Abstract
Aquaporin-1 (AQP1) water channels are expressed in the plasma membrane of dorsal root ganglion (DRG) neurons. We found reduced osmotic water permeability in freshly isolated DRG neurons from AQP1−/− versus AQP1+/+ mice. Behavioral studies showed greatly reduced thermal inflammatory pain perception in AQP1−/− mice evoked by bradykinin, prostaglandin E2, and capsaicin as well as reduced cold pain perception. Patch clamp of freshly isolated DRG neurons showed reduced action potential firing in response to current injections. Single action potentials after pulse current injections showed reduced maximum inward current, suggesting impaired Nav1.8 Na+ function. Whole-cell Nav1.8 Na+ currents in Nav1.8-expressing ND7-23 cells showed slowed frequency-dependent inactivation after AQP1 transfection. Immunoprecipitation studies showed AQP1- Nav1.8 Na+ interaction, which was verified in live cells by single-particle tracking of quantum dot-labeled AQP1. Our results implicate the involvement of AQP1 in DRG neurons for the perception of inflammatory thermal pain and cold pain, whose molecular basis is accounted for, in part, by reduced Nav1.8-dependent membrane Na+ current. AQP1 is, thus, a novel target for pain management.
Keywords: Ion Channels, Membrane Proteins, Neurobiology, Sodium Channels, Water Channel, Aquaporin, Pain, Patch Clamp
Introduction
Aquaporins (AQPs)2 are water-transporting proteins expressed in epithelial, endothelial, and other cell types. In the central nervous system AQP4 is expressed in glial cells, where it plays a role in cerebral edema (1, 2), glial cell migration (3, 4), and neuroexcitation (5, 6). The mechanisms of AQP4 modulation of seizure dynamics (5), cortical spreading depression (6), vision (7), hearing (8), and olfaction (9) remain unclear. AQP4-dependent Kir4.1 K+ channel function has been suggested from delayed K+ reuptake from brain extracellular space after neuroexcitation (6, 10); however, patch clamp analysis showed AQP4-independent Kir4.1 K+ channel function (11). Extracellular space expansion, which has been found AQP4-deficient brain (12, 13), may contribute to the altered neuroexcitation phenotype. Neurons in the central nervous system do not express AQPs.
Water channel AQP1 is abundant in hematopoietic cells and kidney. In normal brain AQP1 expression is restricted to the choroid plexus, where it facilitates the secretion of cerebrospinal fluid (14). In the spinal cord and peripheral nervous system, AQP1 is expressed in sensory neurons in dorsal root ganglia (DRG) that are associated with pain nociception (15–17). These neurons, whose cell bodies reside in the DRG, carry sensory signals from the periphery through small diameter, non-myelinated fibers that synapse in the superficial lamina of the dorsal horn in the spinal cord (18). Two prior studies on pain phenotype in AQP1−/− mice have reported conflicting behavioral findings. Oshio et al. (16) reported mild impairment in pain nociception in AQP1−/− mice after thermal (tail flick) and chemical (capsaicin) stimuli, with no differences in response to mechanical stimuli. Shields et al. (15) confirmed AQP1 expression in DRG neurons and partially colocalization with TRPV1 and substance P; however, they reported no significant differences in behavioral pain tests. The role of AQP1 in neuronal function in the DRG has, thus, remained unclear, as does its role in neurons in trigeminal and nodose ganglia, where it is also expressed.
To clarify the role of AQP1 in pain physiology, we did more extensive behavioral testing as well as immunolocalization, water permeability, and patch clamp studies on freshly isolated, dissociated DRG neurons from wild type (AQP1+/+) and AQP1 null (AQP1−/−) mice. We found greatly reduced behavioral response to inflammatory thermal and cold pain in litter-matched AQP1−/− mice and distinct electrophysiological defects related to impaired Nav1.8 Na+ channel functioning in AQP1-deficient DRG neurons. Patch clamp, immunoprecipitation, and single particle tracking studies in transfected cell models suggested a novel AQP1-Nav1.8 interaction as responsible, in part, for the impairment in pain-sensing in AQP1 deficiency.
EXPERIMENTAL PROCEDURES
Mice
AQP1−/− mice in a CD1 (out-bred) strain were generated by targeted gene disruption as described (19). Mice in a C57/bl6 (inbred) genetic background were generated by >10 back-crosses. 8-to-10-week-old, age-matched mice were used. Investigators were blinded to genotype information in all experiments. Protocols were approved by the University of California San Francisco Committee on Animal Research.
Isolation of DRG Neurons
Adult mice were decapitated after anesthesia. L4–6 DRGs were removed and treated with collagenase (type IA, 1.5 mg/ml, Sigma) and trypsin (1 mg/ml, Sigma) in Dulbecco's modified Eagle's medium at 37 °C for 30 min. After incubation, DRGs were washed five times and gently triturated using fine fire-polished glass pipettes. Dissociated DRG neurons were plated onto poly-d-lysine-coated coverslips and incubated at 4 °C for up to 8 h before electrophysiological measurements.
DRG Cell Line Culture
ND7-23 DRG neuroblastoma cells (20) were obtained from the European Collection of Cell Cultures and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mm glutamine at 37 °C in humidified air with 5% CO2. Cells were stably transfected with cDNA encoding human AQP1 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Stably transfected cells were selected with 800 μg/ml G418 (Invitrogen). Resistant colonies were tested for AQP1 expression and clonally expanded. Rat Nav1.8 (kindly provided by Dr. John Wood, University College London) and green fluorescent protein were co-transiently transfected into non-transfected ND7-23 cells or the AQP1 stably expressing ND7-23 cells. Experiments were done 48 h after transfection.
Electrophysiology
Whole-cell patch clamp recordings were done at room temperature (20–22 °C) using an Axon 200B amplifier (Molecular Devices, Union City, CA). Patch clamp data were acquired using a Digidata 1440A digitizer with Clampex 10.0 software (Molecular Devices). Microelectrodes with a resistance of 3–5 megaohms were fabricated using a P-97 micropipette puller (Sutter Instruments, Novato, CA). Data were sampled at 5 kHz and low pass-filtered at 5 kHz. Series resistance was routinely compensated (85–95%). Capacitive transients were canceled with the amplifier circuitry, and linear leakage currents were digitally subtracted online using the P/4 procedure. Data were analyzed by Clampfit 10.0 and Sigmaplot.
For patch clamp studies, DRG neurons were incubated with 3 μg/ml IB4-Alexa 594 (Invitrogen) for 5 min and washed twice for 2 min. Small diameter (<25 μm) IB4-negative neurons were studied (21). In current-clamp experiments, cells were incubated in the standard external recording solution containing 140 mm NaCl, 5 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 10 mm HEPES, and 10 mm d-glucose, pH 7.4, adjusted with NaOH. The intracellular (pipette) solution contained 5 mm NaCl, 140 mm KCl, 0.5 mm CaCl2, 2 mm MgCl2, 5 mm EGTA, 10 mm HEPES, and 3 mm Na2ATP, pH 7.2, adjusted with NaOH. Depolarizing current ramp (1 s, 1 nA) and depolarizing current pulse (100 pA, 10s) injections were applied to examine action potential (AP) firing. Single APs were evoked by 1-ms depolarizing current pulses. AP threshold was determined from a 1st derivative plot where the dV/dt abruptly increases (22). AP amplitudes were measured from threshold to peak (23). AP duration was measured at half-maximal amplitude. Input resistance (Rin) was determined as the slope of the voltage-current curve measured in the linear region at hyperpolarized potentials. Maximal upstroke velocity (V/s) was determined as dV/dtmax. In some experiments, tetrodotoxin (TTX, 300 nm) (Sigma) was used to block TTX-sensitive (TTX-S) Na+ channels.
