Abstract
Microtubule plus-end tracking proteins (+TIPs) are involved in many microtubule-based processes. End binding (EB) proteins constitute a highly conserved family of +TIPs. They play a pivotal role in regulating microtubule dynamics and in the recruitment of diverse +TIPs to growing microtubule plus ends. Here we used a combination of methods to investigate the dimerization properties of the three human EB proteins EB1, EB2, and EB3. Based on Förster resonance energy transfer, we demonstrate that the C-terminal dimerization domains of EBs (EBc) can readily exchange their chains in solution. We further document that EB1c and EB3c preferentially form heterodimers, whereas EB2c does not participate significantly in the formation of heterotypic complexes. Measurements of the reaction thermodynamics and kinetics, homology modeling, and mutagenesis provide details of the molecular determinants of homo- versus heterodimer formation of EBc domains. Fluorescence spectroscopy and nuclear magnetic resonance studies in the presence of the cytoskeleton-associated protein-glycine-rich domains of either CLIP-170 or p150glued or of a fragment derived from the adenomatous polyposis coli tumor suppressor protein show that chain exchange of EBc domains can be controlled by binding partners. Extension of these studies of the EBc domains to full-length EBs demonstrate that heterodimer formation between EB1 and EB3, but not between EB2 and the other two EBs, occurs both in vitro and in cells as revealed by live cell imaging. Together, our data provide molecular insights for rationalizing the dominant negative control by C-terminal EB domains and form a basis for understanding the functional role of heterotypic chain exchange by EBs in cells.
Keywords: Cytoskeleton/Tubulin, Protein, Protein/Domains, Protein/Protein-Protein Interactions, Protein/Stability, Protein/Structure
Introduction
Microtubules are filamentous structures involved in many vital cellular activities. They contain two structurally and functionally distinct ends, slow-growing minus ends, and fast-growing plus ends and are, thus, intrinsically polar (for review, see Refs. 1 and 2). In cells, the microtubule minus ends are often anchored to the microtubule organizing center and are, thus, stable, whereas microtubule plus ends are highly dynamic and stochastically switch between phases of growing and shortening. The intrinsic dynamic nature of microtubules is central to microtubule function and is tightly regulated both spatially and temporally by so-called microtubule-associated proteins. Microtubule plus-end tracking proteins (+TIPs)4 are a functionally and structurally diverse set of microtubule-associated proteins that specifically accumulate to and track growing microtubule plus ends. They play important roles in controlling microtubule dynamics and in conferring recognition of microtubule ends in all eukaryotes (for review, see Refs. 3 and 4).
Prominent representatives of +TIPs are end-binding proteins (EB). EBs constitute a highly conserved and ubiquitously expressed family of proteins that recently emerged as core components of dynamic +TIP interaction networks at growing microtubule plus ends (4). Besides regulating microtubule plus end dynamics, they are implicated in several vital microtubule-based processes, including maintenance of cell polarity, regulation of chromosome segregation, positioning of the mitotic spindle, and anchoring of microtubules to their nucleation sites (4, 5). EBs autonomously track microtubule tips independent of additional factors, most likely by recognizing a structural feature of the growing microtubule plus end (6–10). Moreover, EBs play an essential role in targeting a diverse range of +TIP binding partners to microtubule ends (4).
EBs comprise about 300 residues and contain highly conserved N- and C-terminal domains that are separated by a more variable linker sequence (4). The globular N-terminal moiety shows the characteristic features of calponin homology (CH) domains and is necessary and sufficient for microtubule binding (11). The C-terminal domain (EBc) contains an α-helical parallel coiled-coil, which mediates the dimerization of EB polypeptide chains (12). It partially overlaps with the unique EB homology (EBH) domain, which contains a four-helix bundle and a disordered C-terminal tail (12, 13). The EBH domain specifically recognizes SXIP motifs embedded within basic and serine-rich sequence regions (14). The phosphorylation-controlled EBH-SXIP interaction is necessary and sufficient for targeting numerous structurally and functionally unrelated +TIPs to microtubule plus ends, including the adenomatous polyposis coli (APC) tumor suppressor protein, the microtubule-actin cross-linking factor (MACF), cytoplasmic linker protein (CLIP-170)-associated proteins (CLASPs), the transmembrane protein stromal interaction molecule-1 (STIM1), and the mitotic centromere-associated kinesin (MCAK). The SXIP motif thus acts as a widespread microtubule tip localization signal (MtLS) (14). The C-terminal 20–30-residue tails of EBs share a highly conserved C-terminal tripeptide, denoted the EE(Y/F) motif. The EE(Y/F) motif with the carboxylate group of the terminal aromatic residue is specifically recognized by the highly conserved GKNDG motif of cytoskeleton-associated protein-glycine-rich (CAP-Gly) domains present in, for example, CLIP-170 and the large dynactin subunit p150glued (15, 16).
In mammals, the EBs are represented by three proteins, EB1, EB2 (RP1), and EB3 (EBF3), which are encoded by separate genes (17). So far, functional studies have been primarily focused on EB1, but it is becoming increasingly clear that despite their high sequence conservation, the individual EBs exhibit different regulatory and functional properties. For example, EB2 displays lower affinity than EB1 and EB3 for several well characterized binding partners, such as APC and mitotic centromere-associated kinesin, and is, thus, less likely to be involved in their targeting to microtubule plus ends (18, 19). EB3 was also found to bind to the F-actin-binding protein drebrin (20) and the E3 ubiquitin ligase SIAH-1 (21), whereas no such interactions were observed for the other EBs. These observations support the view that the individual mammalian EBs can differentially bind to partners. The EBs also exhibit different spatial and temporal expression patterns. In contrast to EB1 and EB2, EB3 is up-regulated in neurons and muscle cells (22–24) and plays an important role in myotube differentiation (23) and neuronal development (20, 22). Furthermore, a phosphorylation controlled interaction between EB3 and SIAH-1 regulates EB3 degradation and, hence, expression levels during the cell cycle, whereas no such regulation was observed for EB1 and EB2 (21).
We have recently documented that mammalian EBs differentially affect microtubule dynamics (9). EB1 and EB3 promote persistent microtubule growth by suppressing catastrophes (a switch from the growing to the shrinkage phase of a microtubule). By comparison, EB2 is much less potent; this functional difference is partially due to amino acid substitutions in the CH domain of EB2. Although both in vivo and in vitro studies revealed that CH domains are sufficient for tracking growing microtubule plus ends, dimerization was shown to be necessary for the anti-catastrophe activity of EB proteins (9). In this context, we reported that the EBs can also form heterodimers through their C-terminal domains (9). Differences or changes in the dimerization properties of the three EBs may, therefore, contribute to the differences in the anti-catastrophe activity. Interestingly, C-terminal EB dimerization domains act as dominant negative mutants and have been used in an array of cellular studies as tools to interfere with the activity of endogenous EB proteins (9, 20, 25–32). They were found to affect all EB species by removing them from microtubule plus ends, which results in an increase in the catastrophe frequency (9).
The molecular basis of EB dimerization and the mechanism underlying the dominant negative effect of EBc domains is poorly understood. Acquiring knowledge of these properties is, however, important to understand the different functions of mammalian EB proteins and to interpret data obtained in the presence of transiently overexpressed EBc domains in cells. Moreover, the dimerization properties of EB proteins are likely to play a role in regulatory mechanisms used by EBs to control microtubule dynamics. Using a combination of biochemical, biophysical, structural, and cell biological methods together with computational modeling, this paper provides molecular insight into the mechanism of EB dimerization and its regulation by +TIPs binding partners.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
For all EB constructs, DNA was PCR-amplified from human EB1, EB2, and EB3 full-length cDNA clones kindly provided by W. Bu and L. Su (18, 19). The insertion of EB1c (Asp-191—Tyr-268) into the bacterial expression vector pET15b (Invitrogen) has been described (33). To create a tag-less version of EB1c, a PCR-amplified EB1c fragment was ligated between the NcoI and BamHI sites of pET15b (Invitrogen). Gateway cloning (Invitrogen) of EB3c (Ala-200—Tyr-281) into a pDEST17 vector has been previously described (9).
N-terminal fluorescent protein-tagged EB1c, EB2c (Thr-242—Tyr-327), and EB3c constructs were generated as follows. First, monomeric variants of enhanced cyan fluorescence protein (mECFP) and enhanced yellow fluorescence protein (mEYFP) were generated using pECFP-C1 and pEYFP-C1 vectors (Clontech) as DNA templates. Next, PCR products of mECFP and mEYFP were inserted by a PCR-splicing method (34) into pET15b between the regions encoding for the hexahistidine tag and the thrombin cleavage site. The thrombin cleavage site and the BamHI site were part of the 11-amino acid linker SGLVPRGSSDP introduced between the fluorescent proteins and EBc domains. The antisense primer used in this step also introduced two stop codons followed by an EcoRI restriction site. Finally, PCR-amplified EBc domains were ligated between the BamHI and EcoRI sites.
