA was isolated from the transformants growing on 40 μg/ml amp selection plates, and the mutations were confirmed by DNA sequencing.
Cloning and Site-directed Mutagenesis of π ORF
Mutation in the π ORF was generated in two steps. In the first step, the N- and C-terminal regions of π ORF were amplified using Nde-Pi F and P106L-F107S R primer and P106L-F107S F with Bam-Pi R primers. The amplified products were purified and mixed in an equimolar ratio. This mixture was used as a template for the second round of PCR where the Nde-PiF and BamPi R primers were added after the first five cycles of PCR reaction. A similar strategy was used to generate π ORF with its own promoter carrying P106L F107S, R75A, and D226A mutations using the primers given in Table 1. Pfu Ultra (Stratagene) was used in these PCR reactions.
Expression and Purification of π Protein
The different forms of the protein were expressed in the vector pTXB1 that contained a self-cleaving intein tag and a chitin binding affinity tag (New England Biolabs, Beverly, MA), under the transcriptional control of the T7 promoter. E. coli BL21(DE3) “codon plus” cells (Stratagene) were used for enhanced protein expression. WT π, π cop, and D226A π ORFs were cloned between NdeI and XhoI sites of pTXB1 plasmid to generate pTXB1-WT, pTXB1-π cop, and pTXB1-D226A expression plasmids. Upon induction with 1 mm isopropyl β-d-thiogalactopyranoside, the π protein was expressed as a fusion with the chitin-binding peptide affinity tag separated from π by an intein sequence, which was cleaved off in the presence of dithiothreitol to generate the original protein. The fusion proteins were purified as described (17) except that the induction was carried out at 18 °C for 18–20 h. The R75A mutant of the π ORF was cloned at the NdeI-BamHI site in pET15b to generate the pET15b-R75A π plasmid, and the His-tagged R75A π protein was purified as described (14).
Gel Mobility Shift Assay
The DNA fragments containing either single α or γ iteron sequences were amplified by PCR, and the purified products were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. 20 fmol of the labeled DNA substrate was mixed with increasing concentrations of π proteins (8, 16, 32, and 64 pmol) in a 20-μl reaction containing 1× π binding buffer (40 mm Hepes-KOH, pH 7.5, 150 mm NaCl, 40 mm potassium glutamate, and 10% glycerol) and 7.5 μg of salmon sperm DNA. After 15 min of incubation at room temperature, the reaction mix was loaded with 1× gel-loading dye on 6% polyacrylamide gel and resolved by electrophoresis for 2 h at 145 V. Gels were analyzed and the data quantified with a phosphorimager.
Ligation Assay
The ligation assay was performed as described (30). Twenty fmol of 32P 5′-end-labeled DNA was mixed with either wt π or monomeric π or the mixture of wild type and monomeric π in a equimolar ratio (64 pmol each) in a buffer containing 40 mm Hepes-KOH, pH 7.5, 150 mm NaCl, 40 mm potassium glutamate, and 10% glycerol. This mixture was incubated for 10 min at room temperature and further for 45 min at 4 °C. To a 75-μl reaction mixture 1 unit of T4 DNA ligase was added, and incubation continued for various times up to 45 min at 16 °C. Finally, the exonuclease reaction was carried out in a 100-μl reaction mix containing 1 unit of T7 exonuclease (NEB 4 buffer) at 25 °C for 6 h. Samples were trichloroacetic acid-precipitated and passed through a GF/C filter. After two washes with ethanol, the filters were dried, and radioactivity retained on the filters was determined by liquid scintillation counting.
ELISA
Protein-protein interactions were performed by immobilizing different π protein on the surface of plastic microtiter plates. After blocking the unoccupied plastic surface with bovine serum albumin, the immobilized proteins were incubated with various concentrations of DnaB, and the extent of interaction was detected with anti-DnaB antibodies and secondary alkaline phosphatase-linked antibodies. The results were quantified with an ELISA plate reader as described previously (25).
