Abstract
Neuronal migration is essential for proper cortical layer formation and brain function, because migration defects result in neurological disorders such as mental retardation and epilepsy. Neuronal migration is divided into several contiguous steps: early phase (multipolar mode), locomotion mode, and terminal translocation mode. The locomotion mode covers most of the migration route and thereby is the main contributor to cortical layer formation. However, analysis of the molecular mechanisms regulating this mode is difficult due to the secondary effects of defects at the early phase of migration. In this study, we established an ex vivo chemical inhibitor screening, allowing us to directly analyze the locomotion mode of migration. Roscovitine and PP2, inhibitors for Cdk5 and Src family kinases, respectively, suppressed the locomotion mode of migration. In line with this, a small percentage of Cdk5- or Src family kinase (Fyn)-knockdown cells exhibited locomoting morphology but retarded migration, although the majority of cells were stalled at the early phase of migration. We also showed that rottlerin, widely used as a specific inhibitor for protein kinase Cδ (PKCδ), suppressed the locomotion mode. Unexpectedly, however, the dominant-negative form as well as RNA interference for PKCδ hardly affected the locomotion, whereas they may disturb terminal translocation. In addition, we found JNK to be a potential downstream target of rottlerin. Taken together, our novel chemical inhibitor screening provides evidence that Cdk5 and Src family kinases regulate the locomotion mode of neuronal migration. It also uncovered roles for Fyn and PKCδ in the early and final phases of migration, respectively.
Keywords: Cell Migration, JNK, Neurodevelopment, Protein Kinase C, Signal Transduction, Cyclin-dependent Kinase 5, Electroporation, Microtubule-associated Protein 1B, Mouse Embryonic Cerebral Cortex, Slice Culture
Introduction
The mammalian cerebral cortex is a six-layered structure, whose formation is largely dependent on regulated neuronal migration during developmental stages. The disruption of cortical layer structure results in several neurological disorders with mental retardation and/or epilepsy, such as lissencephaly and periventricular heterotopia in humans (1–3). In addition, recent reports indicated that schizophrenia and dyslexia are associated with neuronal migration defects (4, 5). Thus, proper regulation of neuronal migration is a pivotal step for the formation of a functional brain with a normal layered structure.
During cortical development, post-mitotic neurons, generated near the ventricle, migrate toward the pial surface exhibiting various morphological changes and neuronal maturation events (3, 6). Neuronal migration is divided into several contiguous steps: the early phase of migration (the multipolar migration mode and some parts of neuronal maturation events), the locomotion mode, and the terminal translocation mode (see Fig. 1A). During the early phase of migration, the majority of post-mitotic neurons display multipolar morphology and undergo several neuronal maturation events, including neuronal polarity and axon formation, in the lower part of the intermediate zone. Subsequently, they transform into bipolar-shaped neurons with a leading process, a future apical dendrite. These bipolar-shaped neurons, called “locomoting neurons,” migrate over a relatively long distance from the intermediate zone to the upper part of the cortical plate (the locomotion mode of migration). At the final phase of migration, these locomoting neurons switch into the terminal translocation mode. While many neuronal maturation events occur at the early phase of migration, the longest part of the neuronal journey is dependent on the locomotion mode of migration. Thus locomotion mode is the principal component of proper layered cortical formation.
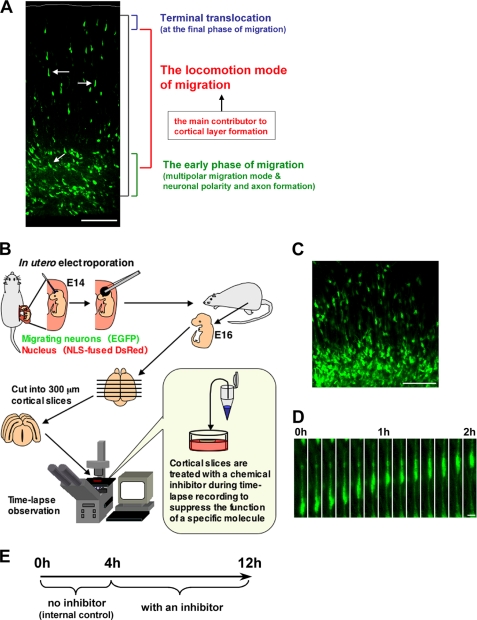
FIGURE 1.
An ex vivo chemical inhibitor screening for the molecules involved in locomotion. A, the cortical section of an electroporated brain, at E17, 3 days after electroporation. Black bracket shows the entire path of the neuronal journey at this stage. In the neuronal migration process, the locomotion mode (white arrow) covers most of the migration (red bracket). In contrast, at the early phase of the migration, neurons exhibit various morphological changes and neuronal maturation events, such as the formation of an axon and a leading process (green bracket). These various cellular events can hinder direct analysis of the mechanisms of the locomotion mode (red bracket) or the final phase (blue bracket) of neuronal migration. B and E, a schematic diagram for an ex vivo chemical inhibitor screening. Embryonic brains were electroporated with EGFP- and NLS-fused DsRed-expressing vectors at E14. After electroporation, the uterus was placed back into the abdominal cavity, allowing embryos to continue developing. 300-μm coronal slices were obtained with a microtome at E16, cultured on the insert membrane under time-lapse microscopy, and treated with chemical inhibitors as indicated. Because a few cells did not migrate even in the control slices, we compared the migration rate of inhibitor-treated locomoting neurons with that of the same one before the inhibitor treatment as an internal control (see “Results” and E). C, a cortical slice at the beginning of the slice tissue culture. At this stage (E16), the EGFP-electroporated cells were just transforming from multipolar neurons into locomoting neurons. D, time-lapse imaging of the locomoting neuron in a cultured tissue slice without any inhibitors. The cell can be observed to continuously migrate toward the pial surface with a locomoting morphology in the culture. Scale bars, 100 μm (A and C) and 10 μm (D).
We and others have identified several molecules involved in neuronal morphological changes and maturation events at the early stage of migration (3, 7). Cyclin-dependent kinase 5 (Cdk5)3 regulates the multipolar cell morphology and leading process formation (8), and its downstream molecules, DCX and Lis1, both of which are causative gene products of lissencephaly (9–11), are required for the transformation from multipolar to locomoting neurons with a leading process (12, 13). c-Jun N-terminal kinase (JNK), a kinase that phosphorylates microtubule-associated protein 1B (MAP1B) and DCX, is also required for the leading process formation (14–16). In addition, many other molecules regulating neuronal polarity have been reported (17). SAD kinase and LKB1 are involved in neuronal polarity and axon formation in the intermediate zone (18–20). These recent findings contribute toward the understanding of the molecular mechanisms regulating the morphological changes and neuronal maturation events during the early phase of neuronal migration.
After the formation of a leading process, neurons enter the locomotion mode of migration. Because the locomotion mode is the critical step for neuronal positioning and cortical layer formation, the molecular analysis of this mode of migration is needed to understand the mechanisms underlying brain construction and related neurological disorders, such as lissencephaly and schizophrenia. However, in contrast to the early phase of migration, it remains difficult to directly analyze the molecular mechanisms for the locomotion. A major hurdle is that no promoter whose activation marks the start of the locomotion mode has yet been identified; thus obstructing in vivo experiments using conditional knockout mice or in utero electroporation-mediated functional suppression technique (21, 22). Thus analysis of the in vivo mechanisms underlying the locomotion mode of migration is usually complicated by various secondary effects of defects arising at the early phase of migration, such as multipolar migration, leading process formation and the acquirement of neuronal polarity and axon.
