Abstract
Although the small GTPase Ran is best known for its roles in nucleocytoplasmic transport, mitotic spindle assembly, and nuclear envelope formation, recent studies have demonstrated the overexpression of Ran in multiple tumor types and that its expression is correlated with a poor patient prognosis, providing evidence for the importance of this GTPase in cell growth regulation. Here we show that Ran is subject to growth factor regulation by demonstrating that it is activated in a serum-dependent manner in human breast cancer cells and, in particular, in response to heregulin, a growth factor that activates the Neu/ErbB2 tyrosine kinase. The heregulin-dependent activation of Ran requires mTOR (mammalian target of rapamycin) and stimulates the capped RNA binding capability of the cap-binding complex in the nucleus, thus influencing gene expression at the level of mRNA processing. We further demonstrate that the excessive activation of Ran has important consequences for cell growth by showing that a novel, activated Ran mutant is sufficient to transform NIH-3T3 cells in an mTOR- and epidermal growth factor receptor-dependent manner and that Ran-transformed cells form tumors in mice.
Keywords: Diseases/Cancer/Transformation, G Proteins/Low Molecular Weight, Hormones/Growth Factors, RNA/Processing, Signal Transduction, Signal Transduction/Protein Kinases
Introduction
Ran is a unique member of the Ras superfamily of GTPases that utilizes a guanine nucleotide exchange factor (the chromatin-associated RCC1 protein), a GTPase-accelerating protein complex (RanGAP/RanBP1), and a single major class of effectors (the importins or karyopherins) to regulate the nucleocytoplasmic transport of various cargo as well as other nuclear functions (for review, see Ref. 1). In interphase cells, a Ran-GTP gradient is formed in response to the nuclear localization of RCC1 together with the cytoplasmic localization of RanGAP. As a result, Ran exists predominantly in an active, GTP-bound state in the nucleus where it is capable of engaging its primary biological effectors, the importins/karyopherins. GTP hydrolysis, catalyzed by RanGAP in the cytoplasm, then results in the dissociation of Ran-GDP from its effector proteins. The nucleocytoplasmic transport of a number of proteins is dependent upon the proper establishment of the Ran-GTP gradient, as best exemplified in the case of classical nuclear import (2). Protein cargo destined for the nucleus is identified by the presence of a nuclear localization sequence. The nuclear localization sequence is recognized by an adapter protein, importin-α, in the cytosol, and upon binding the cargo, importin-α engages importin-β to form a complete import complex that translocates to the nucleus. Within the nucleus, Ran-GTP binds to importin-β, causing it to dissociate from the import complex, which results in the subsequent release of cargo. Thus, the fact that Ran-GTP is predominantly a nuclear species is pivotal in the directional release of import cargo into the nucleus.
Similarly, it has been suggested that Ran plays an essential role in the directional release of capped RNAs in the cytoplasm by regulating the interactions that occur between the nuclear cap-binding complex (CBC)2 and the importins (3). The CBC (comprised of two subunits, CBP20 and CBP80) binds cotranscriptionally to the monomethylated guanosine cap structure (m7G(5′)ppp(5′)N) of RNA polymerase II-transcribed RNAs (4–8). The recognition of the cap by the CBC is important for the stability of these RNAs and to facilitate multiple aspects of RNA metabolism, including the coordinated splicing and export of mRNAs (4, 9–13). One of the protein-binding partners of the CBC is importin-α. The CBC-importin-α complex is able to bind capped RNA with high affinity, whereas the inclusion of importin-β into this complex greatly reduces the affinity of the CBC for capped RNA (3, 14). The interphase Ran-GTP gradient would favor the formation of a nuclear CBC-importin-α complex and a cytosolic CBC-importin-α-importin-β complex, thereby providing a mechanism to promote the directed release of CBC-associated RNAs into the cytoplasm (3, 14). In the case of mRNA, such regulation has especially important consequences, because the release of capped mRNA from the CBC in the cytosol is prerequisite for its subsequent binding to eIF-4E and translation into protein.
We have found that like the case for eIF-4E, the cap binding capability of the CBC is subject to growth factor regulation (15, 16). Moreover, recently we showed that the growth factor regulation of the cap binding capability of the CBC is dependent on its interactions with the importins (14). These findings, when considered together with the role played by Ran-GTP in regulating the interactions between the importins and the CBC, would suggest that the formation of the Ran-GTP gradient in cells is also subject to growth factor regulation. The possibility that Ran is a downstream signaling target of growth factors is especially intriguing, given the growing body of evidence that correlates Ran expression with tumorigenesis (for review, see Ref. 17).
Thus, in the present study, we set out to establish that Ran-GTP levels can be regulated by serum and in particular by the growth factor HRG, because we have shown it to be very effective at activating the CBC in cells (14–16). We used the human breast cancer SKBR3 cell line as a model system because it responds to both HRG and EGF, and it exhibits a specific HRG-stimulated activation of the CBC. Here we show that both serum and HRG treatment are in fact able to activate Ran in an mTOR-dependent manner in the nucleus of these breast cancer cells and that one functional consequence of Ran activation is an increase in capped RNA binding by the CBC. We further examined the consequences of increased Ran activation in cell growth control through the use of a novel, activated Ran mutant (Ran (F35A)). We show that this Ran mutant can induce the transformation of NIH-3T3 cells as read out by a number of assays and that the injection of Ran-transformed cells into mice caused tumor formation. Interestingly, we discovered that increasing the levels of active Ran in cells through the expression of Ran (F35A) triggers the rapamycin-sensitive activation of multiple signaling proteins including the EGFR, Ras, and ERK and that cellular transformation induced by Ran (F35A) was dependent on activated EGFRs. These findings suggest that a potentially important consequence of the activation of Ran and its effects on enhancing CBC-dependent RNA-processing/gene expression is ultimately the production of growth regulatory proteins and/or growth factors (i.e. EGF) that result in excessive signaling through the EGFR. When taken together, these findings highlight Ran as a small GTPase that, like the Ras and Rho proteins, comes under strict growth factor regulation, and the loss of this regulation can have important consequences for cell growth control and give rise to oncogenic transformation.
EXPERIMENTAL PROCEDURES
Materials
V5 and BrdUrd antibodies and Hoechst 33342 were obtained from Invitrogen. The Ran and RanBP1 antibodies were obtained from BD Transduction Laboratories. The anti-phosphotyrosine antibody, as well as antibodies against mTOR, p70 S6K, EGFR, PDGFR, RCC1, and Ras antibodies were from Upstate, as was glutathione S-transferase-Ras. The phospho-ERK (Thr202/Tyr204), phospho-S6K (Thr389), and ERK antibodies were from Cell Signaling. RanGAP1 antibody was from Novus. The His6 and hemagglutinin antibodies were from Covance, and the Oregon Green anti-mouse was from Molecular Probes. [α-32P]GTP was purchased from PerkinElmer Life Sciences. Rapamycin and AG1478 were from Calbiochem.
Cell Culture Conditions
SKBR3 cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum. Prior to growth factor treatment, the cells were switched to serum-free medium for 5 days. Growth factors, including 100 nm heregulin β1 (residues 177–244; a generous gift from Dr. Mark Sliwkowski, Genentech), 100 ng/ml EGF (Invitrogen), or 20% fetal bovine serum, were then added to the serum-free medium for the time periods indicated under “Results” at 37 °C. Following treatment, the growth factor-containing medium was removed, and the cells were washed twice with phosphate-buffered saline and then lysed (see below). Transient transfections were performed using Lipofectamine Plus (Invitrogen) according to the manufacturer's directions.
