Abstract
Several residues are conserved in the transmembrane domains (TMs) of G-protein coupled receptors. Here we demonstrate that a conserved proline, Pro247, in TM6 of platelet-activating factor receptor (PAFR) is required for endoplasmic reticulum (ER) export and trafficking after agonist-induced internalization. Alanine-substituted mutants of the conserved residues of PAFRs, including P247A, were retained in the ER. Because a PAFR antagonist, Y-24180, acted as a pharmacological chaperone to rescue ER retention, this retention is due to misfolding of PAFR. Methylcarbamyl (mc)-PAF, a PAFR agonist, did not increase the cell surface expression of P247A, even though another ER-retained mutant, D63A, was effectively trafficked. Signaling and accumulation of the receptors in the early endosomes were observed in the mc-PAF-treated P247A-expressing cells, suggesting that P247A was trafficked to the cell surface by mc-PAF, and thereafter disappeared from the surface due to aberrant trafficking, e.g. enhanced internalization, deficiency in recycling, and/or accelerated degradation. The aberrant trafficking was confirmed with a sortase-A-mediated method for labeling cell surface proteins. These results demonstrate that the conserved proline in TM6 is crucial for intracellular trafficking of PAFR.
Keywords: Cell/Trafficking, Chaperones, G Proteins/Coupled Receptors (GPCR), Membrane/Proteins, Methods/Fluorescence, Protein/Cell Surface, Protein/Export, Receptors/Recycling
Introduction
Many G-protein coupled receptors (GPCRs)2 classified in the rhodopsin-type family have several common residues located in their seven transmembrane (TM) helices (1, 2). Studies of membrane preparations from cells expressing various GPCRs, including the platelet-activating factor (PAF; 1-O-alkyl-2-acetyl-sn-glycero-3-phosphocholine) receptor (PAFR) have demonstrated the significance of these conserved residues for ligand recognition and/or GTPγS binding (3–6). The three-dimensional structural analysis of some GPCRs (7) suggests that changes in these residues are likely to result in misfolding of the protein during its biosynthesis in the endoplasmic reticulum (ER). However, little is known about the roles of these residues for passage through the ER quality control. Recent information about pharmacological chaperones, which are able to rescue the intracellular retention of several misfolded proteins by stabilizing their conformation and/or enhancing refolding (8–10), raises the possibility that the addition of specific ligands to structurally deficient GPCRs retained in the ER might result in their export to the cell surface.
Although the binding properties of ligands to mutant GPCRs have been examined using membrane preparations (4, 11–15), little is known about intracellular trafficking (for example, the internalization and recycling) of these mutants in living cells, mainly because of their ER retention. In this report, we identified several residues in PAFR that could be crucial for correct folding during its biosynthesis in the ER by mutating the residues and determining which caused a deficiency in PAFR expression at the cell surface. We then used a PAFR antagonist or agonist as a pharmacological chaperone to analyze the structurally defective mutants after they were trafficked to the cell surface. Moreover, we used a new technique, site-specific N-terminal labeling of cell surface proteins on living cells by sortase-A (16, 17), to evaluate the trafficking of mutant PAFR after stimulation with agonist. We found that a conserved proline, Pro247, in TM6 of PAFR is important not only for ER export but also for trafficking, e.g. internalization, recycling, and/or sorting to lysosomes.
EXPERIMENTAL PROCEDURES
Materials
Methylcarbamyl (mc)-PAF C-16 was purchased from Cayman Chemical (Ann Arbor, MI). Chloroquine diphosphate salt was from Sigma. Y-24180 was donated from Yoshitomi Pharmaceutical Industries, Ltd. (Osaka, Japan).
Construction of Mutant GPCRs
N terminally HA-tagged human PAFR (HA-hPAFR), human leukotriene B4 type 2 receptor (HA-hBLT2), or human GPR43 (HA-hGPR43) were used as templates to generate mutant receptors using the QuikChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA) following the manufacturer's instructions. The mutant receptors were inserted into pcDNA3.1 or pCXN2.1. The primer sets utilized are listed under supplemental materials.
Cell Culture and Transfection
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma) supplemented with 10% fetal bovine serum. Chinese hamster ovary-K1 cells were cultured in Ham's F-12 (Sigma) supplemented with 10% fetal bovine serum. PC12h cells were cultured in DMEM supplemented with 10% horse serum and 5% fetal bovine serum. These cells were transfected with a plasmid harboring a wild-type (WT) or mutated receptor using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Stable cell lines with inducible expression of WT or mutant PAFRs were established by transfecting the pTRE plasmid bearing the appropriate PAFRs into a stable HeLa cell line harboring the Tet repressor (HeLa Tet-On Cell Line; Clontech, Palo Alto, CA (18)) using Lipofectamine 2000. Cells were grown under Geneticin (1 mg/ml; Invitrogen) and hygromycin (100 μg/ml; Wako, Osaka, Japan) selection, isolated, expanded, and then tested for doxycycline (Dox; 1 μg/ml; Clontech)-inducible expression of PAFR by Western blotting. The clones used for experiments showed very low basal, but highly inducible, receptor expression. For our experiments, after cells were plated and cultured for 16 h, receptor expression was induced by adding 1 μg/ml of Dox to the culture medium for 24 h.
