Abstract
The transcription factor FoxN1 is essential for differentiation of thymic epithelial cell (TEC) progenitors during thymic organogenesis. However, limited information is available on the postnatal contribution of FoxN1 to thymic maintenance. To address this question, we generated a loxP-floxed FoxN1 (fx) mouse with three different promoter-driven inducible CreERT transgenes. Postnatal ubiquitous deletion of FoxN1 caused dramatic thymic atrophy in 5 days and more severe deterioration in medullary TECs (mTECs) than in cortical TECs (cTECs). Induction of FoxN1 deletion selectively in K5 promoter-driven somatic epithelial cells (mostly mTECs and possibly some adult epithelial stem cells) was sufficient to cause significant thymic atrophy, whereas FoxN1 deletion in K18 promoter-driven somatic epithelial cells (mostly cTECs) was not. Thymic atrophy resulted from increased apoptosis and was associated with activation of the p53 gene in mature mTECs. Although FoxN1 is required for the development of both mTECs and cTECs in thymic organogenesis, it is most important for the maintenance of mTECs in the postnatal thymus, which are in turn necessary to prevent thymic atrophy.
Keywords: Cell/Epithelial, Cell/Stromal, Gene/Knock-out, Genetics/Mouse, Tissue/Organ Systems/Thymocyte, Transcription/Development, Apoptosis, thymic Epithelial Cells
Introduction
The thymus gland is not only required for ontogenesis but is also indispensable for postnatal cellular immune system function (1) through replenishment of the naive T-cell pool and maintenance of diversity of the T-cell receptor repertoire, establishment of central immune tolerance by depleting self-reactive T-cell clones, and generation of natural regulatory T cells to maintain immune balance (2, 3). Many intrinsic and extrinsic factors can induce atrophy of the mature thymus and disrupt thymic functions (4, 5). Thymic atrophy is generally believed to result from deterioration of the interactions between lymphohematopoietic progenitor cells and nonhematopoietic thymic stromal cells (TSCs),3 primarily thymic epithelial cells (TECs) in the thymus (6, 7). Therefore, changes in expression of genes and transcription factors, related to lymphohematopoietic progenitor cell and/or TEC functions may regulate thymic involution.
FoxN1 is an epithelial cell-autonomous gene that encodes a forkhead-box transcription factor related to the immune system (8) and skin epithelial cells (9). FOXN1 in humans and FoxN1 in rodents are highly conserved in their sequence and function. A mutation in FoxN1 generates alymphoid cystic thymic dysgenesis due to defective TECs (10, 11), causing primary T-cell immunodeficiency (12–15), and leads to a hairless “nude” phenotype (9). TECs have two major subsets, medullary and cortical TECs (mTEC and cTEC), based on anatomic regions, expressed molecules, and function. mTECs mediate negative selection of T cells and control the maturation of T cells prior to leaving the thymus, whereas cTECs foster the development of CD4−8− T-cell progenitors and regulate positive selection of T cells. Both mTECs and cTECs are FoxN1-dependent (16) during fetal thymic organogenesis, as demonstrated in mouse models (11, 17, 18). However, it is unclear whether FoxN1 is still required for postnatal TECs and whether postnatal mTECs and cTECs are equally FoxN1-dependent. This is because of the lack of a mouse model in which FoxN1 can be inducibly deleted in postnatal somatic TECs instead of in the germ line.
Recently, the role of FoxN1 in the postnatal thymus was addressed by using a FoxN1-LacZ allele, in which postnatal FoxN1 transcription is disrupted by a LacZ gene inserted in the 3′-untranslated region of the FoxN1 locus, resulting in thymic atrophy (19). However, the mechanism for reduced FoxN1 expression in this mutant is uncertain, because the inserted LacZ gene is not controllable. The LacZ insertion may disrupt FoxN1 in postnatal and prenatal life, and genetic pathways other than FoxN1 may also be affected by the LacZ gene. It is more definitive to study postnatal functions of FoxN1 by using a temporally controllable and keratin type-specific FoxN1 knock-out mouse model. Here, we developed such a mouse, denoted as FoxN1fx (fx). Conditional deletion of FoxN1 in the young adult thymus caused acute thymic atrophy within 5 days, associated with greatest reduction of mTECs, particularly MHC-IIhiUEA-1hi mature mTECs, compared with cTECs and presumptive precursors of mTECs. Conditional FoxN1 deletion in the somatic cells under the control of the keratin-5 (K5) promoter, expressed predominantly in postnatal mTECs in the developed thymus, sufficiently caused acute thymic atrophy, whereas conditional FoxN1 deletion in the somatic cells under the control of the K18 promoter, expressed primarily in postnatal cTECs (20), did not. TEC loss resulted from increased apoptosis and was associated with activation of p53 in mature mTECs. These results indicate that deletion of FoxN1 in the postnatal thymus causes a primary defect in mTECs, and perhaps a secondary defect in other TECs, including cTECs. Therefore, mTECs in the postnatal thymus are essentially FoxN1-dependent, and disrupting the steady state of postnatal mTECs will trigger thymic atrophy.
EXPERIMENTAL PROCEDURES
Mice
All animal experiments were done according to the protocols approved by the Institutional Animal Care and Use Committee of the University of Texas Health Science Center at Tyler, in accordance with guidelines of the National Institutes of Health. The FoxN1fx mice were generated on a C57BL/6/129Sv genetic background as described under “Results.” The pCAGG-CreERTM mice (21), which carry a tamoxifen (TM)-inducible ubiquitous Cre-recombinase (referred to here as uCreERT mice), were purchased from The Jackson Laboratory (catalog No. 004682). FoxN1-null (nude) mice were also purchased from The Jackson Laboratory (catalog No. 000819). Keratin-5-CreERT2 (22) and keratin-18-CreERT2 (23) mice, which we refer to here as K5- or K18-CreERT mice, were kindly provided by Drs. Chambon and Oshima. After cross-breeding with fx/fx mice for >2 generations, we obtained uCreERT-, K5CreERT-, or K18CreERT-driven fx/fx and fx/+ mice. All young mice were genotyped by PCR as described below under “Genotyping FoxN1fx Mice.” The age of the mice used in the experiments was 1–2 months unless indicated otherwise.
Preparation of Tamoxifen and Induction of FoxN1 Deletion
A final concentration of 20 mg/ml stock solution of TM (Sigma T5648) was prepared following the manufacturer's instructions by completely dissolving TM powder into 1 volume of 100% ethanol in 55 °C for about 1 min, adding 9 volumes of warmed (55 °C) corn oil, and mixing them well by vortexing. We have found that this is more effective than sonication for dissolving TM. We induced deletion of exons 5 and 6 of FoxN1 encoding the putative FoxN1 DNA binding domain (24, 25) (denoted as ΔE5&6) by intraperitoneal injection of TM (1 mg/10 g body weight/day) for the indicated number of successive days. This is similar to the total dose used in other reports to treat adult mice (21, 26). Thymus phenotypes and cellular analyses were performed 3–5 days after the last injection. Control mice were WT, fx/fx-only (without Cre-recombinase), and uCreERT-, K5CreERT-, or K18CreERT-fx/+ heterozygous littermates.
Southern Blot
Southern blot was used to screen embryonic stem cells for the introduction of the FoxN1-targeting construct (supplemental Fig. S1B) and to determine the percentage of FoxN1 excision in different tissues (supplemental Fig. S3B). For these studies, a flanking probe corresponding to nucleotides (nt) 35441–35958 of the FoxN1 locus was amplified from FoxN1 genomic DNA by PCR (5′-primer, 5′-gtt tcc tgt ctc agg ctt gc-3′; and 3′-primer, 5′-cag gca gtt ggg tac agc tt-3′) and subcloned into a T-easy vector (Promega). This was labeled with [α-32P]dCTP, and Southern blotting was performed using standard simple salt methods.
Genotyping FoxN1fx Mice
All young mice were genotyped by PCR at 3–4 weeks of age using DNA obtained from the tails. The primers used for the genotypes (indicated by arrows in supplemental Fig. S3A) are primer A, 5′-cca acc tcc tgg gga cat ga-3′; and primer B, 5′-tag gag gag ggg agc gcc ta-3′; these produced 566- and 648-bp amplicons for the wild type (WT) and floxed FoxN1 (fx) loci (82 bp larger than the product of WT DNA because of the inclusion of one FRT and one loxP site), respectively. Primer N is a 3′-primer in the Neor cassette (5′-gcc tac cgg tgg atg tgg aa-3′), which produces a 388-bp amplicon with primer A if Neor has not been deleted (indicated by arrows in supplemental Fig. S2B).