To generate stable cell lines, NIH-3T3 cells were transfected with the various pcDNA3.1-V5-Neo vectors, either alone or incorporating various Ran constructs, using the Lipofectamine transfection kit (Invitrogen). Transfected cells were maintained in DMEM supplemented with 5% calf serum and 750 μg/ml G418 (Invitrogen). After 10–14 days, G418-resistant colonies were selected and subcultured in DMEM supplemented with 10% calf serum and 750–875 μg/ml G418. The clones were then assessed for the expression of Ran.
BrdUrd Incorporation Assays
For analyses of S phase entry, subconfluent cells stably expressing Ran were plated in sterile 60-mm dishes. After 24 h, the cells were maintained in low serum (0.5% calf serum) for an additional 24 h, and then 2–4 × 104 cells were replated onto sterile chamber well slides in 10% calf serum and 200 μm BrdUrd. After 10 h, the cells were processed for immunofluorescence staining with anti-BrdUrd monoclonal antibody, anti-V5 polyclonal antibody, and Hoechst, as described previously (18). The percentage of BrdUrd-positive cells was determined for more than 400 cells from multiple fields for each experiment.
Cell Fractionation and Lysis, Immunoprecipitation, and Western Immunoblotting
Cell fractionation and lysis were performed as previously described (15). When preparing lysates for Ran pulldown assays, dithiothreitol and EDTA were omitted from all lysis buffers.
Immunoprecipitation of EGFRs and PDGFRs
Cell lines stably expressing Ran were serum-starved for 40–48 h in DMEM supplemented with 0.5% calf serum. The serum-starved cells were washed twice in ice-cold phosphate-buffered saline and lysed in MLB buffer (25 mm Hepes, pH 7.5, 150 mm NaCl, 1% Igepal CA-630, 10% glycerol, 25 mm NaF, 10 mm MgCl2, 1 mm Na3VO4, 10 μg/ml leupeptin, and 10 μg/ml aprotonin). The lysates were rotated for 1 h at 4 °C and then centrifuged at 13,200 × g for 10 min at 4 °C. One mg of lysate was then allowed to incubate overnight at 4 °C with an anti-EGFR or anti-PDGFR antibody. Following the primary antibody incubation, 60 μl of protein G-Sepharose beads (slurry) were added to each sample and incubated for 1 h at 4 °C. The samples were centrifuged, and the immunoprecipitated pellets were washed (three times) with MLB buffer. The samples were mixed with 5× Laemmli buffer and boiled. SDS-PAGE was performed using 7.5% acrylamide gels followed by transfer to nitrocellulose filters and Western blotting with anti-phosphotyrosine.
Photoaffinity Labeling of CBP20 with [α-32P]GTP
Photoaffinity labeling of cellular proteins with [α-32P]GTP was performed as described previously (15).
Immunofluorescence
For indirect immunofluorescence, NIH-3T3 cells were seeded at 4 × 104 cells/well in chamber well slides (Nalgene), grown in complete medium for 1 day, and then transiently transfected with V5-tagged Ran constructs. The cells were allowed to grow for an additional 24 h post-transfection and were then washed and processed as described previously (14). Ran expression was visualized using an anti-V5 antibody, and the nuclei were stained using Hoescht dye.
Ras Activation Assay
Activation of cellular Ras was assayed according to the manufacturer's protocol (Ras activation assay kit; Upstate). A Ras antibody (Upstate) was used for Western blotting.
Knockdown Experiments
RNA interference knockdown experiments were performed in SKBR3 cells. The knockdown of mTOR was performed using SignalSilence mTOR siRNA (Cell Signaling). SignalSilence Control siRNA was used as the negative control. The RNA interference oligonucleotides were transiently transfected in SKBR3 cells using Lipofectamine 2000, and the relative knockdown efficiency was determined using antibodies against mTOR. A pKD-p70 S6K construct (Upstate) producing shRNAs directed against human p70 S6K was transiently transfected with FuGENE 6 transfection reagent (Roche Applied Science). A pKD-NegCon-v1 construct (Upstate) producing negative control shRNA was used as the control siRNA expression plasmid. Relative knockdown efficiencies were analyzed by probing the transfected lysates with an antibody directed against p70 S6K.
Cellular Transformation
To determine the growth rates in low serum, 10 × 104 NIH-3T3 cells expressing vector, wild type Ran, or various Ran mutants were cultured in 6-well plates in DMEM supplemented with 10% calf serum. After 5 h, the medium was changed to DMEM supplemented with 0.5% calf serum, which was changed daily. At the indicated times, the cells were trypsinized and counted on a hemacytometer. The data represent the averages of three different experiments.
When assaying the ability of cells to grow to high density, the different cell lines of interest (10 × 104 cells) were cultured in 6-well plates in DMEM supplemented with 5% calf serum, which was replaced every other day. Saturation density was determined by counting the cells, as described above, 2 days after culture confluence (6–8 days total). The data are the averages of at least three independent experiments.
To determine the ability of the different Ran-expressing cell lines to grow in semi-solid agar, 10 × 104 cells from the different clones were mixed with DMEM supplemented with 10% calf serum and 0.3% agarose and plated on a solidified layer of DMEM supplemented with 0.5% agarose and 10% calf serum. The cells were fed weekly by adding 1 ml of DMEM supplemented with 10% calf serum and 0.3% agarose. After 2 weeks, colonies larger than 50 μm were scored under a microscope. The data are the averages of three independent experiments.
Six-week-old NIH-III nude male mice (Charles River Diagnostics) were used to assess the ability of NIH-3T3 cells expressing either wild type Ran or Ran (F35A) to form tumors. All of the study protocols were approved by the Institutional Animal Care and Use Committee of Cornell University and conform to the Guide for the Care and Use of Laboratory Animals from the United States National Institutes of Health. The cells were harvested and resuspended in DMEM and Matrigel (BD Biosciences) at 1 × 106 cells/ml. From this suspension, 200 μl were injected subcutaneously into the right and left flanks of the mouse. After 60 days, the mice were euthanized, and the tumors were excised and weighed. The tumors were then fixed with 10% formalin, and sections of 4 μm were stained with hematoxylin and eosin for routine histopathological examination by light microscopy.
Plasmids
Importin-β was cloned by PCR from untagged importin-β (a generous gift from Dr. Iain Mattaj, EMBL). The 5′- and 3′-primers were designed using the published sequence for human importin-β (GenBankTM accession number AAC41763). Restriction sites (5′ BamHI and 3′ NotI) were introduced to facilitate cloning. The importin-β gene was then amplified using 30 PCR cycles (1 min at 94 °C, 1 min at 58 °C, and 6 min at 72 °C). The PCR product was inserted into a cloning vector (pCR2.1) using a TA cloning kit (Invitrogen) and then subcloned into the Escherichia coli expression vector pet28a immediately downstream of the hexahistidine tag using the rapid DNA ligation kit (Roche Applied Science).
V5-WT Ran was cloned by PCR from pGEX-KG-Ran (a generous gift from Dr. Alfred Wittinghofer, Max-Planck Institut für Molekulare Physiology). The 5′- and 3′-primers were designed from the published sequence for human Ran (GenBankTM accession number NM_006325), and the Ran gene was amplified using PCR and inserted into the pcDNA 3.1D/V5/His-TOPO plasmid according to the manufacturer's directions (Invitrogen). The QuikChange site-directed mutagenesis kit (Stratagene) was used to generate the Ran (F35A) and Ran (T24N) mutants.