For bacterial expression of full-length EB1 and EB3, PCR-amplified DNA was ligated into the pET28a vector (Invitrogen) between the NcoI and BamHI sites, where the intrinsic BamHI site in EB2 was silently mutated. These plasmids were used to generate C-terminal fluorescent protein-tagged EB full-length constructs. PCR-amplified fluorescent protein DNA fragments were derived from pmECFP, pmEYFP, pmEGFP-N1, and pmCherry-N1 (Clontech) and ligated between the BamHI and XhoI sites to provide all constructs with a C-terminal hexahistidine tag. The insertion of p150CG (Met-18—Ser-111 of human p150glued), and the construction of the p150CG(A49M) mutant in pET15b has been described in (33). The Gateway cloning of ClipCG2 (Arg-210—Gly-282 of human CLIP-170) is reported in Ref. 15. All mutagenesis was carried out using the PCR-based QuikChange site-directed mutagenesis protocol from Stratagene.
Protein and Peptide Preparations
The Escherichia coli strain BL21(DE3) (Stratagene) was used for all protein expressions. Bacteria were grown to an A600 of 0.8 in LB medium at 37 °C containing the appropriate antibiotic. Cultures were induced by the addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.5 mm, and protein expression was performed at 22 °C for 16 h. For the preparation of uniformly 15N-labeled EB1c, cells were grown in a minimal medium as previously described (35).
Hexahistidine-tagged fusion proteins were purified by immobilized metal-affinity chromatography on HisTrap Ni2+-Sepharose chelating columns (GE Healthcare) at 4 °C according to the manufacturer's instructions. To remove the His tag, the recombinant proteins were dialyzed against thrombin cleavage buffer (20 mm Tris-HCl, pH 7.4, supplemented with 150 mm NaCl, and 2.5 mm CaCl2), and proteolytic cleavage was carried out for 16 h at 4 °C using human thrombin (Sigma) at a concentration of 3 units/mg of recombinant protein. The processed proteins were reapplied to immobilized metal-affinity chromatography to separate digested proteins from the hexahistidine tag and from uncleaved protein.
Uniformly 15N-labeled EB1c containing no affinity tag was purified by ion exchange chromatography using a self-packed ResourceQTM FastFlow 10/5 column (GE Healthcare) equilibrated in binding buffer (20 mm Bis-Tris, pH 6.0, supplemented with 5 mm NaCl, and 2 mm DTT) at 4 °C. After sample loading, the column was washed with 20 column volumes of binding buffer. Bound proteins were eluted by applying a linear gradient of NaCl (0–0.5 m) in the same buffer. All processed protein samples were gel-filtered on a SEC HiLoadTM Superdex 200 16/60 column (GE Healthcare) equilibrated in PBS, pH 7.5, supplemented with 2 mm DTT, and 1 mm EDTA. The N-acetylated and C-amidated APCp1 (Val-2781—Lys-2819 of human APC) peptide was assembled on an automated continuous-flow synthesizer employing standard methods.
The homogeneity of the proteins and peptides was confirmed by either 15% SDS-PAGE (proteins) or HPLC followed by mass spectral analysis (APCp1). The concentrations of protein and peptide solutions were determined by tyrosine and tryptophan absorbance at 280 nm.
Fluorescence Assay
Förster resonance energy transfer (FRET) measurements at 37 °C were carried out in PBS, pH 7.5, supplemented with 2 mm DTT and 1 mm EDTA. 50-μl protein solutions were analyzed in 384-well plates (black flat-bottom; Greiner no. 781900) covered with an optical clear adhesive seal (ABsolute QPCR Seal (AB-1170), Thermo scientific) in a TECAN Saphir II plate reader. A minimal excitation/emission bandwidth of 5 nm was used. The gain and the Z-position were set manually to 85 and 10,000 μm, respectively. Emission spectra were recorded between 450 and 650 nm, and excitation was performed at 434 nm. For kinetic measurements, FRET fluorescence was recorded at 527 nm. 10 readings per time point with an integration time of 40 μs were collected. For data analysis, three kinetic traces or spectra were averaged. Kinetic half-lives were calculated by single-exponential fitting using MATLAB Version 7.8 (The MathWorks, Inc.). S.D. from the average values were <5%.
Static Light Scattering (SLS)
SLS experiments were performed on a miniDawn TriStar system equipped with an Optilab rEX refractometer (Wyatt Technology Corp.) coupled to a Superdex 200 10/30 (Amersham Biosciences) size-exclusion chromatography (SEC) column run on an Agilent 1100 HPLC. 100 μl of 20 μm protein solutions were injected at a constant flow rate of 0.5 ml/min onto the column equilibrated in PBS supplemented with 2 mm DTT and 1 mm EDTA. The molecular masses of the eluted proteins were determined with the Wyatt Astra Version 5.3.1.5 software package (Wyatt Technology Corp.).
Circular Dichroism (CD) Spectroscopy
CD experiments with EBc domains were performed on a J-715 instrument (Jasco Ltd.) equipped with a computer-controlled water bath using cylindrical jacketed cuvettes of 1-mm optical path length. Thermal melting curves were recorded with 50 μm protein in PBS, pH 7.5, supplemented with 2 mm DTT and 1 mm EDTA by continuous heating at 1 deg min−1. Data points (ellipticity at 222 nm) were collected every 10 s. Reversibility was determined from the recovery of the mean residue ellipticity (MRE222) after cooling and was always found to be better than 95%. Thermal melting curves were analyzed according to a two-state model as described in Ref. 36. For measurements of urea melting curves, 40 μm of protein was incubated overnight at the corresponding urea concentrations. The signal was averaged for 3 min after thermal equilibration. Urea-induced equilibrium unfolding experiments were analyzed by nonlinear least-squares regression according to well established procedures (37).
Stopped-flow Kinetics
Kinetic CD experiments with EBc domains were performed with the π*-180 instrument (Applied Photophysics). The dead time was 1–2 ms, and the optical path length was 10 mm. The detection wavelength was 225 nm, and the slits were set to 4 mm. Refolding was initiated by mixing one volume of buffered protein solution (PBS, pH 7.5, supplemented with 2 mm DTT and 1 mm EDTA) containing 8–9 m urea with 10 or 25 volumes of buffer or with buffer containing various concentrations of the denaturant. Unfolding rates were measured by a 1:10 dilution of the protein into solutions containing final urea concentrations higher than the midpoint of equilibrium urea unfolding. 10–15 mixing experiments were averaged for each kinetic trace. Unfolding traces were modeled with a single-exponential function to extract the rate constants for unfolding (ku) using the software provided by the manufacturer. The rate constants for refolding (kf) in urea were calculated as described previously (36). Linear extrapolation to 0 m denaturant of the corresponding limbs in plots of ln kf,u versus [urea] yielded the kinetic constants in the absence of urea (38).
Structural Modeling
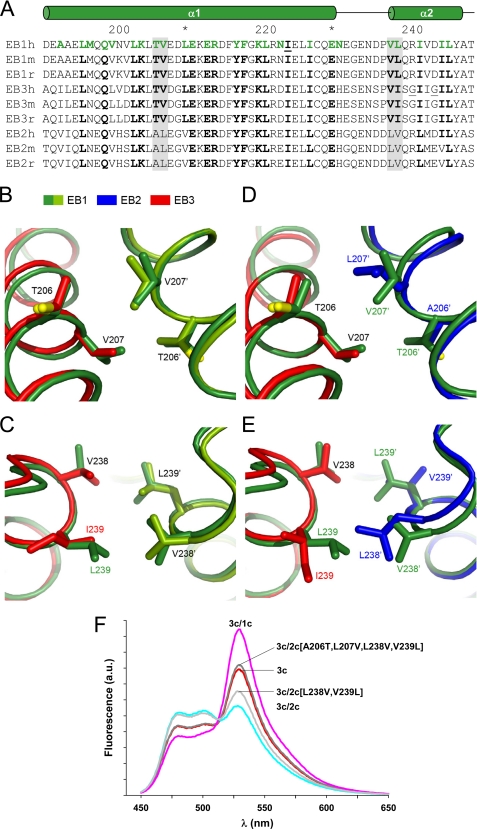
All dimeric forms of human EBc domains (EB2c·EB2c, EB3c·EB3c, EB1c·EB2c, EB1c·EB3c, EB2c·EB3c) were modeled based on the x-ray structure of human EB1c·EB1c at 1.54 Å resolution (PDB code 1WU9). The EB1c homodimer structure was used as a template in Modeler 9v6 (39), allowing for simultaneous energy minimization of both chains in the homo- or heterodimers. A multiple-sequence alignment of human, murine, and rat EB1c, EB2c, and EB3c was used as the basis for the modeling process. The homo- or heterodimeric models, thus, obtained were geometry-regularized with PHENIX 1.4 (40). Fig. 3, B–E, was prepared using PyMOL (41).