Determining FoxN1 Excision by PCR
FoxN1 excision in the FoxN1fx (with Cre) mouse thymus was detected by PCR with 5′-primer C (5′-gtg ggc ttt tca cca tcc ta-3′) and 3′-primer B (sequence in the genotyping section above) to produce a ∼444-bp amplicon in the mutant locus with deletion of exons 5 and 6 of FoxN1 (ΔE5&6). PCR was performed with three primers (A, B, and C (supplemental Fig. S3A)) in one reaction. PCR yielded 648- and ∼444-bp amplicons (for fx and ΔE5&6 loci, respectively) for thymi from FoxN1fx/fx homozygous mice with the CreERT transgene or 648-, 566-, and ∼444-bp amplicons (for fx, WT, and ΔE5&6 loci, respectively) for thymi from FoxN1fx/+ heterozygous mice with the CreERT transgene. PCR products were detected by electrophoresis on 5% polyacrylamide gels.
Measuring FoxN1 mRNA Expression by Real-time RT-PCR and FoxN1+ TECs by Immunofluorescence Staining
Total thymic cells were dissociated by 1 mg/ml collagenase/dispase (Roche Applied Science) plus 5 units/ml DNase I (Sigma), and CD45−MHC-II+ TECs were sorted with a FACSAria cell sorter. Total RNA from the sorted TECs was prepared and reverse transcribed with the SuperScript III cDNA kit (Invitrogen). Real-time PCR was performed by standard techniques in an ABI-7300 thermocycler with TaqMan reagents. The FoxN1 5′-primer was 5′-GGC CAA CGC CGA AGG-3′, the 3′-primer was 5′-TGA AGA TGA GGA TGC TGT AAG AGT AGA-3′, and the probe was 6-FAM-CAC CAG CCA CTC TTC CCA AAG CCC-TAMRA. Samples were normalized to 18s RNA as the internal control. The results were analyzed by the relative gene (RQ) expression ΔΔCT method (27), setting the value for fx/fx-only cDNA control as 1.0. For FoxN1 protein immunohistology staining, cryosections (6 μm) were fixed, blocked, and stained with primary rabbit anti-FoxN1 (gift from Dr. Amagai) based on published methods (28).
Histological and Immunofluorescent Analysis
Thymi were fixed, cut into 5 μm-thick sections, and stained with hematoxylin and eosin; or for immunofluorescent staining, cryosections (6 μm) were cut, fixed in acetone, and blocked with donkey serum in Tris-buffered saline or with an endogenous biotin blocking kit (Invitrogen) if a biotinylated antibody was used. Primary antibodies were rabbit anti-mouse K5 (Covance), rabbit anti-mouse claudin-3,4 (Cld-3,4; Invitrogen, catalog No. 34-1700 and 36-4800), rat anti-mouse K8 (Troma-1 supernatant), rat anti-mouse MHC class II (I-A/I-E; BioLegend), and biotinylated anti-UEA-1 (Vector Laboratories). Secondary reagents included Cy3-donkey anti-rabbit IgG, fluorescein isothiocyanate-donkey anti-rat IgG, Cy3-streptavidin (all from Jackson ImmunoResearch Laboratories), and Alexa Fluor 488-anti-rabbit IgG (Invitrogen). The magnification in Figs. 1, 4, 5, and 6 shows that of the objective lens in a Nikon Eclipse Ti-U fluorescent microscope.
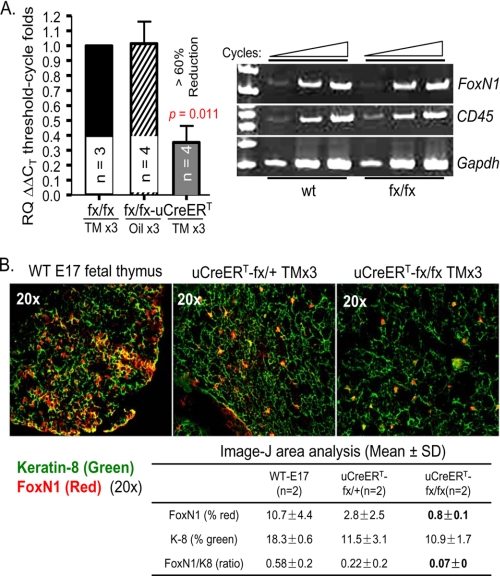
FIGURE 1.
Characterization of uCreERT-driven FoxN1fx mice. A, results of RT-PCR. Left panel, results of real-time RT-PCR from sorted CD45−MHC-II+ TECs of conditional FoxN1-deleted and control mice. Mean values ± S.E. are shown for three distinct sorting experiments; n = biological replicates. Right panel, representative result of semiquantitative RT-PCR from TSE-enriched thymic cells of WT and fx/fx-only mice. Expression of CD45 is an indicator of the extent of hematopoietic cell contamination, and GAPDH was used as an internal control. Data are representative of at least three separate experiments. B, immunofluorescence staining of the thymus with antibodies to keratin-8 (green) and FoxN1 (red). The left panel shows WT E17 fetal thymus (positive control). The middle and right panels show heterozygous fx/+ and homozygous fx/fx mice, both expressing uCreERT and treated with TM x3. A representative result of two experiments is shown with essentially identical results. The table shows the percentage of area occupied by K8+ and FoxN1+ cells as analyzed by NIH ImageJ software.
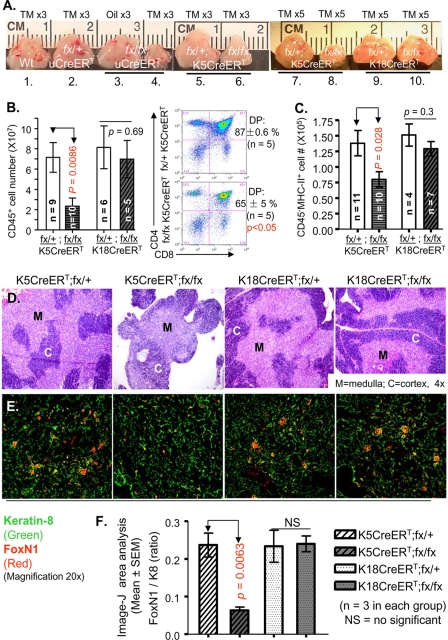
FIGURE 4.
Conditional deletion of FoxN1 in K5 promoter-driven TECs caused acute thymic atrophy, whereas in K18, promoter TECs did not cause obvious changes. A, gross view of thymic size of TM x3-treated WT and uCreERT-fx/+, oil x3-treated uCreERT-fx/fx, and TM x3-treated uCreERT-fx/fx, K5CreERT-fx/+, and K5CreERT-fx/fx, as well as TM x5-treated K5CreERT-fx/+, K5CreERT-fx/fx, K18CreERT-fx/+, and K18CreERT-fx/fx mice (from left to right). B and C, absolute CD45+ thymocyte number and absolute CD45−MHC-II+ TEC number per thymus, respectively, with flow cytometric staining with antibodies to CD45 and MHC-II from mice with the four genotypes (see bar legends) 5 days after treatment with TM x5. Means ± S.E. are shown for a total of 62 animals. The right panel in B shows a representative staining of CD4 versus CD8 plots from mice of K5CreERT-fx/+, K5CreERT-fx/fx, treated with TM x5. D, hematoxylin and eosin staining of thymi from mice with the four genotypes and treatments as shown in B, with two biological replicates in each genotype group. M, medulla; C, cortex. E, immunofluorescence staining of thymi from mice with the four genotypes and treatments as shown in B, with antibodies to keratin-8 (green) and FoxN1 (red). F, bar graph based on E shows ratios of percentage of FoxN1 versus K8-positive TEC area analyzed by NIH ImageJ software (n = mouse numbers).
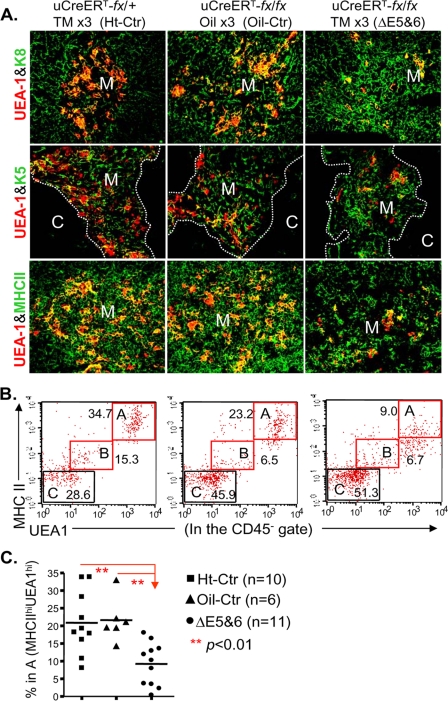
FIGURE 5.