Ran Pulldown Assays
His6-importin-β was expressed in E. coli and affinity-purified on Nickel-chelating Sepharose beads. For the assay, the His6-importin-β proteins were reattached to the beads (15 μl/sample, ∼10 μg of fusion protein) at room temperature for 20 min, and the beads were washed twice with Ran pulldown buffer (equal volumes of cytoplasmic and nuclear lysis buffers). Nuclear lysates (precleared with nickel-chelating Sepharose beads) were adjusted to 500 μl with Ran pulldown buffer and then added to the beads. The lysates were incubated with the His6-importin-β-bound proteins at 4 °C with rocking for 1 h. The nickel-chelating Sepharose beads were pelleted, washed (three times) with 1 ml of Ran pulldown buffer plus 100 mm imidazole to eliminate background binding of nonspecific proteins, and then resuspended in 5× Laemmli buffer for analysis by SDS-PAGE and Western blotting.
Pre-mRNA Splicing Reactions
Splicing extracts were prepared from NIH-3T3 cells stably expressing Ran (F35A) or vector alone that were grown in 1% calf serum for 24 h. The splicing reactions were then carried out as described previously (15).
Quantitative Reverse Transcription-PCR
Total RNA was extracted utilizing the RNeasy kit (Qiagen) from cells that were serum-starved for 20–24 h. Complimentary DNA was synthesized using the SABiosciences RT2 first strand kit. Quantitative PCR was performed in triplicate using primer sets to amplify EGF, the gene of interest (SABiosciences RT2 PCR primer assay for mouse EGF with RT2 SYBR Green/ROX qPCR Master Mix) and mouse GAPDH, the normalizer/housekeeping gene. Controls (i.e. no reverse transcriptase and no template controls) were prepared to exclude the possibility of amplification coming from genomic DNA contamination. Thermal cycling was carried out on an ABI7500 fast real time PCR system starting with one cycle at 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The entire procedure was replicated (three times) on independent RNA isolations. Relative quantification studies were performed with the ABI7500 fast system sequence detection software, and the statistical significance was calculated using Student's t test.
RESULTS
Demonstration of the Growth Factor-dependent Activation of Ran
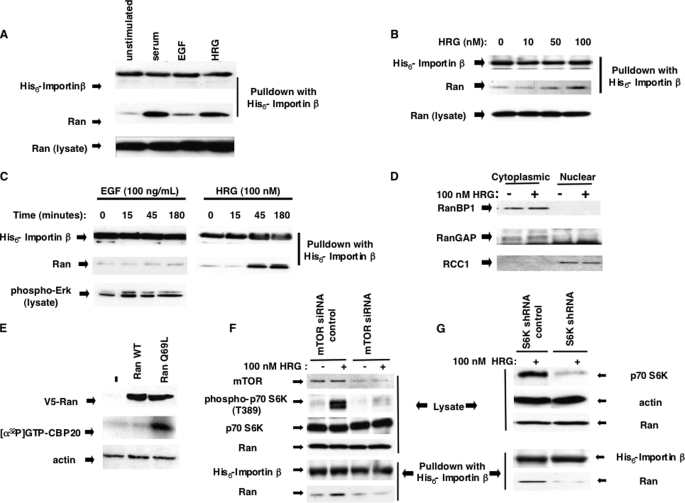
To determine whether Ran is subject to growth factor regulation, we took advantage of an assay that is capable of detecting changes in the activation status of Ran in cells. This involved the use of a His6-importin-β fusion protein that serves as a specific binding partner for activated (GTP-bound) forms of Ran. A similar assay has been used previously to read out Ran activation (19, 20) as well as to assess the activation of a variety of small GTPases including Ras and Rho family members (21, 22). We verified the validity of this assay system for Ran activation in a number of ways. First, we showed that His6-importin-β selectively precipitates a recombinant, constitutively active Ran mutant, Ran (Q69L), as compared with wild type Ran (RanWT) in either its GDP-bound or nucleotide-free form in vitro (supplemental Fig. S1A, bottom panel). Next, we treated nuclear extracts from SKBR3 cells with EDTA to strip guanine nucleotides from the GTPases present in these fractions and then reloaded them with either GDP or GTPγS. As shown in supplemental Fig. S1B (bottom panel), Ran was selectively precipitated by His6-importin-β from nuclear extracts loaded with GTPγS but not from extracts loaded with GDP. Finally, to ensure the integrity of Ran-GTP in the nuclear lysates during the course of the assay, we examined the ability of increasing amounts of recombinant Ran loaded with [γ-32P]GTP to be precipitated with His6-importin-β in the presence or absence of nuclear lysates. The top panel in supplemental Fig. S1C depicts an autoradiogram showing the input levels of Ran-[γ-32P]GTP, whereas the middle and bottom panels are autoradiograms of Ran-[γ-32P]GTP precipitated in the absence or presence of nuclear lysates, respectively. The equal precipitation of Ran- [γ-32P]GTP under these two conditions shows that Ran-GTP is not hydrolyzed during the course of these experiments.
We then examined whether serum-starved SKBR3 cells, a human breast cancer cell line that expresses high levels of EGFR family members when treated with serum or the growth factors HRG or EGF, showed increased cellular levels of Ran-GTP. Following 24 h of stimulation, cells were harvested and fractionated, and the nuclear fractions were assayed for Ran activation using recombinant His6-importin-β as a probe for Ran-GTP as described above. As shown in Fig. 1A (middle panel), Ran-GTP levels were increased significantly in response to treatment with either serum or HRG, whereas there was little change in the amount of activated Ran following the addition of EGF. The HRG-dependent activation of Ran was dose-dependent with maximal effects occurring at 100 nm HRG (Fig. 1B, middle panel) and peaked after 45 min of treatment (Fig. 1C, middle panel), whereas EGF was ineffective through 180 min despite giving rise to a clearly detectable activation of ERK (Fig. 1C, bottom panel). The changes in the levels of Ran-GTP were not a consequence of effects on Ran expression, because the cellular levels of Ran remained constant throughout the time period of the experiment (Fig. 1A, bottom panel). Similarly, these results were not due to changes in either the expression levels or the localization of the key Ran regulatory proteins, RCC1, RanGAP, or RanBP1 (Fig. 1D).
FIGURE 1.