FIGURE 3.
Structural analysis of EBc domains. A, shown is multiple sequence alignment of human (h), murine (m), and rat (r) EB1c, EB2c, and EB3c. Residues of EB1c involved in interchain interactions (based on PDB entry 1WU9) are highlighted in green, and their size conservation among similar residue types is indicated in bold. The locations of the helices α1 and α2 in the structure of the EB1c homodimer and the residue numbering of EB1c are indicated above the alignment. The sites investigated in the mutational study (panel F) are shaded in gray; Gly-241 and Gly-244 in EB3h and Ile-224 in EB1h are underlined. All residue positions are given according to the alignment with the EB1h sequence. B and C, close-up views of the model of the human EB1c·EB3c heterodimer (EB1c and EB3c chains are depicted in light green and red, respectively) superimposed onto the crystal structure of the human EB1c homodimer (depicted in dark green). The main chains are shown as ribbons, and the interacting side chains are in stick representation. The labels are in the color of the corresponding chains or in black when the same residue occurs in both superimposed chains. Panel B shows the dipeptide segment 1, and panel C shows the dipeptide segment 2 (see text). D and E, shown is a close-up view of the model of the human EB2c·EB3c heterodimer (EB2c and EB3c chains depicted in blue and red, respectively) superimposed onto the crystal structure of the human EB1c homodimer (depicted in dark green). Panel D shows dipeptide segment 1, and panel E shows dipeptide segment 2. The residue numbering is according to the EB1 sequence (see panel A). Oxygen atoms of side chains in the panels B to E are indicated in yellow. F, the effect of mutations in EB2c on chain exchange with EB3c is shown. Fluorescence emission spectra (excitation at 434 nm) recorded from equimolar mixtures (10 μm each) of CFP-EB3c with YFP-EB1c (3c/1c, magenta), YFP-EB2c (3c/2c, cyan), YFP-EB3c (3c, red), YFP-EB2c(L238V,V239L) (3c/2c(L238V,V239L), gray) and YFP-EB2c(A206T,L207V,L238V,V239L) (3c/2c(A206T,L207V,L238V,V239L), dark gray). Samples were incubated at 37 °C for 16 h before data acquisition. a.u., arbitrary units.
Nuclear Magnetic Resonance (NMR) Spectroscopy
The previously obtained NMR assignments for the 1H and 15N backbone resonances of EB1c in the free form and in complex with APCp1 at 45 °C in 10 mm potassium phosphate buffer at pH 6.5 (14) were used as a starting point to obtain assignments at 37 °C in 10 mm potassium phosphate buffer at pH 6.5 containing 150 mm NaCl (NMR buffer). A series of 1H,15N heteronuclear single quantum correlation spectra obtained at 45, 42.5, 40, and 37 °C was used to transfer the assignments from 45 to 37 °C. Amide proton exchange experiments were initiated by dissolving the lyophilized protein in 2H2O and measuring a series of 1H,15N heteronuclear single quantum correlation spectra at various time intervals. The first experiment was measured ∼10 min after dissolution in 2H2O. Residues with exchangeable amide signals, which were present in the spectrum obtained in 1H2O but absent in the first spectrum obtained in 2H2O, are identified as “undergoing fast exchange.” A lower limit on the exchange rate of these amide protons was obtained by assuming that 98% of the amide protons had exchanged for 2H before the first experiment could be measured. The intensities of all remaining well resolved signals were fitted to an exponential function,
where I0 is the initial intensity, and kex is the exchange rate. All experiments were performed on Bruker Avance 600 and 750 MHz spectrometers equipped with triple resonance probe heads. Under the conditions where our experiments were conducted, the so-called EX2 mechanism of exchange applies (42, 43). Exchange occurs during breathing motions of the protein, which result in the breaking of hydrogen bonds and proton exposure to the solvent (the “open state”), enabling pH-dependent chemical reaction-controlled exchange with 2H present in the buffer (44, 45). The rate of exchange, kex, then is,
where kopen and kclose are the rate of opening and closing of the exchanging protein state, kintr is the intrinsic rate of exchange for a given amino acid type at a given pH (46), and Keq is the thermodynamic equilibrium constant for the open and closed protein states. Equation 2, thus, shows that amide proton exchange can report on underlying conformational equilibria of the protein. The ratio kintr/kex is commonly referred to as the protection factor P, because it indicates the degree to which exchange is suppressed in the folded protein when compared with an unstructured oligopeptide (46). The program CARA was used for the analysis of NMR spectra (47). Fig. 4C was prepared using Molmol 2.2 (48).
FIGURE 4.
Effect of binding partners on EB1c chain exchange. A, shown is the time-dependent increase of the fluorescence signal at 527 nm (excitation at 434 m) after mixing equimolar amounts (10 μm each) of CFP-EB1c and YFP-EB1c at 37 °C in the absence of a binding partner and in the presence of either 591 μm ClipCG2, 65 μm p150CG, 23 μm p150CG(A49M), or 116 μm APCp1. The concentrations of the EB1c binding partners have been adjusted so that 95% of the EB1c is present in complexed form when taking the Kd values of the individual binding partners into account (15, 33). a.u., arbitrary units. B, shown are amide 1H exchange protection factors of EB1c in the absence (blue) and presence (red) of an equimolar amount of APCp1. No data are shown for residues 207, 211, and 215 in the complex because of resonance overlap and for the residues 237, 256, and 261 in both data sets because these residues are prolines. Residues 191–208 and 231–241 from the free EB1c and residues 191–202, 231–241, and 249–268 from EB1c in complex with APCp1 are not shown because their amide protons have completely exchanged before the first experiment could be started (about 10 min), corresponding to log p values ≤3. The locations of the helices α1 and α2 in the structure of the EB1c homodimer (PDB entry 1WU9) are indicated. C, shown is the location of slowly exchanging amide hydrogens in the structure of the free EB1c dimer (left) and the EB1c dimer in complex with a peptide derived from the microtubule-actin cross-linking factor (right), which was shown to form a closely similar complex to the one with APCp1 (14). The backbone is shown in gray, and selected residue positions are indicated. The exchange data are represented by color-coded spheres at the position of the backbone N atoms: light blue, fast exchanging 1H, not observed in the first exchange spectrum; yellow, log p < 4; orange, log p = 4.0–5.0; red, log p > 5. The locations of the bound peptides are indicated by a green tube representing a spline function through the Cα atom coordinates.
Mammalian Expression Constructs and Plasmid Transfection
For mammalian expression of EB full-length FRET constructs, the cloning cassettes of the bacterial expression vectors (see above) were inserted into the backbone of the pEGFP-N1 mammalian expression vector (Clontech). The PCR products of the C-terminal fluorescent protein-tagged EB constructs were ligated between the HindIII and NotI sites after removal of the GFP cassette. Rescue mutations were introduced for all EB1 and EB3 full-length constructs, resulting in five silent substitutions in the RNAi target site of EB1 or EB3, respectively (9). Plasmids were transfected using FuGENE 6 (Roche Applied Science). Plasmids expressing fluorescent protein-tagged EB proteins were co-transfected with a tandem pSuper short hairpin RNA vector for simultaneous depletion of EB1 and EB3 (9). Imaging was performed at 2–4 days after transfection.
Live Cell Imaging and Image Processing
For live cell imaging we used a Nikon Eclipse Ti-E inverted microscope with perfect focus system (PFS) (Nikon) equipped with a Nikon CFI Apo TIRF 100 × 1.49 NA oil objective (Nikon), QuantEM 512SC EMCCD camera (Roper Scientific), and controlled with MetaMorph 7.5 software (Molecular Devices). For excitation we used a mercury lamp HBO-100W/2 (Osram) or a 491-nm 50-milliwatt Calypso (Cobolt) or a 561-nm 50-milliwatt Jive (Cobolt) laser (for epifluorescence and for total internal reflection fluorescence, respectively). For simultaneous imaging of green and red fluorescent signals, we used the ET-mCherry/GFP filter set (59022, Chroma) together with DualView (DV2, Roper Scientific) equipped with a dichroic filter 565dcxr and a HQ530/30m emission filter (Chroma). To keep cells at 37 °C we used a stage top incubator (model INUG2E-ZILCS, Tokai Hit). 16-bit images were projected onto the charge-coupled device chip at a magnification of 0.065 μm/pixel with an intermediate magnification of 2.5× (Nikon C mount adapter 2.5×). Images were captured using 500-ms exposures with no delay between frames (stream mode). A typical image series comprised 100–500 frames, measured over a period of 50 s. Images were converted to 8-bit format and processed using ImageJ with the plug-in “AColor.”