Ubiquitous postnatal FoxN1 deletion selectively impaired mature mTECs. In all panels: Ht-Ctr, uCreERT-fx/+ mice treated with TM x3; oil-Ctr, uCreERT-fx/fx mice treated with oil vehicle x3; ΔE5&6, uCreERT-fx/fx mice treated with TM x3, which deleted exons 5 and 6. A, immunofluorescence staining of thymi. Thymic cryosections were stained with antibodies to UEA-1 (red) and K8 (green) (top panels), UEA-1 (red) and K5 (green) (middle panels), and UEA-1 (red) and MHC-II (green) (bottom panels). Magnification, 20×. Data are representative of two biological replicates in each group with essentially identical results. M, medulla; C, cortex. B, representative result of flow cytometry analysis shows gating on MHC-IIhiUEA-1hi (double high) mature mTECs (square A) based on gating on the CD45−TSC population. C, summarized results of B showing the percentage of MHC-IIhiUEA-1hi mTECs in mice of three genotypes treated as shown in A.
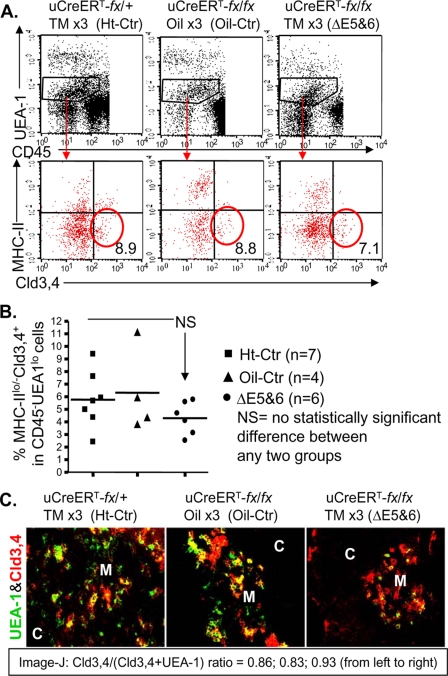
FIGURE 6.
Analysis of mTEC precursors based on the Cld-3,4 marker after ubiquitous postnatal FoxN1 deletion with TM x3 induction. A, representative result of flow cytometry analysis shows the CD45−UEA-1lo and MHC-IIlo/−Cld-3,4+ gates. B, summarized results of the percentage of MHC-IIlo/−Cld-3,4+ TECs in the CD45−UEA-1lo population based on three independent experiments with a total of 17 animals. Lines represent mean values in each group. C, representative result of immunofluorescence staining with antibodies to UEA-1 (green) and Cld-3,4 (red), magnification of 20×, from two biological replicates in each group with essentially identical results. M, medulla; C, cortex.
Flow Cytometry Assay
Cell strainer-isolated thymocytes (29) were stained with fluorochrome-conjugated anti-CD45, anti-CD4, and anti-CD8 (all from BioLegend). Enzymatically (1 mg/ml collagenase and 5 units/ml DNase I) dissociated thymic cells, including TECs (30), were stained with various combinations of fluorochrome-conjugated anti-CD45, anti-MHC-II, anti-Ly51 (all from BioLegend), anti-EpCam (G8.8; eBioscience or BioLegend), and anti-UEA-1 (Vector Laboratories) as well as anti-Cld-3,4, (Invitrogen, catalog No. 34-1700 and 36-4800). Data were acquired with a dual laser FACSCalibur system and analyzed using CellQuest or flowJo software.
Western Blot
As described in our previous publication (30), TSC-enriched total thymic cells were subjected to protein extraction with modified radioimmune precipitation assay buffer. Protein was loaded at ∼50 μg/lane under reducing conditions, and GAPDH was used as an internal loading control. Antibodies to phosphorylated p53 (Ser-15), Puma (catalog No. 4976), and caspase-8 (catalog No. 4927; recognizing 57- and 45-kDa bands, and activated 18- kDa caspase-8) were purchased from Cell Signaling Technology Inc. Antibody to p21 was purchased from BD Biosciences.
TUNEL Assay
This assay was performed as described in our previous publication using TSC-enriched thymic cells (30). The cells were treated with 2.4G2 hybridoma supernatant (from ATCC) to block Fc receptors stained with phycoerythrin/Cy5-anti-CD45, phycoerythrin-anti-MHC class II, and biotin-anti-UEA-1/APC-streptavidin and then fixed with 2% paraformaldehyde/phosphate-buffered saline and permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate, pH 7.2. Terminal transferase reactions were then performed with the in situ cell death detection kit (Roche Applied Science) for the TUNEL assay. A negative control without the terminal deoxynucleotidyl transferase was included with each sample.
Statistics
Groups were compared by the paired Student's t test; p < 0.05 was considered statistically significant. All measurements of variance are presented as S.E.
RESULTS
Generation and Characterization of Conditional FoxN1 Gene Knock-out Mice
The fx mouse was generated by a gene-targeting strategy that utilized the Cre-loxP and Flp-FRT recombinase systems. A DNA fragment corresponding to nt 36067–47099 (XhoI site) of FoxN1 (GenBankTM accession number Y12488) (31) in the pBluescript plasmid was used for the targeting construct. The FRT-Neor-FRT-loxP cassette (supplemental Fig. S1A), used for positive selection of embryonic stem cells, was inserted into the BglII site of the FoxN1 fragment corresponding to nt 40698, yielding a band ∼2 kb larger than the WT band when Southern blot was used to screen embryonic stem cell-targeting clones (supplemental Fig. S1B). The 5′-arm loxP site was inserted into the KpnI site of the FoxN1 fragment corresponding to nt 38339, and the KpnI site was then destroyed. The Neor/Neor homozygous mouse showed a typical FoxN1nu/nu phenotype, including athymia and hairlessness (supplemental Fig. S2A). This suggests that corrupting the germ line FoxN1 DNA at the site we selected resulted in the FoxN1-null phenotype. The Neor insert was deleted by in vivo Flp-mediated excision by cross-breeding with ActFlpe transgenic mice (The Jackson Laboratory; catalog No. 003800) (32). The FoxN1fx locus lost the Neor insert but retained FRT (48 bp) and loxP (34 bp) sites, producing a PCR product that was 82 bp larger than the WT amplicon when amplified with primers A and B (supplemental Fig. S2B). Homozygous (fx/fx-only) mice were phenotypically normal, including their hair, thymic size, and thymocyte number, as discussed below. Their FoxN1 mRNA expression was the same as that of WT mice (data not shown and Fig. 1A, right panel).
Heterozygous and homozygous fx mice carrying the pCAGG-CreERTM transgene (21), which is ubiquitously expressed and TM-inducible (uCreERT), are denoted as uCreERT-fx/+ and uCreERT-fx/fx mice, respectively. Both mice were injected intraperitoneally with TM to yield heterozygous control (Ht-Ctr) and ubiquitously FoxN1-deleted (ΔE5&6) mice (supplemental Fig. S3A), respectively, and induction of FoxN1 deletion was measured in different organs (supplemental Fig. S3, B and C). The percentage of FoxN1 deletion in thymic cells was ∼50% with three-dose TM induction. Because FoxN1 is functional only in TECs in the thymus, we evaluated FoxN1 deletion in TECs. The percentage of FoxN1 deletion in purified TECs (CD45−MHC-II+ cells) with five-dose TM induction was increased to >74% (supplemental Fig. S4A) in the uCreERT-fx/fx mice. Compared with germ line gene excision, induced genomic excision in somatic cells usually causes a variable degree of deletion in different organs depending on the TM dose and duration of administration (26, 33). FoxN1 mRNA expression in FACS-sorted TECs (CD45−MHC-II+ population) in uCreERT-fx/fx mice with three-dose TM treatment was reduced by more than 60% compared with the fx/fx mice without Cre-recombinase and compared with uCreERT-fx/fx mice treated with the oil vehicle (Fig. 1A, left panel). In contrast, there was no significant difference in FoxN1 expression in TEC-enriched thymic cells of WT and fx/fx-only mice without Cre-recombinase (Fig. 1A, right panel). To confirm that the FoxN1 mRNA expression reflected lower protein levels, we performed immunofluorescence staining of the thymi with anti-FoxN1 (28). uCreERT-fx/fx mice treated with three-dose TM showed a ∼70% reduction of FoxN1+ TECs compared with Ht-Ctr mice (Fig. 1B, upper panel and table (lower panel)). Therefore, FoxN1 was substantially deleted from TECs of uCreERT-fx/fx mice with ≥3 doses of TM treatment, but it was not affected in fx/fx-only mice without Cre-recombinase.
