Abstract
Fine-tuning of the Wnt/TCF pathway is crucial for multiple embryological processes, including liver development. Here we describe how the interaction between Hhex (hematopoietically expressed homeobox) and SOX13 (SRY-related high mobility group box transcription factor 13), modulates Wnt/TCF pathway activity. Hhex is a homeodomain factor expressed in multiple endoderm-derived tissues, like the liver, where it is essential for proper development. The pleiotropic expression of Hhex during embryonic development and its dual role as a transcriptional repressor and activator suggest the presence of different tissue-specific partners capable of modulating its activity and function. While searching for developmentally regulated Hhex partners, we set up a yeast two-hybrid screening using an E9.5–10.5 mouse embryo library and the N-terminal domain of Hhex as bait. Among the putative protein interactors, we selected SOX13 for further characterization. We found that SOX13 interacts directly with full-length Hhex, and we delineated the interaction domains within the two proteins. SOX13 is known to repress Wnt/TCF signaling by interacting with TCF1. We show that Hhex is able to block the SOX13-dependent repression of Wnt/TCF activity by displacing SOX13 from the SOX13·TCF1 complex. Moreover, Hhex de-repressed the Wnt/TCF pathway in the ventral foregut endoderm of cultured mouse embryos electroporated with a SOX13-expressing plasmid. We conclude that the interaction between Hhex and SOX13 may contribute to control Wnt/TCF signaling in the early embryo.
Keywords: Development Differentiation, Gene/Regulation, Protein/Protein-Protein Interactions, Signal Transduction, Transcription/Coactivators, Transcription/Homeobox, Transcription/TCF, Transcription/Tissue-specific Factors
Introduction
Wnt signaling is a conserved signaling pathway that plays crucial roles in animal life by controlling the genetic programs of embryonic development and adult homeostasis. Recent reports have particularly highlighted the essential role of Wnt during liver and pancreas development. Wnt repression in the anterior endoderm is required for liver and pancreas specification, whereas active Wnt signaling in the posterior endoderm suppresses these fates (1). However, immediately after the induction of the hepatic program in the endoderm, Wnt signaling is apparently required for the further outgrowth of the endoderm into a liver bud. In fact, the expression of Wnt2b in the lateral plate mesoderm appears essential for liver specification in the endoderm and bud induction (2). In short, Wnt activity is extremely dynamic in the course of liver specification and morphogenesis, suggesting that a tight and fast control of Wnt signaling is essential for the proper development of the organ.
Hhex is a homeobox transcription factor of the Antennapedia/Ftz class. Hhex is expressed in the anterior definitive endoderm (3) that gives rise to the liver and pancreas. After gastrulation, Hhex is also expressed in mesoderm- and endoderm-derived tissues such as hematopoietic and vascular progenitors, endocardium of the heart, liver, thyroid, lung, thymus, gallbladder, and pancreas (4–7). Hhex plays a role in cell proliferation and morphogenesis during organogenesis (5, 8, 9). Loss-of-function of Hhex in mice results in embryonic lethality at E10.5 and shows different degrees of defects in organs derived from the three embryonic germ layers (4, 7, 10, 11). The defects observed in endoderm-derived organs, such as thyroid, liver, and pancreas, are associated with alterations in cell proliferation and cell migration in embryonic progenitors (5, 7, 12, 13). Recently, Hhex locus has been robustly associated in genome-wide association studies with type 2 diabetes (14). Human and mouse Hhex proteins share 95% homology throughout the full sequence and the N-terminal domain.
Hhex is a transcriptional repressor in Xenopus embryos and cultured cells (15–17). The N-terminal proline-rich domain (aa2 1–137) is highly conserved among species and exerts a repressing activity (18). Goosecoid, vascular endothelial growth factor, and endothelial cell-specific molecule 1 are among the best characterized targets transcriptionally repressed by Hhex. But Hhex can also function as a transcriptional activator of the Na+/taurocholate cotransporting polypeptide and sodium iodide symporter promoters (19, 20). The acidic C-terminal domain (aa 197–271) is responsible for the transactivating properties of Hhex (21). Finally, the homeodomain (aa 138–196) is responsible for sequence-specific binding to DNA.
Given the wide expression of Hhex and the complexity of its functions, we speculated that an elevated number of interactors in different tissues contributes to its differential specificity and activity. Therefore, we searched an E9.5-E10.5 mouse embryo library for developmentally regulated proteins that interact with Hhex. Using a yeast two-hybrid screening approach we identified SOX13 as a Hhex interactor. Two SOX13 isoforms of 768 bp (255 aa) and 1815 bp (604 aa) were isolated from human cDNA, but only the longer isoform interacted with Hhex. We mapped the interaction domains in both proteins and showed that Hhex and SOX13 are colocalized in the nucleus. Finally, we addressed the function of the Hhex/SOX13 interaction and show in cultured cells and mouse embryos that it de-represses Wnt activity by disrupting the SOX13·TCF1 complex.
EXPERIMENTAL PROCEDURES
Plasmid Construction
To construct the bait for the yeast two-hybrid screening, different Hhex fragments were amplified by PCR using appropriate primers (supplemental Table S1). After digestion with EcoRI and SalI, fragments were cloned in-frame into the pBD-Gal4-Cam vector (Stratagene), yielding five bait plasmids containing the Hhex N-terminal domain or Hhex-(1–137), HhexΔC or Hhex-(1–196), HhexΔN or Hhex-(138–271), Hhex C-terminal domain or Hhex-(197–271), and full-length Hhex (aa 1–271) fused to the Gal4 DNA binding domain. GST-Hhex fusion constructs were constructed by subcloning the Hhex fragments from pBD-Gal4 Cam vectors into the EcoRI-SalI site in pGex-6P-1 (GE Healthcare). EGFP-Hhex fusion constructs were constructed by subcloning the Hhex BamHI-SalI fragments from the pGex-6P-1 vector series into the BglII-SalI site in pEGFP-C1 (Clontech). The expression plasmids containing HA-tagged Hhex, Hhex-(1–137), and Hhex-(1–196) were obtained by subcloning the EcoRI-SalI fragments from pBD-Gal4-Cam vector into the EcoRI-XhoI site in pIRES-hrGFP-2a. Hhex-(138–271) and Hhex-(197–271) were amplified by PCR to introduce an initiation codon at the 5′ end of the coding sequence and were cloned as above.
The expression plasmids containing FLAG and the Myc-tagged SOX13 isoforms (604 and 255 aa) were obtained by PCR-cloning from a human hepatic cDNA library (Ambion) using primers SOX13clon1–3 (supplemental Table S1). To obtain ΔSOX13, the 1–100-aa fragment was PCR-cloned so that an artificial EcoRI restriction site at the 5′ end and a HindIII restriction site at the 3′ end flanked the product. The 210–604-aa fragment was also PCR-cloned but with an artificial HindIII restriction site at the 5′ end and a SalI restriction site at the 3′ end. The resulting EcoRI-SalI-flanked PCR products were ligated to the EcoRI-SalI fragment of pFLAG-CMV2 (Sigma) and also to the EcoRI-XhoI fragment of the pcDNA3-myc tag to obtain pFLAG-SOX13-255, pFLAG-SOX13-604, pFLAG-ΔSOX13, pcDNA3-myc-SOX13-255, pcDNA3-myc-SOX13-604, and pcDNA3-myc-ΔSOX13. All the constructed plasmids were verified by sequencing.
Cell Lines and Transfection
HeLa cells were maintained as monolayer cultures and grown in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum, 50 units/ml penicillin, and 50 mg/ml streptomycin, whereas 293T cells were also supplemented with 3.5 g/liter of glucose. Plasmid DNAs were purified using Qiagen Maxiprep kit columns (Qiagen). On the day before transfection, 293T or HeLa cells were plated in 24-well plates (reporter assay) or 10-cm plates (protein expression). Plasmids (0.7 μg for the reporter or 10 μg for the protein expression) were transfected using FuGENE 6 following the manufacturer's instructions (Roche Applied Science). To correct the variations in transfection efficiency in the reporter assay, 15 ng of pRL-SV40 (Promega) were cotransfected in parallel. Luciferase activities were assayed at 48 h using Dual Luciferase reporter (Promega).
Adenoviral shRNA Construction
An adenoviral vector expressing shRNA against Hhex was obtained by homologous recombination of pJM17 and pAC-shHhex as described (22). pAC-shHhex was obtained by inserting synthesized Hhex shRNA (targeting sequence in HHex, TGGACAGTTCCTGTGATCAGAG) into the BamHI-EcoRI site in pAC-shRNA plasmid. pAC-shRNA plasmid was obtained by replacing the CMV cassette in pAC-CMV with the CMV expression cassette from pPRIME-CMV-GFP (23). An adenovirus expressing shRNA against luciferase was constructed similarly and used as an infection control.