RESULTS AND DISCUSSION
Dimerization Properties of EBc Domains
We previously speculated that the mechanism underlying EB heterodimer formation involves spontaneous exchange of polypeptide chains (9). To further investigate this hypothesis, we now developed an assay based on FRET. Cyan and yellow fluorescent proteins (CFP and YFP) were fused individually to the N termini of the coiled-coil dimerization domains of the three human EBs (Fig. 1A), and milligram amounts of the recombinantly expressed fusion proteins were purified to homogeneity (referred to as CFP-EB1c, YFP-EB1c, CFP-EB2c, YFP-EB2c, CFP-EB3c, and YFP-EB3c). The oligomerization state of the EBc domains in solution was assessed by SEC-SLS measurements. The apparent molecular masses for the CFP-tagged EB1c, EB2c, and EB3c domains were 76.9, 79.2, and 77.8 kDa, respectively, which is consistent with the formation of dimers (calculated molecular masses for the monomers: CFP-EB1c = 38.4 kDa, CFP-EB2c = 39.6 kDa, CFP-EB3c = 38.9 kDa). This data demonstrate that the N-terminal fluorescent protein tags do not interfere with the formation of EBc homodimers.
FIGURE 1.
Heterodimerization of EBc domains. A, a schematic diagram shows FRET of dimeric EBc domains that are N-terminal-tagged with fluorescence proteins. CFP absorbs light at 434 nm and emits light at 477 nm. YFP absorbs light at 514 nm and emits light at 527 nm. When brought in close proximity, for example, through the coiled-coil domains of EBc dimers, energy absorbed by CFP is transferred to YFP through nonradiative FRET and emitted by YFP. B, shown are fluorescence emission spectra obtained by mixing equimolar amounts of CFP-EB1c and YFP-EB1c (10 μm each) at 37 °C and recorded at 60-min time intervals with excitation at 434 nm. Red and green indicate spectra obtained after 0- and 600-min incubation times, respectively. Note that the fluorescence emission signal at 527 nm observed at time 0 is due to background FRET in the sample (supplemental Fig. 3). a.u., arbitrary units. C, shown is time-dependent increase of the fluorescence signal at 527 nm (excitation at 434 m) after mixing equimolar amounts (10 μm each) of CFP-EB1c and YFP-EB1c (1c), CFP-EB2c and YFP-EB2c (2c), CFP-EB3c and YFP-EB3c (3c), CFP-EB1c and YFP-EB2c (1c/2c), CFP-EB1c and YFP-EB3c (1c/3c), CFP-EB2c and YFP-EB3c (2c/3c), and CFP-EB1c(I224A) and YFP-EB1c(I224A) (1c(I224A)) at 37 °C. D, shown is titration of 10 μm CFP-EB1c with increasing amounts of YFP-EB3c. The data show the loss of intensity of the fluorescence signal at 477 nm due to the dequenching of CFP-EB1c homodimers because of EB1c·EB3c heterodimer formation. Samples were incubated at 37 °C for 16 h before each measurement.
Fig. 1B shows the result of a typical fluorescence experiment in which 10 μm CFP-EB1c was mixed with 10 μm YFP-EB1c and incubated for several hours at 37 °C (throughout, protein concentrations refer to monomer equivalents). The excitation wavelength was set at 434 nm (CFP maximum), and fluorescence spectra were recorded between 450 and 650 nm. The fluorescence emission spectra displayed a characteristic time-dependent transformation that reached an apparent equilibrium after about 360 min. The reduction of CFP donor emission at 477 nm with a concomitant increase in acceptor YFP emission at 527 nm is characteristic of FRET for this fluorescence pair (49). Similar spectral changes were also observed after mixing equimolar amounts of CFP-EB2c and YFP-EB2c or of CFP-EB3c and YFP-EB3c (not shown). In contrast, under the same experimental conditions a CFP-tagged monomeric mutant of EB1c in which Ile-224 was replaced by alanine (denoted EB1c(I224A) (13)) displayed no significant spectral changes after mixing and prolonged incubation at 37 °C with an equimolar amount of YFP-EB1c(I224A) (not shown). The monomeric state of EB1c(I224A) was confirmed by SEC-SLS experiments, which yielded a molecular mass of 38.4 kDa (calculated mass for the monomer, 37.3 kDa). These results provide a reference showing that the fluorescence assay can be used to monitor polypeptide chain exchange of dimeric EBc domains in solution, which occurred on a time scale of minutes to hours under the conditions investigated.
The increase in fluorescence emission at 527 nm was exploited to follow polypeptide chain exchange of EBc dimers as a function of time. Fig. 1C shows the kinetic profiles obtained after mixing equimolar amounts (10 μm each) of CFP-EB1c and YFP-EB1c, CFP-EB2c and YFP-EB2c, CFP-EB3c and YFP-EB3c, or CFP-EB1c(I224A) and YFP-EB1c(I224A) at 37 °C. The traces reveal that although both EB1c and EB2c homodimers exchange their chains on a similar time scale, EB3c homodimers do so much faster. The calculated half-time values, t½, were 81, 84, and 5 min for EB1c, EB2c, and EB3c, respectively. No significant increase in fluorescence emission was detected for the monomeric EB1c(I224A) mutant on the time scale of the experiment (Fig. 1C).
Next we tested the heterodimerization properties of the EBc domains. Mixing equimolar amounts (10 μm each) of CFP-EB1c and YFP-EB3c revealed an increase in fluorescence emission with a t½ = 84 min (Fig. 1C). Under the same experimental conditions only a small increase in fluorescence emission was obtained after mixing equimolar amounts of CFP-EB2c and YFP-EB3c, and no fluorescence signal was observed after mixing CFP-EB1c with YFP-EB2c. These findings suggest that significant heterodimerization occurs only between EB1c and EB3c chains, which is consistent with previous findings based on native PAGE analysis (9).
The ∼2-fold larger fluorescence signal increase observed after mixing an equimolar amount of YFP-EB3c with CFP-EB1c compared with the analogous experiments performed with the homodimers indicates that a larger fraction of the chains form heterodimers than homodimers (Fig. 1C). It, thus, suggests that the EB1c·EB3c heterodimer is significantly more stable than the homodimers. We estimated the fraction of EB1c·EB3c heterodimers formed at various molar ratios of EB1c and EB3c by titrating a 10 μm solution of CFP-EB1c with increasing amounts of YFP-EB3c and measuring the reduction of CFP donor emission at 477 nm after incubating for 16 h at 37 °C. The data in Fig. 1D show that at a 1:1 molar ratio, about 80% of the EBc polypeptide chains form EB1c·EB3c heterodimers. At YFP-EB3c:CFP-EB1c molar ratios above 2, nearly all CFP-EB1c chains are bound as heterodimers (Fig. 1D).
Overall, these experiments demonstrate that the C-terminal domains of human EB proteins readily exchange their chains in solution. They further show that EB1c and EB3c preferentially form heterodimers, whereas EB2c does not significantly participate in heterotypic complexes with its homologues under the conditions investigated.
Thermodynamic and Kinetic Analysis of EBc Domains
In view of the pronounced sequence identity/similarity of 47/65, 60/73, and 50/72% between human EB1c and EB2c, EB1c and EB3c, and EB2c and EB3c, respectively, it came as a surprise that human EBc domains behave differently with respect to their ability to form homo- and heterodimers (Fig. 1C). In search of a rationale for these differences, we determined the thermodynamic stabilities, ΔGU, of EB1c, EB2c, and EB3c, and assessed the rate constants for refolding (kf) and unfolding (ku) by stopped-flow experiments.
For the thermodynamic and kinetic experiments the molar per-residue ellipticity at 222 or 225 nm was used to probe the conformational state of the proteins. Fig. 2A shows the results from urea-induced unfolding at 25 °C with 40 μm protein. For all three proteins, the unfolding reaction can be well described assuming a two-state equilibrium between folded dimer and unfolded monomer (36). The transition midpoints, [urea]½, differ significantly (supplemental Table 1), whereas the steepness of the transitions is comparable, indicating differences in stability with similar cooperativity. The linear extrapolation method adapted to a monomer-dimer equilibrium was used to calculate ΔGU and its dependence on the denaturant concentration. EB2c (ΔGU = 65 kJ mol−1) is slightly more stable than EB1c (ΔGU = 60 kJ mol−1), whereas EB3c (ΔGU = 49 kJ mol−1) is significantly less stable (supplemental Table 1). EB1c, EB2c, and EB3c also undergo reversible thermal unfolding (Fig. 2B). The apparent midpoints of denaturation, Tm, differ and follow the same order as the midpoints of urea denaturation: EB2c > EB1c > EB3c (supplemental Table 1).