Characterization of the stimulus-induced Ran activation. A, nuclear cell lysates were prepared from SKBR3 cells that were starved for 5 days in serum-free medium (lane 1) and subsequently stimulated in serum-free medium supplemented with either 20% serum (lane 2), 100 ng/ml EGF (lane 3) or 100 nm HRG (lane 4) for 24 h. Ran-GTP was assayed in a pulldown experiment with His6-importin-β and analyzed by immunoblotting with anti-His6 antibody (top panel) or anti-Ran antibody (middle panel) as indicated. The bottom panel indicates the relative levels of Ran present in the lysates. B, heregulin activates the Ran GTPase in a dose-dependent manner. SKBR3 cells were serum-starved and then stimulated for 24 h with different concentrations of HRG. The cells were then harvested, and nuclear lysates were assayed for Ran-GTP levels using the His6-importin-α pulldown assay (middle panel). His6-importin-β levels and endogenous Ran levels are shown in the top and bottom panels, respectively. C, a time course for Ran activation in response to either 100 ng/ml EGF or 100 nm HRG was performed using serum-starved SKBR3 cells. The cells were harvested after 15, 45, and 180 min of treatment, and nuclear lysates were generated and assayed for Ran-GTP using the His6-importin-β pulldown assay. Ran-GTP precipitated by His6-importin-β was identified by immunoblot analysis (middle panel), as was the level of His6-importin-β present on the beads (top panel). The efficacy of the EGF treatment was determined by detecting cytosolic levels of phospho-ERK by Western blotting (bottom left panel). D, the cellular localization of Ran regulators in response to HRG treatment was assessed by probing cytoplasmic and nuclear lysates for protein levels of RanBP1 (top panel), RanGAP (middle panel), and RCC1 (bottom panel). E, activation of the CBC by Ran-GTP. SKBR3 cells were transiently transfected with V5-RanWT (wild type) or V5-Ran (Q69L) for 24 h, followed by serum starvation for 5 days. The cells were lysed, and nuclear lysates were assayed for the incorporation of [α-32P]GTP into the cap-binding site on CBP20 (middle panel). The lysates were analyzed for expression of the transfected proteins by Western blotting using an anti-V5 antibody (top panel), and an anti-actin antibody was used to ensure equal protein loading (bottom panel). F, knocking down mTOR blocks HRG signaling to Ran. SKBR3 cells were transfected with mTOR siRNA or control siRNA, serum-starved for 5 days, and stimulated with HRG (100 nm) for 1 h at 37 °C. The cytoplasmic lysates were probed with anti-mTOR, (top panel), anti-phospho-p70 S6K (Thr389) (second panel), and anti-p70 S6K (third panel). The nuclear lysates were assayed for Ran-GTP levels using the His6-importin-β pulldown assay (bottom two panels). The nuclear levels of Ran are shown in the fourth panel from the top. G, knocking down p70 S6K blocks HRG signaling to Ran. SKBR3 cells were transiently transfected with a specific p70 S6K shRNA or a control shRNA and then the cells were treated as in F. The knockdown efficiency was determined using an anti-p70 S6K antibody to probe cytoplasmic lysates (top panel), and equal loading was confirmed by probing with an anti-actin antibody (second panel). Nuclear lysates were used to determine Ran protein levels (third panel) and to assess Ran-GTP levels (bottom two panels).
The finding that HRG was a more potent activator of Ran than EGF was consistent with Ran playing a role in signaling to the CBC, because HRG shows a stronger capability for regulating CBC activity compared with EGF. Indeed, the transient expression of the dominant active, GTP hydrolysis-defective Ran (Q69L) mutant in SKBR3 cells circumvents the need for growth factor stimulation and strongly activates the CBC. This can be read out by the incorporation of [α-32P]GTP (which serves as a radiolabeled cap analog) into the cap-binding site of the CBP20 subunit in nuclear lysates from serum-starved cells (Fig. 1E, middle panel). Because we had previously shown that the ability of HRG to regulate the CBC was mediated by mTOR (16), we examined whether the HRG-dependent activation of Ran was also dependent on mTOR activity. SKBR3 cells were transfected with either a control siRNA or a validated, mTOR-specific siRNA, serum-starved, and then treated with HRG for 1 h. The cells were harvested and fractionated, with the cytoplasmic lysates being used to determine both knockdown efficiency and p70 S6K activity by Western blotting, whereas the nuclear lysates were used to assay Ran activation via the His6-importin-β pulldown assay. As shown in Fig. 1F, the knockdown of mTOR was very effective at abrogating both the HRG-dependent activation of p70 S6K (second panel from the top) and Ran (bottom panel). Likewise, knocking down p70 S6K using an shRNA (Fig. 1G, top panel) blocked HRG signaling to Ran (Fig. 1G, bottom panel).
A Novel Activated Ran Mutant That Stimulates Cell Growth
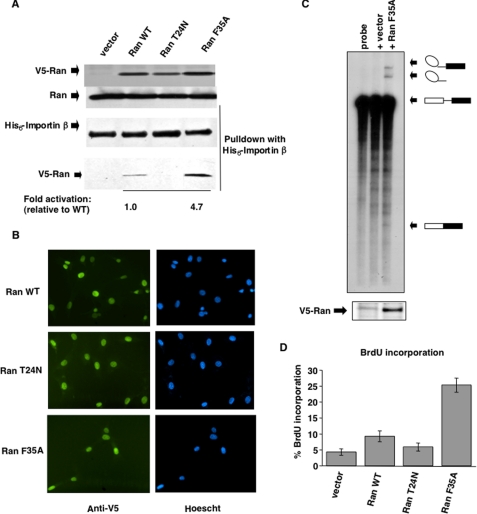
Given that Ran is activated downstream of a HRG signaling pathway in breast cancer cells, we wanted to examine the consequences of losing this regulation, as would occur by introducing an activated Ran mutant into cells. In particular, we were interested in seeing whether such an activated Ran mutant would be capable of stimulating cell proliferation and even inducing cellular transformation when introduced into nontransformed NIH-3T3 cells. An important consideration was that we examine the effects of an activated Ran mutant that was not highly overexpressed in these cells, given that the changes in Ran-GTP levels in response to HRG were likely to represent only a small percentage of the total Ran population. Moreover, a potential complication with these experiments was that it had previously been shown that the overexpression of a constitutively active, GTP hydrolysis-defective form of Ran (Ran (Q69L)) negatively affected cell growth (23–25). In these cases, it was assumed that Ran (Q69L) was acting as a dominant negative inhibitor by binding, but not releasing, effector proteins such as importin-β. This, in turn, would inhibit Ran-dependent functions such as classical nucleo-cytoplasmic trafficking and result in cell cycle arrest. Therefore, to circumvent this potential problem, we screened for Ran mutants that were constitutively active in cells but did not cause any dominant negative effects that would result in cell cycle arrest. One such mutant, Ran (F35A), appeared to be hyperactivated in cells as read out by the His6-importin-β binding assay but was not toxic to cell growth (see below).
We generated NIH-3T3 cell lines stably expressing the Ran (F35A) mutant fused to a V5 tag, as well as wild type Ran and the dominant negative Ran (T24N) mutant. Fig. 2A demonstrates that we were able to select clones that expressed similar levels of each of the different Ran proteins without perturbing the expression of endogenous Ran (top two panels). Although we can detect the Ran fusion proteins using the V5 antibody, we were unable to reliably detect them using a Ran antibody, suggesting that the ectopically expressed Ran proteins represented only a small percentage of the total pool of cellular Ran. Cells expressing the Ran (T24N) mutant were prone to a loss of expression of this Ran construct upon repeated cell passages, possibly because of its potential dominant negative effects. Each of the V5-Ran fusion proteins showed a nuclear localization, as indicated by immunofluorescence using antibodies directed against the V5 tag (Fig. 2B).
FIGURE 2.
Characterization of NIH-3T3 cells stably expressing Ran (F35A). A, stable NIH-3T3 cell lines were generated with V5-tagged versions of WT Ran, Ran (T24N), Ran (F35A), or a vector control. Expression levels of the exogenous proteins were detected using an anti-V5 antibody (top panel), and endogenous Ran was detected using an anti-Ran antibody (second panel). The activation states of the ectopically expressed proteins were determined by using the His6-importin-β pulldown assay and immunoblotting with the anti-V5 antibody (bottom panel). The relative amount of V5-tagged Ran precipitating with His6-importin-β was measured using the National Institutes of Health Image J software. B, cellular localization of V5-fused WT Ran, Ran (T24N), and Ran (F35A) was determined by immunofluorescence using an anti-V5 antibody, and the nuclei were stained with Hoescht dye. C, NIH-3T3 cells stably transfected with Ran (F35A) or vector alone were analyzed for their ability to support the splicing of a specific m7GpppG-capped pre-mRNA probe (upper panel). The mature splice products and intermediates of the splicing reaction are indicated diagrammatically on the right. The lower panel shows the expression of the Ran (F35A) mutant as determined by Western blotting using an anti-V5 antibody. D, BrdUrd incorporation assays were performed on NIH-3T3 cells stably expressing various Ran constructs. The histograms represent the percentages of cells in each experiment incorporating BrdUrd after 10 h of growth. The data are presented as the means ± standard deviation.