In voltage-clamp measurements, TTX-resistant (TTX-R) Na+ currents in DRG neurons were isolated using as extracellular solution 35 mm NaCl, 110 mm choline chloride, 5 mm CsCl, 1 mm MgCl2, 1 mm CaCl2, 0.1 mm CdCl2, 0.3 mm TTX, 10 mm HEPES, 10 mm d-glucose, pH 7.4 (osmolality 320 mosm). Intracellular solution was 140 mm CsF, 1 mm MgCl2, 1 mm EGTA, 5 mm Na2ATP, 10 mm HEPES, pH 7.2 (300 mosm). The external Cd2+ and internal F− block calcium currents (24) and the internal Cs+ inhibits K+ current. Recordings were started 10 min after establishing the whole-cell configuration. Current-voltage curves were generated by application of a series of 100-ms test pulses to voltages that ranged from −50 to +50 mV in 5-mV increments. Peak INa at each depolarized potential was plotted. For current density measurements, membrane currents were normalized to membrane capacitance.
To study the Na+ channels on Nav1.8-transfected ND7-23 cells, the extracellular solution contained 120 mm NaCl, 20 mm tetraethylammonium chloride, 5 mm CsCl, 1 mm MgCl2, 1 mm CaCl2, 0.1 mm CdCl2, 10 mm HEPES, 10 mm d-glucose, pH 7.4 (320 mosm). Tetraethylammonium chloride was used to inhibit K+ current. Current-voltage curves were plotted from −50 to +80 mV in 10-mV increments. Use-dependence of Na+ channels was studied using repetitive 30-ms depolarizing pulses from −80 to 0 mV with different frequencies (5, 10, or 20 Hz). The reduction in peak INa at the 40th pulse compared with the 1st pulse was taken as a measure of use-dependent channel inactivation (25, 26).
Immunohistochemistry and Immunocytochemistry
Mice were perfused with 4% paraformaldehyde, and DRGs were post-fixed for 24 h in 4% paraformaldehyde and processed in paraffin. Sections (7 μm) were deparaffinized in xylene and rehydrated in graded ethanol. After epitope retrieval with citrate buffer (10 mm sodium citrate, 0.05% Tween 20, pH 6, 30 min, 95–100 °C). Staining was also done on isolated cells that were seeded onto poly-d-lysine-coated, 12-mm diameter coverslips and fixed with 4% paraformaldehyde. After blocking with 1% bovine serum albumin, coverslips were incubated with rabbit anti-AQP1 (1:100, Santa Cruz Biotechnology Inc., Santa Cruz, CA), chicken anti-peripherin (1: 200, Millipore), chicken anti-calcitonin gene-related peptide (CGRP; 1:500, Neuromics, Edina, MN) or biotin-conjugated IB4 (Sigma) followed by Texas Red-conjugated anti-rabbit secondary antibody (Sigma) and/or fluorescein isothiocyanate-conjugated anti-chicken secondary antibody (Millipore). Binding of biotinylated IB4 was visualized with fluorescein-conjugated streptavidin (Jackson ImmunoResearch, West Grove, PA). Nuclei were counterstained blue with 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA).
Immunoblot and Immunoprecipitation Analysis
L4–6 DRGs from three mice of each genotype were homogenized in lysis buffer (Cell Signaling, Danvers, MA) and centrifuged at 3000 × g for 10 min, and the supernatant was loaded onto a NOVEX-NuPAGE 4–12% BisTris SDS-PAGE gel (10 μg protein/lane, Invitrogen). Proteins were transferred to a nitrocellulose membrane and incubated with rabbit anti-AQP1 antibody (1:1000), anti-Nav1.8 antibody (1:500; Millipore), or anti-β-actin antibody (1:2000; Cell Signaling) followed by horseradish peroxidase-linked anti-rabbit IgG (1:10.000; GE Healthcare) and visualized by enhanced chemiluminescence (GE Healthcare).
ND7-23 cells were co-transfected with Nav1.8 and full-length AQP1 (AQP1.T120.myc) or a C terminus truncation mutant of AQP1 (AQP1.T120.myc.Δ27 or AQP1.T120.myc.Δ41). Two days after transfection, cell extracts were prepared using radioimmune precipitation assay buffer lysis buffer containing protease inhibitor mixture (Pierce) according to the manufacturer's instructions. Protein concentration was determined using the BCA protein reagent kit (Bio-Rad). The cell lysate was diluted to ∼1 mg/ml of total protein with lysis buffer and mixed with rabbit anti-AQP1 polyclonal antibody (1 g/100 g of total protein, Santa Cruz), rabbit anti-Nav1.8 polyclonal antibody (1 g/100 g of total protein, Millipore), or c-Myc monoclonal antibody (1 g/100 g of total protein, Covance, Emeryville, CA) for 1 h at 4 °C. Equal amounts of samples were mixed with either rabbit IgG or mouse IgG or vehicle as negative controls. Protein A/G beads were then added, and solutions were rocked for 1 h at 4 °C. The protein A/G beads were collected and washed three times with lysis buffer. Proteins were eluted by boiling in elution buffer (Pierce). Samples were resolved by SDS-PAGE and analyzed by immunoblot using mouse anti-AQP1 monoclonal antibody (Santa Cruz, 1:400), rabbit anti-Nav1.8 antibody (Millipore), and rabbit anti-c-Myc antibody (Santa Cruz, 1:400). Clean-Blot IP detection reagent (Pierce) was used as the secondary antibody, and blots were detected using the SuperSignal (Pierce) detection system.
Behavioral Testing
All behavioral testing was done with genotype information blinded. Age-matched male and female mice were used. Several pain behavior studies were done using established procedures as follows. (a) For hotplate testing, mice were placed on a hotplate (model #35100, Ugo Basile, Comerio, Italy), and a ramp stimulus (45–52 °C, 2 °C/min) was applied. The temperature threshold was recorded at which paw-licking or jumping was observed. In some experiments the temperature was fixed at 50 °C, and the latency was determined for paw-licking or jumping (cut-off time 1 min). (b) For capsaicin testing, mice were kept in an empty cage. Total licking time over 5 min was recorded after intraplantar injection of capsaicin (10 μl, 1 or 3 μg). (c) Prostaglandin E2-induced pain hypersensitivity was tested by measuring hotplate latency (50 °C) at specified times after intraplantar injection of PGE2 (10 μl, 300 ng). (d) For bradykinin-induced acute pain testing, total licking time over 10 min was recorded after intraplantar injection of bradykinin (10 μl, 300 ng). In separate experiments hotplate latency (50 °C) was measured at 10 min after bradykinin injection. Paw edema was measured as paw thickness at 10 min. (e) For formalin testing, total licking time in 5-min intervals over 40 min was measured after intraplantar injection of 10 μl of 5% formalin in saline. (f) For cold pain testing, a cold-plate (model #35100, Ugo Basile) maintained at 4 °C was used to assess noxious cold sensitivity of the plantar surface of the hind paws (27). Mice were placed on the cold plate for 5 min, and behavior was videoed and scored in a blinded manner. Severity of cold pain was scored in 5-s intervals using the following scale: 0, standing still; 1, walking; 2, each paw lifting occurrence; 3, each jumping occurrence.
Osmotic Water Permeability
Cell plasma membrane water permeability was measured by a calcein-quenching method (28). DRG cells were isolated after enzymatic digestion as described above, immobilized on a poly-d-lysine-coated cover glass, and loaded with calcein by 30-min incubation with 5 μm calcein-AM (Invitrogen). The cover glass was mounted in a custom perfusion chamber having a solution exchange time of <200 ms at 50 ml/min perfusion rate. The time course of cytoplasmic calcein fluorescence was measured in response to cell swelling produced by a 2-fold dilution of the extracellular bathing solution with water. Single cell calcein fluorescence was measured continuously using a Nikon inverted epifluorescence microscope equipped with 100× oil objective, halogen light source, calcein filter set (480-nm excitation, 490-nm dichroic mirror, 535-nm emission filter), photomultiplier detector, and 14-bit analog-to-digital converter.