Chromatin Immunoprecipitation (ChIP) Assay
E. coli cells containing plasmid γ-α origins were grown to A600 = 0.7, centrifuged, and were snap frozen in liquid nitrogen and washed twice with 50 ml of chilled PBS. Fresh solution of 10 mm DMA (dimethyl adipimidate) in PBS containing 0.25% dimethyl sulfoxide was prepared and was added in solution and incubated with gentle shaking for 1 h at room temperature. Cells were centrifuged, washed once, and resuspended in 50 ml of PBS containing 1% final formaldehyde and were incubated at room temperature for 3–4 h. The reaction was quenched by adding glycine to 100 mm. Cells were washed twice with PBS. ChIP was carried out as described by Ref. 31. After double cross-linking, cells were resuspended in 500 μl of chilled lysis buffer (50 mm Hepes/KOH, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate), and protease inhibitor was added. Cells were lysed, centrifuged at 13,000 rpm for 5 min, and the supernatant was discarded and resuspended in 500 μl of lysis buffer. Chromatin is sheared by sonication on ice. The samples were centrifuged at 4 °C to recover the supernatant. An aliquot of the lysate was set aside as input sample. Lysate was incubated with anti-π polyclonal antibody, overnight at 4 °C on a slow moving rotator. After incubation with the antibody, protein A beads were added and incubated further for 1 h at 4 °C. Samples were centrifuged, supernatant was removed, and beads were washed with 500 mm NaCl containing lysis buffer in 15-min intervals. Beads were pelleted at 3000 rpm (room temperature). Sample beads were resuspended and washed in lysis buffer once and three times with deoxycholate buffer(10 mm Tris-HCl, pH 8.0,0.25 M LiCl, 0.5% Nonidet P-40,0.5% sodium deoxycholate, I mm EDTA. Beads were resuspended in elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1.5% SDS) and incubated at 65 °C for 30 min. After elution, the supernatant was incubated at 65 °C overnight to reverse the formaldehyde cross-linking. After a day, the samples were treated with proteinase K and incubated for 2 h at 55 °C. After adding 25 μg of glycogen as carrier, samples were phenol/chloroform-extracted, DNA recovered by ethanol precipitation, and were dissolved in 50 μl of TE, pH 8.0. Serial dilution of input and immunoprecipitated DNA was made, and PCR was done with appropriate primers to detect the α or the γ origin.
RESULTS
Purification of the Wt and Various Mutant Forms of π and Interaction with α and γ Iterons
To study the possible relative roles of the dimeric and the monomeric forms of π and the various mutants, the proteins were purified as described above, and their interactions with iteron DNA were characterized. It was also considered useful to assign the mutations to the residues displayed in the crystal structure of γ-iteron-π complex. The double mutant P106L F107S of π (π cop), that is known to enhance the copy number of R6K (32, 33) are located in an unstructured region of the complex (Fig. 2A). The N-terminal (blue-green) and the C-terminal (red-brown) recognition helices contained the residue Arg-75 and Asp-226, respectively (Fig. 2A). The N-terminal recognition helix binds to base pairs (bps) at the 3′-half of the iteron, and the red-brown C-terminal recognition helix binds to those on the 5′-half (Fig. 2B). These conclusions on structural aspects of γ-iteron-π contacts are supported by footprinting data (19, 34). We wished to test some of these conclusions further by performing site-directed mutagenesis, purification of the mutant forms of the protein, and performing their biochemical characterization.
We carried out gel mobility shift assays of the proteins bound to labeled α and γ iteron probes. As expected, the wt protein generated a gel shift pattern showing a major more slowly moving band corresponding to the binding of the dimeric form with the probe DNA and a minor, faster moving band corresponding to binding of the DNA probe to the monomers. The gel filtration profile of wt π correspondingly, carried out in a Superose 12 gel filtration column revealed a major dimeric peak and a minor monomeric peak with Stoke's radii indicative of molecular masses of ∼72 and ∼35 kDa, respectively (Fig. 2D, upper panel). The gel filtration profile of π cop also showed a mixture of a majority of dimeric and a minority of monomeric peaks (Fig. 2D, lower panel). However, after binding to the γ iteron DNA, the π-cop-γ complex displayed the characteristic mobility shift of a monomeric π-iteron complex suggesting that iteron binding caused π cop to dissociate into monomer (Fig. 2C). The wt π consistently bound with lower affinity to the iteron probe in comparison with π cop as revealed by the magnitude of depletion of label in the free DNA band and its corresponding enhancement in the DNA-protein complex as a function of increasing concentrations of protein in the DNA binding reaction. Neither the wt nor the π-cop protein bound to the solo α iteron (Fig. 2E). Data are consistent with the interpretation that DNA binding induced the π cop but not the wt form to dissociate almost completely into monomers and that a solo α did not bind to either forms of π protein.
Consistent with the crystal structure (Fig. 2A), the mutant forms R75A and D226A did not bind to the γ iteron DNA (Fig. 2F). We investigated whether these results could be attributed to a localized change in the DNA binding domains caused by mutations or due to global misfolding of the mutant forms of the protein. To distinguish between these alternatives, we measured the relative abilities of the wt and the two mutant forms to bind specifically to DnaB (25). We reasoned that if the R75A and D226A mutations caused global protein misfolding, then the mutant forms of the protein should not retain specific protein-protein interaction with DnaB. To test this expectation, we performed ELISA experiments by immobilizing separately, a fixed and equivalent amount of the wt and the two mutant forms of π to the plastic surfaces of microtiter plates, and challenged the immobilized π proteins with the same increasing range of concentration of purified DnaB in solution. We observed that all 3 forms of π bound almost equally well to DnaB. A negative control with immobilized bovine serum albumin, as expected, did not show any interaction with DnaB. Data are averages of three separate sets of experiments (Fig. 2G). We conclude from the results that the D226A and R75A mutant forms, as expected from the crystal structure, did not bind to DNA. In a control experiment, both the wt π and DnaB did not interact with another replication terminator protein, Reb1 of Schizosaccharomyces pombe (supplemental Fig. S1). The circular dichroism (CD) Spectra of these proteins further revealed that these mutated proteins are folded properly like its wild-type counterpart (supplemental Fig. S2).