Immunofluorescence
HeLa cells growing on coverslips were cotransfected with plasmids expressing EGFP-Hhex and SOX13-myc. After 48 h, cells were fixed for 5 min in 4% paraformaldehyde and permeabilized with PBS containing 0.2% Triton X-100 for a further 5 min. Blocking was carried out with 1% bovine serum albumin in PBS solution for 30 min. Primary mouse anti-Myc antibody (9E10 hybridoma, Santa Cruz Biotechnologies) was incubated using a 1/30 dilution overnight at 4 °C. The secondary anti-mouse Alexa555-conjugated antibody (1/500 dilution, Invitrogen) was incubated for 1 h in PBS containing 1% bovine serum albumin. Cells were washed with PBS, and coverslips were mounted with Mowiol®. Fluorescence was viewed through a Leica TCS-SP2 confocal microscope.
Yeast Two-hybrid Screening
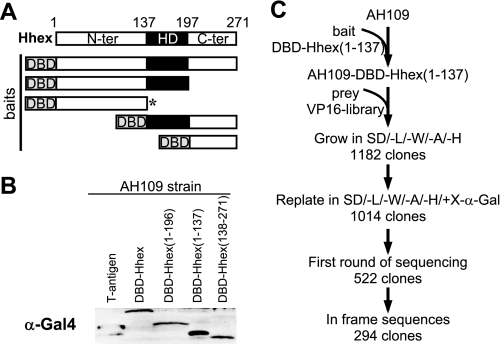
We used a 9.5–10.5-days post-coitum mouse expression library, kindly provided by Dr. Weintraub (Howard Hughes Medical Institute) and Dr. Scambler (Institute of Child Health, UK). In this library, cDNAs are fused to a sequence encoding the activation domain of the herpes simplex virus protein VP16 (24). Hhex-derived bait plasmids (Fig. 1A) were transformed into the AH109 strain (Ade−, His−, Leu−, Trp−; Clontech) following the manufacturer's instructions. AH109 strain expresses α-galactosidase (MEL1) and β-galactosidase (LacZ) upon Gal4 activation. Gal4-Hhex fusion protein expression in yeast was detected by immunoblotting (Fig. 1B). pBD-Gal4-Hhex-(1–137) was selected as bait given the extremely low autoactivating capabilities (probably linked to its function as a repressor) and the biochemical relevance of the N-terminal domain (see the Introduction). For the yeast two-hybrid library, 145 μg of the library (∼106 transformants) were transformed into the AH109 strain expressing Gal4-Hhex-(1–137), plated onto a SD/−Leu/−Trp/−Ala/−His 9-cm plate and grown at 30 °C for 7 days. Plates were checked for growing colonies on a daily basis. Single-growing colonies were transferred to SD/−Leu/−Trp/−Ala/−His/+α-X-gal to reconfirm the interaction. Confirmed positive clones (blue colonies) were stocked in glycerol media at −80 °C. In parallel, all the clones appearing on the first 4 days of selection (a total of 522) were amplified by PCR (colony touching) using the primers flanking the insert: VP16 forward (5′-TCGAGTTTGAGCAGATGTTTACCG-3′), VP16 reverse (5′-TTGTAAAACGCGGCCAGT-3′), and the Biomek FX robot. Translated sequences were blasted against the protein data base. Selected preys were then isolated from yeast using the Wizard® Plus SV Minipreps DNA purification system (Promega). Isolated plasmids were transformed into Escherichia coli, amplified, and retransformed into AH109 yeast expressing the bait plasmids and empty pBD-Gal4 Cam to confirm the interaction. α-Galactosidase-activity was measured using the Beta-Glo Assay System (Promega).
FIGURE 1.
Hhex yeast two-hybrid screening. A, a diagram is shown of Hhex fusion proteins containing the Gal4 DNA binding domain (DBD) in the N terminus. These proteins were tested as putative bait in a yeast two-hybrid assay. An asterisk indicates the protein used as the final bait in the screening. The Hhex protein showing functional domains is also shown above as a reference. N-ter, N-terminal domain; HD, homeodomain; C-ter, C-terminal domain. B, shown is immunoblotting using an anti-Gal4 antibody of extracts from the AH109 yeast transformed with the bait. The yeast-expressing plasmid, pGBKT7-T, expressing the SV40 large antigen T protein was used as a positive control. C, an overview is shown of the Hhex yeast two-hybrid screening by sequential transformation of bait and prey (VP16 library). VP16 library, E9.5-E10.5 mouse embryo library made in the pVP16; SD/−L/−W/−A/−H, synthetic dropout medium lacking leucine, tryptophan, adenine, and histidine; SD/−L/−W/−A/−H/+X-α-gal, synthetic dropout medium lacking leucine, tryptophan, adenine, and histidine, and containing 20 μg/ml of X-α-gal.
In Vitro Glutathione S-Transferase Fusion Protein Pulldown Assays
GST and GST fusion proteins were isolated from the E. coli as described (25). Volumes (15–50 μl) corresponding to approximately even amounts of individual GST fusion proteins were coupled to washed glutathione-Sepharose beads (40 μl suspension) and washed twice with PBS and once with immunoprecipitation buffer (20 mm Tris-HCl, pH 8, 200 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40). In the pulldown experiments using the in vitro translated protein, coupled beads were incubated with 8 μl of [35S]methionine-labeled in vitro translated protein (Promega TNT® T7 Quick Coupled Transcription/Translation system) in immunoprecipitation buffer containing the Halt Protease Inhibitors Mixture (Pierce). Alternatively, when extracts from the cell cultures overexpressing the protein of interest were being pulled down, 500 μg of total protein lysate in M-Per buffer (Pierce) were used instead. In both cases beads were incubated overnight at 4 °C with shaking, washed 6 times with immunoprecipitation buffer, resuspended in 40 μl of electrophoresis sample buffer, and heated (95 °C) for 5 min. After SDS-polyacrylamide gel electrophoresis, the gel was checked by immunoblotting or audioradiography.
Immunoprecipitation and Immunoblotting
Cell protein extracts were obtained by harvesting transfected cells (HeLa, 293T, or HepG2) with M-Per buffer containing the Halt Protease Inhibitors Mixture. Approximately, 0.5–1.0 mg of total protein was incubated with anti-FLAG or anti-HA antibodies bound to protein G-Sepharose beads overnight at 4 °C (Roche Applied Science). After five washes in lysis buffer, immunoprecipitates were resolved by SDS-PAGE and transferred to Immobilon filters (Millipore). Filters were then immunoblotted using a rabbit polyclonal anti-Hhex (26), horseradish peroxidase-conjugated anti-FLAG, or anti-HA antibodies. For the competitive immunoprecipitation assay, 293T cell extracts expressing SOX13-FLAG and TCF1 Myc-tagged proteins were incubated with increasing amounts of GST-Hhex or GST alone (40 and 160 μg). Samples were immunoprecipitated as above using anti-FLAG antibodies bound to protein G-Sepharose. Immunoprecipitates were resolved by SDS-PAGE and transferred to Immobilon filters (Millipore). Filters were immunoblotted using mouse anti-FLAG or anti-Myc antibodies and incubated with a True Blot horseradish peroxidase-conjugated anti-mouse IgG (eBioscience). Immunoreactive proteins were visualized using the ECL detection reagent system (Amersham Biosciences).
Quantitative RT-PCR
DNA-free total RNA from human tissues was obtained from Ambion. Quantification of specific mRNAs was done as described (27) using porphobilinogen deaminase and TATA box-binding proteins as internal controls. We designed specific primer sets for the three putative variants of SOX13 (Table 1). PCR amplicons were specific in terms of size (agarose gel electrophoresis), melting curve analysis, and sequencing. After denaturing for 30 s at 95 °C, amplification was performed in 40 cycles of 1 s at 94 °C, 5 s at 62 °C, and 20 s at 72 °C. The real-time monitoring of the PCR reaction and the precise quantification of the products in the exponential phase of the amplification were performed with the LightCycler quantification software according to the manufacturer's recommendations. Each reaction was done in triplicate.
TABLE 1.