Quantitative Real-time Reverse Transcription-PCR
L4–6 DRGs were collected after euthanasia, total RNA was isolated by a PureLinkTM Micro-to-midi kit (Invitrogen), and cDNA was reverse-transcribed from mRNA with the Super-Script First Strand Synthesis System for reverse transcription-PCR (Invitrogen). Fluorescence-based quantitative real-time reverse transcription-PCR was carried out using the LightCycler 480 and with LightCycler FastStart DNA MasterPLUS SYBR Green I kit (Roche Diagnostics). Primers were as follows: 5′-TGTATGCCTCTGGTCGTACC-3′ (sense) and 5′-CAGGTCCAGACGCAGGATG-3′ (antisense) for β-actin, 5′-CTCCCTAGTCGACAATTCAC-3′ (sense) and 5′-ACAGTACCAGCTGCAGAGTG-3′ (antisense) for AQP1, 5′-ACCGACAATCAGAGCGAGGAG-3′ (sense) and 5′-ACAGACTAGAAATGGACAGAATCACC-3′ (antisense) for Nav1.8, 5′-TGAGGCAACACTACTTCACCAATG-3′ (sense) and 5′-AGCCAGAAACCAAGGTACTAATGATG-3′ (antisense) for Nav1.9, 5′-TCCTTTATTCATAATCCCAGCCTCAC-3′ (sense) and 5′-GATCGGTTCCGTCTCTCTTTGC-3′ (antisense) for Nav1.7, 5′-GTGCATCTCCTGTAAGCGTCGTAG-3′ (sense) and 5′- ATTCTCATAGCGTAGGATCTTGACAA-3′ (antisense) for β1. Data were analyzed by LightCycler software 4.0 (Roche Diagnostics) and reported as calibrated ratios normalized to β-actin. Data were averaged from three mice of each phenotype.
Single Particle Tracking
Single particle tracking measurements were done on confluent cells grown on 18-mm cover glasses that were transfected with plasmid encoding AQP1.T120.myc at 18–24 h before measurements. Nav1.8 or control pcDNA3 vector was co-transfected with AQP1 at a 5:1 ratio. In some experiments Nav1.8 or control pcDNA3 vector was first co-transfected with plasmid encoding green fluorescent protein (10:1) for 36 h and then transfected with the AQP1.T120.myc, allowing measurements to be made selectively on green fluorescent protein-positive cells. Cells were labeled with c-Myc antibody (Covance) and followed by goat F(ab′)2 anti-mouse Qdot 655 (Invitrogen) as described previously (29). Single particle tracking was done on a TE2000S inverted microscope (Nikon, Melville, NY) equipped with a 100× total internal reflection fluorescence oil immersion objective and EM-CCD (Hamamatsu, Bridgewater, NJ) with resolution 80 nm/pixel using continuous 11-ms acquisitions for 6 s (91 Hz). Single particle tracking image sequences were analyzed, and trajectories were constructed using IDL software (Research Systems, Boulder, CO) as described (29). Blinking of individual Qdots was accounted for during trajectory constructions. Only trajectories longer than 200 steps were analyzed with data sets composed of at least 100 trajectories from at least 5 cells. The mean-squared displacement as a function of time 〈r2(t)〉 was computed for each trajectory, and the diffusion coefficient D1–3 and offset (due to positional noise) were determined by a linear fit of the first three time steps of the mean-squared displacement: 〈r2(t)〉1–3 = 4D1–3t + offset. Data are reported as cumulative distributions of D1–3 and ranges at 1 s.
Statistical Analysis
Behavioral data comparing responses in AQP1+/+ versus AQP1−/− mice was done using the Student's t test. Time course data in a formalin test and PGE2 testing were compared by analysis of variance. Electrophysiology data comparing responses in AQP1+/+ versus AQP1−/− DRG neurons was done using Student's t test. Neuron firing time was compared by the rank sum test.
RESULTS
AQP1 Expression and Water Permeability in DRG Neurons
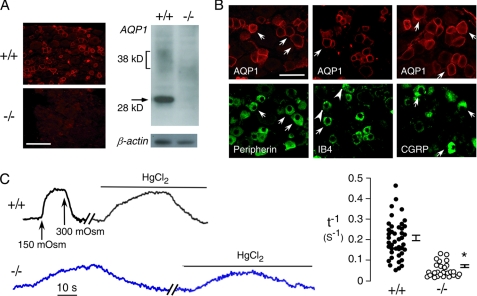
By immunofluorescence, AQP1 immunoreactivity was seen in DRG neurons, mainly in the plasma membranes of small, <25-μm diameter neurons (Fig. 1A, left). Immunoblot analysis showed non-glycosylated AQP1 as a sharp band at 28 kDa and glycosylated AQP1 as a diffuse band migrating at 36–42 kDa (Fig. 1A, right). Specific immunoreactivity was absent in DRGs from AQP1−/− mice. Double immunostaining of DRG sections with AQP1 and peripherin, a small-diameter nociceptor marker (30), showed that 92 ± 2% of AQP1-positive neurons were peripherin-positive, indicating that nearly all AQP1-expressing cells are nociceptors (Fig. 1B). There are two subpopulations of nociceptive neurons, “peptidergic” and “non-peptidergic” types (18). By double immunostaining with AQP1 and IB4, a surface binding lectin specific for non-peptidergic neurons (21), 54 ± 4% of IB4-positive neurons were AQP1-positive. By double immunostaining with AQP1 and CGRP, a neuropeptide specific for peptidergic small DRG neurons (18), 71 ± 1% of CGRP-positive neurons were AQP1-positive. The majority of small-diameter DRG neurons, thus, express AQP1.
FIGURE 1.
AQP1 expression and water permeability in DRG neurons. A, shown is AQP1 immunofluorescence (left) and an immunoblot (right) of freshly isolated DRG neurons from AQP1+/+ and AQP1−/− mice. Bar, 100 μm. B, shown is immunostaining for AQP1 (red) and nociceptor markers peripherin (left), IB4 (center), and calcitonin gene-related peptide (right). Arrows indicate examples of colocalization, and arrowheads indicate examples without colocalization. Bar, 50 μm. C, osmotic water permeability of DRG neurons is shown. Left, time course of calcein fluorescence in response to exchange between isosmolar and hypo-osmolar (150 mosm) solutions is shown. Data are shown for AQP1+/+ and AQP1−/− DRG neurons under control conditions and after application of HgCl2 (100 μm). Right, a summary of deduced reciprocal half-times (t−1) for osmotic equilibration is shown. Filled circles show individual measurements (S.E., t test; *, p < 0.001).
To test whether AQP1 in DRG neurons is functional as a plasma membrane water channel, water permeability was determined in freshly isolated DRG neurons from AQP1+/+ and AQP1−/− mice. Plasma membrane osmotic water permeability was measured by a calcein fluorescence quenching method (28), which quantifies the kinetics of cell volume change in response to osmotic gradients. Two types of responses were seen in small DRG neurons from AQP1+/+ mice, including neurons showing rapid changes in calcein fluorescence that were slowed by the AQP1 inhibitor HgCl2 (Fig. 1C, left). Responses from many AQP1+/+ DRG neurons are summarized in Fig. 1C (right). All DRG neurons from AQP1−/− mice showed slow responses that were insensitive to HgCl2. These measurements indicate that AQP1 is functional in DRG neurons.
Behavioral Analysis Shows Impaired Noxious Thermal Pain Nociception in AQP1−/− Mice
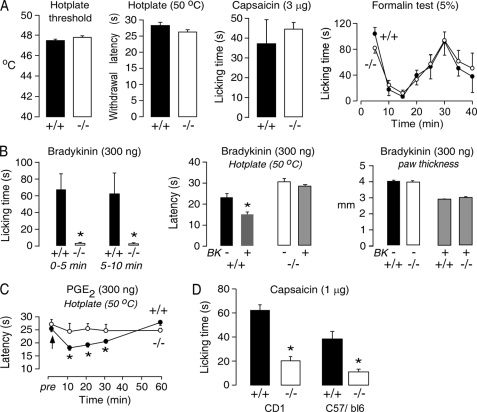
The functional expression of AQP1 in small diameter DRG neurons suggests its involvement in pain nociception. Initial studies of hotplate threshold (temperature ramp), hotplate latency (constant 50 °C temperature), paw licking time after high dose intraplantar injection of capsaicin (3 μg/paw), and paw licking time after intraplantar injection of formalin (5% in saline, 10 μl/paw), as done in the two prior studies (15, 16), showed no significant differences between AQP1+/+ and AQP1−/− mice (Fig. 2A).
FIGURE 2.