As indicated above, the Ran (F35A) mutant is hyperactivated in cells, because its expression results in ∼5-fold greater levels of Ran-GTP compared with that for wild type Ran (Fig. 2A, bottom panel). Biochemical characterization of Ran (F35A) suggests that although this mutant is not capable of spontaneous nucleotide exchange as read out by the decrease in Mant-GDP fluorescence because of its exchange for GTP (supplemental Fig. S2A), it exhibits an increased ability to be activated by substoichiometric levels of the Ran-guanine nucleotide exchange factor RCC1, compared with wild type Ran (supplemental Fig. S2B). The Ran (F35A) mutant also shows little ability to hydrolyze GTP compared with wild type Ran, when assayed in the presence of the mouse RanGAP, FUG1, although its GTP hydrolytic activity can be largely restored by the addition of the RanGAP cofactor RanBP1 (supplemental Fig. S2C). Therefore, the combination of increased responsiveness to RCC1 and a slightly reduced capability for hydrolyzing GTP likely accounts for the increase in the cellular activation of the Ran (F35A) mutant.
We verified that the Ran (F35A) mutant was functional in these stable cell lines by testing its ability to activate the CBC, as assayed by changes in the cap-dependent splicing of RNA (4). Nuclear lysates were prepared from serum-starved cells stably expressing Ran (F35A) or from vector control cells. A radiolabeled precursor mRNA probe was then incubated with the lysates under conditions that are permissive for pre-mRNA splicing in vitro, and the resulting RNA products were resolved on a denaturing polyacrylamide gel. Although cap-dependent splicing was not supported from lysates derived from serum-starved vector control cells, the lysates obtained from serum-starved cells expressing Ran (F35A) exhibited splicing activity as indicated by the generation of splicing intermediates and products from the pre-mRNA (Fig. 2C).
Unlike the Ran (Q69L) mutant that was completely incapable of hydrolyzing GTP and had been shown to have deleterious effects on cell cycle progression (23–25), the Ran (F35A) mutant did not cause cell cycle arrest. This was demonstrated in experiments where we examined the ability of cells expressing Ran (F35A), as well as vector control cells, and cells expressing wild type Ran and the Ran (T24N) mutant, to stimulate DNA synthesis as measured by BrdUrd incorporation (18). The different stable cell lines were synchronized in the G0 phase by serum deprivation, and then 200 μm BrdUrd was added along with 10% calf serum. Ten hours after the addition of serum and BrdUrd, the cells were scored to determine the percentage of cells in the S phase of the cell cycle as read out by the incorporation of BrdUrd into DNA. Approximately twice as many cells that expressed wild type Ran entered the S phase, compared with either the vector control cells or cells expressing the dominant negative Ran (T24N) mutant (Fig. 2D). However, the number of cells in the S phase in the Ran (F35A) stable transfectants was ∼5-fold greater than the number of vector control cells in the S phase, indicating that the Ran (F35A) mutant stimulated cell cycle progression.
Role for Ran-GTP in Cellular Transformation
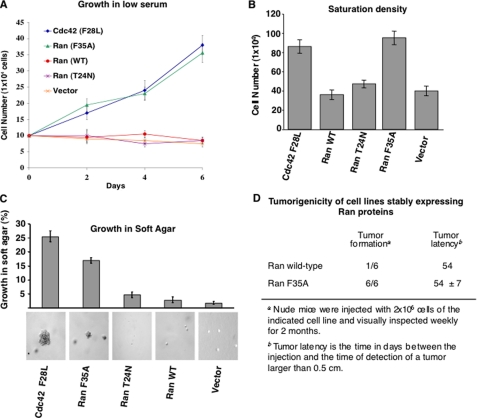
We next examined the capability of the Ran (F35A) mutant to transform cells. We started by examining its ability to stimulate the growth of fibroblasts under low serum conditions. Fig. 3A shows that over the course of 6 days, the cells expressing Ran (F35A) more than tripled in number, similar to cells expressing the constitutively active Cdc42 (F28L) mutant, which has strong transforming activity (26, 27). On the other hand, the vector control cells and cells expressing either wild type Ran or the Ran (T24N) mutant failed to grow under these conditions. Likewise, when we assayed cell growth in 5% calf serum, the cells expressing Ran (F35A) exhibited a loss of contact inhibition and grew to 3-fold higher densities compared with vector control cells or cells expressing either wild type Ran or Ran (T24N), thus matching the growth of cells expressing Cdc42 (F28L) (Fig. 3B).
FIGURE 3.
Growth properties of NIH-3T3 cells stably expressing Ran (F35A). A, fibroblasts stably expressing Cdc42 (F28L) and Ran (F35A) show diminished serum dependence for growth. The indicated cell lines were cultured in DMEM supplemented with 0.5% serum, and at the indicated times, the cells were trypsinized and counted. The data are representative of three experiments. B, fibroblasts stably expressing Ran (F35A) and Cdc42 (F28L) exhibit diminished contact inhibition. Control (vector) NIH-3T3 cells and NIH-3T3 cells stably expressing either Cdc42 (F28L), WT Ran, Ran (T24N), and Ran (F35A) were cultured in DMEM supplemented with 5% calf serum for 6 days, trypsinized, and counted. The data represent the average of three independent experiments. C, fibroblasts stably expressing Ran (F35A) and Cdc42 (F28L) exhibit anchorage-independent growth. NIH-3T3 cells that stably express vector control, WT Ran, Ran (T24N), Cdc42 (F28L), and Ran (F35A) were mixed with medium supplemented with 0.3% agar and 10% calf serum and plated on top of a 0.5% agarose layer. Growing colonies were scored after 14 days for the various cell lines. The values shown are the averages of three independent experiments. The bottom panels are photomicrographs of colony formation of the cell lines in soft agar (40× magnification). D, NIH-3T3 cells stably expressing wild type Ran and Ran (F35A) were injected into the right and left flank of nude mice. Two months after injection, the mice were euthanized, and the tumors were excised. The data are representative of two independent experiments.
An additional parameter that distinguishes normal from transformed cells is the requirement for attachment to an extracellular substratum, so we assessed the extent to which the different cell lines were capable of anchorage-independent growth in soft agar. Constitutive expression of Ran (F35A) stimulated colony formation in soft agar, whereas vector control cells and cells expressing wild type Ran or Ran (T24N) were incapable of forming colonies (Fig. 3C). We then asked whether cells expressing the activated Ran (F35A) mutant exhibit tumorigenic activity in mice. Cells expressing wild type Ran and the Ran (F35A) mutant were injected subcutaneously into immunocompromised (athymic) “nude” mice, and then the mice were monitored for the formation of tumors (>5 mm) over the course of 2 months. The tumors were detected in six of six mice injected with cells expressing the Ran (F35A) mutant, compared with mice injected with cells expressing wild type Ran where only one tumor was detected among the six mice (Fig. 3D). Additionally, tumors formed by Ran (F35A) showed advanced cellular necrosis (not shown), probably because of their difficulty in sustaining an adequate blood supply.