Primers used in this study
FP, forward primer; RP, reverse primer.
| Number | Primer | Target | Sequence |
|---|---|---|---|
| Reverse-transcription quantitative PCR | |||
| 1 | SOX13-FP | SOX13 and S-SOX13 | tgattcagcagcagcataagat |
| 2 | SOX13-RP1 | S-SOX13 | taatggatgagagtggctccag |
| 3 | SOX13-RP2 | SOX13 | cagcggatactccactggtt |
| 4 | L-SOX13-FP | L-SOX13 | cgaagagaagagcgatggga |
| 5 | L-SOX13-RP | L-SOX13 | gtatctgccaagtgcacagc |
| 6 | SOX13-intron-FP | Intron 1 in SOX13 locus | gatcccagcaacaatcctgt |
| 7 | SOX13-intron-RP | Intron 1 in SOX13 locus | cctgcccaagctttgtttag |
| 8 | SOX13-conflict-FP | SOX13 and S-SOX13. Conflict. | aacatgcctgtgatcgtcaa |
| 9 | SOX13-conflict-RP | SOX13 and S-SOX13. Conflict. | cgagttgggacaaggtcttc |
| 10 | DKK1-FPa | DKK1 | tggaatatgtgtgtcttctgatcaaa |
| 11 | DKK1-RPa | DKK1 | aagacaaggtggttcttctggaat |
| Chromatin immunoprecipitation quantitative PCR | |||
| 12 | DKK1-ChIP-FPa | attcaacccttactgccaggc | |
| 13 | DKK1-ChIP-RPa | aaggctaccagcgagcgttat | |
| 14 | SP5-ChIP-FPb | tccagaccaacaaacacacc | |
| 15 | SP5-ChIP-RPb | gcttcaggatcacctccaag | |
| 16 | RPLP0-ChIP-FP | acccagctctggagaagtca | |
| 17 | RPLP0-ChIP-RP | gaggtcctccttggtgaaca | |
Wnt Stimulation and Chromatin Immunoprecipitation
Wnt signaling was activated in 293T cells by treatment with LiCl (28). Cells were grown in 10-cm plates. One-fifth of the cells were used to obtain RNA and protein extract, whereas the rest was used for chromatin immunoprecipitation. Chromatin fixation and immunoprecipitation were performed as previously described (29) with the following modifications. After sonication, the samples were centrifuged at full speed for 10 min at 4 °C. The resultant chromatin was diluted in chromatin immunoprecipitation dilution buffer (1% SDS, 10% Triton X-100, 1.2 mm EDTA, 50 mm Tris-HCl, pH 8, 200 mm NaCl, and protease inhibitors) and precleared for 1 h at 4 °C with salmon sperm DNA/protein G-agarose slurry (Millipore, 16-201). Then 50 μl of diluted chromatin was incubated with 4 μg of mouse monoclonal anti-Myc tag antibody (Upstate Technology, clone 4A6) or monoclonal mouse anti-HisG antibody (negative control; Invitrogen) overnight at 4 °C in a rocking platform. Immunoprecipitated chromatin was eluted from beads by the addition of 250 μl of 0.1 m sodium bicarbonate, 1% SDS and incubation for 15 min by shaking at 42 °C. DNA was recovered using Qiaquick PCR purification kit (Qiagen). Precipitated DNA was analyzed using the LightCycler 480 real-time PCR system (30).
In Vivo Electroporation of Mouse Embryos
The ventral definitive endoderm was electroporated as described in Pierreux et al. (31, 32). After electroporation, embryos were cultured for 24 h at 37 °C in a roller culture system (33). Somites were counted, and only those embryos showing blood circulation and the turning of embryos were selected. The ventral midguts (liver bud region) were dissected, and luciferase activity was measured with the Dual Luciferase Reporter (Promega).
RESULTS
SOX13 Interacts with Hhex
To search for Hhex interactors during embryonic development, we performed a yeast two-hybrid screening using an E9.5-E10.5 mouse embryo library (see “Experimental Procedures”) as prey and Hhex-(1–137) as bait. After the sequential transformation of the AH109 yeast strain (Fig. 1C), we retrieved 1182 clones that grew in stringent media SD/−Leu/−Trp/−Ala/−His. Of these, only 1014 grew and were α-galactosidase-positive when re-plated in SD/−Leu/−Trp/−Ala/−His/+α- X-gal. A first round of sequencing (522 isolated clones) provided 294 in-frame coding sequences with repetitions ranging from 1 to 56 times. TLE-related proteins (AES-1) and members of the Eif4 family (Eif4g1 and Eif4g3) were present in the list, which is in agreement with previous reports (26, 34). Among the different novel sequences, SOX13 was selected based on the high number of clones detected during the screen, expression pattern, and embryological function of Sox family members which activate or repress target genes through interaction with different partner proteins (35). Eight clones of SOX13 were isolated (Fig. 2A), all of which contained the region between aa 126 and 218 comprising the leucine zipper (LZ) and the glutamine-rich (Q-rich) domain. To confirm the interaction and to exclude the auto-activation of the isolated clones, a representative prey plasmid (clone 11.23) was purified from the yeast, amplified in bacteria, and re-transformed in the AH109 yeast strain expressing each pre-selected Hhex bait (see Fig. 1A). Only those transformants expressing bait that included the N-terminal domain (aa 1–137) were able to grow and stain positive in SD/−Leu/−Trp/−Ala/−His α-X-gal media (Fig. 2B). Finally, we quantified the interaction by measuring β-galactosidase activity by luminescence. Once again, only the bait containing the N-terminal domain of Hhex gave β-galactosidase activity above the background level (Fig. 2C). In conclusion, SOX13 (aa 126–218) interacts with Hhex in yeast.
FIGURE 2.
Interaction between Hhex and SOX13 in yeast. A, shown is multiple alignment between mouse Sox13 and the prey clones isolated in the yeast two-hybrid screening. Relevant functional domains in Sox13, the leucine zipper (LZ), and Q-rich are emphasized. Asterisks indicate the position of leucines within the LZ domain. Consensus sequences are in red. B, a representative Sox13 prey plasmid (clone 11.23) was re-transformed into AH109 yeast expressing Hhex bait or empty plasmid as a negative control. The transformed yeast were plated in SD/−Leu/−Trp and SD/−Leu/−Trp/−Ala/−His/+X-α-gal. C, β-galactosidase activity was also measured in yeast cotransformed with each Hhex bait and Sox13 prey plasmid (clone 11.23).
A 1815-bp Open Reading Frame of SOX13 (604 aa) Is Prevalent in Human Tissues
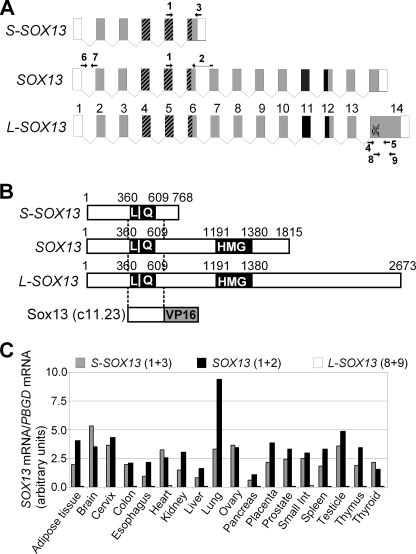
SOX13 is a member of the group D of the SOX transcription factors that contains the characteristic high mobility group (HMG) domain together with a LZ and Q-rich domain. The SOX13 genomic locus contains 14 exons, and three different isoforms have been reported in the literature (Fig. 3A). SOX13 (36) is an open reading frame of 1815 bp encoding a 604-aa protein containing the HMG domain and the LZ and Q-rich domain in the N terminus (Fig. 3B). Long (L)-SOX13 (37) is an open reading frame of 2673 bp encoding an 890-aa protein as a result of a missing G nucleotide at base pair 1950 in the sequence reported by Roose et al. (GenBankTM accession number AF083105). This putative deletion leads to a shift of exon 14 open reading frame, resulting in a longer protein. Finally, short (S)-SOX13 (ECRBrowser (38)) is an open reading frame of 768 bp encoding a 255-aa protein caused by alternative splicing that results in a truncated isoform lacking the C terminus including the HMG domain. The three isoforms contain the putative SOX13 interaction domain detected in the yeast two-hybrid screening (LZ-Q-rich domain, aa 126–218; Fig. 3B). In preparation for mapping the interaction domain of SOX13 with Hhex, we decided to determine which SOX13 isoform prevails in human tissues. We designed three primer pairs capable of binding specifically to each coding sequence (Table 1). These primers were validated by PCR using plasmids containing the three different cDNAs. PCR fragments representative of each PCR reaction were purified and sequenced to confirm identity. The mRNA of the SOX13 isoforms was quantified by qRT-PCR in 18 different tissues from a mixture of at least 3 different human donors (Fig. 3C). Only SOX13 and S-SOX13 were significantly detected in all the tissues tested. SOX13 predominated in most of the tissues, although S-SOX13 mRNA was more abundant in the brain, heart, and thyroid. To confirm the absence of L-SOX13, we designed a pair of primers (numbers 4 and 5; Table 1) to amplify the conflicting region in exon 14. The sequencing of the amplified region revealed no deletion at nucleotide G1950. Based on these results, SOX13 and S-SOX13 coding sequences were amplified by PCR from human hepatic cDNA and cloned into the pFLAG-CMV2 vector. We conclude that isoforms SOX13 and S-SOX13 are prevalently expressed in human tissues and that both contain the LZ and Q-rich domain.
FIGURE 3.
SOX13 isoforms present in human tissues. A, shown is a graphic representation of SOX13 exons as described in ECRBrowser (38). Exons are depicted as boxes. Exon size is not proportional to box size. Gray boxes represent translated exons. The HMG box is represented in black (exons 11–12). Dashed boxes represent the LZ-Q domain (exons 4–6). The conflictive G1950 in exon 14 is also shown. The primers used for qRT-PCR analysis are depicted with a number (1–9). Primer sequences are shown in Table 1. B, alignment of the SOX13 coding sequences are compared with the prey clone 11.23 isolated from the yeast two-hybrid screening. This fragment contains the LZ-Q domain of Sox13. Numbers show base pair positions. C, relative SOX13 mRNA levels in 18 human tissues were determined by qRT-PCR. The numbers in parentheses represent the primers used for the qPCR reaction (see panel A). PBGD, porphobilinogen deaminase.