Inducing a Ubiquitous FoxN1 Deletion in the Postnatal Thymus Caused Acute Thymic Atrophy
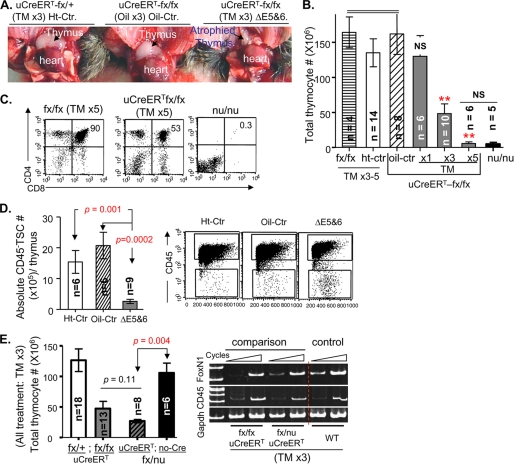
Mice of different genotypes were injected intraperitoneally with TM for 1, 3, or 5 successive days (TM x1, x3, or x5) to induce FoxN1 deletion, and phenotypes were analyzed 3–5 days after the last injection. After three doses of TM, the thymi of uCreERT-fx/fx mice were markedly reduced in size (Fig. 2A), and thymocyte number was significantly decreased to approximately one-third of control values (Fig. 2B, TM x3). After five doses of TM, the thymus almost disappeared, and the cell number decreased by ∼95% (Fig. 2B, TM x5), which was not significantly different from that in congenital FoxN1-null mutant (nu/nu) mice. However, the thymi in TM-treated uCreERT-fx/fx mice contained all mature thymocyte subpopulations, although the percentage of CD4+8+ double positive (DP) cells was reduced compared with findings in fx/fx-only or WT mice (Fig. 2C). In contrast, nu/nu mice did not have any DP or CD4+ or CD8+ single positive (SP) thymocytes (Fig. 2C). The reduced absolute thymocyte number in uCreERT-fx/fx mice was mostly due to reduced numbers of DP cells, which constitute ∼90% of the cells in the normal young adult thymus but only ∼50% in TM-treated uCreERT-fx/fx mice (Fig. 2C). More detailed studies of changes in T-cell developmental checkpoints in fx mice will be addressed in a subsequent report. Because FoxN1 is expressed by TECs, we also examined the total number of TSCs (CD45− population), the majority of which are TECs. The number of TSCs was markedly reduced in ΔE5&6 thymi (Fig. 2D).
FIGURE 2.
Ubiquitous postnatal deletion of FoxN1 caused acute thymic atrophy. In all panels: Ht-Ctr, uCreERT-fx/+ mice treated with TM; oil-Ctr, uCreERT-fx/fx mice treated with oil vehicle; ΔE5&6, uCreERT-fx/fx mice treated with TM, which deleted exons 5 and 6. Analyses were performed 4–5 days after treatment with TM or oil vehicle for 3 successive days (TM x3 or Oil x3) unless indicated otherwise. n = number of animals. A, gross appearance of thymi in mice with different FoxN1 genotypes, treated with TM x3 or oil x3. B, summary of total thymocyte numbers from mice with different genotypes, treated with oil vehicle or TM. Data are based on a total of 53 animals. The three filled bars represent uCreERT-fx/fx mice injected with TM for 1, 3, or 5 successive days. NS (in x1 group), no statistically significant difference versus the fx/fx, Ht-Ctr, or oil-Ctr group. **, p < 0.01 in x3 or x5 groups versus the fx/fx, Ht-Ctr, or oil-Ctr group. The far right dark bar represents FoxN1-null (nu/nu) congenital mutant mice. NS (between x5 and nu/nu groups), no statistically significant difference between these two groups. C, flow cytometry analyses of CD4 versus CD8 from fx/fx-only and uCreERT-fx/fx mice treated with TM for 5 successive days and nu/nu congenital mutant mice. D, mean values ± S.E. of the absolute numbers of CD45− TSCs in thymi with a conditional FoxN1 deletion and controls (left panel) and a representative flow cytometry analysis (right panel) of total thymic cells dissociated with collagenase/DNase I, showing CD45− TSCs (lower, rectangular gates). E, comparison of the efficacy of inactivation of floxed FoxN1 by TM induction in uCreERT-fx/nu and uCreERT-fx/fx mice. Left panel, absolute number of thymocytes in uCreERT-fx/+ (n = 18), uCreERT-fx/fx (n = 13), uCreERT-fx/nu (n = 8), and fx/nu without Cre-recombinase (n = 6) mice (from left to right). Right panel, representative semiquantitative RT-PCR result shows FoxN1 expression in the TSC-enriched thymi of uCreERT-fx/fx, uCreERT-fx/nu, and WT (control) mice treated with TM x3. Expression of CD45 served as a marker of hematopoietic cell contamination, and GAPDH was used as an internal control. Data are representative of at least two separate experiments.
To look at the efficacy of inactivation of floxed FoxN1 by TM induction, we made uCreERT-fx/nu mice by mating uCreERT-fx/fx with FoxN1-null (nude) mice. In uCreERT-fx/nu mice, one copy of the FoxN1 gene is the germ line null mutation from nude mice. When TM x3 was used to delete FoxN1 in uCreERT-fx/nu and uCreERT-fx/fx thymi, both groups had similar numbers of significantly reduced thymocytes compared with uCreERT-fx/+ or fx/nu without Cre-recombinase (no-Cre) mice (Fig. 2E, left panel). In addition, the levels of loss of FoxN1 mRNA were generally the same in TM-induced uCreERT-fx/fx and uCreERT-fx/nu mice (Fig. 2E, right panel). These results suggest that the efficacy of the inactivation of two copies of floxed FoxN1 in uCreERT-fx/fx mice was similar to that of inactivation of one floxed FoxN1 copy in uCreERT-fx/nu mice.
Injection of WT and fx/fx-only (without Cre) mice with TM did not reduce the thymic size, thymocyte numbers, or percentage of DP cells compared with injection of oil vehicle (supplemental Fig. S3D and data not shown), demonstrating that our dosage regimen of TM (x3–5) is not toxic to the thymus. Although three doses of TM can cause one copy of FoxN1 excision in uCreERT-fx/+ mice, these changes did not immediately affect thymic size and thymocyte numbers (Fig. 2, A and B) in young mice.
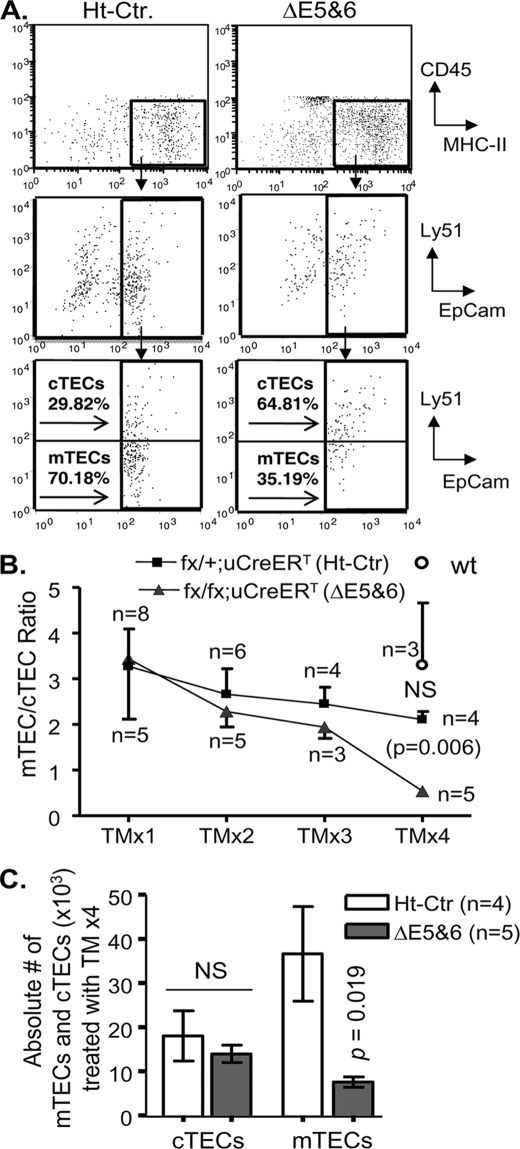
Inducing a Ubiquitous FoxN1 Deletion Caused Dose-related Selective Loss of mTECs
TECs consist of two major subsets, cTECs and mTECs, based on their localization, molecular markers, and functions. The cTECs are generally CD45−, MHC class II+, EpCam+, and Ly51+ (34, 35), whereas the mTECs may include several subpopulations based on shape (stellate or globular), expression of K5, K14, K8 (20, 36), and UEA-1 (37), as well as different levels of MHC class II. The two major phenotypic mTEC subsets are one possessing a stellate morphology with K5+, K14+, and UEA-1− and the other having a globular shape with high levels of MHC-II, K8+K5−K14−, and UEA-1+. Because FoxN1 may not regulate all TEC subpopulations in the adult thymus (28), we used flow cytometry to analyze TEC subpopulations affected by ubiquitous deletion of FoxN1. Because the total number of TSCs was markedly reduced in the ΔE5&6 thymus (Fig. 2D), the reduction in proportion of mTECs (lower gates in Fig. 3A, bottom panels) indicates a substantial decrease in the absolute number of mTECs. In contrast, the proportion of cTECs (Fig. 3A, bottom panels, upper gates) increased in TM-treated uCreERT-fx/fx (ΔE5&6) mice. The ratio of mTECs/cTECs fell steadily as the number of doses of TM treatment increased. After treatment with four doses of tamoxifen (TM x4), the mTEC/cTEC ratio was significantly reduced in ΔE5&6 mice compared with ΔE5&6 mice treated with one dose of TM or compared with WT or Ht-Ctr mice treated with four doses of TM (Fig. 3B), whereas there was no reduction in WT mice compared with TM x1. In Ht-Ctr mice, the mTEC/cTEC ratio fell slightly with increased TM doses, but this decrease was not statistically significant (Fig. 3B). The absolute number of mTECs was also significantly reduced in ΔE5&6 mice but the number of cTECs was not, as compared with heterozygous controls (Fig. 3C). These findings suggest that, in the young adult thymus, mTECs are more susceptible than cTECs to loss of FoxN1.