Link between Ran-induced Cellular Transformation and Protein Expression
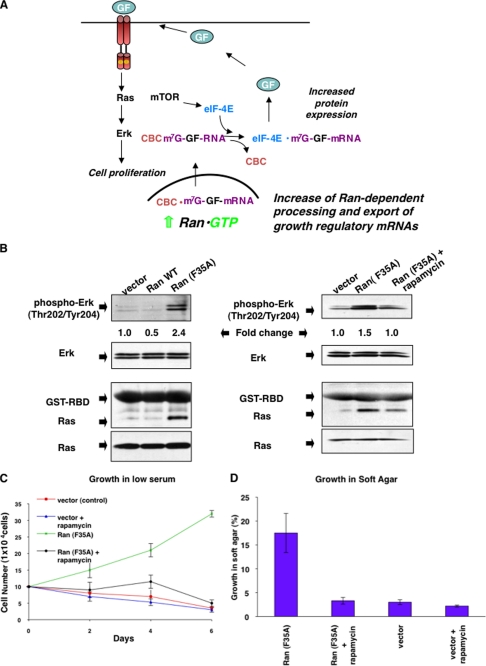
How does the hyperactivation of Ran lead to cellular transformation? The ability of Ran-GTP to stimulate the rates of processing and delivery of mRNAs to eIF-4E could result in the increased expression of growth factors and/or growth regulatory proteins, which in turn stimulate mitogenic signaling events, as depicted in Fig. 4A. This would be analogous to the demonstrated ability of eIF-4E, which binds to mature capped mRNAs in an mTOR-dependent manner (i.e. the rate-limiting step for cap-dependent translation) to cause cellular transformation (28–31). In the model for transformation by eIF-4E, and what we propose for activated Ran, the expression of growth factors and their receptors and/or other growth regulatory proteins would increase when the processing and translation of mRNAs (i.e. through the binding of capped mRNAs by the CBC and/or eIF-4E) are no longer regulated, thus resulting in the stimulation of mitogenic signaling activities. Consistent with this idea is the finding that the overexpression of eIF-4E results in the constitutive activation of Ras (29).
FIGURE 4.
Activation of mitogenic signaling proteins by the stable expression of Ran (F35A). A, model for a mechanism by which Ran (F35A) transforms cells. Increased levels of Ran-GTP in the nucleus (i.e. because of the expression of the Ran (F35A) mutant) leads to increased binding of transcripts encoding growth regulatory proteins (i.e. a growth factor (GF)) to the CBC and ultimately enhanced translation by eIF-4E. The synthesis and secretion of growth factors then result in the activation of the Ras-ERK signaling pathway giving rise to excessive mitogenic signaling. B, stable cell lines expressing WT Ran, Ran (F35A), and a vector control were cultured and serum-starved for 24 h with DMEM supplemented with 0.5% calf serum. Left-hand panels, the lysates were analyzed for the activation of ERK using antibodies against phospho-ERK (Thr202/Tyr204) (top panel) and Ras using the Ras activation assay (third panel from the top). Fold change in ERK activation was quantified with the NIH Image J software and is stated relative to ERK activation in the vector control lane. Total ERK and Ras in the lysates are shown in the second panels from the top and the bottom panels, respectively. Right-hand panels, conditions were identical to those in the left-hand panels except that in some cases rapamycin (50 ng/ml) was added 30 min prior to harvesting the indicated cell lines. C, reversal of Ran transformation by rapamycin. Vector control cells or cells stably expressing Ran (F35A) were grown in 0.5% calf serum, with or without rapamycin (100 nm) every 2 days. The data shown represent the average of three independent experiments. D, the ability of Ran (F35A)-expressing cells and vector control cells to grow in soft agar in the presence of rapamycin was assessed by adding rapamycin (100 nm) every 3 days to the conditions described in Fig. 3C. The results represent an average of three experiments.
From this model, we would make two predictions. First, we would expect to see increases in cellular signaling activities (such as Ras) in cells transformed by Ran (F35A), and we would predict that these activities, as well as Ran-dependent transformation, are sensitive to the mTOR inhibitor, rapamycin. Both of these predictions turned out to be true. NIH-3T3 cells stably expressing wild type Ran, the Ran (F35A) mutant, or vector alone were serum-starved and then harvested without any additional treatment. Whole cell lysates generated from these cells were then used to assess the activation status of Ras by using the Ras activation assay (see “Experimental Procedures”), as well as ERK by the use of phospho-specific antibodies. Fig. 4B shows that both Ras (second panel from the bottom, left column) and ERK (top panel, left column) were indeed activated in cells expressing Ran (F35A) and that the treatment of these cells with rapamycin blocked the Ran (F35A)-dependent activation of ERK (top panel, right column) and Ras (second panel from the bottom, right column). To further test this idea, we examined the effects of rapamycin on the ability of cells expressing Ran (F35A) to grow in low serum and to form colonies in soft agar. Fig. 4 (C and D) shows that treatment with rapamycin completely inhibited the transforming capability of the activated Ran mutant.
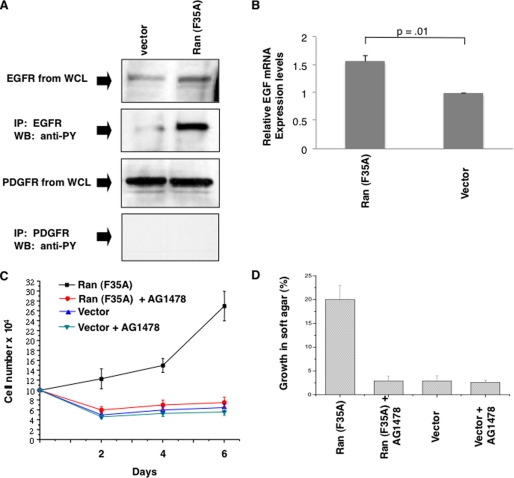
Because it seemed likely that the activation of the Ras/ERK signaling pathways was the outcome of the increased expression of growth factors or growth factor receptors that are known to activate these signaling proteins, we looked at the expression and activation status of two such receptor tyrosine kinases, the EGFR and PDGFR. These receptors were immunoprecipitated from either serum-starved vector control cells or cells expressing Ran (F35A) and then immunoblotted with anti-phosphotyrosine to determine their activation status. Although the expression levels for the EGFR and PDGFR did not appear to be significantly different between the two cell lines (the top panel of Fig. 5A shows an example for the EGFR), the EGFRs were highly activated in Ran (F35A)-expressing cells (Fig. 5A, second panel), whereas we did not detect PDGFR activation under these conditions (Fig. 5A, bottom panel).
FIGURE 5.
Ran (F35A)-induced transformation requires EGF signaling. A, activation of the EGFR in Ran (F35A) cells. NIH-3T3 cells expressing either vector or Ran (F35A) were serum-starved for 48 h and then harvested. Relative EGFR levels were assessed by probing whole cell lysates with anti-EGFR (top panel). EGFR was also immunoprecipitated (IP) from whole cell lysates and analyzed for activation by probing with an anti-phosphotyrosine antibody (second panel). Relative PDGFR levels were determined by probing whole cell lysates with an anti-PDGFR antibody (third panel), and their activation state was determined by immunoprecipitating PDGFRs from whole cell lysates and then probing with an anti-phosphotyrosine antibody (bottom panel). B, the expression levels of EGF mRNA from NIH-3T3 cells expressing either Ran (F35A) or vector alone were determined using quantitative PCR. The results shown are the averages of three independent experiments with standard deviation. The data were further analyzed using Student's t test. C, vector control and Ran (F35A)-expressing cells were grown in low serum with and without the addition of AG1478 (3 μm) every 2 days. The data shown are representative of three independent experiments. D, the effect of AG1478 on the ability of vector control or Ran (F35A)-expressing cells to grow in soft agar was assessed by adding AG1478 (3 μm) to the soft agar growth conditions every 3 days. The values that are shown are the averages of three independent experiments. WB, Western blot.