Mapping Hhex and SOX13 Interaction Domains
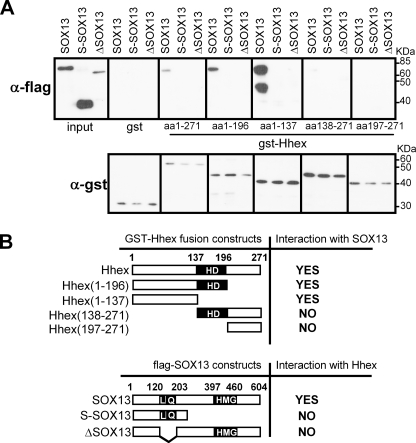
We investigated which Hhex domains are required for the interaction with SOX13 and vice versa by pulldown assays using GST-Hhex fusion constructs. We purified GST-Hhex deletion mutants from bacteria. We also obtained total protein extracts from the HeLa cells ectopically expressing FLAG-tagged full-length SOX13 (604 aa), S-SOX13 (255 aa), and ΔSOX13, a mutant SOX13 lacking the LZ and Q-rich domain (lacking aa 126–218, the interaction domain). As Fig. 4A illustrates and as Fig. 4B summarizes, GST-Hhex and all the fragments containing the N-terminal domain (aa 1–137) were able to co-precipitate full-length SOX13. When only the N-terminal domain of Hhex was expressed as a single fragment, it was capable of pulling down full-length SOX13 more efficiently. This result clearly establishes the N terminus of Hhex as the binding interface with SOX13 and confirms the result obtained in the yeast two-hybrid assay (Fig. 2). On the one hand, ΔSOX13, which specifically lacks the LZ-Q domain of SOX13, was unable to bind Hhex or any of its fragments. This result is also in agreement with the yeast two-hybrid assay. On the other hand, we found that the N terminus of SOX13 alone (the S-SOX13 isoform), which contains the LZ-Q domain, did not interact with Hhex or any of its deletion mutants. In short, the N terminus, or more specifically the LZ and Q-rich domain, is essential for SOX13 interaction with Hhex, but the domain alone is not sufficient for the interaction in mammalian cells. We conclude that the N terminus of Hhex is essential for SOX13 interaction, whereas the LZ and Q-rich domain of SOX13 is necessary, but not sufficient, for Hhex interaction.
FIGURE 4.
Hhex and SOX13 interaction domains. A, shown is an in vitro binding assay between purified GST, GST-Hhex-(1–271), GST-Hhex-(1–196), GST-Hhex-(1–137), GST-Hhex-(138–271), or GST- Hhex-(197–271) from an E. coli lysate and HeLa cell extract expressing FLAG-tagged SOX13, S-SOX13, or ΔSOX13. HeLa total protein extracts expressing SOX13-related proteins were incubated with GST-Hhex proteins. Glutathione-Sepharose was used to pull down the GST-Hhex fusion proteins, and anti-FLAG antibody was used in Western blots. One-twentieth of the extract was loaded as the input. The lower band in the SOX13 incubation was routinely observed after overnight incubation and is a C-terminal degradation product of SOX13. Lower panel, shown is an immunoblot using anti-GST antibodies in 1% of the beads. B, shown are diagrams summarizing the results obtained in the GST pulldown assays. Numbers show amino acid positions. HD, homeodomain; L, leucine zipper; Q, glutamine-rich; HMG, high mobility group domain.
SOX13 Directly Interacts with Hhex
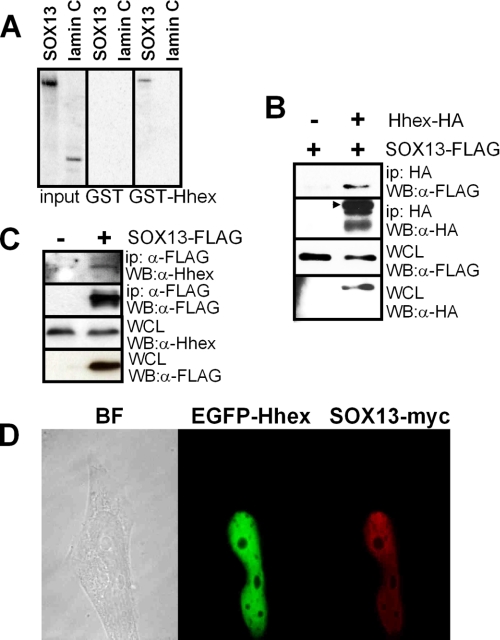
The results illustrated in Fig. 4 are compatible with the existence of a direct association between SOX13 and Hhex. These interactions were studied by assessing the binding of 35S-labeled in vitro translated SOX13 to GST-Hhex immobilized on glutathione-agarose beads. The GST protein alone did not interact with SOX13 (Fig. 5A). However, GST-Hhex was able to interact with full-length SOX13 and L-SOX5 (another member of the group D family of SOX; data not shown). Neither GST nor GST-Hhex pulled down the unrelated protein Lamin C. These results show that a direct interaction takes place between SOX13 and Hhex. To further corroborate the interaction between Hhex and SOX13, we performed immunoprecipitation experiments with HeLa cells that ectopically express both transcription factors. Full-length SOX13-FLAG co-immunoprecipitated with full-length Hhex-HA (Fig. 5B). No co-precipitation was detected in either mock-transfected HeLa cells or cells transfected with a plasmid expressing SOX9-FLAG (data not shown). Endogenous Hhex was also immunoprecipitated with SOX13 after transfection of HepG2 cells with a plasmid expressing SOX13-FLAG (Fig. 5C). Co-localization studies in transfected HeLa cells showed nuclear localization of the proteins and are in agreement with both co-immunoprecipitation and the pulldown analysis (Fig. 5D).
FIGURE 5.
Interaction nature and co-localization of Hhex and SOX13. A, 35S-SOX13 and 35S-lamin C (negative control) were synthesized in an in vitro transcription and translation system and incubated with glutathione-Sepharose beads coated with GST-Hhex or GST. Bound proteins were run in SDS/PAGE and visualized using phosphorimaging. B, immunoblot (WB) analysis of 293T cell lysates transfected with SOX13-FLAG and Hhex-HA detects SOX13-FLAG after immunoprecipitation (ip) with anti-HA antibody and immunoblotting against the FLAG tag. The whole cell lysate (WCL) was immunoblotted using anti-FLAG or anti-HA to show the expression levels of SOX13-FLAG and Hhex-HA in the assay. C, immunoblot analysis of HepG2 cell lysates transfected with SOX13-FLAG detects endogenous Hhex after immunoprecipitation with anti-FLAG antibody and immunoblotting against a primary antibody against Hhex. The whole cell lysate was immunoblotted using anti-FLAG or anti-Hhex to show the expression levels of SOX13-FLAG and Hhex in the assay. D, co-localization of ectopically expressed Hhex and SOX13 is shown. HeLa cells were co-transfected with plasmids expressing the fusion protein EGFP-Hhex (green) and a SOX13-myc construct and labeled with anti-Myc antibody (red). Images were acquired with a Leica TCS-SP2 confocal microscope. BF, bright field.
Hhex Modulates SOX13-dependent Wnt Signaling in 293T Cells
Because Hhex can act as a transcriptional repressor or activator, we tested the possibility that SOX13 could be a co-repressor or co-activator of Hhex. To look for SOX13 co-repressing activity, we used the plasmid Gsc-LUC, a Hhex-specific reporter system (15). SOX13 did not significantly change the transcriptional activity of Hhex toward the Goosecoid promoter (data not shown). Similarly, we tested the possible role of SOX13 as a transcriptional co-activator of Hhex using the Na+/taurocholate cotransporting polypeptide promoter (19). Again, SOX13 was not able to change Hhex transcriptional activity. Alternatively, we tested whether Hhex could alter SOX13 transcriptional activity using the general SOX reporter 3xSx (39). SOX13 did not transactivated the 3xSx luciferase reporter, although other SOX proteins, such as SOX9, effectively activated this reporter more than 30-fold (data not shown). The addition of Hhex did not change SOX13 response (data not shown). In short, neither Hhex nor SOX13 altered the transcriptional capabilities of each other in specific luciferase reporter plasmids.