Measurement of π-Mediated DNA Looping
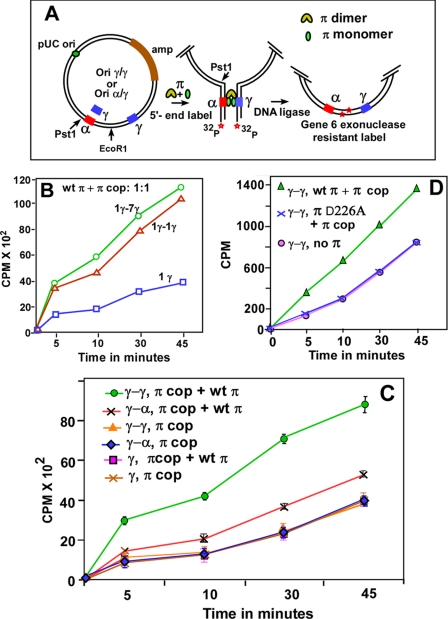
We then proceeded to investigate the roles of the wt and the mutant forms of π in promoting DNA looping with the following questions in mind: (i) do two γ iterons separated by ∼3-kb interact with each other in vitro by π-mediated iteron pairing?; (ii) which forms of the protein are required for the looping interaction? The technique used to measure looping interactions is based on the γ-γ and α-γ plasmid DNA substrates shown in Fig. 3A (30). The substrate DNA, that contained the ori of pUC19 and the R6K iteron(s), was linearized by cutting at the single EcoR1 site, 5′-end-labeled, incubated either with the wt π, with π cop or with a 1:1 mixture of the 2 forms on ice, after which the mixture was further incubated with T4 DNA ligase at 16 °C for various periods of time. The reaction products were digested with T7 exonuclease, and measurements of π-mediated enhancement of circle formation were carried out by counting the exonuclease-resistant label. The enzyme hydrolyzes dsDNA with 5′-protruding ends in a 5′ to 3′ direction. The magnitude of the exo-resistant radioactivity, measured as a function of time, was directly proportional to the rate of DNA circularization, which in turn was a measure of the extent of DNA looping.
FIGURE 3.
Determination of the frequency of DNA looping by enhancement of ligase-catalyzed DNA circularization. A, schematic depiction of the circularization assay showing the pUC19-based substrate that contained γ-γ or α-γ iterons cloned at the indicated locations on either side of a unique EcoR1 site. A similar plasmid having a single γ iteron on one side and the 7 γ iterons on the other was also constructed. The assay steps including linearization by EcoR1, 5′-end labeling, incubation with either π cop, or with a 1:1 mixture of wt π and π cop, ligation with T4 DNA ligase at 16 °C for various periods of time, and digestion with T7 gene 6 exonuclease are shown; 20 fmol of DNA were incubated with a total of 128 pmol of π. B, DNA circularization kinetics showing that there was a small difference between γ-γ and γ-7 γ iteron interactions; the control experiment with a solo γ iteron is also shown. C, DNA circularization kinetics with various combinations of DNA substrates and π proteins as indicated. The α-γ plasmid incubated with a 1:1 mixture of wt π and π cop had a small but distinct and reproducible enhancement in ligation kinetics over the same incubated with π cop only. The γ-γ plasmid incubated with π monomer-dimer mixture had a much higher rate of DNA circularization over that when only π cop was used. Data were collected from six separate sets of experiments and are presented with the S.D. as error bars. D, ligation experiments showing that π D226A (non-DNA binding mutant dimers) did not complement π cop to loop two γ-γ iterons. Controls show that under identical conditions, wt π complemented π cop in promoting γ-γ interaction, and there was no interaction as expected when π was withheld from the reaction.
Because ori γ naturally contains 7 iterons, it was necessary to determine whether the full set of iterons placed on one side of the EcoR1-cut DNA substrate enhanced interaction with a single γ iteron placed on the other side of the cut (Fig. 3B) in comparison with a similar DNA substrate that contained one γ iteron on either side of the EcoRI site. Data showed that there was only a small difference between looping promoted by two γ iterons versus that in which one γ iteron and 7 γ iterons flanked the EcoR1 site (Fig. 3B). Therefore in all subsequent experiments, for the sake of simplicity, we used DNA substrates that either contained one pair of single γ iterons or a pair α-γ iterons. Substrates containing a single γ iteron or no iteron were used as negative controls. Implicit in the DNA circularization experiments is the notion that when 2 binding sites located near the ends of a linear DNA substrate with complementary sticky ends are held together by a protein(s), the rate of ligase-catalyzed circularization of the substrate tends to be directly proportional to the efficiency with which the two ends are brought together causing enhanced ligation by the binding protein (16, 30). The presence of a single binding site should not loop DNA, and the data showed no enhancement of ligation rate with such a substrate after incubation with the wt π-π cop mixture (Fig. 3B). A DNA substrate, without preincubation with π, as expected, showed only background levels of end-joining at 16 °C (not shown).