FIGURE 3.
Reduction of the mTEC/cTEC ratio in young mice with a ubiquitous FoxN1 deletion was associated with the dose of TM induction. A, representative flow cytometry plots of thymic cells from uCreERT-fx/+ (Ht-Ctr) and uCreERT-fx/fx (ΔE5&6) mice treated with four doses of TM. The top panels show the results of gating on CD45−MHC-II+ cells. The middle and bottom panels show the results of gating on CD45−MHC-II+ cells and staining with Ly51 versus EpCam. B, summarized ratios of mTEC/cTEC in Ht-Ctr mice (filled squares), ΔE5&6 mice (triangles), and WT mice (circle) based on flow cytometry analyses shown in A. We gated on CD45−MHC-II+ cell and measured the percentages of EpCam+Ly51+ cells (cTECs) and EpCam+Ly51− cells (mTECs). Means ± S.E. are shown for a total of 22 Ht-Ctr mice, 18 ΔE5&6 mice, and 3 WT mice; n = number of animals treated with the indicated TM dose. NS, not significant between WT and Ht-Ctr. C, summarized absolute cell numbers of cTECs (CD45−MHC-II+EpCam+Ly51+ population) and mTECs (CD45−MHC-II+EpCam+Ly51− population) from four Ht-Ctr and five ΔE5&6 mice treated with TM x4. Means ± S.E. are shown.
Acute Thymic Atrophy Was Induced by K5-CreERT but Not by K18-CreERT, although Both CreERT Transgenes Caused Thymic FoxN1 Deletion
The results shown above suggest that postnatal mTECs are more susceptible than cTECs to loss of FoxN1. As an alternative means of evaluating this finding, we produced mice in which we could selectively delete FoxN1 from major cTEC and mTEC subpopulations at the somatic cell level. fx mice were cross-bred with K5-CreERT or K18-CreERT mice to obtain K5-/K18-CreERT-fx/fx or -fx/+ mice. In the mature thymus, K5+ TECs are distributed primarily in the medulla, although K5+K8+ double positive TECs are located at the corticomedullary junction (CMJ), and some K5+ TECs are considered TEC progenitors. K18+ TECs, similar to K8+ TECs, are located mainly in the cortical region, although a few are present in the medulla (20). Therefore, treatment of K5- and K18-CreERT-fx/fx mice with TM should selectively delete FoxN1 from major mTEC and cTEC subpopulations in somatic cells rather than in the germ line of the postnatal thymus, respectively.
Mice with four different genotypes were injected intraperitoneally with TM for 3–5 successive days (TM x3–5) to induce FoxN1 deletion in keratin-specific TECs, and phenotypes were analyzed 5 days after the last injection. K5CreERT-fx/fx mice showed a slight reduction in thymic size after three doses of TM and a more marked reduction after five doses of TM (Fig. 4A). Thymic atrophy caused by conditional FoxN1 deletion did not change for 6 months.4 This atrophy was associated with a significantly reduced absolute number of total thymocytes (Fig. 4B, left panel), particularly the percentage and absolute number of DP cells (Fig. 4B, right panel), as well as the number of CD45−MHC-II+ TECs (Fig. 4C). In addition, the atrophy was associated with architectural changes in the medullary regions and/or the CMJ, which became indistinct (Fig. 4D, second panel). Furthermore, the percentage of FoxN1+ TECs, which reflects the expression of FoxN1 protein, was reduced by ∼70% in the K5CreERT-fx/fx mice compared with the K5- and K18-CreERT-fx/+ mice (Fig. 4, E and F). In contrast, TM-treated K18CreERT-fx/fx mice showed no change in thymic size (Fig. 4A), thymocyte number (Fig. 4B), profile of CD4 versus CD8 cells (data not shown), TEC number (Fig. 4C), thymus architecture (Fig. 4D), and number of FoxN1+ TECs (Fig. 4F). This was not due to failure to induce deletion of FoxN1 genomic DNA in 5x TM-treated K18CreERT-fx/fx mice, as there was substantial deletion of FoxN1 genomic DNA (∼40%), more than that (∼20%) in the K5CreERT-fx/fx mice, in sorted CD45−MHC-II+ TECs in these mice (supplemental Fig. S4A). The ubiquitous promoter-driven CreERT showed the greatest degree of FoxN1 deletion in TECs, the K18 promoter showed intermediate levels of FoxN1 deletion, and the K5 promoter showed the lowest degree of FoxN1 deletion (supplemental Fig. S4A). Expression of FoxN1 mRNA in sorted CD45−MHC-II+ TECs was lowest in TM-treated uCreERT-fx/fx mice, intermediate in TM-treated K5CreERT-fx/fx mice, and greatest in TM-treated K18CreERT-fx/fx mice (supplemental Fig. S4B). This correlated directly with the extent of thymic atrophy in these three genotypes after TM treatment, which was most severe in TM-treated uCreERT-fx/fx mice, intermediate in TM-treated K5CreERT-fx/fx mice, and least in TM-treated K18CreERT-fx/fx mice (Fig. 4A). These findings support the conclusion that mTECs, which are the major subpopulation that expresses K5+, are highly FoxN1-dependent, whereas cTECs, which are the dominant K18+ TEC subpopulation, are less FoxN1-dependent in the postnatal thymus. This is significantly different from the fetal thymus, in which mTECs and cTECs are equally FoxN1-dependent (16).
Inducing a Ubiquitous FoxN1 Deletion Affected Mature mTECs More Than Presumptive mTEC Precursors
We next wished to evaluate the effect of FoxN1 loss on mature mTECs, which express high levels of UEA-1 and MHC-II. When we induced a ubiquitous FoxN1 deletion in the uCreERT-fx/fx mice, the overall percentage of mTECs that expressed both high levels of MHC-II and UEA-1 was significantly decreased as assessed by immunohistology (Fig. 5A, right panels) and flow cytometry (Fig. 5, B (gate A) and C). This result is similar to that in a recently published study of mice with a non-inducible FoxN1 mutation bearing the FoxN1-LacZ allele (19).
To determine whether postnatal FoxN1 deletion affects precursors of mature mTECs, we measured the percentage of CD45−UEA-1loMHC-IIlo/−Cld-3,4+ TECs, which are presumptive mTEC precursors (38). They were not substantially reduced by conditional FoxN1 deletion as assessed by flow cytometry (Fig. 6, A and B). Immunohistochemistry confirmed that the medulla in TM-treated uCreERT-fx/fx mice (ΔE5&6) showed no reduction in Cld-3,4+ TECs (red staining in Fig. 6C) but had a marked decrease in UEA-1+ TECs (green staining in Fig. 6C) per unit area compared with Ht-Ctr and oil-Ctr mice (Fig. 6C).