Impaired peripheral pain nociception in AQP1−/− mice. A, shown are behavioral responses to thermal stimuli (hotplate threshold and withdrawal latency) (S.E., 27 AQP1+/+ mice, 20 AQP1−/− mice; t test; *, p = 0.15 and 0.6), intraplantar injection of 3 μg/paw capsaicin (S.E., 5 mice per genotype; t test; p = 0.22), and intraplantar injection of formalin (S.E., 6 mice per genotype, analysis of variance, p = 0.33). B, left, shown is licking time at 0–5 min and 5–10 min after intraplantar injection of 300 ng bradykinin (S.E., 10 mice per genotype; t test; *, p < 0.001). Middle, shown is hotplate latency at 10 min after bradykinin injection (S.E., 6 mice per genotype; t test; *, p = 0.001). Right, shown is paw thickness at 10 min after bradykinin injection (S.E., n = 10 mice per genotype, differences are not significant). C, shown is hotplate latency after intraplantar injection of 300 ng of PGE2 (S.E., 6 mice per genotype; t test; *, p = 0.008 at 10 min, p = 0.01 at 20 min, p = 0.03 at 30 min). D, shown is paw licking time 5 min after intraplantar injection of 1 μg/paw capsaicin (S.E., 7 mice per genotype in CD1 background; t test; *, p < 0.001, 8 mice per genotype in C57/b6 background; t test; *, p < 0.001).
Behavioral studies involving acute inflammatory pain were done in an attempt to discover differences in pain nociception in AQP1−/− mice that were not observed using the protocols in Fig. 2A. Bradykinin responses were tested as a measure of acute inflammatory pain (31). Intraplantar bradykinin injection produced an immediate paw licking response, which was greatly reduced in AQP1−/− mice (Fig. 2B, left). Also, after bradykinin injection, the paw withdrawal latency on a 50 °C hotplate was significantly reduced in AQP1+/+ mice but not changed significantly in AQP1−/− mice (Fig. 2B, middle). As a control, we found that paw edema, which is mediated after bradykinin injection by mast cell B1 receptors (31), was comparable in AQP1+/+ and AQP1−/− mice (Fig. 2B, right). Studies were also done using a different inflammatory mediator, PGE2, which is involved in the development of acute inflammatory pain (32). Although intraplantar injection of PGE2 did not itself produce a paw licking response, it produced a transient thermal hypersensitivity in AQP1+/+ mice which was absent in AQP1−/− mice (Fig. 2C).
After discovery of these significant phenotype differences in bradykinin and PGE2 testing, we re-examined capsaicin responses but using a lower dose (1 μg/paw) of capsaicin, which produces less nerve damage but a similar licking response (33, 34); 3 μg of capsaicin has been reported to destroy C fibers (35). Intraplantar injection of 1 μg of capsaicin produced a greatly reduced licking time in AQP1−/− mice in both out-bred (CD1) and inbred (C57/bl6) mouse stains (Fig. 2D).
Altered Electrophysiological Responses of AQP1-deficient DRG Neurons
To investigate possible mechanisms to account for the greatly reduced pain nociception in AQP1−/− mice, whole-cell patch clamp was done to compare electrophysiological responses from isolated, small DRG neurons (diameter <25 μm). Of 45 and 50 DRG neurons studied from AQP1+/+ and AQP1−/− mice, respectively, there were no differences in cell size, resting membrane potential, or input resistance (Table 1). Because of the distinct electrophysiological properties of IB4-positive and IB4-negative small DRG neurons (21, 36), in subsequent patch clamp studies we labeled live cells with IB4-Alexa 594 and only studied IB4-negative small DRG neurons.
TABLE 1.
Electrophysiological properties of AQP1+/+ and AQP1−/− small DRG neurons
Differences are not significant. AHP, after-hyperpolarization.
| Membrane potential | Input resistance | Cell capacitance | Injection current threshold | AP 50% width | AHP | |
|---|---|---|---|---|---|---|
| mV | Gigaohms | Picofarads | nA | ms | mV | |
| AQP1+/+ | −62.7 ± 1.0 (n = 45) | 1.3 ± 0.1 (n = 45) | 18.7 ± 0.7 (n = 45) | 0.6 ± 0.1 (n = 26) | 3.4 ± 0.4 (n = 26) | 12.3 ± 1.2 (n = 26) |
| AQP1−/− | −62.9 ± 1.4 (n = 50) | 1.0 ± 0.1 (n = 50) | 17.6 ± 0.8 (n = 50) | 0.6 ± 0.1 (n = 28) | 3.3 ± 0.4 (n = 28) | 11.3 ± 1.0 (n = 28) |
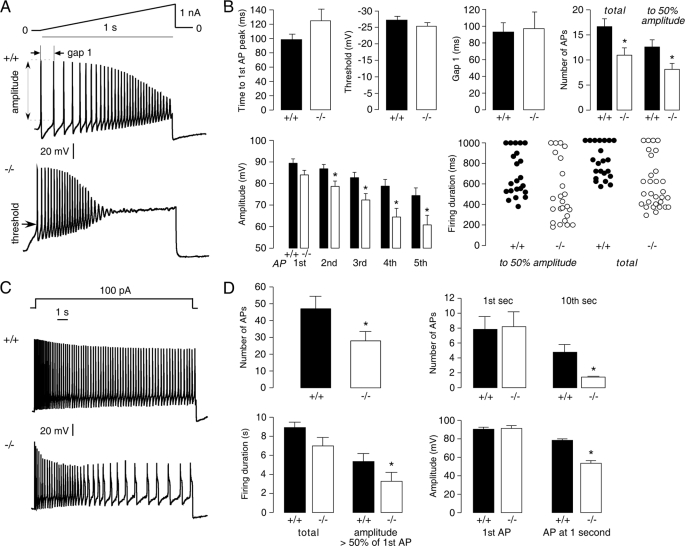
A first set of studies was done under current clamp conditions with 1-s current injections with a linear ramp current from 0–1 nA (Fig. 3A). The ramp current injection elicited a series of spikes over the 1-s injection with progressively reduced amplitude. Spikes with peak voltage >0 mV were defined as APs (23). The threshold was defined as the potential when the curve slope abruptly increased, as determined by derivative analysis, and the amplitude was defined as the potential change from the threshold to the peak. Of 26 small DRG neurons studied from AQP1+/+ mice, 22 responded, with the number of APs in response to the current ramp ranging from 6 to 28, with an average 16 ± 1 (S.E.). Of 28 small DRG neurons studied from AQP1−/− mice, 24 responded, with a significantly reduced average number of APs of 11 ± 1 (S.E.). Fig. 3B summarizes the analyses done on data as in Fig. 3A. The threshold, initial latency (to the first AP peak), and the gap time (between the first two APs) showed no differences. The amplitude of the first AP showed no difference, but with continue firing the amplitudes of the fourth and fifth APs of AQP1−/− neurons were significantly decreased. The total firing duration was reduced in AQP1−/− DRG neurons. Counting of APs with an amplitude greater than 50% that of the first AP indicated remarkably fewer APs and decreased the firing duration of AQP1−/− DRG neurons.
FIGURE 3.
Impaired firing of AQP1−/− DRG neurons after sustained stimulation. A, examples are shown of small DRG neuron responses to a 1-s linear current ramp (0–1 nA). B, shown is analysis of data as in A (S.E., 22 AQP1+/+ neurons and 24 AQP1−/− neurons). Differences were not significant by t test, except as indicated by the asterisks (p < 0.05). C, examples are shown of small DRG neuron responses to 10-s constant current injection (100 pA). D, shown is analysis of data as in C (S.E., 22 AQP1+/+ neurons and 21 AQP1−/− neurons). Differences were not significant by t test or rank sum test for firing duration, except as indicated by the asterisks (p < 0.05).