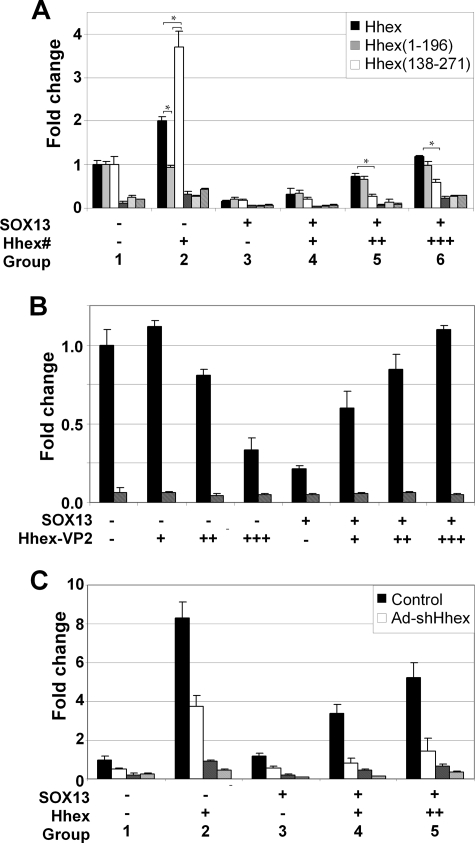
SOX13 was recently described as a key regulator of T-lymphocyte differentiation by the inhibition of Wnt signaling (40). To test whether Hhex could modulate the function of SOX13 in Wnt signaling, we used a luciferase reporter controlled by multiple wild-type (TOPflash) or mutated (FOPflash) TCF binding sites (41). Wnt pathway was constitutively activated by co-transfecting an expression vector containing activated β-catenin-S37Y. We used 293T cells because they express TCF1 and are fully competent to activate transcription of a luciferase reporter gene controlled by TCF binding sites (41). Full-length Hhex caused a 2-fold induction of the TCF/β-cat luciferase reporter, whereas Hhex-(1–196) had no effect (Fig. 6A, compare group 1 and 2), suggesting that the activating domain of Hhex located in the C terminus is necessary for this induction. In fact, when a deletion mutant of Hhex containing only the activating domain and the homeodomain was used Hhex-(138–271), Wnt reporter activity increased by 4-fold. Full-length SOX13 caused a dose-dependent inhibition of the Wnt reporter in 293T cells (group 3). A concentration of SOX13 causing an 80% reduction of TOPflash reporter activity was selected for co-transfection studies. The same concentration of ΔSOX13 and S-SOX13 did not change Wnt reporter activity (supplemental Fig. S1A), suggesting that the LZ-Q domain is necessary, but not sufficient, for modulation of Wnt activity. The addition of Hhex to SOX13 abrogated the repressive effect of the latter on Wnt activity in a dose-dependent manner (Fig. 6A, black bars, compare group 4–6). We speculated that the resulting TOPflash activity may be either the simple addition of the repressing and activating features of SOX13 and Hhex, respectively, or the result of the interference caused by Hhex in the SOX13·TCF1 complex on the TOPflash reporter. To explore these hypotheses and establish the direction of this functional interaction (does Hhex rescue SOX13 inhibition or does SOX13 block Hhex induction?), we used Hhex-(1–196), a fragment of Hhex that interacts with SOX13 (Fig. 4), but it was unable to induce Wnt reporter activity on its own. Nevertheless, Hhex-(1–196) was still able to restore TOPflash activity (Fig. 6A). Conversely, Hhex-(138–271), a fragment of Hhex that cannot interact with SOX13 (Fig. 4) but is an enhancer of TOPflash reporter activity on its own, only induced Wnt reporter activity at higher doses and significantly less efficiently than Hhex. Next, our aim was to definitively address whether the ability of Hhex to block the SOX13 repression of Wnt activity was directly dependent on SOX13 interaction or if it was indirect by co-repressing a factor that would ultimately alter Wnt reporter. Hhex is a transcriptional repressor that can be converted into an activator by fusing two copies of the minimal transcriptional activation domain of VP16 (Hhex-VP2 (15)). Retention of the entire Hhex open reading frame allows the examination of Hhex function without disrupting potential protein-protein interactions. As previously reported (42), Hhex-VP2 alone caused a dose-dependent decrease in the activity of the TOPflash reporter, which is in agreement with the conversion of Hhex into an activator (Fig. 6B). Paradoxically, when increasing amounts of Hhex-VP2 were added to SOX13, Wnt activity increased to basal levels. This indicates that abolishment of the inhibitory activity of SOX13 by Hhex does not reside in the repressing capabilities of the latter. Rather, it strongly suggests that Hhex antagonizes SOX13 activity on the Wnt signaling pathway by displacing and sequestering SOX13 and not indirectly repressing other genes ultimately involved or linked to Wnt activity.
FIGURE 6.
Effect of Hhex and SOX13 interaction on Wnt activity. A, shown are functional domains of Hhex involved in Wnt regulation. HEK 293T cells were transfected with the Wnt reporter plasmid TOPflash (filled bars) and the mutated version, FOPflash (dashed bars), as a negative control. An expression vector containing activated β-catenin-S37Y was cotransfected in each condition. Those plasmids expressing SOX13 or Hhex derivatives were transfected as indicated below the graph. The means and S.E. of the relative firefly/renilla ratios and -fold change over the control of at least three experiments are shown. The total firefly/renilla ratio of untreated cells transfected with activated β-catenin (S37Y) was set as 1.0. Hhex#, Hhex-(1–271), Hhex-(1–196), or Hhex-(138–271). B, shown is dose-dependent regulation of Wnt by HhexVP2, a Hhex fusion protein containing two copies of the VP16 activator domain that converts Hhex into a transcriptional activator with or without SOX13 modulation. The same experiment settings as in panel A were used. C, Wnt activity is shown in HepG2 cells after knocking down endogenous Hhex by adenovirus-mediated expression of shRNA (Ad-shHhex). An adenovirus expressing shRNA against luciferase was used in HepG2 control cells. A plasmid expressing Hhex is represented at the bottom of the panel as Hhex. The same experiment settings as in panel A were used. *, p < 0.05, paired Student's t test.
Next, we conducted loss-of-function experiments in Hhex-expressing HepG2 cells by means of an adenoviral vector expressing shRNA against Hhex. Knockdown of Hhex was validated by immunoblotting using specific Hhex primary antibodies (supplemental Fig. S2). Knockdown of Hhex resulted in inhibition of Wnt activity in agreement with the role of Hhex as a Wnt inducer (Fig. 6C). SOX13 only caused inhibition of Wnt activity when Hhex was knocked down simultaneously (Fig. 6C, group 3). The addition of increasing doses of Hhex slowly increased Wnt activity in shRNA-treated HepG2 cells, although at a significantly lower rate than control HepG2 cells. In summary, endogenous Hhex in HepG2 cells abolish the repression of Wnt induced by SOX13.
Hhex Competes with TCF1 to Bind SOX13 in 293T Cells
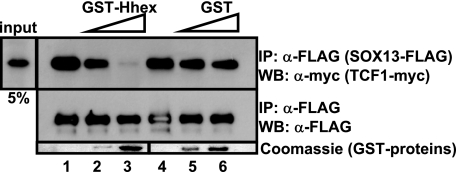
SOX13 is a potent repressor of the Wnt pathway by interacting and sequestering TCF1 from the Wnt transcriptionally active complex (40). As shown above, the co-expression of Hhex together with SOX13 is able to restore Wnt activity to basal levels. One hypothesis to explain Hhex activity is that Hhex interacts with SOX13 to release TCF1 that is then restored into the Wnt activating complex. To test this hypothesis, we performed a competitive immunoprecipitation assay using purified GST-Hhex as a competitor. As reported, SOX13 binds full-length TCF-1 (Fig. 7, upper panel, lanes 1 and 4). The addition of Hhex was able to abolish the formation of the SOX13·TCF1 complex in a dose-dependent manner (Fig. 7; lanes 2 and 3). Concomitantly, the addition of Hhex restores the Wnt activity in transfected 293T cells (Fig. 6A). Basically, Hhex is not only able to interact with SOX13 but is also capable of withdrawing SOX13 from the SOX13·TCF1 complex and releasing back TCF1 and restoring Wnt activity to basal levels.
FIGURE 7.
Hhex and SOX13 competitive immunoprecipitation assay. A lysate from 293T cells co-transfected with SOX13-FLAG- and TCF1-myc-tagged-expressing plasmids was immunoprecipitated (IP) using anti-FLAG antibody in the absence of GST proteins (lanes 1 and 4) or with increasing amounts of purified GST-Hhex (lanes 2 and 3) or GST (lanes 5 and 6). The immunoprecipitated material was immunoblotted (WB) with anti-Myc antibody. The presence of increasing amounts of GST-proteins is shown with Coomassie staining of a gel run in parallel.
Hhex Influences SOX13-dependent Wnt Activity in Mouse Embryos
To validate the results obtained in the cell culture, we designed an in vivo reporter assay. We electroporated mouse embryos with reporter and expressing plasmids, cultured the embryos for 24 h, isolated the presumptive liver domain, and measured luciferase activity on the isolated embryonic tissue extract (Fig. 8). The method was developed to target the definitive ventral endoderm where the liver and ventral pancreas develop (31). Once again, electroporation of SOX13 resulted in a severe (>80%) reduction in the Wnt reporter activity. Unlike our observation in 293T cells, Hhex electroporation also caused a profound inhibition of the Wnt pathway in mouse ventral targeted endoderm. The different behavior of Hhex alone in this system and in cultured cells may result from tissue-specific differences in expression of other Wnt regulators. Co-electroporation of Hhex and SOX13 restored TOPflash reporter activity at the control levels, thus confirming our cell culture experiments. In short, in vivo experiments corroborated that Hhex can revert Wnt inhibition triggered by SOX13.