In this study, we established a time-lapse imaging-based ex vivo chemical inhibitor screening method, allowing us to analyze the molecular mechanisms for the locomotion mode of neuronal migration in cortical tissues. Using this technique, we revealed that treatment with roscovitine, PP2, and bisindolylmaleimide I (BIM), inhibitors for Cdk5, Src family kinases, and protein kinase C (PKC), respectively, suppressed the migration rate of the locomoting neurons in cortical tissues. Consistently, a small percentage of the knockdown cells for Cdk5 or Fyn (an Src family kinase), exhibited locomoting morphology, but their migration in the cortical plate was retarded. Furthermore, treatment of primary cortical neurons with PP2 increased Ser-732 phosphorylation on Focal adhesion kinase (FAK), which is known to be phosphorylated by Cdk5, suggesting a cooperative role for Cdk5 and Src family kinases in cortical neurons. We also found that rottlerin (widely used as a specific inhibitor for PKCδ), but not safingol (an inhibitor for PKCα), suppressed the locomotion mode of migration. Surprisingly, use of RNA interference (RNAi) or dominant-negative PKCδ hardly affected the locomotion mode of migration, suggesting that this inhibitory effect of rottlerin occurs in a PKCδ-independent manner. Instead, the functional suppression of PKCδ seemed to disturb the terminal translocation mode at the final phase of migration.
Taken together, this study proposes a novel method for direct analysis of the locomotion mode of neuronal migration and describes several regulatory molecules at each step of neuronal migration, particularly the locomotion mode. Our results suggest the involvement of Cdk5, PKC, Src family kinases, and a non-PKCδ rottlerin target (a candidate downstream molecule is JNK) in the locomotion mode of migration. Novel roles for Fyn and PKCδ in the early phase of migration and terminal translocation mode, respectively, are also reported.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids were prepared using the EndoFree plasmid purification kit (Qiagen, Valencia, CA). DN-PKCδ (23) and nuclear localizing signal (NLS)-fused DsRed (Clontech, Mountain View, CA) cDNAs were inserted into pCAG = MCS2 vector (24) to generate CAG = DN-PKCδ and CAG = DsRed-NLS, respectively. CAG = EGFP was described previously (14). To construct shRNA-expressing vectors, oligonucleotides targeting two distinct regions in the PKCδ coding sequence (PKCδ-sh284: 5′-GCAAGAAGAACAACGGCAA-3′ and PKCδ-sh911: 5′-CAACAGAGTCTGTCGGAAT-3′); one region in the Cdk5 coding sequence (Cdk5-sh47: 5′-GAACTGTGTTCAAGGCTAA-3′); two distinct regions in the Fyn coding sequence (Fyn-sh465: 5′-CCGCAAAGATGCTGAGAGA-3′ and Fyn-sh631: 5′-GGTGGATACTATATCACAA-3′); a control scrambled sequence (sh-scr2: 5′-GAAGATTCTTCTTCGCAAA-3′) and their complementary sequences were inserted into the mU6pro vector (25). All contain a nine-base hairpin loop sequence (5′-TTCAAGAGA-3′). These sequences were designed based on information from shRNA sequence analyses (B-Bridge International, Sunnyvale, CA).
Antibodies and Chemical Regents
Primary antibodies used in this study were anti-PKCδ (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phosphorylated PKCδ (Cell Signaling Technology, Danvers, MA), anti-JNK (BD Biosciences, Franklin Lakes, NJ), anti-phosphorylated JNK (Promega, Madison, WI), anti-MAP1B (Santa Cruz Biotechnology), SMI31 (Sternberger Monoclonal Inc., Baltimore, MD), anti-Cdk5 (Santa Cruz Biotechnology), anti-Fyn (Santa Cruz Biotechnology), anti-β-actin (Abcam, Cambridge, UK), anti-S732 phosphorylated FAK (Sigma), and anti-FAK (Santa Cruz Biotechnology) antibodies. For nuclear staining, 0.1 μm quinolinium, 4-[3-(3-methyl-2(3H)-benzothiazolylidene)-1-propenyl]-1-[3-(trimethylammonio)propyl]diiodide (TO-PRO-3, Molecular Probes, Eugene, OR) was used. Roscovitine and safingol were purchased from Sigma; BIM, rottlerin, KT5823, and PP2 were purchased from Calbiochem; H89 was purchased from Seikagaku (Tokyo); and SP600125 was purchased from BIOMOL (Plymouth Meeting, PA).
In Utero Electroporation and Slice Culture of Embryonic Cerebral Cortex
Pregnant ICR mice were purchased from SLC Japan (Shizuoka, Japan). Animals were handled in accordance with guidelines established by Kyoto University and Keio University. In utero electroporation was performed as described previously (14) on embryonic day 14 (E14) embryos. For slice tissue cultures, at E16, electroporated brains were cut into 300-μm coronal slices with a microtome in Dulbecco's modified Eagle's medium/F-12 1:1 media (Invitrogen). In this study, we used brain slices at the levels from the rostral edge of the corpus callosum to the middle part of the hippocampus. Cortical slices were incubated on the insert membrane (Millipore, Bedford, MA) in 2 ml of enriched media (26) in a CO2/O2 incubator (37 °C, 5% CO2, 40 or 60% O2), and cultured for 12 h (Fig. 1B). The following inhibitors were added into culture media 4 h after the start of time-lapse observation; control DMSO (0.1%), roscovitine (100 μm), H89 (20 μm), BIM (5 μm), KT5823 (2 μm), PP2 (10 μm), safingol (30 μm), rottlerin (1 or 5 μm), and SP600125 (50 μm). The concentrations of inhibitors were determined by referring to previous in vitro studies that used cultured cells (27–34).
Immunoblot Analysis, Primary Culture, and Transfection
For preparing cell lysates, primary cultured neurons were washed with 2 ml of ice-cold phosphate-buffered saline and harvested with a cell scraper. The harvested cells were centrifuged at 2300 rpm for 5 min and lysed in buffer containing 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Triton X-100, EDTA-free Complete protease inhibitor mixture (Roche Applied Science), 10 mm β-glycerophosphate, 50 mm sodium fluoride, and 1 mm sodium orthovanadate. After 1-h incubation on ice, the lysates were sonicated and centrifuged at 5000 rpm for 5 min at 4 °C to remove cell debris. The supernatants were mixed with SDS sample buffer (50 mm Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 100 mm dithiothreitol, or 5% 2-mercaptoethanol and bromophenol blue). 5 μg of cell lysates in SDS sample buffer was separated with SDS-PAGE and electrophoretically transferred onto polyvinylidene difluoride membranes. Membranes were blocked with 5% skim milk in TBST (20 mm Tris-HCl (pH 8.0), 150 mm NaCl, and 0.05% Tween 20) for 1 h and probed with primary antibodies in 5% skim milk in TBST or Can Get Signal reagents (Toyobo, Osaka, Japan), followed by treatment with horseradish peroxidase-conjugated secondary antibodies and ECL Plus Western blotting detection reagents (Amersham Biosciences). Signals were detected and measured with a cooled charge-coupled device camera (LAS-3000mini or LAS-4000mini, Fujifilm, Tokyo). Primary culture of embryonic cerebral cortices and transfection using a nucleofector kit (Amaxa Biosystems, Cologne, Germany) were performed as described previously (8).