A second set of studies was done with a greater duration (10 s) of current injection using 100 pA constant current. Of 26 small DRG neurons studied from AQP1+/+ mice, 22 responded with more than 1 AP, with 21 of 28 responders from AQP1−/− mice. Most DRG neurons from AQP1+/+ mice fired throughout the depolarization, with minimal decay in the amplitude and frequency of APs; a substantially greater decay in APs was seen in DRG neurons from AQP1−/− mice (Fig. 3C). Analysis of all responding small DRG neurons showed significantly fewer APs in DRG neurons from AQP1−/− mice (47 ± 7 versus 28 ± 5 in 10 s) (Fig. 3D). Although the number of APs was comparable over the first second in the AQP1−/− neurons, the number of APs was much lower over the last second. Also, AP amplitudes in AQP1−/− DRG neurons decayed faster, taking 2.9 ± 0.8 s to decrease by 50% compared with 5.7 ± 0.5 s in AQP1+/+ DRG neurons. At 1 s, the amplitude of APs in AQP1−/− DRG neurons was significantly decreased. The reduced AP firing in AQP1−/− DRGs, seen in two different current injection protocols, may account for the impaired pain nociception in AQP1−/− mice.
Evidence for Nav1.8 as Responsible for the AQP1-sensitive Na+ Current
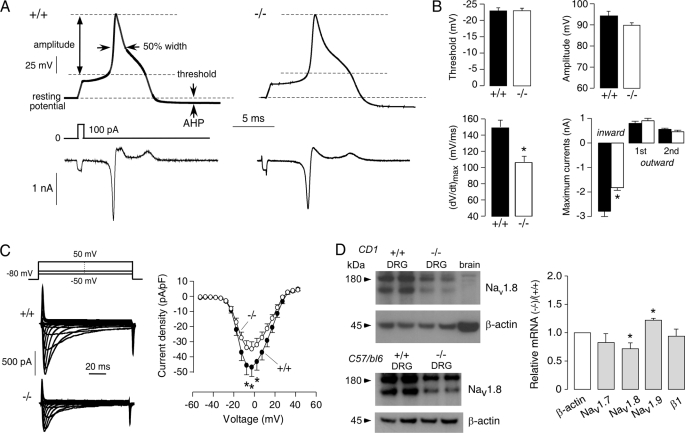
To identify the membrane channels responsible for the different responses in DRG neurons from AQP1+/+ and AQP1−/− mice, we measured single APs after a short (1-ms) step-depolarizing current, leaving most of each AP free of the effect of injected current (22). Nociceptive DRG neurons generally show a long duration AP with a shoulder on the falling phase of the AP (21), as seen in the representative single APs in Fig. 4A. Comparing single APs from 26 and 28 small DRG neurons from AQP1+/+ and AQP1−/− mice, respectively, no significant differences were found in AP threshold, AP amplitude, width at 50% amplitude, or maximum after-hyperpolarization (AHP) amplitude (Fig. 4B and Table 1). However, analysis of the time derivative of the voltage response (dV/dt) showed reduced maximum dV/dt in AQP1−/− neurons, giving a significant reduced maximum ionic current (Iionic = −C.dV/dt) in the inward direction. The maximum inward current represents a Na+ current with little contribution from other ion channels (22).
FIGURE 4.
Impaired inward Na+ current in AQP1−/− DRG neurons. A, top, examples are shown of single APs from small DRG neurons elicited by injection of a 1-ms depolarizing current. Threshold, amplitude, 50% width, resting potential, and after-hyperpolarization (AHP) indices are indicated. Bottom, shown is ion current computed from the voltage waveform as −C.dV/dt, where C is the cell capacitance. B, analysis of data is as in A (S.E., 26 AQP1+/+ neurons and 28 AQP1−/− neurons). Differences not are significant by t test, except as indicated by the asterisks (p < 0.05). C, left, examples are shown of whole-cell TTX-R Na+ currents. Right, shown is the current-voltage relationship of TTX-R Na+ currents (normalized by cell capacitance) (S.E., 22 AQP1+/+ neurons and 17 AQP1−/− neurons, paired t test; *, p < 0.05). pF, picofarads. D, left, shown is a Nav1.8 immunoblot of DRG in AQP1+/+ and AQP1+/+ mice (CD1 and C57/b6 genetic backgrounds), with β-actin immunoblot for the same samples. Right, shown is relative mRNA expression of AQP1, Nav1.8, Nav1.7, Nav1.9, and sodium channel β1 subunit quantified by real-time PCR (S.E., 3 mice per genotype). *, p < 0.05.
Both TTX-S and TTX-R Na+ channels are expressed on small DRG neurons. Several TTX-S Na+ channels are expressed on all DRG neurons, including Nav1.1, Nav1.2, and Nav1.7 (37). TTX-R Na+ channels Nav1.8 and Nav1.9 are expressed mainly in small DRG neurons (38). In small DRG neurons >80% of the peak inward current during an AP upstroke is carried by Nav1.8 (39, 40), whereas the TTX-S Na+ current contributes mainly to the AP initial threshold. We found that 300 nm TTX did not significantly change the maximal inward currents in AQP1+/+ or AQP1−/− small DRG neurons (data not shown). To isolate Nav1.8 Na+ currents, whole-cell currents were measured in the presence of TTX and reduced extracellular Na+ concentration of 35 mm. We studied 22 and 17 small DRG neurons from AQP1+/+ and AQP1−/− mice, respectively. Although the voltage dependence of Nav1.8 Na+ activation was similar in DRG neurons from AQP1+/+ and AQP1−/− mice (Fig. 4C), the maximum current amplitude was significantly reduced in AQP1−/− DRG neurons.
The reduced Nav1.8 Na+ current in AQP1−/− could be a consequence of reduced Nav1.8 Na+ channel expression and/or a functional effect of AQP1 expression on Nav1.8 Na+ current. Both possibilities were investigated. By immunoblot analysis, the expression of Nav1.8 protein in DRGs from AQP1−/− mice was reduced compared with that from AQP1+/+ mice, as seen in both out-bred (CD1) and inbred (C57/bl6) mouse strains (Fig. 4D). By quantitative reverse transcription-PCR, Nav1.8 transcript expression was also significantly lower in AQP1−/− mice, whereas that of Nav1.7 and the sodium channel β1 subunit, which is widely expressed in all DRG neurons, were not changed (Fig. 4D). Transcript encoding Nav1.9 was significantly increased, which might represent a compensatory effect. Reduced Nav1.8 expression in AQP1−/− mice might account, in part, for the pain phenotype findings.
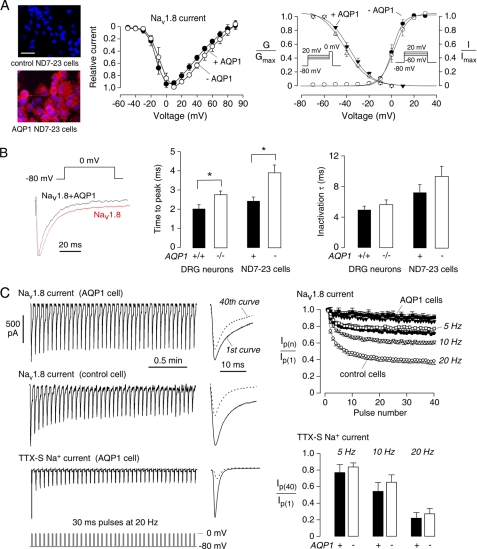
AQP1 Modulates the Kinetic Properties of Nav1.8 Na+ Currents in ND7-23 Cells
The data above suggest impaired Nav1.8 current in AQP1−/− DRG neurons. To further study the involvement of AQP1 in Nav1.8 function, we measured Nav1.8 Na+ current in ND7-23 cells, an immortalized rat DRG/mouse neuroblastoma cell line, which is the only cell line that forms functional Nav1.8 channels after transfection without the need to transfect accessory subunits (41). These cells do not express AQP1 as seen by immunostaining (Fig. 5A, left) and immunoblot analysis (data not shown). For these studies Nav1.8 was transiently transfected (along with green fluorescent protein to identify transfected cells) into control and AQP1-expressing ND7-23 cells. The current-voltage relation of the Nav1.8 Na+ current was similar in the control and AQP1-expressing ND7-23 cells (Fig. 5A, middle). Analysis of the voltage-dependent activation and steady-state inactivation also showed no differences between control and AQP1-expressing cells. However, analysis of activation and inactivation kinetics of Nav1.8 current at 0 mV did reveal differences (Fig. 5B). The activation time (the time to peak current) was significantly shorter in the presence of AQP1, which was seen both in freshly isolated DRG neurons and transfected ND7-23 cells. The inactivation time did not differ significantly.
FIGURE 5.