We then proceeded to measure the extent of looping as a function of time using the γ-γ and α-γ iteron sets in the presence of solo π cop, or just dimeric π or an equimolar mixture of both forms. It should be noted that wt π has a significantly lower affinity of binding for iteron DNA even though it contained ∼10% of monomers, presumably generated by intracellular chaperone activity. Control experiments consistently showed that the extent of DNA circularization promoted by wt π alone (as measured by cpm protected after 45 min of ligation at 16 °C) approached ∼15% of that obtained when an equimolar mixture of the monomeric and dimeric proteins was used (supplemental Fig. S3).
The ligation kinetics obtained from six separate experiments (shown with standard error bars) revealed that a pair of γ iterons interacted with each other in the presence of a 1:1 mixture of wt π and π cop to loop DNA in vitro but not when only π cop, was supplied (Fig. 3C). A single γ iteron, as expected, did not promote ligation enhancement when incubated with either the π cop or an equimolar mixture of the wt and the mutant form. Furthermore, the data showed that an α iteron interacted with a γ iteron although at a lower rate than that generated by γ-γ interaction. However, the magnitude of the former interaction was clearly above the control, i.e. α-γ interaction in the presence of the solo π cop protein (Fig. 3C). We have also performed control experiments without the addition of any π to the reaction mixture and consistently observed only background levels of ligation (not shown). The main conclusion derived from the above-mentioned experiments was that π-mediated DNA looping between α-γ or γ-γ iteron pairs required both the monomeric and the dimeric forms of π.
Plasmid Maintenance in Vivo by Ori α Also Required Both Forms of π
We wished to investigate whether the in vitro data on DNA looping presented above showing the requirement for both wt π and π cop was also reflected in a similar protein requirement for plasmid replication/plasmid maintenance in vivo directed by ori α. We investigated this possibility by constructing various replicons containing a solo γ and a replicon with both α and γ origins (Fig. 1). The latter construct was needed because of the obligatory requirement of a γ sequence present in cis for ori α activity (9, 11). We also made use of the observation that ori γ (but not ori α) requires the DNA-bending protein IHF to be functional and consequently is maintained poorly in an ihfΔ strain. In contrast, the presence of an α sequence in cis in the replicon in the proper sequence context rescues plasmid maintenance (23) (see Fig. 4A).
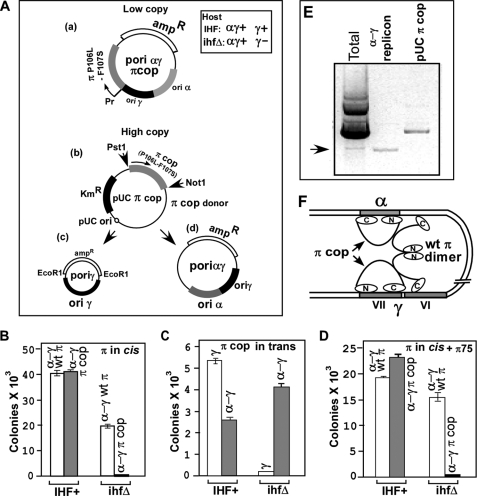
FIGURE 4.
Data showing that both the dimeric and monomeric forms of π are needed for promoting ori-α-dependent plasmid maintenance (in ihfΔ cells) and that the dimeric mutant forms (R75A) that failed to bind to DNA, also failed to support (along with monomeric π) ori α activity. A, a, schematic representation of α-γ plasmid DNA substrate; b, high copy number plasmid that supplied π cop in trans; c, γ and α-γ reporter replicons. B, propagation of α-γ plasmids carrying wt π and π cop in normal to low concentrations, in both IHF+ and ihfΔ cells. C, propagation of γ and α-γ plasmids in IHF+ and ihfΔ cells when π cop was supplied in trans from the high copy pUC19 plasmid. D, failure of the dimeric π containing the R75A mutation that prevented iteron binding to complement π cop in sustaining plasmid propagation. E, image of an agarose gel showing that cells that contained high concentration of π cop in trans did propagate the α-γ reporter as a plasmid. F, model showing a looped structure in which two iteron-bound monomeric π are bridged by a single dimer that not only requires protein-protein interaction with π monomers but also interaction with DNA.
We constructed two types of ori α-γ and ori γ reporter plasmids for these experiments. One class of constructs included the π ORF in cis. The other just contained the cis components of the replicons along with an amp-resistant marker but no π; such a plasmid was maintained by supplying π in trans from a π donor plasmid. The first class of the plasmids, because of their relatively low copy number and because of their autoregulated natural promoter produced moderate to low levels of π protein that were adequate for plasmid maintenance (Fig. 4A, top). The protein expressed in a high copy pUC19 vector from a constitutive lac promoter and supplied in trans (Fig. 4A, middle) naturally resulted in a much higher intracellular level of the protein in the cell milieu (as verified by SDS gel electrophoresis that produced a major band of the protein after staining with Coomassie Blue, not shown). The π protein supplied in trans supported the maintenance of the ampR ori γ and ori α-γ replicons that did not contain the π ORF in cis (Fig. 4A, bottom). First we tested whether the α-γ, ampR mini-replicon required both the dimeric wt π and the monomeric π cop for maintenance in an ihfΔ strain. We transformed equal amounts of the α-γ reporter DNA, one containing the wt π and the other π cop into isogenic IHF+ and ihfΔ host cells and plated the cells on LB plates containing 40 μg of amp/ml. We performed control transformation with pUC19 DNA to calculate the relative plating efficiencies of the two strains and used the data to normalize the transformation efficiencies of the α-γ plasmids. The data showed, as expected, that the replicon carrying the wt π was maintained in both the IHF+ and the ihfΔ strains. In contrast the same replicon containing π cop failed to propagate in the ihfΔ host strain at the specified antibiotic concentration (Fig. 4B).