RESULTS
Chemical Inhibitor Screening for Locomotion Regulatory Molecules
To identify molecules that regulate the locomotion mode of neuronal migration (Fig. 1A), we established an ex vivo chemical inhibitor screening method (Fig. 1B). Using in utero electroporation (14, 35, 36), EGFP- and NLS-fused DsRed-expressing vectors were transferred into embryonic cerebral cortices at E14. The brains were dissected and sliced for tissue culture at E16, when most of the electroporated cells were just transforming into locomoting neurons (Fig. 1, C and D). The prepared cortical slices were cultured on an insert membrane under time-lapse microscopy and treated with several chemical inhibitors. Initially, the migration speeds of individual locomoting neurons were measured in the absence of inhibitors as an internal control. After 4-h observation, a chemical inhibitor was added to the culture media, and the migration speeds of the same abovementioned locomoting neurons were measured in succession (Fig. 1E). Addition of a control solvent (DMSO) did not affect the migration of locomoting neurons toward the pial surface (Figs. 2A and 3A and supplemental movie 1). The migration rates of neurons treated with DMSO (average speed = 21.7 μm/h) were similar to those of non-treated neurons (average speed = 20.1 μm/h) (Fig. 4), consistent with reported time-lapse observations in cultured cortical slices (37–39).
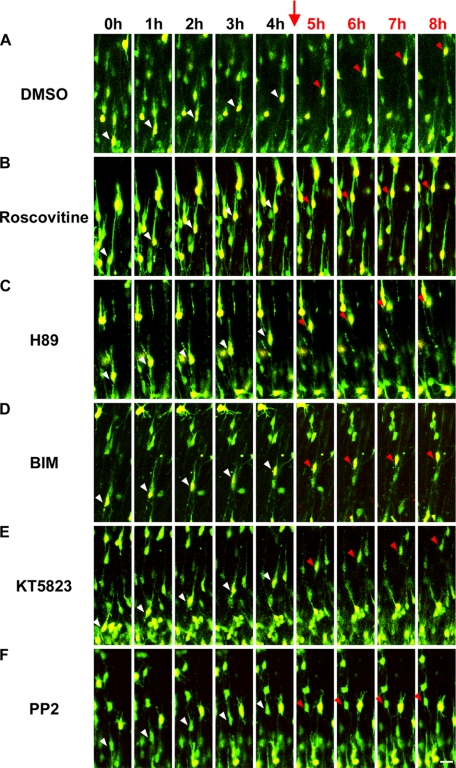
FIGURE 2.
Inhibitors for Cdk5, PKC, and Src family kinases suppressed the migration rate of the locomoting neurons. A–F, time-lapse observation of the locomoting neurons in cultured cortical tissues with or without the indicated inhibitors. Cortical slices were cultured without any inhibitors for the first 4 h as an internal control (white arrowheads). Subsequently, we added the indicated inhibitors into cultured media (red arrows) and continued to observe the same locomoting neurons for an additional 4 h (red arrowheads). Green, EGFP; red, DsRed with NLS; scale bar, 20 μm.
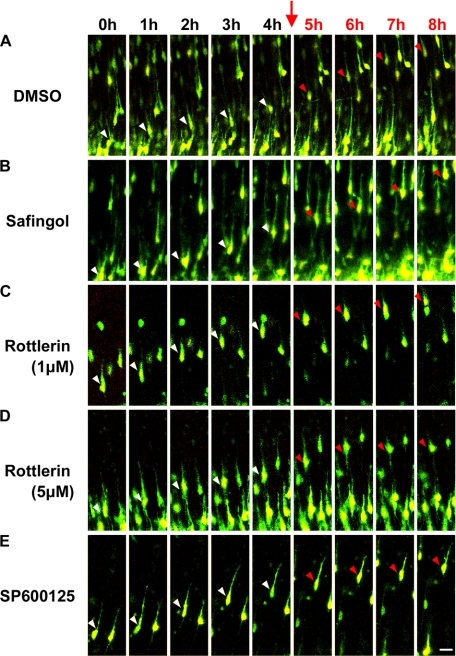
FIGURE 3.
Rottlerin and SP600125 decreased the migration rate of locomoting neurons. A–E, time-lapse observation of locomoting neurons in cultured cortical tissues with or without rottlerin (1 or 5 μm), safingol, or SP600125. Cortical slices were cultured without any inhibitors for the first 4 h as an internal control (white arrowheads). Subsequently, we added the indicated inhibitors into cultured media (red arrows), and continued to observe the same locomoting neurons for an additional 4 h (red arrowheads). Green, EGFP; red, DsRed with NLS; scale bar, 20 μm.
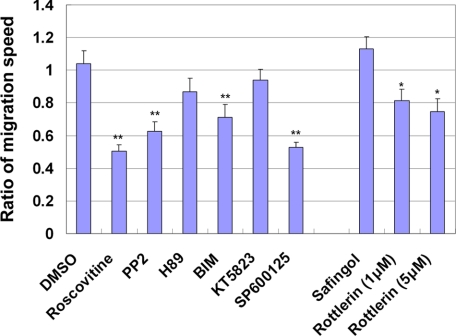
FIGURE 4.
The ratio of the migration speed of the locomoting neurons after inhibitor treatment to before treatment. The migration speeds of the locomoting neurons were measured in cortical slices before and after treatment with the indicated inhibitors. The ratios of their migration speeds after inhibitor treatment to before inhibitor treatment were calculated. More than 3 slices and 10 cells were examined for each inhibitor experiment. To omit extreme exceptions, the cells exhibiting the maximum or minimum speed were excluded. Each score represents the mean of ratios ± S.E., significant differences between control and each inhibitor treatment were investigated by the use of Welch's t test: *, p < 0.05; **, p < 0.01.
Previous mouse knockout (40) and dominant-negative experiments (14) revealed that Cdk5 has essential roles in the early phase of neuronal migration, including the formation of multipolar morphology (8), neuronal polarity (41), and leading process (8). These multiple roles of Cdk5 during the early phase of migration complicate the analyses of Cdk5 function in the locomotion mode of migration. Because treatment of the cortical slices with roscovitine (a Cdk5 inhibitor), when most of the electroporated cells are in the early phase of migration, causes a phenotype similar to that of Cdk5-knockout mice or in utero electroporation-mediated functional suppression of Cdk5 (24), we used roscovitine to examine the role of Cdk5 in the locomotion. We treated cultured cortical tissues with roscovitine while observing individual locomoting neurons. Roscovitine treatment decreased the migration speed of the locomoting neurons (average speed = 9.5 μm/h) (Fig. 2B and supplemental movie 2), indicating an additional role for Cdk5 during neuronal migration.
We next examined the effects of several chemical inhibitors for serine/threonine or tyrosine kinases on the locomotion mode of migration by means of our ex vivo chemical inhibitor screening method. Treatment with BIM, an inhibitor for all subtypes of PKC when used at 5 μm of final concentration, suppressed the locomotion mode of migration (Fig. 2D), whereas H89 and KT5823, inhibitors for protein kinase A and protein kinase G, respectively, did not affect the locomotion (BIM: average speed = 10.7 μm/h, KT5823: average speed = 18.3 μm/h, H89: average speed = 15.9 μm/h) (Fig. 2, C–E and supplemental movies 3–5). In addition, PP2, an inhibitor for Src family tyrosine kinases, suppressed the migration of the locomoting neurons (average speed = 8.5 μm/h) (Fig. 2F and supplemental movie 6). Quantitative analysis indicated that BIM and PP2 resulted in ∼30% or 40% reduction of the migration rate of locomoting neurons, respectively (Fig. 4). It has been reported that both PKC and Src family kinases are involved in the preplate splitting by the earliest-born cortical neurons around E11–13 (30). Our data suggest novel roles for PKC and Src family kinases in the locomotion mode of migration at mid to late corticogenesis.