FIGURE 2.
Thermodynamic and kinetic properties of EBc domains. A, shown is isothermal urea-induced unfolding at 25 °C with 40 μm protein. Experimental data are represented by circles. Continuous lines show the best fits obtained assuming a monomer-dimer equilibrium model. B, shown is thermal unfolding with 50 μm protein. This is the same presentation as in A. C, urea dependence of the rate constants for refolding (solid symbols) and unfolding (open symbols) at 25 °C was measured by stopped-flow CD experiments. The rate constants under native conditions were calculated by extrapolation of the linear regression lines to 0 m urea. In all panels the data are color-coded: green, EB1c (1c); blue, EB2c (2c); red, EB3c (3c); brown, EB3c(G241R,G244D). The data shown correspond to the mean residue ellipticities at 222 nm (A and B) or at 225 nm (C).
The rate of folding (unfolding) of the three EBc proteins was studied by following the time course of secondary structure formation (disruption) after rapid dilution from (into) urea at 25 °C. All unfolding traces could be fitted as single exponential reactions (not shown). The refolding traces were fitted by a model assuming bimolecular formation of dimers from monomers (not shown). Both the refolding and unfolding rate constants exponentially depend on the urea concentration in the range studied. The Chevron-type plots of the data are shown in Fig. 2C, and the rate constants extrapolated to 0 m urea are listed in supplemental Table 1. The refolding constants determined for EB1c (kf = 1.5 × 105 m−1 s−1) and EB3c (kf = 1.3 × 105 m−1 s−1) are identical within the precision of this approach and are 7–8 times lower than that of EB2c (kf = 9.9 × 105 m−1 s−1). EB2c and EB3c unfold with comparable rates (ku = 0.019 and 0.013 s−1, respectively) and are 30–40 times faster than EB1c (ku = 0.0005 s−1; supplemental Table 1). These data reveal that under the conditions investigated, EB1c is kinetically the most stable protein.
The thermodynamic data suggest that EB3c is significantly less stable than EB1c and EB2c despite the high sequence similarity of the core residues (Fig. 3A). Visual inspection of the interactions involved at the protein-protein interface of an EB3c dimer model (see below) did not reveal any peculiarities that might explain the decreased stability of EB3c. However, it is noteworthy that the two glycine residues, 241 and 244 (residue numbering is according to the sequence alignment with human EB1c throughout), occur in the α2 helix of EB3c and are not located at the predicted interchain interface. Because glycines are well known to destabilize α-helices (50), we replaced Gly-241 and Gly-244 in EB3c simultaneously by arginine and aspartate, which corresponds to the residues in human EB1c and EB2c (Fig. 3A, EB3c(G241R,G244D)). The shape of the thermal unfolding profile as well as the Tm of 65.1 °C obtained for EB3c(G241R,G244D) are very similar to that obtained for EB1c (Fig. 2B). Concomitant with the increase in Tm of 11.1 °C for EB3c(G241R,G244D) when compared with EB3c, the t½ value of chain exchange increased by a factor of 51 to 255 min (not shown). These findings show that Gly-241 and Gly-244 contribute significantly to the decreased stability and faster rate of chain exchange of EB3c when compared with EB1c and EB2c.
Model Building
To extend our thermodynamic and kinetic studies, we generated molecular models for the presently studied dimers based on the crystal structure of the human EB1c homodimer (see “Experimental Procedure”). Analysis of a sequence alignment of mammalian EB1c, EB2c and EB3c domains (Fig. 3A) coupled with visual inspection and stereochemical analysis of the interchain interfaces of the models did not provide clues to rationalize why EB1c preferentially forms heterodimers with EB3c. However, in the case of EB2c, which does not significantly form heterodimers with EB1c and EB3c, the models allowed us to identify two non-conserved dipeptide segments of interest, i.e. Thr-206—Val-207 and Val-238—Leu-239 (Fig. 3A). These two dipeptide segments display favorable interchain interactions in the x-ray structure of the EB1c homodimer and in the modeled EB1c·EB3c heterodimer (Fig. 3, B and C), but they exhibit unfavorable packing in the EB2c·EB1c/EB2c·EB3c heterodimers (Fig. 3, D and E). Fig. 3D shows that in the EB2c·EB1c/EB2c·EB3c interfaces, Ala-206 of EB2c is too short for an optimal van der Waals contact with Val-207 of EB1c/EB3c, whereas Leu-207 of EB2c is too bulky and sterically clashes with Thr-206 of EB1c/EB3c. In Fig. 3E, Leu-238 of EB2c forces Leu/Ile-239 of EB1c/EB3c to adopt unusual rotamer states, whereas Val-239 of EB2c is too short for an optimal interaction with Val-238 of EB1c/EB3c. By comparison, optimal packing is represented in Fig. 3E by the similarly positioned Val-238 and Leu-239 residues in the EB1c homodimer.
To test whether the two non-conserved dipeptide segments in the EBc domains are responsible for preventing heterodimer formation by EB2c, we generated two EB2c mutants and tested their ability to form heterodimers with EB3c using our fluorescence assay. In the first mutant we replaced Leu-238 and Val-239 of EB2c by the corresponding EB1c residues valine and leucine, respectively (YFP-EB2c(L238V,V239L)). As shown in Fig. 3F, at equilibrium an equimolar mixture (10 μm each) of CFP-EB3c with YFP-EB2c(L238V,V239L) displayed a stronger fluorescence emission at 527 nm when compared with an equimolar mixture of CFP-EB3c with YFP-EB2c, which is indicative of heterodimer formation. The fluorescence intensity was in between the signals obtained for a sample containing 10 μm each of CFP-EB3c and YFP-EB2c (no chain exchange) and a sample containing 10 μm each of CFP-EB3c and YFP-EB3c (Fig. 3F), suggesting that the homodimers are favored over the heterodimer. In a second mutant, Ala-206 and Leu-207 of YFP-EB2c(L238V,V239L) were substituted with the corresponding EB1c residues threonine and valine, respectively (YFP-EB2c(A206T,L207V,L238V,V239L)). When a 10 μm concentration of this mutant was incubated with 10 μm CFP-EB3c, it displayed a fluorescence emission comparable with the one obtained for 10 μm concentration each of CFP-EB3c and YFP-EB3c, suggesting that the homo- and heterodimers are equally stable (Fig. 3F). These findings indicate that multiple elements dispersed along the coiled coil and four-helix bundle contribute to homo- versus heterotypic chain selection.
In combination with the thermodynamic and kinetic data (see above), these findings show that despite their high sequence similarities, mammalian EBc domains exhibit different biophysical properties and suggest that substitutions within the EBH domain contribute to the differential stabilities and rate of chain exchange of the EBc variants. They further show that variable packing interactions at the dimer interface determine whether EBc domains preferentially form homo- or heterodimers.
Effects of EB1c Binding Partners on Chain Exchange
To investigate the possible impact of binding partners on chain exchange of EBc domains, we executed our fluorescence assay in the presence of several different +TIP domains. The first set of experiments was performed with CAP-Gly domains, which target the C terminus of EB proteins. Prominent members of CAP-Gly +TIPs are CLIP-170 and p150glued. We recombinantly expressed and purified the second CAP-Gly domain of CLIP-170 (ClipCG2), the CAP-Gly domain of p150glued (p150CG), and a mutant of p150CG in which Ala-49 was mutated to methionine (p150CG(A49M)). The affinities of the three CAP-Gly domains for EB1c have been determined previously by isothermal titration calorimetry (15, 33). The dissociation constants, Kd, were 29.7 μm for ClipCG2, 2.7 μm for p150CG, and 0.17 μm for p150CG(A49M), with the stoichiometries for all complexes corresponding to 2 mol of CAP-Gly domain/1 mol of EB1c dimer. CFP-EB1c and YFP-EB1c (10 μm each) were mixed separately with different CAP-Gly domains, and the increase in fluorescence emission at 527 nm (excitation at 434 nm) was monitored at 37 °C during an incubation time of 600 min. The amount of CAP-Gly domain added was adjusted such that 95% of the EB1c molecules are complexed.