We then proceeded to test whether the failure of the α-γ plasmid to be maintained in an ihfΔ host when π cop protein was being supplied from the low copy plasmid in cis, and limited amounts can be overcome by supplying the protein at higher concentrations from a pUC19 vector (copy number ∼200 per cell). This experiment was prompted by our observations3 that at high intracellular levels of the protein, when present in many fold molar excess over the concentration of the binding site DNA, the π cop exists as a mixture of dimers and monomers presumably because a significant amount of the protein was not iteron-bound. Alternatively or in addition, the high concentrations of π cop present in the cell milieu could promote even the mutant form to generate some dimers. We further expected that at high intracellular concentration of π cop, α-γ replicon should be maintained in an ihfΔ host even in the absence of wt π. This expectation was borne out by the data (Fig. 4C). We confirmed that the α-γ was present in the episomal form when π cop was supplied in trans from a high copy vector and was not integrated into the host DNA by extracting total plasmid DNA from the cells and resolving these in an agarose gel that showed a faint α-γ replicon band in addition to the much more intense π donor plasmid band (Fig. 4E).
There are at least 2 alternative models with regard to protein-protein and DNA-protein interactions by the dimer and the monomers needed to form a productive looped structure generated by iteron pairing. First, the structure might involve a pair of iterons, each bound to a monomer that are bridged together by a dimeric π (the remaining 6 iterons of ori γ probably wrapped around the π monomers) (35) (Fig. 1, middle). Alternatively, the looped structure could require not only iteron binding by the monomers but also by a dimeric bridge that possibly stabilized the structure strictly by protein-protein interaction without itself binding to DNA. If the first model were correct, then the R75A and D226A mutant forms (Fig. 2, A and F), should still complement monomeric π cop to promote looping-dependent plasmid maintenance in vivo. If on the other hand, interaction of the dimer with DNA is also obligatory, the mutant dimers should fail to complement π monomers required to form a looped structure in vitro and plasmid maintenance in an ihfΔ host in vivo. Data in Fig. 4D showed that the α-γ replicon that also included the ORF of π cop was not propagated in the ihfΔ host even though the mutant form R75A was supplied in trans from the high copy plasmid. We have also repeated the experiment with D226A protein with identical results (supplemental Fig. S4). In control experiments, wt π supplied from the same expression vector in trans supported the in vivo propagation of the α-γ reporter in the ihfΔ host (supplemental Fig. S4). The in vitro data described in a preceding section are also fully consistent with the in vivo data that π dimers needed to contact DNA on the one hand and the monomeric π on the other to promote productive iteron-iteron contacts (Fig. 3D). Fig. 4F schematically presents a model of the looped structure that includes a dimeric π that bridges each of the iteron-bound monomers, not only by protein-protein contacts, but also by contacting the DNA (Fig. 4F).
Plasmid Maintenance by Ori α Requires Weak and Transient Interactions between the α and the γ Iterons
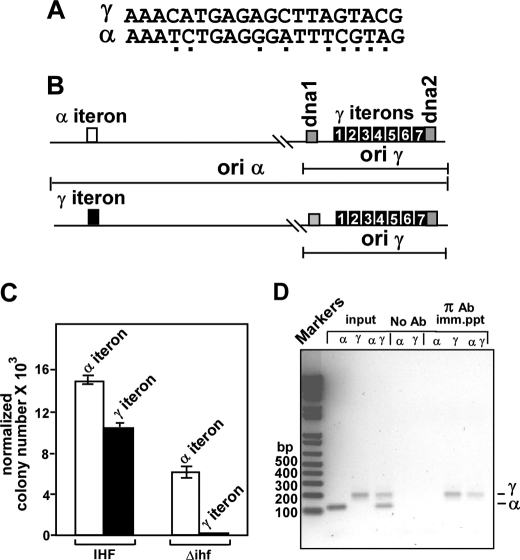
We wished to investigate why π-mediated α-γ iteron pairing supported ori α-driven plasmid maintenance whereas in general, iteron pairing is known to negatively impact plasmid replication in vivo (5, 6, 36, 37) and in vitro by blocking origin melting (30). We wished to approach this problem by an iteron swap experiment in which we precisely replaced the single iteron of ori α by a consensus γ iteron, without changing in any way the sequence context. We then measured the ability of the mutated ori α to support plasmid maintenance in the ihfΔ strain. The differences in nucleotide sequences between the α and γ iterons are shown in Fig. 5A. The normal and mutated replicons are shown schematically in Fig. 5B. Equal moles of the ampR normal and mutated α-γ replicons were separately transformed into the IHF+ and the isogenic ihfΔ host cells. We also transformed equal amounts of pUC19 DNA into both types of host cells to determine the plating efficiency, and the ratio of transformants in IHF+/ihfΔ was used to normalize the data. The results shown in Fig. 5C suggest that the wt α-γ replicon was able to propagate in the ihfΔ host but not the mutant form in which the original α iteron was replaced by a γ iteron. In fact, even in the IHF+ cells, there was some reduction in the propagation of the plasmid (Fig. 5C). We concluded from the results that whereas low affinity interaction between an α and a γ iteron (Fig. 3C) promoted plasmid propagation in the ihfΔ cells, enhanced iteron-iteron interaction promoted by a γ to α exchange significantly inhibited plasmid maintenance.