Rottlerin Decreases the Migration Rate of Locomoting Neurons
PKC kinases are classified into three subfamilies; conventional PKC (PKCα, -βI, -βII, and -γ), novel PKC (PKCδ, -ϵ, -η, and -θ), and atypical PKC (PKCζ and -λ) (42). Because BIM decreased the migration rate of the locomoting neurons, we repeated the experiment using safingol and rottlerin, which are widely used as specific inhibitors for PKCα or -δ, respectively. Although treatment with safingol did not affect the locomotion mode of migration (average speed = 17.2 μm/h), rottlerin (5 μm) resulted in decreased migration speeds (average speed = 10.0 μm/h) (Fig. 3, B and D, and supplemental movies 7 and 9).
Suppression of PKCδ Had Little Effect on the Locomotion
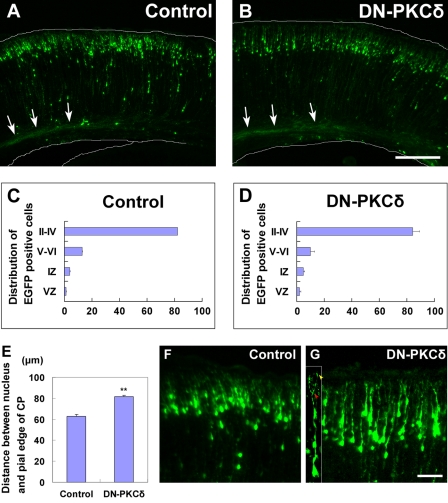
To elucidate the requirement of PKCδ in the locomotion mode of migration, we first performed dominant-negative experiments using in utero electroporation. A dominant-negative PKCδ (DN-PKCδ)-expressing vector was electroporated into E14 embryonic cerebral cortices, and the electroporated brains were fixed at postnatal day 0 (P0), 5 days after electroporation. The electroporated cells were visualized with co-expressed EGFP. Consistent with previous reports (14), control cells migrated to reach the superficial layer of the cortical plate at P0 (Fig. 5, A and C). To our surprise, DN-PKCδ-expressing cells also reached the upper part of the cortical plate at P0, suggesting that PKCδ function is not required for the locomotion mode of neuronal migration (Fig. 5, B and D). It is possible that rottlerin disturbs locomotion in a non-cell autonomous manner. For example, disorganized axon bundles from superior neurons may affect neuronal migration. We found that cells, in which PKCδ was suppressed, were observed to form axon bundles normally at P0 (Fig. 5, A and B), although this possibility cannot be excluded.
FIGURE 5.
DN-PKCδ had little effect on the locomotion mode of neuronal migration. A and B, embryonic brains were electroporated with control (A) or DN-PKCδ expressing vector (B) plus EGFP-expressing vector at E14, followed by fixation at P0. White arrows show the axon bundles from the electroporated neurons. C and D, estimation of neuronal migration in cerebral cortices electroporated with the indicated plasmids. Numbers of cells in each layer were counted in control and DN-PKCδ electroporated brains, respectively. Layers of the cortical plate are indicated in Roman numerals; IZ, intermediate zone; VZ, ventricular zone. Each score represents the mean percentage of numbers of cells ± S.E., n = 3. E, DN-PKCδ-electroporated cells exhibited an increased distance between the nucleus and the upper edge of the cortical plate. Each score represents mean distance ± S.E. More than 3 slices and 200 cells were analyzed, and the significant difference was examined by the use of Student's t test: **, p < 0.01. F and G, high magnification of A and B. The tips of the leading processes in many DN-PKCδ expressing cells reached the marginal zone (yellow arrowhead in the inset of G), and some of them exhibited branched morphologies (red arrowhead in the inset of G). Scale bars, 200 μm (A and B) and 50 μm (F and G).
To confirm the dominant-negative experiments, we performed RNAi experiments. We constructed two shRNA-expressing vectors for PKCδ, both of which efficiently reduced the expression levels of PKCδ (supplemental Fig. S1A). These shRNAs for PKCδ were electroporated into E14 cerebral cortices, which were subsequently fixed for histological observation at P0. Consistent with the dominant-negative experiments, most of the PKCδ-knockdown cells migrated to the upper part of the cortical plate at 5 days after electroporation, similar to control (scrambled non-targeted) shRNA-electroporated cells (supplemental Fig. S1, B–G). Our findings are consistent with previous reports indicating that PKCδ-deficient mice are viable with no increase of other PKC isoforms (43) and showed no gross anatomical abnormalities and evidence of abnormal neuronal migration (44).
PKCδ Regulates the Terminal Translocation Mode, Rather Than the Locomotion
Although our in vivo analyses indicated that PKCδ-suppressing cells migrate to reach the upper part of the cortical plate, similar to control cells (Fig. 5, A–D), the positioning of PKCδ-suppressing cells seemed to be slightly lower than that of control cells at P0 (Fig. 5B and supplemental Fig. S1, C and D). Measurement of the distance from the nucleus of the electroporated cells to the pial edge of the cortical plate revealed that both DN-PKCδ-electroporated cells and PKCδ-knockdown cells showed the significantly increased distance between the nucleus and the pial edge of the cortical plate (Fig. 5E and supplemental Fig. S1H). This phenotype is similar to that of Dab1 knockdown, a cytoplasmic adaptor protein involved in the Reelin signaling pathway (45, 46). During the final phase of migration, neurons do not use the locomotion mode, but a specific migration mode, called terminal translocation (38), which is thought to be dependent on Dab1 (47). The phenotypic similarity between the suppression of PKCδ and Dab1 suggests that PKCδ may regulate the terminal translocation mode, rather than the locomotion.
It has been reported that the terminal translocation mode occurs after the leading process reaches the marginal zone, whereas the locomotion mode consists of repetitive cycles comprised of elongation of the leading process followed by somal movement (38, 48). We observed that the tips of the leading processes in many DN-PKCδ-expressing or PKCδ-knockdown cells reached the marginal zone, and that some of them displayed branched morphologies: another feature of terminal translocating neurons (Fig. 5G and supplemental Fig. S1I). These results further support the idea that PKCδ is involved in the terminal translocation mode, although we cannot exclude the possibility that the locomotion mode or early phase of migration may also be slightly affected in PKCδ-suppressing cells (49). Interestingly, both Dab1 and PKC are reported to be involved in the preplate splitting (30, 45, 46). Although it is unclear whether PKCδ is required for the preplate splitting, our results implicate that PKCδ is involved in the terminal translocation mode, in cooperation with the Reelin-Dab1 signaling pathway.
Rottlerin Suppressed JNK Activity
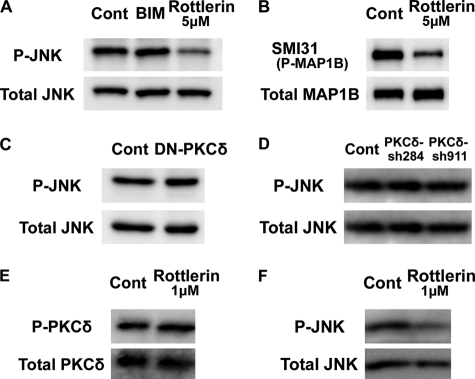
Our findings suggest that the observed rottlerin inhibition on the locomotion mode is occurring through a PKCδ-independent pathway. Consistently, there are recent reports indicating that rottlerin exhibits certain side-effects (50), although rottlerin is still widely used as a PKCδ-specific inhibitor. Therefore, we next tried to identify a downstream target molecule of rottlerin, other than PKCδ. Previously, we have reported that suppression of JNK disturbed neuronal migration and leading process morphology (14). Because a leading process is thought to have important roles in the locomotion mode of migration, we focused on JNK. Primary cortical neurons were treated with rottlerin or BIM, and subjected to immunoblot analysis with anti-activated JNK antibody. While suppression of all subtypes of PKC, including PKCδ, by BIM had little effect on JNK phosphorylation, treatment with rottlerin decreased the activated phosphorylation of JNK (Fig. 6A). Furthermore, rottlerin treatment also decreased the mode I phosphorylation of MAP1B (Fig. 6B), which is a downstream molecule of JNK to control neuronal migration (14, 24).