Fig. 4A documents that ClipCG2 increased the t½ of chain exchange of EB1c about 2-fold. In contrast, p150CG caused a 10–20-fold increase in t½ of chain exchange and for the duration of the experiment of 600 min, p150CG(A49M) suppressed chain exchange completely. The differences in potency of the various CAP-Gly domains can be explained based on their binding mechanisms. ClipCG2 primarily targets the C-terminal EEY tripeptide of EB1c (Fig. 1A) using its highly conserved GKNDG motif (33). Binding to the two C-terminal tail regions of EB1c, which are flexibly disordered in solution (14), is not expected to interfere significantly with EB1c chain exchange, consistent with the experiment (Fig. 4A). In contrast, each p150CG molecule uses its GKNDG motif to bind the C-terminal EEY tripeptide of one EB1c dimer and its β2-β3 loop to interact with the hydrophobic cavity and the polar rim of the EBH domain formed by highly conserved residues stemming from both monomers of the dimer (Fig. 1A) (33). The simultaneous binding of p150CG to both chains of EB1c explains the inhibition of chain exchange. This effect is enhanced for p150CG(A49M), which displays a 15-fold higher affinity for EB1c when compared with the wild-type domain. The increased affinity can be rationalized by the improved packing of the methionine side chain against the hydrophobic cavity of the EBH domain when compared with the alanine present in the wild-type CAP-Gly domain (33).
Another prominent +TIP member that binds to the C terminus of EB1 is APC. A fragmentation study revealed that a 39-residue polypeptide segment derived from the C terminus of APC is sufficient for binding to EB1c with a Kd of 5.1 μm (12, 14, 18). A peptide encompassing the EB1-binding site of APC (denoted APCp1) completely inhibited EB1c chain exchange on the time scale of the experiment (Fig. 4A). Structural analysis of the EB1c·APCp1 complex revealed that APCp1 tightly binds with its SXIP motif to the interface formed between the two EB1c monomers (14), which is consistent with its strong inhibitory effect on chain exchange. Compared with p150CG, which displays a comparable affinity for EB1c as APCp1, the binding mechanism used by APCp1 appears more efficient in suppressing chain exchange (Fig. 4A). This difference may be explained by the observation that APCp1 interacts more extensively with the EBH domain than p150CG and possibly also because the binding of APCp1 induces structure in the C-terminal tail segment of EB1c (14, 33). Overall, these experiments show that chain exchange of mammalian EBc domains can be differentially controlled by binding partners.
NMR Analysis of the EB1c·APCp1 Complex
We characterized the influence of APCp1 on the stability of the EB1c homodimer in more detail using amide hydrogen exchange monitored by NMR. In this method the uniformly 15N-labeled protein is lyophilized from an 1H2O buffer and then redissolved in 2H2O. Two-dimensional 1H,15N correlation NMR experiments recorded with 1H2O and 2H2O solutions of the protein then reveal the extent of 1HN → 2HN exchange, as 2HN signals do not contribute to the signal intensities. Within the dead time of about 10 min for the presently used 37 °C exchange experiment, a large number of amide protons of EB1c have exchanged completely and are no longer detected in the first spectrum of the exchange series (compare supplemental Fig. 1, A and C), so that we can only estimate an upper bound on the protection factors (see “Experimental Procedure”). These rapidly exchanging residues are in positions 191–208 of the N-terminal part of the coiled coil, 231–241, of the loop connecting the helices α1 and α2, and 247–268 of the flexible C-terminal tail of EB1c (Fig. 4B). A second set of amide group signals was visible in the first exchange spectrum (supplemental Fig. 1C), and fits to exponential decays yielded exchange rates of 0.05–0.14 min−1, with corresponding protection factors of 1,600–57,000 (log p = 3.2–4.7); these signals are assigned to residues 209–230 and 242–246, which form the core of the highly conserved EBH domain. These data indicate that the EBH domain is the most stable part of EBc, explaining why even conservative substitutions in this part of the protein can have a strong impact on stability as seen, for example, for the monomeric single-site mutant EB1c(I224A) (see above).
When the amide proton exchange experiment was performed in the presence of an equimolar amount of APCp1 and otherwise identical experimental conditions, the residues 191–202, 231–241, and 249–268 were again completely exchanged within 10 min (supplemental Fig. 1, B and D), whereas the residues 203–230 and 242–248 showed a large reduction of the exchange rates to 0.00015–0.049 min−1, with corresponding protection factors of 2700–3.2·106 (log p = 3.4–6.5). Two regions show a plateau with high protection factors, i.e. residues 212–228 of the helix α1 and residues 242–246 of the helix α2 (Fig. 4B). These helical segments form the core of the EBH domain structure (Fig. 4C). Both of these regions show a significant increase in protection of 1.5–2 orders of magnitude in the presence of APCp1 (Fig. 4, B and C). Although both helices show a large increase in protection, helix α2 stands out because its residues show a uniform set of protection factors in the complex, suggesting that the exchange of all these amide protons is governed by the same molecular process. Possibly this feature in the exchange profile reflects a separation of helix α2 from the pair of α1 helices, which could represent a first step in the release of APCp1 that would precede monomer exchange. We found that the glycine residues at positions 241 and 244 in EB3c account for most of the difference in stability and rate of chain exchange between EB1c and EB3c (see above). Both of these substitutions introduce a residue that is unfavorable for the formation of α-helices, suggesting that chain exchange of EB3c may be facilitated by a destabilizing effect of these substitutions in helix α2.
The amide proton exchange data of EB1c obtained in the presence and absence of APCp1 show that the intramolecular hydrogen bond network is stabilized by the bound peptide. Because this network includes residues from the entire EBH domain (Fig. 4C), it appears likely that the increase of protection indicates enhanced stability of the dimer, so that a smaller fraction of monomer would be available for chain exchange when APCp1 is bound. The largest increase in protection factors upon binding APCp1 is at the central interface between the two monomers formed by the helices α1 and α2. The implicated stabilization of this interface region in the EB1c·APCp1 complex is consistent with the greatly reduced extent of chain exchange observed in the fluorescence assay (Fig. 4A) and may explain how APCp1 achieves efficient suppression of chain exchange despite its comparably modest affinity for EB1c. Overall, these findings indicate that suppression of chain exchange of EB1c by APCp1 is accomplished primarily by preventing the release of helix α2 from the EBH domain.
Dimerization Properties of Full-length EB1 and EB3 in Vitro
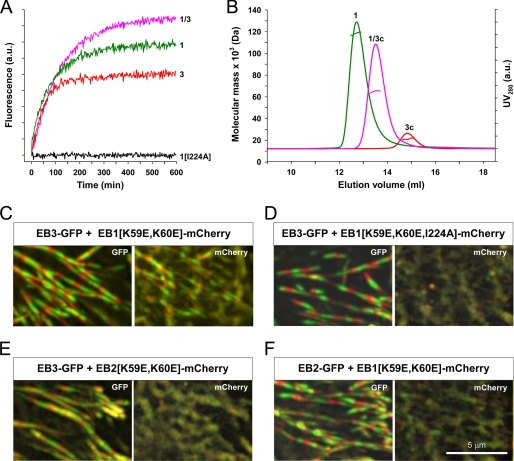
To extend the studies of chain exchange on the C-terminal domains of EB1 and EB3 (Fig. 1C) to the full-length proteins, we individually fused CFP and YFP to the C termini of EB1 and EB3 (referred to as EB1-CFP, EB1-YFP, EB3-CFP, and EB3-YFP) and assessed chain exchange with the same fluorescence assay as for the EBc domains. The oligomerization state of EB1-CFP in solution was found to be dimeric by SEC-SLS measurements, which yielded a molecular mass of 118.2 kDa (calculated molecular mass for the monomer, 58.3 kDa). This finding demonstrates that C-terminal tagging of full-length EBs with a fluorescent protein does not interfere with formation of the native dimeric structure.
Upon mixing equimolar amounts (10 μm each) of EB1-CFP with EB1-YFP or EB3-CFP with EB3-YFP at 37 °C we observed spontaneous chain exchange of both homodimers, as evidenced by a time-dependent increase of fluorescence emission at 527 nm (excitation at 434 nm) (Fig. 5A). The significantly stronger signal obtained at equilibrium for EB1 when compared with EB3 could be due to the four-residue-shorter C-terminal tails of the EB1 dimer, which reduces the average distance between the two fluorescent protein pairs. Evaluation of the kinetic profiles yielded t½ values of 63 and 38 min for EB1 and EB3, respectively. In contrast, no significant increase in fluorescence was detected for the monomeric fluorescent protein-tagged EB1 mutant, where Ile-224 was replaced by alanine (denoted EB1(I224A)-CFP and EB1(I224A)-YFP). The monomeric state of EB1(I224A)-CFP was confirmed by SEC-SLS experiments, which yielded a molecular mass of 60.0 kDa (calculated molecular mass for the monomer, 58.3 kDa). The t½ value for chain exchange of EB1 (63 min) is comparable with the one obtained for EB1c (81 min); the significant increase of the t½ value for EB3 (38 min) compared with EB3c (5 min) indicates that the full-length EB3 dimer is stabilized relative to the dimeric EB3c fragment.