Flow Cytometry
For staining, cells were incubated with anti-HA antibody (clone 3F10; Roche Applied Science) in phosphate-buffered saline (PBS) containing 2% goat serum at room temperature for 30 min, followed by staining with phycoerythrin-conjugated anti-rat IgG (Beckman Coulter, Fullerton, CA) at room temperature for 30 min. An EPICS XL (Beckman Coulter) was used for flow cytometry.
Western Blotting
Two days after transfection, cells were harvested with PBS containing 2 mm EDTA. Cells were disrupted in ice-cold sonication buffer (25 mm HEPES-NaOH, pH 7.4, 0.25 m sucrose, 10 mm MgCl2) plus protease inhibitor mixture (Roche, one tablet in 50 ml) by sonication. The cell debris was removed by centrifugation at 8,000 × g for 10 min at 4 °C, and the resultant supernatants were used as protein samples. The protein concentration was determined by the Bradford method (19) using a Protein Assay Kit (Bio-Rad) with bovine serum albumin (BSA, Sigma) as a standard. For Western blot analyses, protein samples were separated on SDS-10% polyacrylamide gels and transferred to a nitrocellulose membrane. After a blocking step using 5% skim milk in TBS-T (20 mm Tris-buffered saline (pH 7.4), 0.1% Tween 20), blots were probed with the primary antibody for 1 h. The membrane was washed with TBS-T and incubated with a biotin-conjugated antibody (Vector Laboratories, Burlingame, CA) or horseradish peroxidase-conjugated anti-rat IgG antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h. In the case of the biotin-conjugated antibody, the membrane was then incubated with horseradish peroxidase-conjugated streptavidin (GE Healthcare) for 0.5 h. The signal was visualized using an ECL Western blotting detection system (GE Healthcare).
Endoglycosidase Treatment of PAFRs
Protein samples were obtained from the 8,000 × g supernatant described above. They were treated with endoglycosidase-H (Endo-H; 0.005 units; Roche Applied Science) in 50 μl of buffer (11.7 mm Na2HPO4, 168.3 mm NaH2PO4, 0.4% SDS, 20 mm EDTA, 2% 2-mercaptethanol) or with N-glycosidase F (PNGase-F; 1 unit; Roche Applied Science) in 50 μl of buffer (139.2 mm Na2HPO4, 40.8 mm NaH2PO4, 0.4% SDS, 20 mm EDTA, 2% 2-mercaptoethanol) for 16 h at 4 °C. The resultant protein samples were suspended in sampling buffer (25 mm Tris-HCl (pH 6.5), 5% glycerol, 1% SDS, and 0.05% bromphenol blue) and subjected to Western blot analyses.
Immunoprecipitation
All operations were carried out at 4 °C. Cells were lysed with lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Nonidet P-40 (Nacalai Tesque, Kyoto, Japan), 0.5% sodium deoxycholate, protease inhibitor mixture (one tablet in 25 ml)). Total cellular lysates were subjected to immunoreaction with the anti-HA antibody (3F10) and Protein A/G Plus-agarose (Santa Cruz Biotechnology) for 16 h. After centrifugation at 10,000 × g for 10 min, the pellets were washed with wash buffer 1 (50 mm Tris-HCl (pH 7.5), 500 mm NaCl, 0.1% Nonidet P-40, 0.05% sodium deoxycholate) and then with wash buffer 2 (50 mm Tris-HCl (pH 7.5), 0.1% Nonidet P-40, 0.05% sodium deoxycholate). The resulting pellets were suspended in sampling buffer and subjected to Western blot analyses using anti-ubiquitin antibody (P4D1) and horseradish peroxidase-conjugated anti-mouse IgG whole antibody (Santa Cruz Biotechnology).
Immunofluorescence Confocal Microscopy Using Anti-calreticulin and Anti-golgin-97 Antibodies
Cells (5 × 105) were seeded into collagen-coated glass-bottomed 35-mm dishes (MatTek Corporation, Ashland, MA). After transfection and incubation for 48 h at 37 °C, the cells were fixed with 2% paraformaldehyde for 10 min at room temperature and rinsed twice with ice-cold PBS. Subsequently, the cells were incubated with ¼× permeabilization reagent (Beckman Coulter) for 10 min at room temperature. Then, primary antibodies (anti-HA (1:100, 3F10) and anti-calreticulin (1:200, Stressgen Bioreagents, Ann Arbor, MI) or anti-golgin-97 (1:50; Invitrogen) were added, and the mixture was incubated for 1 h. After the cells were washed with ice-cold PBS, secondary antibodies (Alexa Fluor 488-conjugated anti-rat IgG (1:200; Invitrogen) and Alexa Fluor 546-conjugated anti-rabbit IgG (1:200; Invitrogen) or Alexa Fluor 546-conjugated anti-mouse IgG (1:200; Invitrogen)) were added, and incubation was continued for 1 h. Images were obtained using an LSM510 Laser Scanning Confocal Microscope (Carl Zeiss, Jena, Germany) equipped with argon and helium/neon lasers and ×100 oil immersion objective lens.