FIGURE 5.
Effect of a α to γ iteron swap on propagation of a α-γ plasmid in an ihfΔ host. A, sequences of α and γ iterons. The sequence differences between the two are shown as underlying dots. B, schematic representations of the wt α-γ replicon and one with the α iteron replaced with a γ iteron. C, propagation of a normal and the iteron-exchanged α-γ replicon in IHF+ and ihfΔ hosts. D, ChiP data showing that in exponentially growing cells π protein remains bound to the iterons of γ but not to the α iteron; the lanes labeled marker, input, no Ab, and π Ab imm. ppt show a DNA ladder of different base pairs, PCR products of the α- and γ-specific primer pairs using a part of the input DNA as the template. no Ab., PCR products from mock-precipitated DNA; π Ab. imm. ppt, PCR products from cross-linked DNA immunoprecipitated with anti π Ab, respectively. The subcolumns corresponding to the input, mock immunoprecipitation, and π Ab immunoprecipitation, labeled as α, γ, and αγ, refer to primer pairs used to amplify either each iteron region separately or together.
ChIP Analyses Showed Occupation of γ Iterons by π but Not at the α Iteron
Is low affinity interaction between α-γ iterons observed in vitro revealed by the DNA circularization experiments transitory in vivo? To address this question, we investigated the frequency of occupation of the α and γ iterons by π in vivo during vigorous, exponential cell growth. We performed chromatin immunoprecipitations to detect the presence of π on the iterons using monospecific polyclonal anti-π antibodies (Ab). We cross-linked protein to protein and protein to DNA by incubating the cells sequentially with DMA and HCHO and prepared cross-linked chromatin samples, immunoprecipitated the material with π-specific Ab, reversed the cross-link, recovered the DNA, and amplified it by PCR using α-specific and separately γ-specific primer pairs flanking the respective iteron sequences (Table 1). The reactions were carried out with the input DNA (without cross-linking) as a control (Fig. 5D, input lanes). Additional controls were done with mock-precipitated samples (Fig. 5D, no Ab). PCR amplification using separately the α- and γ-specific primer pair or both pairs together showed consistently the occupation of only the γ iterons not the α iteron by π (Fig. 5D, lanes under the subheading π Ab imm. ppt). Increasing the number of PCR amplification cycles by a factor of 5–10 did not produce any evidence of α-specific signal (not shown). Data are consistent with the interpretation that the γ iterons, but not the α iteron, were occupied in vivo by π protein or that the interaction of π with α was so transient and weak that it escaped capture by ChIP. We favor the second interpretation because of two pieces of in vitro evidence: (i) DNase footprinting data showed unequivocal α-γ interaction (9, 14) and (ii) enhancement of ligation-catalyzed DNA circularization data shown in Fig. 3C also supported the same conclusion.
DISCUSSION
It is known that a regulatory protein such as a prokaryotic transcriptional repressor (e.g. gal repressor, λ repressor) can also act as a transcriptional activator depending on the relative location of the operator with respect to the RNA polymerase binding site (38, 39). If the regulatory protein binding site overlaps the promoter and blocks its access to RNA polymerase, it is likely to act as a repressor of transcription. If on the other hand, the repressor binding site is located in such a way that its binding to the cognate site allows it to touch the RNA polymerase bound to the promoter and stabilizes the enzyme binding, then it can act as a transcriptional activator (40, 41). Therefore, it is the sequence context rather than the repressor-operator binding affinity that determines whether such a protein would act as a repressor or an activator of transcription. In contrast, as shown in this work, a replication initiator protein can also serve either as an activator or a repressor of replication (e.g. at ori α) not because of the sequence context of its binding site(s) at the ori but apparently by the relative affinity of the initiator for the iteron (9) and the avidity of iteron pairing.