FIGURE 5.
Heterodimerization of full-length EB1 and EB3. A, shown is the time-dependent increase of the fluorescence signal at 527 nm (excitation at 434 m) after mixing equimolar amounts (10 μm each) of EB1-CFP and EB1-YFP (1), EB3-CFP and EB3-YFP (3), EB1-CFP and EB3-YFP (1/3), and EB1(I224A)-CFP and EB1(I224A)-YFP (1(I224A)) incubated at 37 °C. B, shown are SLS-SEC experiments of EB1-CFP (1), EB3c (3c), and a mixture of EB1-CFP and EB3c (1/3c) that has been incubated for 16 h at 37 °C. Molecular mass determination (colored horizontal lines located below the maximum of each peak) yielded values of 118.2 kDa for EB1-CFP, 18.2 kDa for EB3c, and 68.0 kDa for EB1-CFP/EB3c. a.u., arbitrary units. C–F, heterodimerization of full-length EB proteins in cells is shown. Chinese hamster ovary cells were co-transfected with short hairpin RNA plasmids to deplete endogenous EB1 and EB3 and co-transfected with the indicated combination of fluorescent protein-tagged EBs. GFP and mCherry fluorescence were imaged simultaneously using total internal reflection fluorescence (TIRF) microscopy. Images were obtained at 0.5-s intervals, and 5 consecutive frames were averaged. Each panel shows a maximum intensity projection of 20 averaged frames, where each odd frame is shown in green, and each even frame is in red. Growing microtubule ends appear in this representation as rows of alternating green and red dashes. Note that EB3-GFP and EB2-GFP display robust plus-end tracking. EB1(K59E,K60E)-mCherry is recruited to plus ends by EB3-GFP (C). The I224A mutation, which interferes with dimerization of EB1(K59E,K60E,I224A)-mCherry, abrogates recruitment by EB3-GFP (D). EB3-GFP does not recruit EB2(K59E,K60E)-mCherry (E). In contrast to EB3-GFP(C), EB2-GFP fails to recruit EB1(K59E,K60E)-mCherry to growing microtubule ends (F).
Mixing equimolar amounts (10 μm each) of EB1-CFP and EB3-YFP also revealed a time-dependent increase in fluorescence emission that is indicative of heterodimer formation with t½ = 83 min (Fig. 5A). The ability of full-length EB1 and EB3 to form heterotypic complexes is consistent with previous findings based on immunoprecipitation experiments (9). A stronger fluorescence signal at steady state was observed after mixing EB1-CFP with EB3-YFP than in the solutions of the homodimers, indicating that similar to the EBc domains, the heterodimer of the full-length proteins is more stable than the homodimers.
As mentioned in the introduction, C-terminal domains of EBs are frequently used in cellular studies as dominant negative mutants that interfere with the activity of endogenous EB proteins. To probe the ability of EB3c to form heterodimers with full-length EB1, we performed SEC-SLS experiments. As shown in Fig. 5B, EB1-CFP and EB3c applied separately onto a SEC column eluted as single peaks at 12.8 and 14.9 ml, respectively, and the molecular masses determined for both proteins were consistent with the formation of dimers (Fig. 5B). Injection of an equimolar mixture of EB1-CFP and EB3c (10 μm each), which had been preincubated for 16 h at 37 °C, revealed a single peak that displayed an increase in elution volume of 0.8 ml with respect to EB1-CFP (Fig. 5B). Molecular mass determination by SEC-SLS yielded a value of 68.0 kDa, which corresponds to an EB1-CFP·EB3c heterodimer (the calculated molecular mass is 58.3 (EB1-CFP monomer) + 9.7 (EB3c monomer) = 68.0 kDa).
Collectively, these data show that just as for the C-terminal domains of EB1 and EB3, the full-length proteins also readily exchange their chains in solution. They further reveal that EB1 forms heterodimers with EB3, and that a single polypeptide chain of EB3c can displace an EB1 monomer in the full-length EB1 dimer.
Dimerization Properties of Full-length EBs in Cells
Here we extend the in vitro studies by investigations of the ability of full-length EBs to heterodimerize in cells. We have previously shown that in an EB1/EB3 knockdown background, a monomeric CH domain construct of EB3 can, although weakly, autonomously track growing microtubule plus-ends in cells (9). Therefore, if EB1 and EB3 do efficiently heterodimerize in cells, then EB3 should be able to target an EB1 version with a mutated CH domain, which is incapable of plus-end tracking on its own, to growing microtubule ends. Lys-59 and Lys-60 of EB1 were shown to be essential for microtubule binding (51), and mutation of these two residues to glutamic acids completely abolished plus-end tracking of the EB1 C terminal tagged with the red fluorescent protein mCherry (EB1(K59E,K60E)-mCherry) in an EB1/EB3 knockdown background (supplemental Fig. 2). However, EB1(K59E,K60E)-mCherry was recruited to the growing microtubule plus ends when co-expressed together with EB3-GFP in cells depleted in EB1 and EB3 (Fig. 5C; supplemental Movie 1). As a control, we used the monomeric version of EB1(K59E,K60E)-mCherry containing the I224A mutation (EB1(K59E,K60E,I224A)-mCherry), and as expected, this protein was not recruited to the growing microtubule ends upon coexpression with EB3-GFP (Fig. 5D; supplemental Movie 2). Similarly, EB2(K59E,K60E)-mCherry and EB1(K59E,K60E)-mCherry were not relocalized to growing microtubule ends by EB3-GFP (Fig. 5E; supplemental Movie 3) and EB2-GFP (Fig. 5F; supplemental Movie 4), respectively, which is in agreement with lack of significant heterodimerization between EB2c and the other two EBc domains in vitro (Fig. 1C).
The combined results of the above experiments support the view that in cells, full-length EB1 heterodimerizes with EB3 but not with EB2. It should be noted, however, that we previously observed a dominant negative effect on the endogenous EB2 pool when the EB1c domain was overexpressed in cells (9). A possible explanation of this observation could be that under overexpression conditions, EB1c is potent enough to sequester the much lower cellular levels of EB2.
Conclusions
This study provides a detailed description of the dimerization properties of the three human EB proteins, which is controlled by their C-terminal dimerization domains. EB dimers were found to readily exchange their chains on a time scale of minutes to hours. The data reveal that EB1 preferentially heterodimerizes with EB3, whereas EB2 does not significantly form heterotypic complexes. They further suggest that chain exchange of EBs can be differentially suppressed by +TIP binding partners.
Our findings have important functional implications. Based on the present data, we anticipate that in cells expressing lower levels of EB3 than of EB1, a large fraction of the EB3 pool will be tied up into EB1·EB3 heterodimers. Heterotypic complex formation between EB1 and EB3, thus, generates an additional EB variant which is expected to display yet a different functional profile when compared with its homotypic counterparts. Our findings also suggest that EBs are not present in separate pools but, rather, form a common pool undergoing continuous exchange within the cytoplasm. A consequence of this consideration is that in cells that co-express different EB species, their functions cannot be contemplated and analyzed separately from each other. Our observation that +TIP binding partners suppress chain exchange further indicates that this stabilization mechanism could have regulatory consequences for EBs, for example, in their spatial and temporal degradation (21). The dimerization of EBs is necessary to preserve their function of promoting persistent microtubule growth by suppressing catastrophes (9). Modulation of dimerization of EBs is, thus, an additional route to regulation of microtubule dynamics and function, an aspect that adds exciting prospects for future studies addressing the molecular mechanisms of EBs.
An important finding of our study is that the C-terminal domain of EB3 can interact with full-length EB1, resulting in a truncated heterodimer with only one CH domain. Transient overexpression of EB1 or EB3 C-terminal domains has been used as a tool in many studies with the intent to interfere with the binding of +TIPs with endogenous full-length EBs in different cell types (9, 20, 25–32). We recently showed that a very similar dominant negative effect is also obtained with an EB1 C-terminal domain lacking the tail region, which is essential for partner binding (9), indicating that sequestration of +TIP binding partners is not the prevailing mode of interference. The analysis presented here strongly suggests that the primary mechanism of the dominant negative effect of coexpressed C-terminal EB domains is to knock down all EB species by heterodimerization with the endogenous full-length proteins. This results in a strong reduction in the localization of EBs at microtubule plus ends due to the presence of only one functional CH domain in EBc·EB heterodimers.
Acknowledgments
We are grateful to Drs. I. Smal and E. Meijering for generating the ImageJ plugin AColor, to Dr. J. Missimer for help with the data analysis, and to Dr. I. Vakonakis for careful reading of the manuscript.