FIGURE 8.
Wnt activity in mouse embryos electroporated with SOX13 and Hhex. A, a diagram shows the orientation of the embryo in the electroporation cuvette. E8.5 mouse embryos were orientated with a 45° angle to specifically transfer DNA to the pre-hepatic and pre-pancreatic endoderm. The region targeted by DNA is highlighted in red. After electroporation, embryos were incubated for 24 h in vitro. Turned-embryos, with the E9.5 developmental stage, were then dissected to isolate the liver bud region, and the reporter activity was measured. B, luciferase activity is shown of isolated liver buds from non-electroporated (first chart) or electroporated embryos with TOPflash reporter, SOX13, and/or Hhex. The total firefly/renilla ratio of embryos electroporated with TOPflash alone was set at 1.0. Each condition was done at least three times, and the total number of embryos recovered and assayed was at least six.
DKK1, a Target of the β-Catenin-TCF Pathway Is Regulated by Hhex·SOX13
We addressed the question of whether the modulation of the Wnt activity by the Hhex·SOX13 complex observed in reporter assay could also be detected in endogenous Wnt target. We selected DKK1, a well known Wnt target previously validated by chromatin immunoprecipitation and qRT-PCR in cultured cells (28). mRNA levels of DKK1 behaved in parallel to reporter activity (compare Fig. 9B with Fig. 6A, group 3 and 4). After activation of Wnt signaling by LiCl, SOX13 caused a down-regulation of DKK1 mRNA. This repression was accompanied by a reduction of TCF1 bound to the DKK1 promoter (Fig. 9D). Hhex coexpression increased the level of DKK1 mRNA, obliterating the inhibition caused by SOX13. Again, the derepression caused by Hhex was followed by a recovery in the amount of TCF1 bound to the DKK1 promoter. Similar results were obtained with another Wnt target, SP5 (43) (supplemental Fig. S4).
FIGURE 9.
Regulation of DKK1, a target of the β-catenin-TCF pathway, by Hhex. A, shown is a schematic representation of the human DKK1 gene. TCF binding sites are depicted as black boxes. B, 293T cells were transfected with plasmids expressing TCF1-myc, SOX13, or Hhex, as shown below the graph. The expression of DKK1 was assessed by quantitative RT-PCR. C, shown is immunoblot analysis of the transfected 293T cells. D, 293T cells transfected as in B were cross-linked and lysed. The relative amounts of TCF1-myc on DKK1 promoter were analyzed by real-time qPCR of immunoprecipitated chromatin using anti-Myc antibody. Binding of TCF1 to the RPLP0 gene was used as a negative control. The image of a representative qPCR stopped at the exponential phase is also shown. Data are the mean of three experiments. PBGD, porphobilinogen deaminase.
DISCUSSION
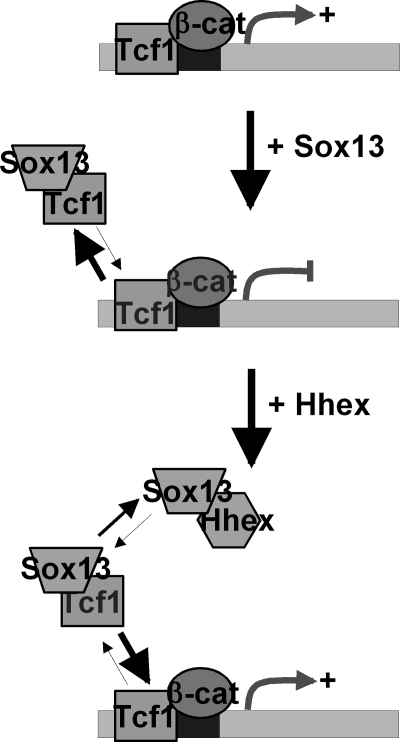
We have shown that binary interactions between Hhex, SOX13, and TCF regulate Wnt activity in vitro and in vivo. The three genes are co-expressed in several embryonic tissues, like the liver and pancreas (44, 45). The β-catenin-TCF complex is a downstream effector of the Wnt signaling pathway (Fig. 10). SOX13 binds the transcription factor, TCF1, possibly through the LZ-Q domain to disrupt the TCF1-β-catenin complex and inhibit Wnt activity (40). The addition of Hhex results in the formation of the Hhex·SOX13 complex (Fig. 4) to dislodge SOX13 from the SOX13·TCF1 complex (Fig. 7) and results in a restoration/elevation of the Wnt levels (Fig. 6A, 8, and 9). In short, Hhex and SOX13 act as a pair of antagonists of the Wnt activity to achieve the right level in different biological contexts.
FIGURE 10.
Model depicting the role of Hhex in Wnt activity modulation. In this model Wnt-responsive genes are up-regulated upon the binding of a complex containing TCF1-β-catenin to consensus TCF1 binding sites. Wnt activity is inhibited in the presence of SOX13 because of its interaction with the TCF1 protein, which disrupts the TCF1-β-catenin complex. The addition of Hhex results in the formation of a Hhex·SOX13 complex, displacing SOX13 from TCF1 and allowing TCF1 to form the effector complex in sensitive promoters once more.
Through the screening for Hhex-binding proteins with an E9.5-E10.5 mouse embryo library (24), we isolated cDNA for a HMG box protein, SOX13. In the only previous yeast two-hybrid screening using Hhex as bait, two interactors were isolated; that is, the HC8 subunit of the proteasome (46) and the CK2β (47). In our study we have used an embryo-derived library instead of a chronic myelogenous leukemia cell line (K562)-derived cDNA library, which lacks any developmental context. Other studies using different bait have identified Hhex as an interacting partner of Jun (48), GATA2 (49), Sox10 (50), TLE1 (34), or PML (51). Noteworthy, our screening confirmed the interaction of Hhex with TLE1. Besides Sox13, we obtained other 68 candidates, 24 of them with 2 repetitions or more. Sox13 was isolated 8 times with a common spanning region of ∼100 aa (Fig. 2).
Among the different SOX13 mRNAs described in the literature, we identify SOX13 (1815 bp) and S-SOX13 (768 bp) as the isoforms present in human tissues. Our identification is in conflict with Roose et al. (37) who described a longer open reading frame of 2673 bp isolated from embryonic thymic cDNA. We did not find a significant amount of this longer mRNA in any of the 18 tissues tested, including adult thymus, which is also in agreement with a later report from Harley and co-workers (36).
SOX transcription factors are involved in the differentiation of multiple cell types and developmental processes from the endoderm. Sox17 plays an essential role in definitive endoderm formation in the mouse (52). Sox9 is expressed in the pancreas where it plays a role in the maintenance of a progenitor cell pool (53). It is also expressed asymmetrically in the ductal plate, and it seems to control the timing of biliary tubulogenesis (54). Sox4 is essential for the normal formation of endocrine pancreas (55). SOX13 is expressed in the pancreas and liver, in the visceral mesoderm of the extraembryonic yolk sac, and also in the spongiotrophoblast layer of the placenta (45, 56). At the biochemical level, SOX13 contains a leucine zipper and glutamine-rich domain at the C terminus and the HMG box, an 80-amino acid sequence that mediates DNA binding (37). SOX13 spans 14 exons and belongs to the group D of the SOX family together with SOX5 and SOX6. They are characterized by the absence of the transactivation domain (57). SOX13 is able to bind the consensus sequence (A/T)(A/T)CAA(A/T)G as a monomer or homodimer, probably through the LZ domain (58). Which domains are responsible for Hhex/SOX13 interaction? The N terminus of Hhex (aa 1–137) is apparently necessary and sufficient for SOX13 interaction. When used as bait, it interacts with the LZ-Q domain of SOX13 to transactivate MEL1 and LacZ reporter genes under the control of a heterologous GAL4-responsive upstream activation sequence and promoter elements in yeast (Fig. 2). In a GST pulldown assay, only Hhex itself and its deletion mutants containing the N-terminal domain, i.e. Hhex-(1–137) and Hhex-(1–196), achieve a significant amount of SOX13 pulldown (Fig. 4). On the other hand, the LZ-Q domain in SOX13 is necessary, but not sufficient, for interaction in cells. S-SOX13 contains the LZ-Q domain but does not interact with any fragment of Hhex. ΔSOX13, which lacks only the LZ-Q domain, is not able to interact either. If Hhex interacts through the LZ-Q, then we may expect its effects to be mediated by a disruption of the putative SOX13 homodimer. However, SOX13 might act as a monomer, as suggested by the fact that co-transfection with S-SOX13 (containing the LZ-Q domain but not the HMG domain) did not block the inhibitory effect of SOX13 on Wnt activity (supplemental Fig. S1B). In short, the N-terminal domain of Hhex mediates the binding to SOX13 in mammalian cells, whereas the LZ-Q domain in SOX13 is necessary, but not sufficient, for the interaction. The interaction between Hhex and SOX13 seems to be specific as SOX9 did not interact with Hhex in an immunoprecipitation assay. Finally, we showed that SOX13 and Hhex interact in a cell system and can do so directly without a bridging protein (Fig. 5).