FIGURE 6.
Rottlerin treatment reduced JNK activity. A and B, influence of rottlerin on the phosphorylation of JNK1. Primary cultures of E15 mouse embryonic cerebral cortices (2 days in vitro) were treated with 5 μm rottlerin or 5 μm BIM for 3 h, and subjected to immunoblot analyses with anti-phospho-JNK (P-JNK) and anti-JNK (Total JNK) or SMI31 mAb (P-MAP1B) and anti-MAP1B (Total MAP1B) antibodies. C and D, suppression of PKCδ had little effect on JNK1 phosphorylation in primary cortical neurons. Primary cortical neurons from E15 cortices were transfected with the indicated plasmids, and subjected to the immunoblot analysis with anti-phospho-JNK (P-JNK) and anti-JNK (Total JNK) antibodies at 2 days in vitro. The ratios of P-JNK1/Total JNK1 were 106.6% (DN-PKCδ), 77.6% (PKCδ-sh284), and 80.2% (PKCδ-sh911), compared with each control sample. E and F, low concentrations of rottlerin suppressed the activated phosphorylation of JNK1, but not PKCδ. Primary cortical neurons from E15 cortices were treated with 1 μm rottlerin for 3 h and subjected to the immunoblot analysis with anti-autophosphorylated PKCδ (P-PKCδ) and anti-PKCδ (Total PKCδ) or anti-phospho-JNK (P-JNK) and anti-JNK (Total JNK) antibodies at 2 days in vitro.
It has been reported that JNK is activated by many extracellular stimuli. Whereas acute hypoxic stress-induced JNK activation is mediated by PKCδ (51), platelet-derived growth factor-induced JNK activation is independent of PKCδ in primary human dermal fibroblasts (52). This suggests that whether PKCδ action occurs upstream of JNK is context-dependent. Therefore, we investigated the relationship between PKCδ and JNK in embryonic cortical neurons. The transfection of a DN-PKCδ-expressing vector had little effect on JNK activation in primary cortical neurons (Fig. 6C). In addition, the effect of PKCδ-shRNAs on the phosphorylation of JNK was very small (Fig. 6D).
To confirm these observations, we next analyzed the effect of a low concentration (1 μm) of rottlerin on JNK activation, because a previous report showed that it did not affect the activity of PKCδ (53). Consistent with this, treatment with 1 μm rottlerin did not change the activated autophosphorylation of PKCδ in primary cortical neurons (Fig. 6E). Under the same condition, however, the phosphorylation of JNK continued to be reduced (Fig. 6F). These data suggest that a large part of JNK activation is independent of PKCδ in cortical neurons.
We next examined whether the rottlerin-induced migration defect of locomoting neurons is caused by PKCδ-independent JNK activation. In cortical slices, treatment with 1 μm rottlerin suppressed the locomotion mode of migration (average speed = 11.9 μm/h) (Fig. 3C and supplemental movie 8), similar to that of 5 μm rottlerin. Furthermore, treatment with SP600125, a JNK inhibitor, also reduced the migration speed of locomoting neurons (average speed = 7.9 μm/h) (Fig. 3E and supplemental movie 10), consistent with previous reports (14, 54). Because 1 μm rottlerin did not affect PKCδ activity, JNK is a good candidate for a rottlerin-downstream molecule involved in the locomotion mode of migration.
Knockdown of Cdk5 or Fyn Caused the Defects of Neuronal Migration
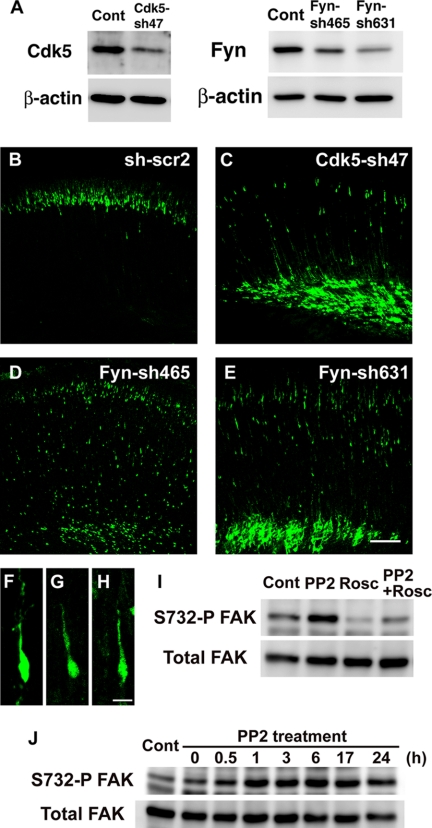
Recent widespread analyses of inhibitor specificities revealed that, unlike rottlerin, roscovitine, and PP2 are reliable specific inhibitors for CDK and Src family kinases, respectively (55, 56). To confirm this, we examined the effects of Cdk5 and Fyn (Src family kinase) knockdown on neuronal migration. shRNAs for Cdk5 and Fyn, which were shown to efficiently reduce cellular levels of their respective target protein (Fig. 7A) (8), were electroporated into E14 cerebral cortices. The electroporated brains were fixed at P0, 5 days after electroporation. In contrast to control cells, which migrated to reach the superficial layer of the cortical plate, Cdk5-knockdown cells displayed migration defects (Fig. 7, B and C). Although the majority of electroporated cells were stalled in the intermediate zone, a few cells displayed retarded migration and exhibited locomoting morphology in the cortical plate (Fig. 7F).
FIGURE 7.
Knockdown of Cdk5 and Fyn caused defects in neuronal migration. A, amount of Cdk5 and Fyn proteins in primary cortical neurons transfected with the indicated shRNA vectors. Primary cortical neurons from E15 cerebral cortices were transfected with the indicated plasmids and incubated for 2 days in vitro. The cell lysates were subjected to the immunoblot analysis with the indicated antibodies to evaluate the efficiency for RNAi. B–E, embryonic brains electroporated with control scrambled shRNA (sh-scr2) (B), Cdk5-sh47 (C), Fyn-sh465 (D), or Fyn-sh631 (E) plus EGFP-expressing vectors at E14, followed by fixation at P0. F–H, Cdk5 or Fyn-knockdown cells in the cortical plate exhibited locomoting morphology with a leading process. F, Cdk5-sh47; G, Fyn-sh465; H, Fyn-sh631; and I and J, primary cortical neurons from E15 cerebral cortices were treated with PP2 for 3 h (I) or the indicated time (J), and subjected to immunoblot analysis with the indicated antibodies. Control (Cont) is cell lysates treated with DMSO for 3 h (I) or 6 h (J). Because Ser-732 phosphorylation on FAK is known to be a good and specific substrate of Cdk5, we used the FAK phosphorylation as an indicator for Cdk5 activity. Scale bars, 100 μm (B–E) and 10 μm (F–H).