AQP1-sensitive Nav1.8 current in transfected ND7-23 cells. A, left, AQP1 immunofluorescence (red) in non-transfected and AQP1 stably transfected ND7-23 cells is shown. Nuclei were counterstained blue with 4′,6-diamidino-2-phenylindole. Bar, 50 μm. Middle, shown is the current-voltage relationship of Nav1.8 currents in control and AQP1 stably expressing ND7-23 cells. Current was normalized to the current at 0 mV (analysis of variance, p = 0.52). Right, shown is voltage-dependent activation (G/Gmax) of Nav1.8 with fitted parameters: AQP1-expressing ND7-23 cells, V1/2 = −4 ± 1 mV, k = 5 ± 1 mV, n = 8; control ND7-23 cells, V1/2 = −3 ± 1 mV, k = 5 ± 1 mV, n = 8. Shown is steady-state inactivation (I/Imax) with fitted parameters: AQP1-expressing ND7-23 cells, V1/2 = −38 ± 2 mV, k = −10 ± 1 mV, n = 9; control ND7-23 cells, V1/2 = −41 ± 2 mV, k = −10 ± 1 mV, n = 8 (t test, p = 0.35 and 0.45). B, left, examples of Nav1.8 current evoked by 100-ms pulses from −80 to 0 mV from control and AQP1-expressing ND7-23 cells; middle, activation time of Nav1.8 at 0 mV was significantly lower in AQP1-expressing ND7-23 cells and AQP1+/+ DRG neurons (S.E., t test; *, p = 0.001 for the ND7-23 cell, 0.004 for DRG cells); right, inactivation time at 0 mV shows no differences (p = 0.26 for ND7-23 cells and 0.33 for DRG cells). C, left, examples are shown of Nav1.8 current evoked by 30-ms pulses from −80 to 0 mV at 20 Hz and example of TTX-S Na+ current in AQP1-expressing cell. Na+ currents elicited by the 1st and 40th pulses are shown at the right. Right, top, peak Nav1.8 currents from control and AQP1-expressing ND7-23 cells (normalized to peak current from the 1st pulse) are shown as a function of pulse number and pulse simulation frequencies of 5, 10, and 20 Hz (S.E., 6 control and 6 AQP1 cells). Right, bottom, shown is normalized peak TTX-S Na+ currents at the 40th pulse at 5, 10, and 20 Hz frequencies in control and AQP1-expressing cells (S.E., t test, 4 control and 4 AQP1 cells).
Patch clamp of DRG neurons in Fig. 3 showed decreased AP amplitude upon sustained stimulation, with a greater decrease seen in the AQP1−/− neurons. Decreased AP amplitude with repetitive firing is produced by slow inactivation of TTX-R Na+ current (26). We measured Nav1.8 Na+ channel slow inactivation in Nav1.8-expressing ND7-23 cells after repeated depolarizations to 0 mV from a −80 mV holding potential, with frequencies of 5, 10, and 20 Hz. Nav1.8 Na+ current amplitudes decreased progressively as a consequence of slow inactivation (Fig. 5C, top left). The reduced current at the 40th step compared with the first step was taken as a measure of slow inactivation. At 20 Hz, ∼60% reduction in Nav1.8 current was seen in control cells compared with ∼25% in AQP1-expressing cells, with the percentage reduction depending on frequency (Fig. 5C, top right). As a control, we studied intrinsic TTX-S Na+ current in ND7-23 cells, which has similar kinetics and activation/inactivation voltage dependence as the TTX-S current in DRG neurons (41). The TTX-S Na+ current was more sensitive to the slow inactivation protocol, with only ∼20% current remaining at the 40th pulse (Fig. 5C, bottom). However, in contrast to the TTX-R Na+ current, no significant differences in TTX-S current were found in control versus AQP1-expressing cells.
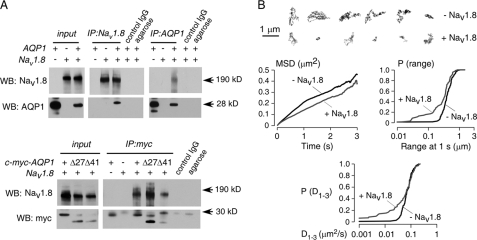
Evidence for Physical Interaction between Nav1.8 and AQP1
The above data indicate AQP1-modulation of Nav1.8 function. Both AQP1 and Nav1.8 are expressed in small DRG neurons. To test for physical association between Nav1.8 and AQP1, immunoprecipitations were done in ND7-23 cells at 48 h after co-transfection. As shown in Fig. 6A, top, Nav1.8 was detected in an AQP1 immunoprecipitate, and AQP1 was also detected in a Nav1.8 immunoprecipitate. Negative IgG and agarose controls indicated assay specificity. To test for involvement of the AQP1 C terminus in the AQP1-Nav1.8 interaction, immunoprecipitations were done using a series of AQP1 C terminus truncation mutants in which a c-Myc epitope was engineered into an extracellular site on AQP1. Fig. 6A, bottom, shows Nav1.8 in the c-Myc immunoprecipitates, even with the fully C terminus-truncated AQP1.T120.myc.Δ41, indicating that the AQP1 C terminus is not required for Nav1.8-AQP1 association. Similar results were obtained in a separate set of co-immunoprecipitation studies done using co-transfected HEK293T cells (data not shown).
FIGURE 6.
Evidence for physical interaction between Nav1.8 and AQP1. A, top, immunoprecipitations (IP) from ND7-23 cell expressing AQP1 and Nav1.8 are shown. Input shows AQP1 and Nav1.8 protein expression. Pulldown with anti-AQP1 antibody (against AQP1 C terminus) shows co-precipitated Nav1.8 protein (control IgG and agarose beads negative). Pulldown with anti-Nav1.8 antibody shows co-precipitated AQP1 protein (control IgG and agarose beads negative). WB, Western blot. Bottom, AQP1-Nav1.8 interaction does not involve the AQP1 C terminus. Pulldown done with anti-Myc antibody (against engineered extracellular c-myc epitope on AQP1) shows co-precipitated Nav1.8 protein with full-length AQP1.T120.myc and C-terminal AQP1 truncations. B, shown is single particle quantum dot tracking of AQP1 in the plasma membrane of ND7-23 cells. Top, shown is representative trajectories for AQP1 diffusion over 6 s in the absence (black) or presence (red) of co-expressed Nav1.8. Bar, 1 μm. Bottom, data summary is shown as a mean-squared displacement (MSD) plot and cumulative probability distributions of AQP1 diffusion coefficient, D1–3, and range at 1 s. Data are summarized for ∼200 trajectories on >15 cells.
To further investigate the AQP1-Nav1.8 protein interaction demonstrated biochemically, we determined whether Nav1.8 expression would slow plasma membrane diffusion of co-expressed AQP1 in live cells. AQP1 diffusion was found previously to be rapid and unrestricted in multiple non-neural cell types (29). Single particle tracking was done to quantify the diffusion of Qdot-labeled AQP1 in ND7-23 cells. Examples of Qdot trajectories are shown in Fig. 6B, top, and analysis of many trajectories is summarized in Fig. 6B, bottom. Nav1.8 expression produced significant slowing of AQP1 diffusion in a subpopulation of AQP1 molecules, as shown in mean-squared displacement plots by a small reduction in average AQP1 diffusion coefficient and in cumulative probability distribution plots of diffusion coefficients and range at 1 s, where a distinct subpopulation of slowly moving AQP1 molecules was seen corresponding to ∼30% of trajectories. These results provide evidence for AQP1-Nav1.8 interaction in live cells.
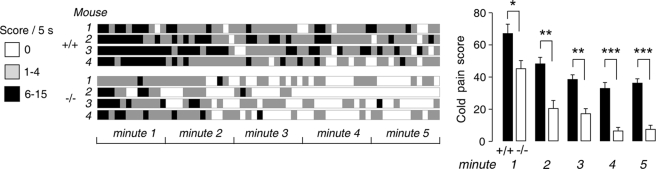
Reduced Cold Pain Perception in AQP1−/− Mice
The above data implicate Nav1.8 as an important determinant of AQP1-dependent DRG neuron function and pain perception. Based on a report showing that Nav1.8 is essential for the cold pain perception (27), we tested whether AQP1 deletion in mice affected cold pain perception. As summarized in Fig. 7, AQP1−/− mice showed remarkably reduced sensitivity to cold pain. During 5 min of observation on a 4 °C cold plate, the AQP1+/+ litter-matched mice showed frequent jumping and lifting the whole time, whereas the AQP1−/− mice mainly sensed the cold pain within the first 1 min and showed little jumping/lifting behavior after 3 min.