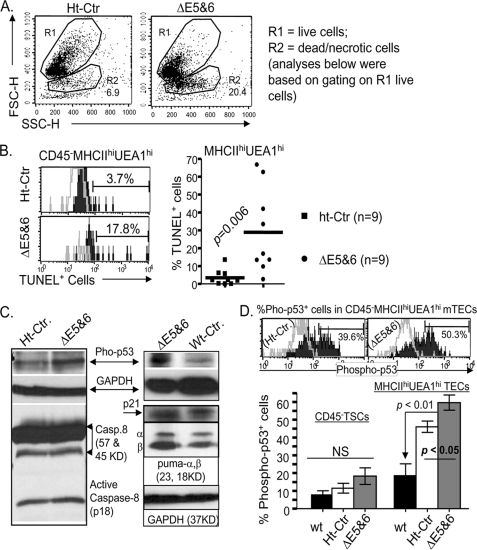
Inducing a Ubiquitous FoxN1 Deletion Increased Apoptosis and Activation of p53 in Mature mTECs
We next sought to determine whether the acutely reduced numbers of TECs after conditional FoxN1 deletion resulted from altered cell death. We first found a 3-fold increase in the number of dead cells in the conditional FoxN1-deleted mice compared with heterozygous controls, based on flow cytometry analysis of forward scatter and side scatter (Fig. 7A, R2 gate). When we gated on live cells (Fig. 7A, R1 gate), apoptosis, measured by staining with TUNEL (30), was increased 3-fold in total TSCs (data not shown) and ∼10-fold in CD45−MHC-IIhiUEA-1hi mature mTECs (Fig. 7B). These findings indicate that deleting FoxN1 may lead to the withdrawal of TEC survival signals and/or eliciting of TEC apoptotic signals. To identify possible mechanisms of apoptosis induced by FoxN1 deletion, we examined the activation of p53 and caspase-8, which are representative molecules that regulate intrinsic and extrinsic apoptotic pathways, respectively. Internal signals of cellular stress generally activate p53, whereas engagement of pro-apoptotic cell surface receptors, such as Fas ligand, activates the caspase-8 pathway (39). After conditional FoxN1 deletion, the thymi showed increased expression of phosphorylated p53 but almost no changes in activated caspase-8 (p18) expression (Fig. 7C). The p53 gene targets several downstream genes, in which Puma (p53-up-regulated modulator of apoptosis) is associated with the p53-activated apoptosis function, whereas p21 is related to p53-activated cell cycle arrest function. We found that FoxN1 deletion caused increased expression of phospho-p53 and Puma but not p21 (Fig. 7C, right panel). This finding correlated with a previous report that Puma is required for p53-dependent cell death in the thymus (40). Because FoxN1 is expressed in TECs, which are a small subpopulation of total thymic cells, we next quantified the expression of phosphorylated p53 in different TEC subsets. Using FACS intracellular staining, expression of phosphorylated p53 was increased specifically in MHC-IIhiUEA-1hi mature mTECs compared with total TSCs (Fig. 7D). These results suggest that conditional FoxN1 deletion causes TEC apoptosis by activation of the p53-dependent intrinsic pathway and the p53 response gene, Puma.
FIGURE 7.
Ubiquitous postnatal FoxN1 deletion caused thymic cell death and increased apoptosis in mature mTECs. In all panels: Ht-Ctr, uCreERT-fx/+ mice treated with TM x3; ΔE5&6, uCreERT-fx/fx mice treated with TM x3, which deleted exons 5 and 6. A, representative flow cytometric dot plots (FSC versus SSC) showing gating on live (R1) and dead and necrotic cells (R2) from ht-Ctr and ΔE5&6 mice. B, left panel shows a representative flow cytometry result of staining for TUNEL (filled histogram) and a negative control, i.e. without the terminal deoxynucleotidyl transferase enzyme (open histogram). The right panel shows summarized results of TUNEL+ apoptotic cells in two groups of nine mice each. Data were obtained by gating on live (R1) CD45−MHC-IIhiUEA-1hi mature mTECs. The numbers show the percentage of positively stained cells. Horizontal lines represent mean values. C, Western blotting analysis of activation of p53 and caspase-8, and the p53 targets, p21 and Puma. All mice were analyzed ∼16 h after treatment with TM for 3 successive days. Total thymic cells were subjected to SDS-PAGE and blotting with antibodies to phosphorylated p53 (Ser-15), active caspase-8 (p18), p21, and Puma. GAPDH served as a loading control. Data are representative of 2–3 biological replicates in each group with essentially identical results. D, flow cytometry analysis of phosphorylated p53 (Ser-15) in CD45−TSC and CD45−MHC-IIhiUEA-1hi mature mTEC subpopulations. The top panels show a representative result of staining CD45−MHC-IIhiUEA-1hi mature mTECs with anti-phosphorylated p53 (open histograms). The unfilled histograms show staining with secondary antibody (APC-conjugated anti-rabbit IgG) only. The bar graph (bottom) shows summarized results of the percentage of phospho-p53+ cells in CD45−TSCs (left group) and in mature mTECs (MHC-IIhiUEA-1hi (right group)) from six WT, seven Ht-Ctr, and seven ΔE5&6 mice. The percentage of phospho-p53+ mTECs in ΔE5&6 was compared with that in Ht-Ctr and WT mice by t test; the p values are shown in the graph. NS, not significant between the groups.
DISCUSSION
In this study, we found that deletion of FoxN1 in the postnatal thymus in an inducible FoxN1 gene knock-out mouse model caused acute thymic atrophy, characterized by more severe effects on mTECs, particularly MHC-IIhiUEA-1hi mature mTECs, with less marked reductions in Cld-3,4+ presumptive mTEC precursors and cTECs. These changes were due to increased cell death and apoptosis associated with activation of the p53 gene. We speculate that the loss of FoxN1 increases cell stress in postnatal mTECs, which triggers stress-induced intrinsic apoptotic pathways mediated by activation of the p53 gene. Because FoxN1 may not be involved directly in the signaling pathway of the apoptosis, we will investigate in future work the apoptotic mechanisms through a determination of whether thymus atrophy mediated by deletion of FoxN1 can be attenuated by lack of p53 expression, as when using p53 knock-out mouse.
Identification of cTECs and mTECs in histological sections was initially based on anatomic location with hematoxylin and eosin staining. Now anti-keratin (K) immunofluorescent antibodies (20) have been widely used. K8 and K18 are expressed in cTECs, whereas K5 and K14 are expressed in mTECs, and K5+K8+ co-stained TECs are localized in CMJ (20, 41) in the mature thymus. A K5 promoter-driven IKKα transgene shows selective activity in the thymic medulla in K5+ and UEA-1+ mTECs but is not involved in cTECs (42), implying that the K5 promoter specifically (or mainly) drives mTECs. Furthermore, a FoxN1-LacZ mouse with reduced postnatal FoxN1 expression has also been found to have decreased numbers of mature mTECs (MHC-IIhiUEA-1+ subset) but not cTECs (19).
The K5 promoter exhibits activity in epithelial progenitor/stem cells on the basal layer of two-dimensional epithelial cells in the skin and mammary gland (43–45). It may also exhibit activity in thymic epithelial progenitor/stem cells, at least in the fetal stage, but the persistence of TEC progenitors in an organized three-dimensional adult thymus remains controversial (46). However, it is clear that mature TECs are constantly regenerated from TEC progenitor/stem cells in situ (47). Furthermore, it has been suggested that K5+K8+ co-staining TECs at the CMJ (20, 48) or certain TECs in the postnatal thymic medulla (49–51), which mainly contains K5+ mTECs (20, 36), are potential TEC progenitor cells. We favor the hypothesis that in the adult thymus, some K5 promoter-driven TECs are progenitors at the CMJ, where, without long distance migration, they can easily differentiate into either cTECs or mTECs; such a feature of bipotential TEC progenitors has been described elsewhere (18, 52). Indeed, the defect in the CMJ in TM-induced K5CreERT-fx/fx thymus (Fig. 4D, second panel) further supports this concept. If TEC progenitors are present in the adult thymus, the defects in our conditional FoxN1 knock-out mice could be caused in part by blocking differentiation from TEC progenitor cells to mature mTECs.
Although mTECs and cTECs are equally FoxN1-dependent during fetal thymic organogenesis (16), our current data have established that mTECs are more FoxN1-dependent than cTECs in the postnatal thymus (Table 1). Given a totally different CD4 versus CD8 thymocyte developmental profile in FoxN1-null nude mice and our ubiquitous and K5 promoter-driven FoxN1-deleted mice (Figs. 2C and 4B), it is clear that elimination of FoxN1 in the germ line during fetal life (nude mice) and in somatic cells postnatally (our conditional FoxN1 gene knock-out mice) has markedly different effects. Others have also reported differences in TEC FoxN1 dependence in fetal and adult thymi, showing that TECs expressing the chemokine CCL25 and the Notch ligand Dll4 are FoxN1-dependent in the fetal thymus but are FoxN1-independent postnatally (28).