To elucidate whether these Cdk5-knockdown cells with locomoting morphologies showed delayed migration, we measured their migration speed in cortical slices. Their migration speeds were significantly reduced compared with control scrambled shRNA-electroporated cells (supplemental Fig. S2 and supplemental movies 11 and 12). Furthermore, when Cdk5-shRNA-electroporated cortical slices were treated with roscovitine, Cdk5-knockdown cells barely exhibited any additional migration delays (supplemental Fig. S2 and supplemental movie 12), suggesting that the effect of roscovitine on the locomotion is largely dependent on Cdk5 activity. These observations support our chemical inhibitor experiments using roscovitine (Fig. 2B) as well as previous findings revealing the essential roles of Cdk5 in the early phase of migration (8). It also suggests that roscovitine cell-autonomously suppressed the locomotion mode of neuronal migration.
Knockdown of Fyn disturbed the early phase of neuronal migration, although like Cdk5 knockdown, some cells exhibited retarded migration with locomoting morphology in the cortical plate (Fig. 7, D, E, G, and H). In addition, the migration speeds of Fyn-knockdown locomoting neurons were reduced in cortical slice tissues (supplemental Fig. S2). Treatment of Fyn-shRNA-electroporated cortices with PP2 did not further affect their migration speeds (supplemental Fig. S2 and supplemental movie 13), suggesting that, among several Src family kinases, Fyn is a major target of PP2 for controlling the locomotion mode of migration. These data are consistent with our chemical inhibitor experiments.
It is believed that Fyn is involved in the Reelin-Dab1 signaling pathway, which regulates the preplate splitting and terminal translocation mode of migration, rather than the locomotion mode and early phase of migration (30, 45–47). However, our findings suggest a novel role for Fyn in the early phase of migration as well as the locomotion.
Because our data suggest that both Cdk5 and Src family kinases are required for the locomotion mode and early phase of migration probably in a cell-autonomous manner, we next analyzed the relationship between Cdk5 and Src family kinases. Although Fyn is known to phosphorylate Cdk5 at Tyr-15 and to activate it in dendrites (57), Tyr-15 phosphorylation on endogenous Cdk5 could hardly be detected in whole cell lysates from primary cortical immature neurons (2-day in vitro) before synaptogenesis (data not shown). Primary cortical neurons were treated with PP2 and subjected to immunoblot analysis with antibodies recognizing Ser-732-phosphorylated FAK, a good Cdk5 substrate (58). Interestingly, treatment with PP2 increased the phosphorylation of FAK at Ser-732, which was suppressed by cotreatment of roscovitine with PP2, suggesting that Src family kinases negatively regulate Cdk5 activity (Fig. 7I). The maximal effects on FAK phosphorylation were observed at 1–3 h after treatment with PP2, with effects tapering with time (Fig. 7J). Such a transient effect of PP2 may explain why other functional suppression experiments for Src family kinases do not increase the activity of Cdk5.
DISCUSSION
Neurons utilize the locomotion mode of migration during most of the journey to their final destination, so that the locomotion mode is the major contributor to proper layer formation of cerebral cortex and thereby to construction of a functional brain (Fig. 1A). However, the analysis of the molecular mechanisms for the locomotion mode of migration without secondary effects of various cellular events at the early phase of migration has remained difficult. Here, we have established a novel ex vivo chemical inhibitor screen to identify molecules involved in the locomotion mode of migration directly. Our results reveal that Cdk5 and Src family kinases are required for the migration of locomoting neurons (Fig. 8). Because the knockdown of Cdk5 or Fyn, using in utero electroporation, strongly inhibited the early phase of neuronal migration, it is apparent that conventional methods would not be useful for analyzing the molecular mechanisms underlying the locomotion mode. Our ex vivo chemical inhibitor screen represents a powerful method for identifying the molecules that regulate the locomotion mode of migration. However, in interpreting the results of the chemical inhibitor experiments, we should be careful of the following points. One is the side-effects of inhibitors. With regard to this issue, previous reports analyzing the specificities of various inhibitors are very useful (55, 56, 59). For example, roscovitine is reportedly a reliable specific inhibitor for CDK kinases but rottlerin is not a good specific inhibitor for PKCδ. Another problem is that the ex vivo chemical inhibitor experiments cannot discriminate between cell-autonomous and non-cell-autonomous effects. Therefore, the chemical inhibitor analysis should be used in combination with RNAi or dominant-negative experiments for confirmation (e.g. Fig. 7 and supplemental Fig. S2).
FIGURE 8.
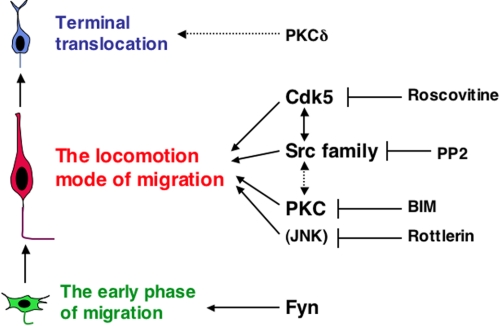
Molecules involved in each phase of neuronal migration. Our ex vivo chemical inhibitor experiments provide evidence that Cdk5, PKC, and Src family kinases as well as one or more unidentified rottlerin-target molecules (a candidate molecule is an upstream regulator for JNK) are required for the migration of locomoting neurons. Cdk5 and Src family kinases may function in similar signaling cascades to regulate neuronal migration. Although it was believed that PKCδ was the target molecule of rottlerin, PKCδ may be involved in the terminal translocation at the final phase of neuronal migration, rather than the locomotion. In addition, our in vivo RNAi data showed that Fyn is required for the early phase of neuronal migration.
Several reports indicated that Cdk5 is required for many cellular events at the early phase of neuronal migration. Cdk5 regulates multipolar cell morphology through the phosphorylation and stabilization of p27kip1, but leading process formation in a p27kip1-independent manner (8), suggesting that Cdk5 phosphorylates different substrates to control separate steps of neuronal migration. Actually, Cdk5 phosphorylates many signaling and cytoskeletal proteins, including FAK, DCX, Ndel1, p21-activated kinase, Paxillin, and Neurabin I (58, 60–64). It is important to identify the downstream pathway of Cdk5 in the regulation of the locomotion mode of migration, which may be achieved by means of our ex vivo chemical inhibitor screening method.
We also found that BIM, an inhibitor for all subtypes of PKC, decreased the migration rate of locomoting neurons (Fig. 8), whereas the suppression of PKCδ (a novel PKC subfamily member) or treatment with safingol, an inhibitor for PKCα (a conventional PKC subfamily member), did not. It suggests that other PKC subtype(s) may regulate the locomotion mode of migration. Because Par6, which forms a complex with atypical PKC, is involved in neuronal migration in the developing cerebellum (65), BIM treatment might suppress the locomotion at least in part through the inhibition of atypical PKC activities.
In addition to PKC, we found that Src family kinases are also involved in the locomotion mode of migration. Inhibitors for both PKC and Src family kinases are reported to disturb the preplate splitting (30), implicating that the cooperation between PKC and Src family kinases may play some important roles. Src family kinases, Fyn and Src, phosphorylate Dab1, whose deficiency also causes a failure to split the preplate (45, 46). On the other hand, knockdown of Dab1 does not affect the locomotion mode of migration at mid- or late-corticogenesis (47, 66), suggesting that Src family kinases regulate the locomotion mode independent of Dab1, unlike the terminal translocation and preplate splitting. Furthermore, we found that the knockdown of Fyn inhibited the early phase of neuronal migration (Fig. 8). These findings provide novel Dab1-independent functions of Fyn and other Src family kinases in the locomotion and/or early phase of neuronal migration. Together with the observation that there is some relationship between Cdk5 and Src family kinases in primary cortical neurons, Src family kinases may act with Cdk5 in the locomotion mode and/or early phase of migration, but with Reelin and Dab1 in the terminal translocation mode.