FIGURE 7.
Reduced cold pain perception in AQP1−/− mice. Real-time scoring of cold pain sensitivity in litter-matched AQP1+/+ and AQP1 −/− mice is shown. A, scores are shown in 5-s intervals over 5 min for 4 mice of each genotype. B, average scores are shown over 5 min (S.E., 8 AQP1+/+ mice and 7 AQP1−/− mice; t test; *, p = 0.03 for 1 min; **, p = 0.002 for 2 min; p = 0.001 for 3 min; ***, p < 0.001 for 4 and 5 min).
DISCUSSION
Our results establish a significant pain phenotype in AQP1−/− mice that was seen in assessments of acute thermal inflammatory, chemical, and cold pain. Evidence by patch clamp, immunoprecipitation, and single particle tracking analysis suggested that AQP1-Nav1.8 interaction is in part responsible for the pain phenotype and, thus, is involved in the tuning of action potential firing in dorsal root ganglion neurons in response to certain types of pain.
Two prior studies on pain phenotype in AQP1−/− mice have reported conflicting behavioral findings. Oshio et al. (16) reported mild impairment in nociceptive response in AQP1−/− mice after thermal (tail-flick) and chemical (capsaicin) stimuli, with no differences in response to mechanical stimuli. Shields et al. (15) reported no significant differences in a series of acute and persistent behavioral pain tests. In this paper we carefully re-tested nociceptive behavior related to acute thermal and cold pain and inflammation-induced thermal pain. In agreement with the two prior studies (15, 16), we found no differences in hotplate threshold or latency or the response to formalin. We also found no differences in response to high dose capsaicin, in agreement with Shields et al (15). However, it was recently reported by the same group (35) that high dose capsaicin produces mainly mechanical rather than thermal hyperalgesia, probably by C-fiber destruction. C-fibers are involved in nociceptive thermosensation (both hot and cold), whereas mechanosensation is mediated by both non-myelinated C-fibers and myelinated A-type fibers (33). High dose capsaicin, thus, sensitizes a different subtype of DRG neurons. We found here that low dose capsaicin, which evokes a near-pure thermal hyperalgesia without C-fiber destruction (27, 33), produced much reduced pain perception in the AQP1−/− mice. Remarkable phenotype differences were also found upon challenge with bradykinin and PGE2, which are major components of the inflammatory soup and sensitize small DRG neurons through G protein-coupled receptors targeting TTX-R and TRP channels (43). Regarding PGE2 testing, Shields et al. (15) reported no differences for a hypoosmotic challenge in the setting of PGE2-induced inflammation. This response is mediated primarily by the TRPV4 (44), a channel expressed on both small and large DRG neurons (45). We used a noxious thermal challenge in the setting of PGE2-induced inflammation, which is mediated exclusively by TRPV1 and Nav1.8 in small DRG neurons. The different results in two papers may, thus, reflect probing of different subtype of DRG neurons.
Nociception involves detection of noxious chemical or thermal stimuli by specific receptors and generation of APs. The impaired pain phenotype in AQP1−/− mice might be related to the impaired function of sensory receptors, such as TRPV1, or to ion channels involved in the generation and transduction of APs along the fibers. In a preliminary study, in comparing the function of TRPV1 in small DRG neurons of AQP1+/+ and AQP1−/− mice, we found no significant difference in the percentage of responding neurons or the averaged evoked current density (data not shown). Furthermore, we tested several different stimuli, including hot and cold, which activate different receptors (46), providing evidence the impaired pain perception in the AQP1−/− mice is not receptor-specific.
We, therefore, focused on ion channels involved in the generation of APs. Nociceptive fibers respond with AP firing over several seconds (26, 47). Less AP firing was found in the AQP1−/− neurons in response to the same level of electrical stimulation. The Nav1.8 channel produces the majority of inward current during an AP and is critical for generation of multiple APs during high frequency repetitive simulation (39, 40). Nav1.8−/− DRG neurons showed intermittent APs and failed to sustain high frequency firing (40). The impairment in repetitive AP firing in AQP1-deficient DRG neurons and their accelerated adaptation provides a link between AQP1 expression, Nav1.8 function, and pain nociception. Electrophysiological analysis of ND7-23 cells indicated accelerated slow inactivation in the absence AQP1, which occurred after prolonged depolarization in the seconds-to-minutes range. Slow inactivation has been reported in various mammalian neuronal cells including rat hippocampal neurons (48), tetrodotoxin-resistant DRG neurons (23), and cultured neuroblastoma cells (49) and is important to firing adaptation. Several molecular partners and membrane proteins have been reported to modulate Na+ channel slow inactivation, including β1–4 subunits, ankyrin G (50), and calmodulin (25). Our data suggest selective AQP1 modulation of Nav1.8 but not Nav1.7.
Na+ channels Nav1.1 to Nav1.9 are composed of a large α subunit containing the pore- and voltage-sensing machinery and several auxiliary β subunits (50). Only a few other proteins have been reported to interact directly with Nav1.8, including annexin II light chain (51), calmodulin (25), and clathrin-associated protein-1A (42). The immunoprecipitation and single molecule tracking indicated a physical interaction between AQP1 and Nav1.8. Impairment in cold pain sensing in the AQP1−/− mice provides further evidence for involvement of AQP1 in the Nav1.8 cold pain signaling pathway (27, 33).
In conclusion, we have discovered a significant pain phenotype in AQP1−/− mice that at the cell level appears to involve impairment in the firing of action potentials in small DRG neurons in AQP1 deficiency and at the molecular level appears to involve accelerated Nav1.8 Na+ channel inactivation. One clinical consequence of our results is the potential utility of AQP1 inhibitors to reduce pain nociception, which may provide a novel strategy to achieve analgesia at the presynaptic spinal level.
Acknowledgments
We thank Drs. J. Crane and P. Haggie for technical assistance and discussions on quantum dot single particle tracking and L. Qian for mouse breeding and genotype analysis. The University of California, San Francisco Neurobehavioral Core for Rehabilitation Research provided instrumentation and expertise for behavioral studies.
This work was supported, in whole or in part, by National Institutes of Health Grants DK35124, EY13574, EB00415, DK86125, DK72517, and HL73856. This work was also supported by a grant from the Guthy-Jackson Charitable Foundation.
- AQP
- aquaporin
- AP
- action potential
- CGRP
- calcitonin gene-related peptide
- DRG
- dorsal root ganglion
- IB4
- isolectin B4
- PGE2
- prostaglandin E2
- TTX
- tetrodotoxin
- TTX-S
- TTX-sensitive
- TTX-R
- TTX-resistant.