TABLE 1.
TEC subset sensitivity to loss of FoxN1 in the postnatal thymus
| TEC subset | Sensitivity to loss of FoxN1 | As identified in this manuscript | Anatomic location | TEC categorya |
|---|---|---|---|---|
| MHC-IIhiUEA-1hi | Sensitive | uCreERT-mediated FoxN1 recombination + IF staining and FACS analysis | Medullary region | mTECs (19, 37) |
| K5 promoter-driven TECs (at somatic cell level) | Sensitive | K5 promoter-driven CreERT-mediated FoxN1 recombination + H&E staining and FACS analysis | Medullary region | mTECs (20, 36, 42) |
| CD45−MHC-II+ EpCam+Ly51− | Sensitive | uCreERT-mediated FoxN1 recombination + FACS analysis | Not confirmed | mTECs (34, 35) |
| Cld3,4+ TECs (CD45−UEA-1lo MHC-IIlo/+ Cla3,4+) | Not sensitive | uCreERT-mediated FoxN1 recombination + IF staining and FACS analysis | Medullary region | Presumptive mTEC precursors (38) |
| CD45−MHC-II+ EpCam+Ly51+ | Not sensitive | uCreERT-mediated FoxN1 recombination + FACS analysis | Cortical region | cTECs (34, 35) |
| K18 promoter-driven TECs (at somatic cell level) | Not sensitive | K18 promoter-driven CreERT-mediated FoxN1 recombination + H&E staining and FACS | Cortical region | cTECs (20, 36) |
a Based on published reports.
Our findings show that in the postnatal thymus, UEA-1+ globular mTECs and K5 promoter-driven TECs (stellate mTECs and/or TEC progenitors in the adult thymus) are sensitive to the loss of FoxN1 (Figs. 4 and 5). By using FACS analysis with a combination of cell surface markers, we also determined that CD45-negative, MHC-II+ EpCam+Ly51− TECs (reported to be mTECs (34, 35)), but not CD45-negative, MHC-II+EpCam+Ly51+ TECs (reported to be cTECs (34, 35)), are FoxN1-sensitive to the ubiquitous loss of FoxN1 (Fig. 3). In the postnatal thymus, mTECs can also be subdivided into immature and mature mTECs based on CD80 and Cld-3,4 markers (38, 53). Immature TECs (MHC-IIloCD80− and/or MHC-IIloCld-3,4+ TECs) in the medullary region are presumptive mTEC precursors (38). Our results show that loss of FoxN1 does not affect these mTEC precursors (Fig. 6). As for cTECs, they express K8 and its pairing partner, K18, markers. We found that K18 promoter-driven TECs (mostly cTECs but not TEC progenitor/stem cells) are not FoxN1-dependent in the postnatal thymus, as K18 promoter-driven CreERT mice did not exhibit thymic epithelial defects (Fig. 4) despite effective FoxN1 deletion (supplemental Fig. S4A). We have summarized our results on the FoxN1 dependence of postnatal TEC subsets in Table 1.
One puzzle is why the deletion of FoxN1 resulted in the loss of DP thymocytes in the ubiquitous or K5 promoter-driven FoxN1 knock-out (Figs. 2C and 4B), because DP cells reside in the cortex. There are at least two potential explanations. First, deterioration of mTECs may affect the integration of the entire thymic epithelial network and alter the cortical thymic microenvironment, resulting in reduced volume and dramatic shrinking of the thymus. Second, the loss of FoxN1 may cause a loss or defect in differentiation of common TEC progenitors, as noted above, resulting in reduced cTEC number, because a common TEC progenitor population has the bipotential to differentiate into either cortical or medullary TECs (18, 52). Therefore, loss of FoxN1 possibly causes a passive or FoxN1-independent defect in cTECs.
The floxed FoxN1 site that we targeted for disruption was essential for FoxN1 function, because disrupting this site in the germ line with a Neor cassette caused a typical FoxN1-null phenotype (supplemental Fig. S2A). Treatment of uCreERT-fx/fx mice with 3–5 doses of TM resulted in a 50–75% FoxN1 gene deletion in somatic cells of different organs (supplemental Figs. S3C and S4A and data not shown), consistent with most reported conditional gene knock-out models, in which the levels of deletion in somatic cells range from 30 to 80% depending on dose, induction time, and the organ studied (26, 33).
We chose to delete FoxN1 from fx/fx mice rather than fx/nu mice, with one copy of a FoxN1 germ line null mutation, because the FoxN1 gene is considered to exhibit haploinsufficiency. Young +/nu heterozygous mice have mildly reduced thymic size (54, 55) and may have premature TEC dysfunction in the fetal and postnatal stage due to the presence of one copy of FoxN1 carrying a germ line null mutation. This would make it difficult to determine whether TEC changes result from induced postnatal loss of FoxN1 or from prenatal haploinsufficiency. Furthermore, we confirmed that the efficacy of inactivation of two copies of floxed FoxN1 in uCreERT-fx/fx mice was similar to that of inactivation of one floxed FoxN1 copy in uCreERT-fx/nu mice (Fig. 2E).
Aging causes thymic involution, but the molecular bases of genetic pathways that control this process are poorly understood. The decline of FoxN1 expression is associated with aging and has been hypothesized to cause age-related thymic involution (56). However, it is also possible that reduced FoxN1 expression may be an effect of aging-induced TEC deterioration. Our inducible FoxN1 deletion mouse model will be useful in distinguishing these possibilities and determining the cause-and-effect relationship between the decline of FoxN1 expression and age-related TEC deterioration.
Together, our current findings suggest that FoxN1 is essential for the maintenance of the steady-state mTEC microenvironment to prevent acute thymic atrophy through regulating apoptosis and promoting epithelial differentiation in the postnatal thymus. Investigation of the molecular mechanisms by which FoxN1 controls thymic homeostasis will facilitate the development of strategies to restore thymic function by reversing the immunologic changes resulting from thymic atrophy and involution (57).
Acknowledgments
We thank Dr. Nancy R. Manley (University of Georgia) for providing FoxN1 genomic DNA and for advice on initial gene-targeting design; Dr. Jason Liu (University of California San Diego) for technical advice; Dr. Eva Zsigmond (University of Texas Health Science Center at Houston) for technical assistance with embryonic stem cell transfection and microinjection of targeted embryonic stem cell clones; Dr. Amy Tvinnereim (University of Texas Health Science Center at Tyler (UTHSCT)) for thymic epithelial cell sorting; and Sandra E. Nash (UTHSCT) for cryosectioning.
This work was supported, in whole or in part, by National Institutes of Health Grants R21AI064939, R01AI081995, and R21AI079747 (to D.-M. S.) from NIAID. This work was also supported by the University of Texas Health Science Center at Tyler.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
L. Sun, J. Guo, R. Brown, T. Amagai, Y. Zhao, and D.-M. Su, unpublished data.
- TSC
- thymic stromal cell
- TEC
- thymic epithelial cell
- mTEC
- medullary TEC
- cTEC
- cortical TEC
- TM
- tamoxifen
- UEA-1
- Ulex europaeus agglutinin-1
- WT
- wild type
- K5
- keratin-5
- nt
- nucleotide(s)
- FACS
- fluorescence-activated cell sorter
- Cld-3,4
- claudin-3,4
- MHC
- major histocompatibility complex
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TUNEL
- terminal dUTP nick-end labeling
- Ht-Ctr
- heterozygous control
- DP
- double positive
- CMJ
- corticomedullary junction
- RT
- reverse transcription
- FRT
- Flippase recombination target.