Although it hardly disturbed the locomotion mode, the functional suppression of PKCδ caused abnormal cell positioning near the pial edge of the cortical plate, probably due to a defect in terminal translocation at the final phase of migration (Fig. 8). Our findings, using the dominant-negative form and RNAi, are consistent with the observation that abnormal neuronal migration was not detected in PKCδ-knockout mice (44). However, we could not exclude the possibility that PKCδ is also involved in the locomotion mode or early phase of migration, in addition to the final phase of migration, because knockdown or dominant-negative experiments may not completely inhibit all PKCδ activity in migrating neurons.
We found that rottlerin suppressed JNK activity. Because a low concentration of rottlerin, which did not affect PKCδ activity, also reduced JNK phosphorylation in primary cortical neurons and the migration rate of the locomoting neurons in cortical slices, JNK may partly account for rottlerin-induced suppression of the locomotion mode of migration. However, a previous report, using an in vitro kinase assay, indicates that direct effect of rottlerin on the activities of JNK1, -2, and -3 are not substantial (56), suggesting that rottlerin may reduce JNK activity indirectly.
In summary, our novel ex vivo chemical inhibitor screening has revealed important molecules that regulate the locomotion mode of neuronal migration. We show that Cdk5, PKC, and Src family kinases as well as a yet to be identified rottlerin-target molecule(s) (a candidate molecule is an upstream regulator for JNK) are required for the migration of locomoting neurons. Furthermore, we showed that Fyn is required for the early phase of neuronal migration, and that PKCδ may be involved in the terminal translocation at the final phase of neuronal migration. These findings provide some critical pieces toward our understanding of the molecular machinery for neuronal migration and cortical layer formation, which are closely associated with neurological disorders.
Acknowledgments
We greatly appreciate the gift of CAG vector from Dr. Jun-ichi Miyazaki (Osaka University, Osaka, Japan), mU6pro vector from Dr. David L. Turner (University of Michigan), and PKCδ plasmid from Dr. Jae-Won Soh (Inha University, Nam-gu Incheon, Korea). We give our thanks to Dr. Ruth Yu for critical reading of the manuscript, and to Dr. Mima Shikanai for technical assistance.
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M. H., T. K., Y. N., and K. N.), by grants from the Uehara Memorial Foundation (to T. K.), Takeda Science Foundation (to T. K.), Global Center of Excellence (to K. N. and T. K.), and by Keio Gijuku Academic Development Funds (to Y. V. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental movies 1–13 and Figs. S1 and S2.
- Cdk5
- cyclin-dependent kinase 5
- JNK
- c-Jun N-terminal kinase
- MAP1B
- microtubule-associated protein 1B
- BIM
- bisindolylmaleimide I
- PKC
- protein kinase C
- RNAi
- RNA interference
- NLS
- nuclear localizing signal
- E
- embryonic day
- DN
- dominant-negative
- P
- postnatal day
- shRNA
- short hairpin RNA
- EGFP
- enhanced green fluorescent protein
- PP2
- 4-amino-5-(4-chlorophenyl)-7-(±-butyl)pyrazolo[3,4-d]pyrimidine.
REFERENCES
- 1.Gleeson J. G., Walsh C. A. (2000) Trends Neurosci. 23, 352–359 [DOI] [PubMed] [Google Scholar]
- 2.Bielas S., Higginbotham H., Koizumi H., Tanaka T., Gleeson J. G. (2004) Annu. Rev. Cell Dev. Biol. 20, 593–618 [DOI] [PubMed] [Google Scholar]
- 3.Kawauchi T., Hoshino M. (2008) Dev. Neurosci. 30, 36–46 [DOI] [PubMed] [Google Scholar]
- 4.Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., Ross C. A., Hatten M. E., Nakajima K., Sawa A. (2005) Nat. Cell Biol. 7, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 5.Galaburda A. M., LoTurco J., Ramus F., Fitch R. H., Rosen G. D. (2006) Nat. Neurosci. 9, 1213–1217 [DOI] [PubMed] [Google Scholar]
- 6.Rakic P. (2006) Cereb. Cortex 16, Suppl. 1, i3–17 [DOI] [PubMed] [Google Scholar]
- 7.Ayala R., Shu T., Tsai L. H. (2007) Cell 128, 29–43 [DOI] [PubMed] [Google Scholar]
- 8.Kawauchi T., Chihama K., Nabeshima Y., Hoshino M. (2006) Nat. Cell Biol. 8, 17–26 [DOI] [PubMed] [Google Scholar]
- 9.des Portes V., Pinard J. M., Billuart P., Vinet M. C., Koulakoff A., Carrié A., Gelot A., Dupuis E., Motte J., Berwald-Netter Y., Catala M., Kahn A., Beldjord C., Chelly J. (1998) Cell 92, 51–61 [DOI] [PubMed] [Google Scholar]
- 10.Gleeson J. G., Allen K. M., Fox J. W., Lamperti E. D., Berkovic S., Scheffer I., Cooper E. C., Dobyns W. B., Minnerath S. R., Ross M. E., Walsh C. A. (1998) Cell 92, 63–72 [DOI] [PubMed] [Google Scholar]
- 11.Reiner O., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W. B., Caskey C. T., Ledbetter D. H. (1993) Nature 364, 717–721 [DOI] [PubMed] [Google Scholar]
- 12.Bai J., Ramos R. L., Ackman J. B., Thomas A. M., Lee R. V., LoTurco J. J. (2003) Nat. Neurosci. 6, 1277–1283 [DOI] [PubMed] [Google Scholar]
- 13.Tsai J. W., Chen Y., Kriegstein A. R., Vallee R. B. (2005) J. Cell Biol. 170, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawauchi T., Chihama K., Nabeshima Y., Hoshino M. (2003) EMBO J. 22, 4190–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gdalyahu A., Ghosh I., Levy T., Sapir T., Sapoznik S., Fishler Y., Azoulai D., Reiner O. (2004) EMBO J. 23, 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang L., Jones Y., Ellisman M. H., Goldstein L. S., Karin M. (2003) Dev. Cell 4, 521–533 [DOI] [PubMed] [Google Scholar]
- 17.Arimura N., Kaibuchi K. (2007) Nat. Rev. Neurosci. 8, 194–205 [DOI] [PubMed] [Google Scholar]
- 18.Kishi M., Pan Y. A., Crump J. G., Sanes J. R. (2005) Science 307, 929–932 [DOI] [PubMed] [Google Scholar]
- 19.Barnes A. P., Lilley B. N., Pan Y. A., Plummer L. J., Powell A. W., Raines A. N., Sanes J. R., Polleux F. (2007) Cell 129, 549–563 [DOI] [PubMed] [Google Scholar]
- 20.Shelly M., Cancedda L., Heilshorn S., Sumbre G., Poo M. M. (2007) Cell 129, 565–577 [DOI] [PubMed] [Google Scholar]
- 21.LoTurco J., Manent J. B., Sidiqi F. (2009) Cereb. Cortex 19, (Suppl. 1) i120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikolic M. (2004) Trends Cell Biol. 14, 1–5 [DOI] [PubMed] [Google Scholar]