Our work also uncovered the independent structural requirements of Hhex and SOX13 to effectively modulate Wnt activity. The Wnt signaling pathway describes a complex network of proteins most well known for their roles in embryogenesis and cancer but also involved in normal physiological processes in adult animals. Hhex has been recently described as an inducer of Wnt by repressing the expression of TLE4 (42). However, Hhex-(1–196), which retains the repressor domain but lacks the transactivator domain, did not induce Wnt activity (Fig. 6A). In parallel, Hhex-(138–271), which lacks the repressor domain, induced Wnt activity by almost 4-fold, suggesting that Wnt activation depends on the C terminus of Hhex. It is feasible that the assigned transcriptional function of specific Hhex domains might be more plastic than initially thought. Alternatively, TLE4 might be a secondary target of Hhex. Fragments of Hhex lacking the homeodomain, i.e. Hhex-(197–271) and Hhex-(1–137), did not alter Wnt activity (data not shown). This is possibly linked to their extranuclear localization (supplemental Fig. S3) and the inability to bind DNA. On the other hand, SOX13 integrity seems to be crucial for Wnt repression. A specific deletion of the LZ-Q domain in SOX13 (ΔSOX13) completely abolished Wnt repression. S-SOX13, which lacks the C-terminal domain of SOX13, was also unable to inhibit Wnt activity (supplemental Fig. S1). An inspection of the S-SOX13 structure suggests that it might function as a SOX13 inhibitor. However, it did not interfere with the SOX13-dependent regulation of Wnt. Full-length Hhex is able to restore Wnt inhibition after SOX13 co-transfection in 293T cells. Hhex domains have a statistically significant difference to restore SOX13-dependent Wnt repression. Hhex-(1–196) was as efficient as the full-length protein, whereas Hhex-(138–271) only caused a certain degree of Wnt restoration at the highest dose, which was significantly less pronounced (Fig. 6A). Given that Hhex-(138–271) lacks the N terminus involved in SOX13 interaction, this result suggests that Wnt activity restoration is caused by direct SOX13/Hhex interaction. In agreement with this hypothesis, Hhex-VP2, a modified form of Hhex containing the N terminus, behaves as a transcriptional activator and is still able to reinstate Wnt activity (Fig. 6B). Thus, Hhex influences Wnt signaling in the presence of SOX13 and does not reside in Hhex transcriptional activity. Reporter assay analysis in HepG2 cells confirmed that Hhex is able to block the inhibitory effect of SOX13 either by endogenous expression in HepG2 cells (Fig. 6C) or by overexpression in 293T cells (Fig. 6A, group 6, black bar). Moreover, knockdown of endogenous HHEX in HepG2 restored the inhibitory activity of SOX13 (Fig. 6C, group 3). In summary, HepG2 cells behave as 293T cells in terms of SOX13-dependent Wnt inhibition, when endogenous Hhex is knocked down.
We validated in cultured mouse embryos the results obtained in cell cultures (Fig. 8). It is somewhat striking that ectopic expression of Hhex cDNA in 293T cells induces Wnt activity, whereas in the embryo it inhibits Wnt activity. We can best explain this discrepancy by suggesting that Wnt induction is a cell-autonomous effect of Hhex. In the embryo, non-cell autonomous effects, such as interaction between Hhex expressing endoderm and mesoderm during patterning, may induce expression of the Wnt antagonist that ultimately accounts for a general inhibition of Wnt activity. In fact, phenotypes observed in response to overexpression of Hhex resemble those obtained after expression of Wnt antagonists (15). Loss-of-function of Hhex in the mouse causes a dorsalization of the embryo, where most of the ventral structures are missing, i.e. forehead, thyroid, liver, or ventral pancreas (5, 7), which is compatible with extended Wnt activity (59).
Keeping in mind that our reporter assay is based on an heterologous promoter, we wanted to confirm our results in an endogenous promoter. We chose DKK1 because it is a relevant developmentally regulated gene and it has been validated extensively as a β-catenin-TCF1 target (28). The mRNA levels of DKK1 reproduced the expression pattern obtained with the TOPflash reporter plasmid. Moreover, DKK1 mRNA expression profile was paralleled by TCF1 occupancy of the DKK1 promoter. In summary, the results obtained in reporter assay were validated with endogenous TCF1-regulated promoter by qRT-PCR and a chromatin immunoprecipitation assay (Fig. 9).
There seems to be an intense tissue-specific cross-regulation between Hhex and Wnt during development. Activation of Wnt/β-catenin signaling in zebrafish (61) induces Hhex expression in the dorsal yolk syncytial layer. In agreement, β-catenin-deficient mouse embryos do not express Hhex in the anterior-posterior axis on embryonic day 5.5 (62). However, inhibition of the canonical Wnt pathway in Xenopus induces Hhex in the underlying endoderm of the heart field (63). Do modulators like SOX13 determine the output of Hhex and Wnt interrelationship in different tissues? Wnt regulation is particularly essential for the specification of the liver and pancreas from the ventral foregut endoderm. Wnt down-regulation in the anterior endoderm is shown to be crucial for liver and pancreas specification in the ventral foregut endoderm. But immediately after inducing the hepatic program in the endoderm, Wnt signaling is apparently required for the endoderm to outgrow further into a liver bud (1). In zebrafish, the expression of Wnt2b in the lateral plate mesoderm, acting through the β-catenin canonical pathway, appears essential for liver specification in the endoderm and bud induction (2). Briefly, there must be fast and sharp control of Wnt in the early steps of liver and pancreas specification. Part of this control is exerted by the expression of the Wnt inhibitors (15). In this study we propose that the reciprocal interactions between the triad Hhex, SOX13, and TCF1 together with the autoregulatory loop Hhex⇔Wnt contribute to achieve the correct Wnt intensity in the appropriate spatiotemporal dimension.
Acknowledgments
We thank Drs. J. M. Brickman, A. Garcia de Herreros, G. Gil, and M. Wegner for kindly providing plasmids. We also thank Marina Blazquez for discussions and assistance in some experiments. CIBEREHD (CIBER de Enfermedades Hepaticas y Digestivas) is funded by the Instituto de Salud Carlos III, Spain.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
- aa
- amino acids
- GST
- glutathione S-transferase
- EGFP
- enhanced green fluorescence protein
- HA
- hemagglutinin
- shRNA
- short hairpin RNA
- CMV
- cytomegalovirus
- PBS
- phosphate-buffered saline
- RT
- reverse transcription
- qRT
- quantitative real-time
- LZ
- leucine zipper
- HMG
- high mobility group
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- ChIP
- chromatin immunoprecipitation
- TCF
- T cell factor.