REFERENCES
- 1.Papadopoulos M. C., Manley G. T., Krishna S., Verkman A. S. (2004) FASEB J. 18, 1291–1293 [DOI] [PubMed] [Google Scholar]
- 2.Manley G. T., Fujimura M., Ma T., Noshita N., Filiz F., Bollen A. W., Chan P., Verkman A. S. (2000) Nat. Med. 6, 159–163 [DOI] [PubMed] [Google Scholar]
- 3.Auguste K. I., Jin S., Uchida K., Yan D., Manley G. T., Papadopoulos M. C., Verkman A. S. (2007) FASEB J. 21, 108–116 [DOI] [PubMed] [Google Scholar]
- 4.Saadoun S., Papadopoulos M. C., Watanabe H., Yan D., Manley G. T., Verkman A. S. (2005) J. Cell Sci. 118, 5691–5698 [DOI] [PubMed] [Google Scholar]
- 5.Binder D. K., Yao X., Zador Z., Sick T. J., Verkman A. S., Manley G. T. (2006) Glia 53, 631–636 [DOI] [PubMed] [Google Scholar]
- 6.Padmawar P., Yao X., Bloch O., Manley G. T., Verkman A. S. (2005) Nat. Methods 2, 825–827 [DOI] [PubMed] [Google Scholar]
- 7.Li J., Patil R. V., Verkman A. S. (2002) Invest. Ophthalmol. Vis. Sci. 43, 573–579 [PubMed] [Google Scholar]
- 8.Li J., Verkman A. S. (2001) J. Biol. Chem. 276, 31233–31237 [DOI] [PubMed] [Google Scholar]
- 9.Lu D. C., Zhang H., Zador Z., Verkman A. S. (2008) FASEB J. 22, 3216–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagelhus E. A., Mathiisen T. M., Ottersen O. P. (2004) Neuroscience 129, 905–913 [DOI] [PubMed] [Google Scholar]
- 11.Zhang H., Verkman A. S. (2008) Mol. Cell. Neurosci. 37, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zador Z., Magzoub M., Jin S., Manley G. T., Papadopoulos M. C., Verkman A. S. (2008) FASEB J. 22, 870–879 [DOI] [PubMed] [Google Scholar]
- 13.Binder D. K., Papadopoulos M. C., Haggie P. M., Verkman A. S. (2004) J. Neurosci. 24, 8049–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshio K., Watanabe H., Song Y., Verkman A. S., Manley G. T. (2005) FASEB J. 19, 76–78 [DOI] [PubMed] [Google Scholar]
- 15.Shields S. D., Mazario J., Skinner K., Basbaum A. I. (2007) Pain 131, 8–20 [DOI] [PubMed] [Google Scholar]
- 16.Oshio K., Watanabe H., Yan D., Verkman A. S., Manley G. T. (2006) Biochem. Biophys. Res. Commun. 341, 1022–1028 [DOI] [PubMed] [Google Scholar]
- 17.Solenov E. I., Vetrivel L., Oshio K., Manley G. T., Verkman A. S. (2002) J. Neurosci. Methods 113, 85–90 [DOI] [PubMed] [Google Scholar]
- 18.Julius D., Basbaum A. I. (2001) Nature 413, 203–210 [DOI] [PubMed] [Google Scholar]
- 19.Ma T., Yang B., Gillespie A., Carlson E. J., Epstein C. J., Verkman A. S. (1998) J. Biol. Chem. 273, 4296–4299 [DOI] [PubMed] [Google Scholar]
- 20.Wood J. N., Bevan S. J., Coote P. R., Dunn P. M., Harmar A., Hogan P., Latchman D. S., Morrison C., Rougon G., Theveniau M. (1990) Proc. Biol. Sci. 241, 187–194 [DOI] [PubMed] [Google Scholar]
- 21.Stucky C. L., Lewin G. R. (1999) J. Neurosci. 19, 6497–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bean B. P. (2007) Nat. Rev. Neurosci. 8, 451–465 [DOI] [PubMed] [Google Scholar]
- 23.Choi J. S., Dib-Hajj S. D., Waxman S. G. (2007) J. Neurophysiol. 97, 1258–1265 [DOI] [PubMed] [Google Scholar]
- 24.Kostyuk P. G., Krishtal O. A., Pidoplichko V. I. (1977) Nature 267, 70–72 [DOI] [PubMed] [Google Scholar]
- 25.Choi J. S., Hudmon A., Waxman S. G., Dib-Hajj S. D. (2006) J. Neurophysiol. 96, 97–108 [DOI] [PubMed] [Google Scholar]
- 26.Blair N. T., Bean B. P. (2003) J. Neurosci. 23, 10338–10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann K., Leffler A., Babes A., Cendan C. M., Carr R. W., Kobayashi J., Nau C., Wood J. N., Reeh P. W. (2007) Nature 447, 855–858 [DOI] [PubMed] [Google Scholar]
- 28.Solenov E., Watanabe H., Manley G. T., Verkman A. S. (2004) Am. J. Physiol. Cell Physiol. 286, C426–C432 [DOI] [PubMed] [Google Scholar]
- 29.Crane J. M., Verkman A. S. (2008) Biophys. J. 94, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larivière R. C., Nguyen M. D., Ribeiro-da-Silva A., Julien J. P. (2002) J. Neurochem. 81, 525–532 [DOI] [PubMed] [Google Scholar]
- 31.Dray A., Perkins M. (1993) Trends Neurosci. 16, 99–104 [DOI] [PubMed] [Google Scholar]
- 32.Narumiya S., Sugimoto Y., Ushikubi F. (1999) Physiol. Rev. 79, 1193–1226 [DOI] [PubMed] [Google Scholar]
- 33.Abrahamsen B., Zhao J., Asante C. O., Cendan C. M., Marsh S., Martinez-Barbera J. P., Nassar M. A., Dickenson A. H., Wood J. N. (2008) Science 321, 702–705 [DOI] [PubMed] [Google Scholar]
- 34.Amaya F., Wang H., Costigan M., Allchorne A. J., Hatcher J. P., Egerton J., Stean T., Morisset V., Grose D., Gunthorpe M. J., Chessell I. P., Tate S., Green P. J., Woolf C. J. (2006) J. Neurosci. 26, 12852–12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Ratnam J., Zou B., England P. M., Basbaum A. I. (2009) J. Neurosci. 29, 5508–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang X., Djouhri L., McMullan S., Berry C., Waxman S. G., Okuse K., Lawson S. N. (2006) J. Neurosci. 26, 7281–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nassar M. A., Stirling L. C., Forlani G., Baker M. D., Matthews E. A., Dickenson A. H., Wood J. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12706–12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akopian A. N., Souslova V., England S., Okuse K., Ogata N., Ure J., Smith A., Kerr B. J., McMahon S. B., Boyce S., Hill R., Stanfa L. C., Dickenson A. H., Wood J. N. (1999) Nat. Neurosci. 2, 541–548 [DOI] [PubMed] [Google Scholar]
- 39.Blair N. T., Bean B. P. (2002) J. Neurosci. 22, 10277–10290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renganathan M., Cummins T. R., Waxman S. G. (2001) J. Neurophysiol. 86, 629–640 [DOI] [PubMed] [Google Scholar]
- 41.John V. H., Main M. J., Powell A. J., Gladwell Z. M., Hick C., Sidhu H. S., Clare J. J., Tate S., Trezise D. J. (2004) Neuropharmacology 46, 425–438 [DOI] [PubMed] [Google Scholar]
- 42.Liu C., Cummins T. R., Tyrrell L., Black J. A., Waxman S. G., Dib-Hajj S. D. (2005) Mol. Cell. Neurosci. 28, 636–649 [DOI] [PubMed] [Google Scholar]
- 43.Woolf C. J., Ma Q. (2007) Neuron 55, 353–364 [DOI] [PubMed] [Google Scholar]
- 44.Alessandri-Haber N., Yeh J. J., Boyd A. E., Parada C. A., Chen X., Reichling D. B., Levine J. D. (2003) Neuron 39, 497–511 [DOI] [PubMed] [Google Scholar]
- 45.Grant A. D., Cottrell G. S., Amadesi S., Trevisani M., Nicoletti P., Materazzi S., Altier C., Cenac N., Zamponi G. W., Bautista-Cruz F., Lopez C. B., Joseph E. K., Levine J. D., Liedtke W., Vanner S., Vergnolle N., Geppetti P., Bunnett N. W. (2007) J. Physiol. 578, 715–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foulkes T., Wood J. N. (2007) Channels 1, 154–160 [DOI] [PubMed] [Google Scholar]
- 47.Rush A. M., Bräu M. E., Elliott A. A., Elliott J. R. (1998) J. Physiol. 511, 771–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mickus T., Jung H., Spruston N. (1999) Biophys. J. 76, 846–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quandt F. N. (1988) Mol. Pharmacol. 34, 557–565 [PubMed] [Google Scholar]
- 50.Chahine M., Ziane R., Vijayaragavan K., Okamura Y. (2005) Trends Pharmacol. Sci. 26, 496–502 [DOI] [PubMed] [Google Scholar]
- 51.Okuse K., Malik-Hall M., Baker M. D., Poon W. Y., Kong H., Chao M. V., Wood J. N. (2002) Nature 417, 653–656 [DOI] [PubMed] [Google Scholar]