- 23.Soh J. W., Weinstein I. B. (2003) J. Biol. Chem. 278, 34709–34716 [DOI] [PubMed] [Google Scholar]
- 24.Kawauchi T., Chihama K., Nishimura Y. V., Nabeshima Y., Hoshino M. (2005) Biochem. Biophys. Res. Commun. 331, 50–55 [DOI] [PubMed] [Google Scholar]
- 25.Yu J. Y., DeRuiter S. L., Turner D. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6047–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyata T., Kawaguchi A., Saito K., Kuramochi H., Ogawa M. (2002) J. Neurosci. Res. 69, 861–868 [DOI] [PubMed] [Google Scholar]
- 27.Kwon Y. T., Gupta A., Zhou Y., Nikolic M., Tsai L. H. (2000) Curr. Biol. 10, 363–372 [DOI] [PubMed] [Google Scholar]
- 28.Maa M. C., Chang M. Y., Chen Y. J., Lin C. H., Yu C. J., Yang Y. L., Li J., Chen P. R., Tang C. H., Lei H. Y., Leu T. H. (2008) J. Biol. Chem. 283, 31408–31416 [DOI] [PubMed] [Google Scholar]
- 29.Mishra S., Fujita T., Lama V. N., Nam D., Liao H., Okada M., Minamoto K., Yoshikawa Y., Harada H., Pinsky D. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5191–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jossin Y., Ogawa M., Metin C., Tissir F., Goffinet A. M. (2003) J. Neurosci. 23, 9953–9959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takuma K., Phuagphong P., Lee E., Mori K., Baba A., Matsuda T. (2001) J. Biol. Chem. 276, 48093–48099 [DOI] [PubMed] [Google Scholar]
- 32.Huang C., Rajfur Z., Borchers C., Schaller M. D., Jacobson K. (2003) Nature 424, 219–223 [DOI] [PubMed] [Google Scholar]
- 33.Sachs C. W., Safa A. R., Harrison S. D., Fine R. L. (1995) J. Biol. Chem. 270, 26639–26648 [DOI] [PubMed] [Google Scholar]
- 34.Chen Y., Cantrell A. R., Messing R. O., Scheuer T., Catterall W. A. (2005) J. Neurosci. 25, 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabata H., Nakajima K. (2001) Neuroscience 103, 865–872 [DOI] [PubMed] [Google Scholar]
- 36.Saito T., Nakatsuji N. (2001) Dev. Biol. 240, 237–246 [DOI] [PubMed] [Google Scholar]
- 37.O'Rourke N. A., Dailey M. E., Smith S. J., McConnell S. K. (1992) Science 258, 299–302 [DOI] [PubMed] [Google Scholar]
- 38.Nadarajah B., Brunstrom J. E., Grutzendler J., Wong R. O., Pearlman A. L. (2001) Nat. Neurosci. 4, 143–150 [DOI] [PubMed] [Google Scholar]
- 39.Tabata H., Nakajima K. (2003) J. Neurosci. 23, 9996–10001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilmore E. C., Ohshima T., Goffinet A. M., Kulkarni A. B., Herrup K. (1998) J. Neurosci. 18, 6370–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohshima T., Hirasawa M., Tabata H., Mutoh T., Adachi T., Suzuki H., Saruta K., Iwasato T., Itohara S., Hashimoto M., Nakajima K., Ogawa M., Kulkarni A. B., Mikoshiba K. (2007) Development 134, 2273–2282 [DOI] [PubMed] [Google Scholar]
- 42.Newton A. C. (2001) Chem. Rev. 101, 2353–2364 [DOI] [PubMed] [Google Scholar]
- 43.Miyamoto A., Nakayama K., Imaki H., Hirose S., Jiang Y., Abe M., Tsukiyama T., Nagahama H., Ohno S., Hatakeyama S., Nakayama K. I. (2002) Nature 416, 865–869 [DOI] [PubMed] [Google Scholar]
- 44.Choi D. S., Wei W., Deitchman J. K., Kharazia V. N., Lesscher H. M., McMahon T., Wang D., Qi Z. H., Sieghart W., Zhang C., Shokat K. M., Mody I., Messing R. O. (2008) J. Neurosci. 28, 11890–11899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice D. S., Curran T. (2001) Annu. Rev. Neurosci. 24, 1005–1039 [DOI] [PubMed] [Google Scholar]
- 46.Tissir F., Goffinet A. M. (2003) Nat. Rev. Neurosci. 4, 496–505 [DOI] [PubMed] [Google Scholar]
- 47.Olson E. C., Kim S., Walsh C. A. (2006) J. Neurosci. 26, 1767–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakic P. (1972) J. Comp. Neurol. 145, 61–83 [DOI] [PubMed] [Google Scholar]
- 49.Zhao C. T., Li K., Li J. T., Zheng W., Liang X. J., Geng A. Q., Li N., Yuan X. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21353–21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soltoff S. P. (2007) Trends Pharmacol. Sci. 28, 453–458 [DOI] [PubMed] [Google Scholar]
- 51.Chen J. L., Lin H. H., Kim K. J., Lin A., Forman H. J., Ann D. K. (2008) J. Biol. Chem. 283, 34432–34444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan J., Guan S., Cheng C. F., Cho M., Fields J. W., Chen M., Denning M. F., Woodley D. T., Li W. (2006) J. Invest. Dermatol. 126, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 53.Gschwendt M., Müller H. J., Kielbassa K., Zang R., Kittstein W., Rincke G., Marks F. (1994) Biochem. Biophys. Res. Commun. 199, 93–98 [DOI] [PubMed] [Google Scholar]
- 54.Hirai S. I., Cui de F., Miyata T., Ogawa M., Kiyonari H., Suda Y., Aizawa S., Banba Y., Ohno S. (2006) J. Neurosci. 26, 11992–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bain J., McLauchlan H., Elliott M., Cohen P. (2003) Biochem. J. 371, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sasaki Y., Cheng C., Uchida Y., Nakajima O., Ohshima T., Yagi T., Taniguchi M., Nakayama T., Kishida R., Kudo Y., Ohno S., Nakamura F., Goshima Y. (2002) Neuron 35, 907–920 [DOI] [PubMed] [Google Scholar]
- 58.Xie Z., Sanada K., Samuels B. A., Shih H., Tsai L. H. (2003) Cell 114, 469–482 [DOI] [PubMed] [Google Scholar]
- 59.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka T., Serneo F. F., Tseng H. C., Kulkarni A. B., Tsai L. H., Gleeson J. G. (2004) Neuron 41, 215–227 [DOI] [PubMed] [Google Scholar]
- 61.Niethammer M., Smith D. S., Ayala R., Peng J., Ko J., Lee M. S., Morabito M., Tsai L. H. (2000) Neuron 28, 697–711 [DOI] [PubMed] [Google Scholar]
- 62.Rashid T., Banerjee M., Nikolic M. (2001) J. Biol. Chem. 276, 49043–49052 [DOI] [PubMed] [Google Scholar]
- 63.Miyamoto Y., Yamauchi J., Chan J. R., Okada A., Tomooka Y., Hisanaga S., Tanoue A. (2007) J. Cell Sci. 120, 4355–4366 [DOI] [PubMed] [Google Scholar]
- 64.Causeret F., Jacobs T., Terao M., Heath O., Hoshino M., Nikolic M. (2007) Mol. Biol. Cell 18, 4327–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Solecki D. J., Model L., Gaetz J., Kapoor T. M., Hatten M. E. (2004) Nat. Neurosci. 7, 1195–1203 [DOI] [PubMed] [Google Scholar]
- 66.Tabata H., Nakajima K. (2002) J. Neurosci. Res. 69, 723–730 [DOI] [PubMed] [Google Scholar]