Precisely, how does iteron-iteron pairing either activate or repress origin activity? From our work with another plasmid system, namely F (30), it appears that iteron pairing leads to abolition of unwinding of the AT-rich region of the ori thereby blocking replication initiation. To uncover such information in R6K, the α-γ replicon needs to be studied using a reconstituted in vitro replication system (21, 33). It would be useful to determine, by using a partial reaction catalyzed by purified wt π, DnaJ-DnaK-GrpE chaperone system, IHF, DnaA, and ATP, precisely where in the vicinity of the genetically defined oriα, DNA melting occurs, and whether a stronger iteron-iteron interaction, such as that caused by the α-γ iteron swap, would lead to abolition of the ori melting at α. It should also be instructive to determine whether there is an inverse relationship between ori melting at α versus that at γ in a α-γ replicon. These experiments might explain in the future (i) a molecular basis of ori activation versus repression and (ii) also might illuminate the mechanism(s) that ensures that only one origin fires at any given time in a multi-origin prokaryotic replicon (11). What is the stoichiometry of a productive looped complex at the ori? Although we have postulated that it could be a monomer bound to each one of the paired iterons that are bridged by a single dimer, this model needs to be tested by further work.
Although, the present work shows that not only π cop monomers but also wt dimers are both needed their DNA binding activities to form an activating looping complex; in another plasmid system, called RK2, it was reported that runaway replication caused by monomerization of the initiator protein called TrfA could be damped down by intracellular co-expression of mutant forms of the dimeric initiator that itself could not bind to DNA (42). R6K dimeric π is somewhat different from other plasmid-encoded initiator dimers in that π dimers bind to iteron DNA albeit with much lower affinity in comparison with the monomeric form. In other plasmid systems, dimeric initiators mostly do not bind to DNA (43). It appears that π dimers need to contact DNA while bridging a pair of iteron-bound monomers to form a stable looped structure at or near the replication origins of R6K.
Are there separate domains in π for dimerization on one hand and for dimer-monomer or dimer-dimer interactions on the other? Previous work shows that mutations in the unstructured area of π (18, 32) (Fig. 2A) such as amino acid residues 106–108 cause high copy phenotype and here we have shown that the mutant forms of the protein but not wt π are induced to monomerize by iteron binding. The residues 106–108 mostly likely affect the dimerization domain of π. We have previously reported that a specific selection screen for the isolation of nonlooping mutants of π yielded the mutation P42L that prevented DNA looping (9). Clearly, the latter site is well separated, in a linear dimension from the residues 106–108. However, the crystal structure reveals that the residues are not far apart in the folded three-dimensional structure of the protein (18). It is possible that the Pro-42 residue is an indicator of a domain for dimer-monomer interaction that is separate from the dimerization domain. The repressor of phage λ appears to contain domains for both dimer-dimer interaction and dimerization that seem to be located very close to each other (44).
Do host-encoded proteins modulate π-mediated looping interaction? We have previously reported that π interacts with a number of replisomal proteins such as DnaA, DnaB, DnaG, etc. (25, 26). A short range looping interaction that promoted plasmid-encoded initiator and DnaA interaction was promoted by host-encoded DNA-bending protein IHF. Furthermore, interaction between the plasmid-encoded initiator RepA with host-encoded DnaA enhances short range DNA looping at the plasmid ori (45). Therefore it is conceivable that interaction of π with host replisomal protein(s) could modulate π-mediated DNA looping.
In conclusion, the availability of a crystal structure, a reconstituted in vitro replication system, and abundant biological information (including numerous mutants) recommend R6K as an outstanding model system to further investigate the roles of long range DNA-protein interactions in replication control. Further molecular analysis of this mechanism might provide useful pointers to the study of parallel mechanisms in eukaryotes. The advent of the techniques such as chromosome conformation capture (3C) (46) and circular chromosome conformation capture analyses (4C) (47) have uncovered long range intra and interchromosomal interactions in eukaryotes that appear to play important roles in effecting and controlling transcription, immunoglobulin switching, and other important DNA transactions (27). Perhaps it is just a matter of time before similar intrachromosomal and interchromosomal interactions controlling DNA replication might be revealed by further work.
Acknowledgments
We thank Dr. Chris Davies for help with Fig. 2 and Dr. D. K. Chattoraj of the NCI, National Institutes of Health for many useful discussions.
This work was supported, in whole or in part, by Grant GM049264 from the National Institutes of Health (to D. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