The hyperactivation of EGFRs in cells expressing the Ran (F35A) mutant, in the absence of increased receptor expression, suggested that the production and/or secretion of EGF and/or other ligands that activate EGFRs might be elevated in these transformed cells. In fact, we have found that EGF message levels are significantly increased in cells expressing the Ran (F35A) mutant compared with vector control cells (Fig. 5B), although we do not yet know whether these changes fully account for the significant differences in EGFR activation status. However, it does appear that EGFR signaling is a necessary component of transformation by Ran (F35A), because treatment with the EGFR tyrosine kinase inhibitor, AG1478, completely reversed the ability of cells expressing Ran (F35A) to grow in low serum (Fig. 5C) or to form colonies in soft agar (Fig. 5D).
DISCUSSION
The demonstration that Ran activation is under growth factor control would not necessarily have been predicted, given the usual cellular roles that have been attributed to this small GTPase. Ran is a critical player in the regulation of multiple nuclear processes that have not typically been thought of as being susceptible to growth factor regulation including nucleocytoplasmic transport, the formation of the mitotic spindle, and the nuclear envelope (2, 32). However, the finding that growth factors work through the importins to regulate the cap binding capability of the CBC (14), coupled with the role played by Ran-GTP in regulating CBC-importin interactions, raised the intriguing possibility that the activation status of Ran might come under growth factor control, and indeed, we have found this to be the case. Although it was recently reported that growth factors promote the phosphorylation of RanBP3 (33), which was suggested to increase Ran-GTP levels either by binding and stabilizing Ran-GTP or by enhancing the guanine nucleotide exchange factor activity of RCC1 toward Ran, it seems unlikely that this accounts for the HRG-promoted activation of Ran that we describe here. In particular, the phosphorylation of RanBP3 was shown to be downstream of Ras/ERK/RSK signaling, with no role being assigned to p70 S6K, whereas the converse is true for the HRG-dependent activation of Ran that we see in breast cancer cells, where knockdowns of mTOR and p70 S6K block this HRG signaling outcome.
The ability of growth factors to activate Ran leads to an important question, namely how might small changes to the Ran-GTP gradient, such as those that occur in response to HRG or that accompany the introduction of low levels of an activated Ran mutant into cells, result in a change in the growth response of a cell? The Ran-GTP gradient provides a possible mechanism for stimulating the release of capped RNAs from the CBC into the cytoplasm, through the regulation of differential complex formation between the CBC and the importins. Thus, increases in the levels of GTP-bound Ran as induced by growth factors would have the effect of stimulating the capped RNA binding cycle of the CBC and increasing nucleocytoplasmic transport rates. This would have the potential of increasing the import of transcription factors into the nucleus while also accelerating the Ran-dependent processing and export by the CBC of mRNAs encoding growth factors and other growth regulatory proteins. As a consequence, the expression of these proteins may be enhanced and thus trigger mitogenic signaling pathways that can result in cellular transformation. In fact, we show here that the activated Ran (F35A) mutant can induce the transformation of NIH-3T3 cells and that cells expressing this mutant promote tumor formation in mice.
Taken together, our findings are in accordance with the working model presented in Fig. 4A. Quiescent cells may possess background levels of nascent transcripts encoding growth factors, growth factor receptors, and possibly other growth regulatory proteins that normally would not be processed further. In the presence of increased Ran activity, these transcripts would be processed and exported to the cytoplasm in a CBC-dependent fashion, ultimately to be transferred to eIF-4E for translation. The production of these growth factors and growth regulatory proteins would then activate signaling pathways that stimulate cell growth and might even trigger a positive feedback loop by further activating the RNA-processing and translation machinery. Such a model would fit our data with regard to the increased message levels for EGF detected in cells expressing the Ran (F35A) mutant, compared with vector control cells, and the constitutive activation of EGFR/Ras signaling activities, as well as the ability of both rapamycin and the EGFR tyrosine kinase inhibitor, AG1478, to inhibit cellular transformation by this activated Ran mutant. Although increases in EGF levels in response to Ran (F35A) appear to be one necessary factor that contributes to the transformation of these cells, there may very well be other activities which are up-regulated and contribute to the Ran (F35A) transformed phenotype. A proteomics analysis of genes whose expression is altered in response to Ran (F35A) would further aid in understanding the mechanisms underlying Ran transformation.
It has been reported that the activity of both the c-Met tyrosine kinase and Akt is up-regulated in rat mammary cells overexpressing wild type Ran, with this being attributed to an adapter complex containing a putative Ran-binding partner, RanBPM (34). We feel that such a mechanism would not explain the transforming activity we observed for the activated Ran (F35A) mutant. RanBPM is a scaffold protein known to interact with different receptors including c-Met and to facilitate their activation (35), but it has also been reported to be incapable of interacting with the EGFR (36). In addition, although RanBPM was originally identified in a yeast two-hybrid screen using Ran as bait (37), thus far there is little evidence in the literature to support the idea that RanBPM is a bona fide Ran target, and we have not been able to detect an interaction between Ran and RanBPM in our stable cell lines (data not shown).
The findings that we describe here regarding the HRG-dependent activation of Ran and the effects of hyperactivated Ran on cell growth and transformation may be related to and shed some interesting light on the growing body of evidence that correlates Ran expression with tumorigenesis (for review, see Ref. 17). For example, in a study of epithelial ovarian cancer, Ran was found to be a specific marker that defined the invasiveness of the tumor, and its high expression was significantly correlated with poor patient survival (38). Ran was also shown to be highly expressed in gastric, colon, pancreatic, and lung cancer tissues, but not in normal cells within the same tumor sample (39). Additionally, in an RNA interference-based screen, the knockdown of Ran and a protein under Ran regulation in mitotic spindle assembly, TPX2, was found to significantly reduce the survival of multiple human tumor cell lines (40). More recently, it was shown that the overexpression of wild type Ran was sufficient to transform rat mammary cells (34). We would suspect that in these different examples, the overexpression of wild type Ran might lead to a sufficient increase in the cellular levels of Ran-GTP so as to tip the balance from normal cell growth to malignant transformation, particularly given our findings that modest increases in Ran activation status are able to induce transforming phenotypes.
In conclusion, we have provided to our knowledge the first direct demonstration that Ran-GTP levels can be modulated in response to serum and a specific growth factor, HRG, and have presented evidence that low levels of activated Ran have oncogenic potential. These findings lend credence to previous correlations between high Ran expression and the growth of multiple tumor types, point to previously unappreciated Ran-related nuclear functions (in particular, the regulation of RNA processing via the CBC) for possible points of therapeutic intervention, and underscore the importance of Ran both in normal and tumorigenic cell growth.
Acknowledgments
We thank Dr. Alfred Wittinghofer (Max-Planck Institut für Molekulare Physiology) for pGEX-KG-Ran, Dr. Iain Mattaj (EMBL) for the importin-β construct, Dr. Ian Macara (University of Virginia) for the mouse RanGAP pGEX-FUG1, and Dr. Gino Cingolani (SUNY Upstate Medical Center) for pET28a-Ran-BP1. We also thank Dr. Sandra Dias for helpful discussions and Cindy Westmiller for excellent secretarial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM040654.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- CBC
- nuclear cap-binding complex
- HRG
- heregulin
- S6K
- S6 kinase
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- eIF
- eukaryotic translation initiation factor
- ERK
- extracellular signal-regulated kinase
- BrdUrd
- bromodeoxyuridine
- DMEM
- Dulbecco's modified Eagle's medium
- WT
- wild type
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- shRNA
- small hairpin RNA
- PDGFR
- platelet-derived growth factor receptor
- mTOR
- mammalian target of rapamycin.