This work was supported by the Swiss National Science Foundation (to M. O. S. and I. J.), by Netherlands Organization for Scientific Research NWO-ALW-VICI and ZonMW-TOP grants (to A. A.), by a Federation of European Biochemical Societies fellowship (to R. M. B.), and by the Swiss National Science Foundation and the ETH Zürich through the NCCR Structural Biology (to I. J. and K. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. 1–3, and Movies 1–4.
- +TIP
- microtubule plus-end tracking protein
- EB
- end-binding protein
- CH
- calponin homology
- EBH
- EB homology
- EBc
- C-terminal domain of EB
- APC
- adenomatous polyposis coli
- HPLC
- high performance liquid chromatography
- SEC
- size exclusion chromatography
- FRET
- Förster resonance energy transfer
- CFP
- cyan fluorescent proteins
- ECFP
- enhanced CFP
- YFP
- yellow fluorescent proteins
- EYFP
- enhanced YFP
- SLS
- static light scattering
- CAP-Gly
- cytoskeleton-associated protein-glycine-rich
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PBS
- phosphate-buffered saline
- DTT
- dithiothreitol
- GFP
- green fluorescent protein.
REFERENCES
- 1.Desai A., Mitchison T. J. (1997) Annu. Rev. Cell Dev. Biol. 13, 83–117 [DOI] [PubMed] [Google Scholar]
- 2.Howard J., Hyman A. A. (2003) Nature 422, 753–758 [DOI] [PubMed] [Google Scholar]
- 3.Schuyler S. C., Pellman D. (2001) Cell 105, 421–424 [DOI] [PubMed] [Google Scholar]
- 4.Akhmanova A., Steinmetz M. O. (2008) Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 5.Tirnauer J. S., Bierer B. E. (2000) J. Cell Biol. 149, 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D., Surrey T. (2007) Nature 450, 1100–1105 [DOI] [PubMed] [Google Scholar]
- 7.Vitre B., Coquelle F. M., Heichette C., Garnier C., Chrétien D., Arnal I. (2008) Nat. Cell Biol. 10, 415–421 [DOI] [PubMed] [Google Scholar]
- 8.Bieling P., Kandels-Lewis S., Telley I. A., van Dijk J., Janke C., Surrey T. (2008) J. Cell Biol. 183, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komarova Y., De Groot C. O., Grigoriev I., Gouveia S. M., Munteanu E. L., Schober J. M., Honnappa S., Buey R. M., Hoogenraad C. C., Dogterom M., Borisy G. G., Steinmetz M. O., Akhmanova A. (2009) J. Cell Biol. 184, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandblad L., Busch K. E., Tittmann P., Gross H., Brunner D., Hoenger A. (2006) Cell 127, 1415–1424 [DOI] [PubMed] [Google Scholar]
- 11.Hayashi I., Ikura M. (2003) J. Biol. Chem. 278, 36430–36434 [DOI] [PubMed] [Google Scholar]
- 12.Honnappa S., John C. M., Kostrewa D., Winkler F. K., Steinmetz M. O. (2005) EMBO J. 24, 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slep K. C., Rogers S. L., Elliott S. L., Ohkura H., Kolodziej P. A., Vale R. D. (2005) J. Cell Biol. 168, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J., Buey R. M., Lawera A., Jelesarov I., Winkler F. K., Wüthrich K., Akhmanova A., Steinmetz M. O. (2009) Cell 138, 366–376 [DOI] [PubMed] [Google Scholar]
- 15.Weisbrich A., Honnappa S., Jaussi R., Okhrimenko O., Frey D., Jelesarov I., Akhmanova A., Steinmetz M. O. (2007) Nat. Struct. Mol. Biol. 14, 959–967 [DOI] [PubMed] [Google Scholar]
- 16.Steinmetz M. O., Akhmanova A. (2008) Trends Biochem. Sci. 33, 535–545 [DOI] [PubMed] [Google Scholar]
- 17.Su L. K., Qi Y. (2001) Genomics 71, 142–149 [DOI] [PubMed] [Google Scholar]
- 18.Bu W., Su L. K. (2003) J. Biol. Chem. 278, 49721–49731 [DOI] [PubMed] [Google Scholar]
- 19.Lee T., Langford K. J., Askham J. M., Brüning-Richardson A., Morrison E. E. (2008) Oncogene 27, 2494–2500 [DOI] [PubMed] [Google Scholar]
- 20.Geraldo S., Khanzada U. K., Parsons M., Chilton J. K., Gordon-Weeks P. R. (2008) Nat. Cell Biol. 10, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 21.Ban R., Matsuzaki H., Akashi T., Sakashita G., Taniguchi H., Park S. Y., Tanaka H., Furukawa K., Urano T. (2009) J. Biol. Chem. 284, 28367–28381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaworski J., Kapitein L. C., Gouveia S. M., Dortland B. R., Wulf P. S., Grigoriev I., Camera P., Spangler S. A., Di Stefano P., Demmers J., Krugers H., Defilippi P., Akhmanova A., Hoogenraad C. C. (2009) Neuron 61, 85–100 [DOI] [PubMed] [Google Scholar]
- 23.Straube A., Merdes A. (2007) Curr. Biol. 17, 1318–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa H., Koyama K., Murata Y., Morito M., Akiyama T., Nakamura Y. (2000) Oncogene 19, 210–216 [DOI] [PubMed] [Google Scholar]
- 25.Askham J. M., Vaughan K. T., Goodson H. V., Morrison E. E. (2002) Mol. Biol. Cell 13, 3627–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., Gundersen G. G. (2004) Nat. Cell Biol. 6, 820–830 [DOI] [PubMed] [Google Scholar]
- 27.Wang F., Zhang Q., Cao J., Huang Q., Zhu X. (2008) Exp. Cell Res. 314, 213–226 [DOI] [PubMed] [Google Scholar]
- 28.Schrøder J. M., Schneider L., Christensen S. T., Pedersen L. B. (2007) Curr. Biol. 17, 1134–1139 [DOI] [PubMed] [Google Scholar]
- 29.Zhou F. Q., Zhou J., Dedhar S., Wu Y. H., Snider W. D. (2004) Neuron 42, 897–912 [DOI] [PubMed] [Google Scholar]
- 30.Etienne-Manneville S., Manneville J. B., Nicholls S., Ferenczi M. A., Hall A. (2005) J. Cell Biol. 170, 895–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niethammer P., Kronja I., Kandels-Lewis S., Rybina S., Bastiaens P., Karsenti E. (2007) PLoS. Biol. 5, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kronja I., Kruljac-Letunic A., Caudron-Herger M., Bieling P., Karsenti E. (2009) Mol. Biol. Cell 20, 2684–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honnappa S., Okhrimenko O., Jaussi R., Jawhari H., Jelesarov I., Winkler F. K., Steinmetz M. O. (2006) Mol. Cell 23, 663–671 [DOI] [PubMed] [Google Scholar]
- 34.Geiser M., Cèbe R., Drewello D., Schmitz R. (2001) Biotechniques 31, 88–92 [DOI] [PubMed] [Google Scholar]
- 35.Honnappa S., Cutting B., Jahnke W., Seelig J., Steinmetz M. O. (2003) J. Biol. Chem. 278, 38926–38934 [DOI] [PubMed] [Google Scholar]
- 36.Dürr E., Jelesarov I., Bosshard H. R. (1999) Biochemistry 38, 870–880 [DOI] [PubMed] [Google Scholar]
- 37.Santoro M. M., Bolen D. W. (1988) Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 38.Fersht A. R. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme and Protein Folding, W. H. Freeman and Company, New York [Google Scholar]
- 39.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 40.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 41.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 42.Englander S. W., Downer N. W., Teitelbaum H. (1972) Annu. Rev. Biochem. 41, 903–924 [DOI] [PubMed] [Google Scholar]
- 43.Hvidt A., Nielsen S. O. (1966) Adv. Protein Chem. 21, 287–386 [DOI] [PubMed] [Google Scholar]
- 44.Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York [Google Scholar]
- 45.Wagner G., Wüthrich K. (1979) J. Mol. Biol. 134, 75–94 [DOI] [PubMed] [Google Scholar]
- 46.Bai Y., Milne J. S., Mayne L., Englander S. W. (1993) Proteins 17, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller R. (2004) The Computer-aided Resonance Assignment Tutorial, 1st Ed., Cantina Verlag, Goldau, Switzerland [Google Scholar]
- 48.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 49.Piston D. W., Kremers G. J. (2007) Trends Biochem. Sci. 32, 407–414 [DOI] [PubMed] [Google Scholar]
- 50.Chakrabartty A., Baldwin R. L. (1995) Adv. Protein Chem. 46, 141–176 [PubMed] [Google Scholar]
- 51.Slep K. C., Vale R. D. (2007) Mol. Cell 27, 976–991 [DOI] [PMC free article] [PubMed] [Google Scholar]