REFERENCES
- 1.Spits H. (2002) Nat. Rev. Immunol. 2, 760–772 [DOI] [PubMed] [Google Scholar]
- 2.Petrie H. T., Kincade P. W. (2005) J. Exp. Med. 202, 11–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kronenberg M., Rudensky A. (2005) Nature 435, 598–604 [DOI] [PubMed] [Google Scholar]
- 4.Petrie H. T. (2002) Immunol. Rev. 189, 8–19 [DOI] [PubMed] [Google Scholar]
- 5.Gruver A. L., Sempowski G. D. (2008) J. Leukoc. Biol. 84, 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taub D. D., Longo D. L. (2005) Immunol. Rev. 205, 72–93 [DOI] [PubMed] [Google Scholar]
- 7.Ladi E., Yin X., Chtanova T., Robey E. A. (2006) Nat. Immunol. 7, 338–343 [DOI] [PubMed] [Google Scholar]
- 8.Coffer P. J., Burgering B. M. (2004) Nat. Rev. Immunol. 4, 889–899 [DOI] [PubMed] [Google Scholar]
- 9.Mecklenburg L., Tychsen B., Paus R. (2005) Exp. Dermatol. 14, 797–810 [DOI] [PubMed] [Google Scholar]
- 10.Nehls M., Pfeifer D., Schorpp M., Hedrich H., Boehm T. (1994) Nature 372, 103–107 [DOI] [PubMed] [Google Scholar]
- 11.Nehls M., Kyewski B., Messerle M., Waldschütz R., Schüddekopf K., Smith A. J., Boehm T. (1996) Science 272, 886–889 [DOI] [PubMed] [Google Scholar]
- 12.Cunningham-Rundles C., Ponda P. P. (2005) Nat. Rev. Immunol. 5, 880–892 [DOI] [PubMed] [Google Scholar]
- 13.Adriani M., Martinez-Mir A., Fusco F., Busiello R., Frank J., Telese S., Matrecano E., Ursini M. V., Christiano A. M., Pignata C. (2004) Ann. Hum. Genet. 68, 265–268 [DOI] [PubMed] [Google Scholar]
- 14.Frank J., Pignata C., Panteleyev A. A., Prowse D. M., Baden H., Weiner L., Gaetaniello L., Ahmad W., Pozzi N., Cserhalmi-Friedman P. B., Aita V. M., Uyttendaele H., Gordon D., Ott J., Brissette J. L., Christiano A. M. (1999) Nature 398, 473–474 [DOI] [PubMed] [Google Scholar]
- 15.Amorosi S., D'Armiento M., Calcagno G., Russo I., Adriani M., Christiano A. M., Weiner L., Brissette J. L., Pignata C. (2008) Clin. Genet. 73, 380–384 [DOI] [PubMed] [Google Scholar]
- 16.Shakib S., Desanti G. E., Jenkinson W. E., Parnell S. M., Jenkinson E. J., Anderson G. (2009) J. Immunol. 182, 130–137 [DOI] [PubMed] [Google Scholar]
- 17.Su D. M., Navarre S., Oh W. J., Condie B. G., Manley N. R. (2003) Nat. Immunol. 4, 1128–1135 [DOI] [PubMed] [Google Scholar]
- 18.Bleul C. C., Corbeaux T., Reuter A., Fisch P., Mönting J. S., Boehm T. (2006) Nature 441, 992–996 [DOI] [PubMed] [Google Scholar]
- 19.Chen L., Xiao S., Manley N. R. (2009) Blood 113, 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klug D. B., Carter C., Crouch E., Roop D., Conti C. J., Richie E. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11822–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi S., McMahon A. P. (2002) Dev. Biol. 244, 305–318 [DOI] [PubMed] [Google Scholar]
- 22.Indra A. K., Warot X., Brocard J., Bornert J. M., Xiao J. H., Chambon P., Metzger D. (1999) Nucleic Acids Res. 27, 4324–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen F., Cecena G., Munoz-Ritchie V., Fuchs E., Chambon P., Oshima R. G. (2003) Genesis 35, 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schüddekopf K., Schorpp M., Boehm T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9661–9664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brissette J. L., Li J., Kamimura J., Lee D., Dotto G. P. (1996) Genes Dev. 10, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 26.Schwenk F., Kuhn R., Angrand P. O., Rajewsky K., Stewart A. F. (1998) Nucleic Acids Res. 26, 1427–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 28.Itoi M., Tsukamoto N., Amagai T. (2007) Int. Immunol. 19, 127–132 [DOI] [PubMed] [Google Scholar]
- 29.Zhu X., Gui J., Dohkan J., Cheng L., Barnes P. F., Su D. M. (2007) Aging Cell 6, 663–672 [DOI] [PubMed] [Google Scholar]
- 30.Gui J., Zhu X., Dohkan J., Cheng L., Barnes P. F., Su D. M. (2007) Int. Immunol. 19, 1201–1211 [DOI] [PubMed] [Google Scholar]
- 31.Schorpp M., Hofmann M., Dear T. N., Boehm T. (1997) Immunogenetics 46, 509–515 [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez C. I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A. F., Dymecki S. M. (2000) Nat. Genet. 25, 139–140 [DOI] [PubMed] [Google Scholar]
- 33.Seibler J., Zevnik B., Küter-Luks B., Andreas S., Kern H., Hennek T., Rode A., Heimann C., Faust N., Kauselmann G., Schoor M., Jaenisch R., Rajewsky K., Kühn R., Schwenk F. (2003) Nucleic Acids Res. 31, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray D. H., Chidgey A. P., Boyd R. L. (2002) J. Immunol. Methods 260, 15–28 [DOI] [PubMed] [Google Scholar]
- 35.Gray D. H., Seach N., Ueno T., Milton M. K., Liston A., Lew A. M., Goodnow C. C., Boyd R. L. (2006) Blood 108, 3777–3785 [DOI] [PubMed] [Google Scholar]
- 36.Surh C. D., Gao E. K., Kosaka H., Lo D., Ahn C., Murphy D. B., Karlsson L., Peterson P., Sprent J. (1992) J. Exp. Med. 176, 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farr A. G., Anderson S. K. (1985) J. Immunol. 134, 2971–2977 [PubMed] [Google Scholar]
- 38.Hamazaki Y., Fujita H., Kobayashi T., Choi Y., Scott H. S., Matsumoto M., Minato N. (2007) Nat. Immunol. 8, 304–311 [DOI] [PubMed] [Google Scholar]
- 39.Hsu H. C., Scott D. K., Mountz J. D. (2005) Immunol. Rev. 205, 130–146 [DOI] [PubMed] [Google Scholar]
- 40.Jeffers J. R., Parganas E., Lee Y., Yang C., Wang J., Brennan J., MacLean K. H., Han J., Chittenden T., Ihle J. N., McKinnon P. J., Cleveland J. L., Zambetti G. P. (2003) Cancer Cell 4, 321–328 [DOI] [PubMed] [Google Scholar]
- 41.Klug D. B., Carter C., Gimenez-Conti I. B., Richie E. R. (2002) J. Immunol. 169, 2842–2845 [DOI] [PubMed] [Google Scholar]
- 42.Lomada D., Liu B., Coghlan L., Hu Y., Richie E. R. (2007) J. Immunol. 178, 829–837 [DOI] [PubMed] [Google Scholar]
- 43.Vassar R., Rosenberg M., Ross S., Tyner A., Fuchs E. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1563–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuraguchi M., Ohene-Baah N. Y., Sonkin D., Bronson R. T., Kucherlapati R. (2009) PLoS Genet. 5, e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanpain C., Lowry W. E., Geoghegan A., Polak L., Fuchs E. (2004) Cell 118, 635–648 [DOI] [PubMed] [Google Scholar]
- 46.Swann J. B., Boehm T. (2007) Eur. J. Immunol. 37, 2364–2366 [DOI] [PubMed] [Google Scholar]
- 47.Anderson G., Jenkinson E. J., Rodewald H. R. (2009) Eur. J. Immunol. 39, 1694–1699 [DOI] [PubMed] [Google Scholar]
- 48.Klug D. B., Crouch E., Carter C., Coghlan L., Conti C. J., Richie E. R. (2000) J. Immunol. 164, 1881–1888 [DOI] [PubMed] [Google Scholar]
- 49.Rodewald H. R., Paul S., Haller C., Bluethmann H., Blum C. (2001) Nature 414, 763–768 [DOI] [PubMed] [Google Scholar]
- 50.Farr A. G., Dooley J. L., Erickson M. (2002) Immunol. Rev. 189, 20–27 [DOI] [PubMed] [Google Scholar]
- 51.Gillard G. O., Farr A. G. (2006) J. Immunol. 176, 5815–5824 [DOI] [PubMed] [Google Scholar]
- 52.Rossi S. W., Jenkinson W. E., Anderson G., Jenkinson E. J. (2006) Nature 441, 988–991 [DOI] [PubMed] [Google Scholar]
- 53.Kyewski B., Klein L. (2006) Annu. Rev. Immunol. 24, 571–606 [DOI] [PubMed] [Google Scholar]
- 54.Scheiff J. M., Cordier A. C., Haumont S. (1978) Anat. Embryol. 153, 115–122 [DOI] [PubMed] [Google Scholar]
- 55.Kojima A., Saito M., Hioki K., Shimanura K., Habu S. (1984) Exp. Cell Biol. 52, 107–110 [PubMed] [Google Scholar]
- 56.Ortman C. L., Dittmar K. A., Witte P. L., Le P. T. (2002) Int. Immunol. 14, 813–822 [DOI] [PubMed] [Google Scholar]
- 57.Goldberg G. L., Zakrzewski J. L., Perales M. A., van den Brink M. R. (2007) Semin. Immunol. 19, 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]