REFERENCES
- 1.Macara I. G. (2001) Microbiol. Mol. Biol. Rev. 65, 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart M. (2007) Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 3.Görlich D., Kraft R., Kostka S., Vogel F., Hartmann E., Laskey R. A., Mattaj I. W., Izaurralde E. (1996) Cell 87, 21–32 [DOI] [PubMed] [Google Scholar]
- 4.Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I. W. (1994) Cell 78, 657–668 [DOI] [PubMed] [Google Scholar]
- 5.Izaurralde E., Stepinski J., Darzynkiewicz E., Mattaj I. W. (1992) J. Cell Biol. 118, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kataoka N., Ohno M., Kangawa K., Tokoro Y., Shimura Y. (1994) Nucleic Acids Res. 22, 3861–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kataoka N., Ohno M., Moda I., Shimura Y. (1995) Nucleic Acids Res. 23, 3638–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visa N., Izaurralde E., Ferreira J., Daneholt B., Mattaj I. W. (1996) J. Cell Biol. 133, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng H., Dufu K., Lee C. S., Hsu J. L., Dias A., Reed R. (2006) Cell 127, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 10.Izaurralde E., Lewis J., Gamberi C., Jarmolowski A., McGuigan C., Mattaj I. W. (1995) Nature 376, 709–712 [DOI] [PubMed] [Google Scholar]
- 11.Lewis J. D., Görlich D., Mattaj I. W. (1996) Nucleic Acids Res. 24, 3332–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis J. D., Izaurralde E., Jarmolowski A., McGuigan C., Mattaj I. W. (1996) Genes Dev. 10, 1683–1698 [DOI] [PubMed] [Google Scholar]
- 13.Ohno M., Sakamoto H., Shimura Y. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5187–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dias S. M., Wilson K. F., Rojas K. S., Ambrosio A. L., Cerione R. A. (2009) Nat. Struct. Mol. Biol. 16, 930–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson K. F., Fortes P., Singh U. S., Ohno M., Mattaj I. W., Cerione R. A. (1999) J. Biol. Chem. 274, 4166–4173 [DOI] [PubMed] [Google Scholar]
- 16.Wilson K. F., Wu W. J., Cerione R. A. (2000) J. Biol. Chem. 275, 37307–37310 [DOI] [PubMed] [Google Scholar]
- 17.Rensen W. M., Mangiacasale R., Ciciarello M., Lavia P. (2008) Front. Biosci. 13, 4097–4121 [DOI] [PubMed] [Google Scholar]
- 18.Melkoumian Z. K., Peng X., Gan B., Wu X., Guan J. L. (2005) Cancer Res. 65, 6676–6684 [DOI] [PubMed] [Google Scholar]
- 19.Yasuda Y., Miyamoto Y., Saiwaki T., Yoneda Y. (2006) Exp. Cell Res. 312, 512–520 [DOI] [PubMed] [Google Scholar]
- 20.Yudin D., Hanz S., Yoo S., Iavnilovitch E., Willis D., Gradus T., Vuppalanchi D., Segal-Ruder Y., Ben-Yaakov K., Hieda M., Yoneda Y., Twiss J. L., Fainzilber M. (2008) Neuron 59, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagrodia S., Taylor S. J., Jordon K. A., Van Aelst L., Cerione R. A. (1998) J. Biol. Chem. 273, 23633–23636 [DOI] [PubMed] [Google Scholar]
- 22.Taylor S. J., Shalloway D. (1996) Curr. Biol. 6, 1621–1627 [DOI] [PubMed] [Google Scholar]
- 23.Clarke P. R., Klebe C., Wittinghofer A., Karsenti E. (1995) J. Cell Sci. 108, 1217–1225 [DOI] [PubMed] [Google Scholar]
- 24.Dasso M., Seki T., Azuma Y., Ohba T., Nishimoto T. (1994) EMBO J. 13, 5732–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren M., Coutavas E., D'Eustachio P., Rush M. G. (1994) Mol. Cell Biol. 14, 4216–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin R., Bagrodia S., Cerione R., Manor D. (1997) Curr. Biol. 7, 794–797 [DOI] [PubMed] [Google Scholar]
- 27.Wu W. J., Tu S., Cerione R. A. (2003) Cell 114, 715–725 [DOI] [PubMed] [Google Scholar]
- 28.Lazaris-Karatzas A., Montine K. S., Sonenberg N. (1990) Nature 345, 544–547 [DOI] [PubMed] [Google Scholar]
- 29.Lazaris-Karatzas A., Smith M. R., Frederickson R. M., Jaramillo M. L., Liu Y. L., Kung H. F., Sonenberg N. (1992) Genes Dev. 6, 1631–1642 [DOI] [PubMed] [Google Scholar]
- 30.Mamane Y., Petroulakis E., Rong L., Yoshida K., Ler L. W., Sonenberg N. (2004) Oncogene 23, 3172–3179 [DOI] [PubMed] [Google Scholar]
- 31.Smith M. R., Jaramillo M., Liu Y. L., Dever T. E., Merrick W. C., Kung H. F., Sonenberg N. (1990) New Biol. 2, 648–654 [PubMed] [Google Scholar]
- 32.Clarke P. R., Zhang C. (2008) Nat. Rev. Mol. Cell Biol. 9, 464–477 [DOI] [PubMed] [Google Scholar]
- 33.Yoon S. O., Shin S., Liu Y., Ballif B. A., Woo M. S., Gygi S. P., Blenis J. (2008) Mol. Cell 29, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurisetty V. V., Johnston P. G., Johnston N., Erwin P., Crowe P., Fernig D. G., Campbell F. C., Anderson I. P., Rudland P. S., El-Tanani M. K. (2008) Oncogene 27, 7139–7149 [DOI] [PubMed] [Google Scholar]
- 35.Wang D., Li Z., Messing E. M., Wu G. (2002) J. Biol. Chem. 277, 36216–36222 [DOI] [PubMed] [Google Scholar]
- 36.Yuan Y., Fu C., Chen H., Wang X., Deng W., Huang B. R. (2006) Neurosci. Lett. 407, 26–31 [DOI] [PubMed] [Google Scholar]
- 37.Nakamura M., Masuda H., Horii J., Kuma K., Yokoyama N., Ohba T., Nishitani H., Miyata T., Tanaka M., Nishimoto T. (1998) J. Cell Biol. 143, 1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouellet V., Guyot M. C., Le Page C., Filali-Mouhim A., Lussier C., Tonin P. N., Provencher D. M., Mes-Masson A. M. (2006) Int. J. Cancer 119, 599–607 [DOI] [PubMed] [Google Scholar]
- 39.Azuma K., Sasada T., Takedatsu H., Shomura H., Koga M., Maeda Y., Yao A., Hirai T., Takabayashi A., Shichijo S., Itoh K. (2004) Clin. Cancer Res. 10, 6695–6702 [DOI] [PubMed] [Google Scholar]
- 40.Morgan-Lappe S. E., Tucker L. A., Huang X., Zhang Q., Sarthy A. V., Zakula D., Vernetti L., Schurdak M., Wang J., Fesik S. W. (2007) Cancer Res. 67, 4390–4398 [DOI] [PubMed] [Google Scholar]