D. Bastia, unpublished data.
- IHF
- integration host factor
- ORF
- open-reading frame
- wt
- wild type
- ELISA
- enzyme-linked immunosorbent assay
- ChIP
- chromatin immunoprecipitation assay
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Chattoraj D. K., Snyder K. M., Abeles A. L. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 2588–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helinski D. R., Toukdarian A. E., Novick R. P. (1996) in Escherichia coli and Salmonella, 2nd Ed. (Neidhardt F. C., Curtiss R., III, Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Schaechter M., Umbarger H. E. eds), American Society of Microbiology, Washington, D. C. [Google Scholar]
- 3.Bastia D., Mohanty B. K. (1996) in DNA Replication in Eukaryotic Cells (DePamphilis M. ed), Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 4.Tomizawa J. (1990) J. Mol. Biol. 212, 683–694 [DOI] [PubMed] [Google Scholar]
- 5.Pal S. K., Chattoraj D. K. (1988) J. Bacteriol. 170, 3554–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McEachern M. J., Bott M. A., Tooker P. A., Helinski D. R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 7942–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filutowicz M., McEachern M. J., Mukhopadhyay P., Greener A., Yang S. L., Helinski D. R. (1987) J. Cell Sci. Suppl.7, 15–31 [DOI] [PubMed] [Google Scholar]
- 8.Khan S. A., Murray R. W., Koepsel R. R. (1988) Biochim. Biophys. Acta 951, 375–381 [DOI] [PubMed] [Google Scholar]
- 9.Miron A., Mukherjee S., Bastia D. (1992) EMBO J. 11, 1205–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inuzuka N., Inuzuka M., Helinski D. R. (1980) J. Biol. Chem. 255, 11071–11074 [PubMed] [Google Scholar]
- 11.Crosa J. H., Luttropp L. K., Falkow S. (1978) J. Mol. Biol. 124, 443–468 [DOI] [PubMed] [Google Scholar]
- 12.Lovett M., Sparks R. B., Helinski D. R. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 2905–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee S., Erickson H., Bastia D. (1988) Cell 52, 375–383 [DOI] [PubMed] [Google Scholar]
- 14.Miron A., Patel I., Bastia D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6438–6442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosa J. H., Luttropp L. K., Falkow S. (1976) J. Bacteriol. 126, 454–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee S., Erickson H., Bastia D. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 6287–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abhyankar M. M., Zzaman S., Bastia D. (2003) J. Biol. Chem. 278, 45476–45484 [DOI] [PubMed] [Google Scholar]
- 18.Swan M. K., Bastia D., Davies C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18481–18486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Germino J., Bastia D. (1983) Cell 34, 125–134 [DOI] [PubMed] [Google Scholar]
- 20.Kunnimalaiyaan S., Rakowski S. A., Filutowicz M. (2007) J. Bacteriol. 189, 4953–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zzaman S., Reddy J. M., Bastia D. (2004) J. Biol. Chem. 279, 50886–50894 [DOI] [PubMed] [Google Scholar]
- 22.Filutowicz M., Appelt K. (1988) Nucleic Acids Res. 16, 3829–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley W. L., Bastia D. (1991) J. Biol. Chem. 266, 15924–15937 [PubMed] [Google Scholar]
- 24.Kelley W. L., Patel I., Bastia D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5078–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratnakar P. V., Mohanty B. K., Lobert M., Bastia D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5522–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y. B., Datta H. J., Bastia D. (1998) EMBO J. 17, 5192–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jhunjhunwala S., van Zelm M. C., Peak M. M., Cutchin S., Riblet R., van Dongen J. J., Grosveld F. G., Knoch T. A., Murre C. (2008) Cell 133, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley W. L., Bastia D. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 2574–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filutowicz M., Davis G., Greener A., Helinski D. R. (1985) Nucleic Acids Res. 13, 103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zzaman S., Bastia D. (2005) Mol. Cell 20, 833–843 [DOI] [PubMed] [Google Scholar]
- 31.Kurdistani S. K., Grunstein M. (2003) Methods 31, 90–95 [DOI] [PubMed] [Google Scholar]
- 32.Krüger R., Konieczny I., Filutowicz M. (2001) J. Mol. Biol. 306, 945–955 [DOI] [PubMed] [Google Scholar]
- 33.Abhyankar M. M., Reddy J. M., Sharma R., Büllesbach E., Bastia D. (2004) J. Biol. Chem. 279, 6711–6719 [DOI] [PubMed] [Google Scholar]
- 34.Kunnimalaiyaan S., Krüger R., Ross W., Rakowski S. A., Filutowicz M. (2004) J. Biol. Chem. 279, 41058–41066 [DOI] [PubMed] [Google Scholar]
- 35.Mukherjee S., Patel I., Bastia D. (1985) Cell 43, 189–197 [DOI] [PubMed] [Google Scholar]
- 36.Blasina A., Kittel B. L., Toukdarian A. E., Helinski D. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3559–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ables A. L., Austin S. J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9011–9015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ptashne M. (2004) Genetic Switch, 3rd Ed., Cold Spring Harbor Press, NY [Google Scholar]
- 39.Choy H. E., Hanger R. R., Aki T., Mahoney M., Murakami K., Ishihama A., Adhya S. (1997) J. Mol. Biol. 272, 293–300 [DOI] [PubMed] [Google Scholar]
- 40.Ptashne M., Gann A. A. (1990) Nature 346, 329–331 [DOI] [PubMed] [Google Scholar]
- 41.Bushman F. D., Ptashne M. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 9353–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toukdarian A. E., Helinski D. R. (1998) Gene 223, 205–211 [DOI] [PubMed] [Google Scholar]
- 43.Zzaman S., Abhyankar M. M., Bastia D. (2004) J. Biol. Chem. 279, 17404–17410 [DOI] [PubMed] [Google Scholar]
- 44.Koudelka G. B. (2000) Curr. Biol. 10, R704–R707 [DOI] [PubMed] [Google Scholar]
- 45.Stenzel T. T., MacAllister T., Bastia D. (1991) Genes Dev. 5, 1453–1463 [DOI] [PubMed] [Google Scholar]
- 46.Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 47.Ohlsson R., Göndör A. (2007) Curr. Opin. Cell Biol. 19, 321–325 [DOI] [PubMed] [Google Scholar]