REFERENCES
- 1.McLin V. A., Rankin S. A., Zorn A. M. (2007) Development 134, 2207–2217 [DOI] [PubMed] [Google Scholar]
- 2.Ober E. A., Verkade H., Field H. A., Stainier D. Y. (2006) Nature 442, 688–691 [DOI] [PubMed] [Google Scholar]
- 3.Thomas P. Q., Brown A., Beddington R. S. (1998) Development 125, 85–94 [DOI] [PubMed] [Google Scholar]
- 4.Bogue C. W., Zhang P. X., McGrath J., Jacobs H. C., Fuleihan R. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 556–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bort R., Martinez-Barbera J. P., Beddington R. S., Zaret K. S. (2004) Development 131, 797–806 [DOI] [PubMed] [Google Scholar]
- 6.Crompton M. R., Bartlett T. J., MacGregor A. D., Manfioletti G., Buratti E., Giancotti V., Goodwin G. H. (1992) Nucleic Acids Res. 20, 5661–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez Barbera J. P., Clements M., Thomas P., Rodriguez T., Meloy D., Kioussis D., Beddington R. S. (2000) Development 127, 2433–2445 [DOI] [PubMed] [Google Scholar]
- 8.Kubo A., Chen V., Kennedy M., Zahradka E., Daley G. Q., Keller G. (2005) Blood 105, 4590–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obinata A., Akimoto Y., Omoto Y., Hirano H. (2002) Dev. Growth Differ. 44, 281–292 [DOI] [PubMed] [Google Scholar]
- 10.Keng V. W., Yagi H., Ikawa M., Nagano T., Myint Z., Yamada K., Tanaka T., Sato A., Muramatsu I., Okabe M., Sato M., Noguchi T. (2000) Biochem. Biophys. Res. Commun. 276, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 11.Hallaq H., Pinter E., Enciso J., McGrath J., Zeiss C., Brueckner M., Madri J., Jacobs H. C., Wilson C. M., Vasavada H., Jiang X., Bogue C. W. (2004) Development 131, 5197–5209 [DOI] [PubMed] [Google Scholar]
- 12.Bort R., Signore M., Tremblay K., Martinez Barbera J. P., Zaret K. S. (2006) Dev. Biol. 290, 44–56 [DOI] [PubMed] [Google Scholar]
- 13.Hunter M. P., Wilson C. M., Jiang X., Cong R., Vasavada H., Kaestner K. H., Bogue C. W. (2007) Dev. Biol. 308, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Balkau B., Heude B., Charpentier G., Hudson T. J., Montpetit A., Pshezhetsky A. V., Prentki M., Posner B. I., Balding D. J., Meyre D., Polychronakos C., Froguel P. (2007) Nature 445, 881–885 [DOI] [PubMed] [Google Scholar]
- 15.Brickman J. M., Jones C. M., Clements M., Smith J. C., Beddington R. S. (2000) Development 127, 2303–2315 [DOI] [PubMed] [Google Scholar]
- 16.Cong R., Jiang X., Wilson C. M., Hunter M. P., Vasavada H., Bogue C. W. (2006) Biochem. Biophys. Res. Commun. 346, 535–545 [DOI] [PubMed] [Google Scholar]
- 17.Guo Y., Chan R., Ramsey H., Li W., Xie X., Shelley W. C., Martinez-Barbera J. P., Bort B., Zaret K., Yoder M., Hromas R. (2003) Blood 102, 2428–2435 [DOI] [PubMed] [Google Scholar]
- 18.Tanaka T., Inazu T., Yamada K., Myint Z., Keng V. W., Inoue Y., Taniguchi N., Noguchi T. (1999) Biochem. J. 339, 111–117 [PMC free article] [PubMed] [Google Scholar]
- 19.Denson L. A., Karpen S. J., Bogue C. W., Jacobs H. C. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279, G347–G355 [DOI] [PubMed] [Google Scholar]
- 20.Puppin C., Puglisi F., Pellizzari L., Manfioletti G., Pestrin M., Pandolfi M., Piga A., Di Loreto C., Damante G. (2006) BMC Cancer 6, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasamatsu S., Sato A., Yamamoto T., Keng V. W., Yoshida H., Yamazaki Y., Shimoda M., Miyazaki J., Noguchi T. (2004) J. Biochem. 135, 217–223 [DOI] [PubMed] [Google Scholar]
- 22.Gómez-Foix A. M., Coats W. S., Baqué S., Alam T., Gerard R. D., Newgard C. B. (1992) J. Biol. Chem. 267, 25129–25134 [PubMed] [Google Scholar]
- 23.Stegmeier F., Hu G., Rickles R. J., Hannon G. J., Elledge S. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13212–13217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollenberg S. M., Sternglanz R., Cheng P. F., Weintraub H. (1995) Mol. Cell. Biol. 15, 3813–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frangioni J. V., Neel B. G. (1993) Anal. Biochem. 210, 179–187 [DOI] [PubMed] [Google Scholar]
- 26.Topisirovic I., Guzman M. L., McConnell M. J., Licht J. D., Culjkovic B., Neering S. J., Jordan C. T., Borden K. L. (2003) Mol. Cell. Biol. 23, 8992–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez-Jiménez C. P., Gómez-Lechón M. J., Castell J. V., Jover R. (2006) J. Biol. Chem. 281, 29840–29849 [DOI] [PubMed] [Google Scholar]
- 28.Niida A., Hiroko T., Kasai M., Furukawa Y., Nakamura Y., Suzuki Y., Sugano S., Akiyama T. (2004) Oncogene 23, 8520–8526 [DOI] [PubMed] [Google Scholar]
- 29.Weinmann A. S., Bartley S. M., Zhang T., Zhang M. Q., Farnham P. J. (2001) Mol. Cell. Biol. 21, 6820–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-Jiménez C. P., Castell J. V., Gómez-Lechón M. J., Jover R. (2006) Mol. Pharmacol. 70, 1681–1692 [DOI] [PubMed] [Google Scholar]
- 31.Pierreux C. E., Vanhorenbeeck V., Jacquemin P., Lemaigre F. P., Rousseau G. G. (2004) J. Biol. Chem. 279, 51298–51304 [DOI] [PubMed] [Google Scholar]
- 32.Pierreux C. E., Poll A. V., Jacquemin P., Lemaigre F. P., Rousseau G. G. (2005) JOP 6, 128–135 [PubMed] [Google Scholar]
- 33.Tremblay K. D., Zaret K. S. (2005) Dev. Biol. 280, 87–99 [DOI] [PubMed] [Google Scholar]
- 34.Swingler T. E., Bess K. L., Yao J., Stifani S., Jayaraman P. S. (2004) J. Biol. Chem. 279, 34938–34947 [DOI] [PubMed] [Google Scholar]
- 35.Wilson M., Koopman P. (2002) Curr. Opin. Genet. Dev. 12, 441–446 [DOI] [PubMed] [Google Scholar]
- 36.Kasimiotis H., Myers M. A., Argentaro A., Mertin S., Fida S., Ferraro T., Olsson J., Rowley M. J., Harley V. R. (2000) Diabetes 49, 555–561 [DOI] [PubMed] [Google Scholar]
- 37.Roose J., Korver W., de Boer R., Kuipers J., Hurenkamp J., Clevers H. (1999) Genomics 57, 301–305 [DOI] [PubMed] [Google Scholar]
- 38.Ovcharenko I., Nobrega M. A., Loots G. G., Stubbs L. (2004) Nucleic Acids Res. 32, W280–W286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber J., Sock E., Wegner M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4739–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melichar H. J., Narayan K., Der S. D., Hiraoka Y., Gardiol N., Jeannet G., Held W., Chambers C. A., Kang J. (2007) Science 315, 230–233 [DOI] [PubMed] [Google Scholar]
- 41.Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 42.Zamparini A. L., Watts T., Gardner C. E., Tomlinson S. R., Johnston G. I., Brickman J. M. (2006) Development 133, 3709–3722 [DOI] [PubMed] [Google Scholar]
- 43.Valenta T., Lukas J., Doubravska L., Fafilek B., Korinek V. (2006) EMBO J. 25, 2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bogue C. W., Ganea G. R., Sturm E., Ianucci R., Jacobs H. C. (2000) Dev. Dyn. 219, 84–89 [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Ristevski S., Harley V. R. (2006) J. Histochem. Cytochem. 54, 1327–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bess K. L., Swingler T. E., Rivett A. J., Gaston K., Jayaraman P. S. (2003) Biochem. J. 374, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soufi A., Noy P., Buckle M., Sawasdichai A., Gaston K., Jayaraman P. S. (2009) Nucleic Acids Res. 37, 3288–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaefer L. K., Wang S., Schaefer T. S. (2001) J. Biol. Chem. 276, 43074–43082 [DOI] [PubMed] [Google Scholar]
- 49.Minami T., Murakami T., Horiuchi K., Miura M., Noguchi T., Miyazaki J., Hamakubo T., Aird W. C., Kodama T. (2004) J. Biol. Chem. 279, 20626–20635 [DOI] [PubMed] [Google Scholar]
- 50.Wissmüller S., Kosian T., Wolf M., Finzsch M., Wegner M. (2006) Nucleic Acids Res. 34, 1735–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Topcu Z., Mack D. L., Hromas R. A., Borden K. L. (1999) Oncogene 18, 7091–7100 [DOI] [PubMed] [Google Scholar]
- 52.Kanai-Azuma M., Kanai Y., Gad J. M., Tajima Y., Taya C., Kurohmaru M., Sanai Y., Yonekawa H., Yazaki K., Tam P. P., Hayashi Y. (2002) Development 129, 2367–2379 [DOI] [PubMed] [Google Scholar]
- 53.Seymour P. A., Freude K. K., Tran M. N., Mayes E. E., Jensen J., Kist R., Scherer G., Sander M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antoniou A., Raynaud P., Cordi S., Zong Y., Tronche F., Stanger B. Z., Jacquemin P., Pierreux C. E., Clotman F., Lemaigre F. P. (2009) Gastroenterology 136, 2325–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson M. E., Yang K. Y., Kalousova A., Lau J., Kosaka Y., Lynn F. C., Wang J., Mrejen C., Episkopou V., Clevers H. C., German M. S. (2005) Diabetes 54, 3402–3409 [DOI] [PubMed] [Google Scholar]
- 56.Lioubinski O., Müller M., Wegner M., Sander M. (2003) Dev. Dyn. 227, 402–408 [DOI] [PubMed] [Google Scholar]
- 57.Kiefer J. C. (2007) Dev. Dyn. 236, 2356–2366 [DOI] [PubMed] [Google Scholar]
- 58.Roose J., Korver W., Oving E., Wilson A., Wagenaa r G., Markman M., Lamers W., Clevers H. (1998) Nucleic Acids Res. 26, 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fagotto F., Guger K., Gumbiner B. M. (1997) Development 124, 453–460 [DOI] [PubMed] [Google Scholar]
- 60.Thompson B. A., Tremblay V., Lin G., Bochar D. A. (2008) Mol. Cell. Biol. 28, 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bischof J., Driever W. (2004) Dev. Biol. 276, 552–562 [DOI] [PubMed] [Google Scholar]
- 62.Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C., Birchmeier W. (2000) J. Cell Biol. 148, 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foley A. C., Mercola M. (2005) Genes Dev. 19, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]