Immunofluorescence Confocal Microscopy Using Transferrin Conjugate
Twenty-four hours after transfection, HeLa cells were cultured in DMEM containing 0.1% BSA with or without 1 μm mc-PAF C-16 for 24 h. Thereafter, the cells were incubated with Alexa Fluor 546-conjugated transferrin (50 μg/ml; Invitrogen (20) for 1 h. After these cells were fixed and permeabilized, they were incubated with primary (anti-HA) and secondary (Alexa Fluor 488-conjugated anti-rat IgG) antibodies. To detect the plasma membranes, cells were stained with CellMaskTM Orange Plasma Membrane Marker (2.5 μg/ml, Invitrogen) (21) for 5 min, fixed, and incubated with the above antibodies under non-permeabilized conditions. Images were obtained as described above.
Calcium Mobilization Assay
Transiently transfected HeLa cells were plated in a 96-well plate (4 × 104 cells/well) and incubated for 16 h. Then, Y-24180 (1 μm) was added to the culture medium, and the cells were incubated for a further 24 h at 37 °C. The cells were then incubated with loading buffer (buffer A (1× HBSS and 2.5 mm probenecid; Sigma, 20 mm HEPES, 1 mm CaCl2, 1 mm MgCl2, and 0.01% BSA)) containing 4 μm Fluo 3-AM (Dojindo, Kumamoto, Japan) and 0.04% pluronic acid (Molecular Probes, Eugene, OR) at 37 °C for 1 h. Then they were washed twice with buffer A. Intracellular Ca2+ mobilization was monitored with a scanning fluorometer (FlexStation, Molecular Devices Corp.) by measuring emission fluorescence at 525 nm in response to excitation at 485 nm. Relative fluorescence units (maximum − minimum) are indicated.
Reporter Gene Assay
1.5 × 105 PC12h cells were transfected with 450 ng of HA-hPAFR or empty vector (pcDNA3.1), 480 ng of zif 268-firefly luciferase-pGL2 (a kind gift from Japan Tobacco Co., Tokyo, Japan), 20 ng of cytomegalovirus promoter-driven Renilla luciferase-pRL (Promega, Madison, WI), and 50 ng of expression vector for C3 exoenzyme (pEF-C3; a kind gift from Dr. S. Narumiya (Kyoto University)) or empty vector (pEF) using SuperFect (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Transfected cells were cultured on a collagen-coated 24-well plate for 36 h. Cells were then stimulated for 12 h with DMEM, 0.1% BSA containing 1 μm mc-PAF or vehicle only under 5% CO2 at 37 °C. Thereafter, firefly and Renilla luciferase activities were measured using PICAGENE Dual Seapansy (Toyo Ink, Tokyo, Japan) and a MiniLumat LB 9506 luminometer (Berthold, Bundoora, Australia). The relative luciferase activity is represented as the firefly luciferase value/Renilla luciferase (F/R) value (22).
Site-specific N-terminal Labeling of Cell Surface Receptors
A sortase-A recognition sequence (MLPETGGGGG) was generated at the N termini of the HA-tagged human PAFRs by PCR, and the resulting constructs were inserted into pCXN2.1. The primer sets utilized are listed under supplemental materials. Preparation of His6-sortase-A and Alexa Fluor 488-labeled peptide and labeling of cell surface receptors by sortase-A were performed as described (16). Briefly, the transpeptidation reaction was performed by incubation of the transfected HeLa cells in DMEM (0.1% BSA) containing 30 μm His6-sortase-A and 1 mm triglycine at 37 °C for 30 min. After the cells were washed with PBS, they were further incubated in fresh DMEM (0.1% BSA) containing 30 μm His6-sortase-A and 10 μm Alexa Fluor 488-labeled LPETGG peptide at 37 °C for 15 min. Thereafter, the cells were washed with PBS and observed with a confocal laser microscope.
Statistical Analysis
Data were analyzed for statistical significance using Prism 4 software (GraphPad Software, Inc., San Diego, CA). Statistical significance was analyzed using analysis of variance with Dunnett post hoc pairwise comparisons or unpaired t test. Differences were considered significant at p < 0.05, 0.01, and 0.001 as indicated.
RESULTS
Requirement of Conserved Residues for Cell Surface Expression of PAFR
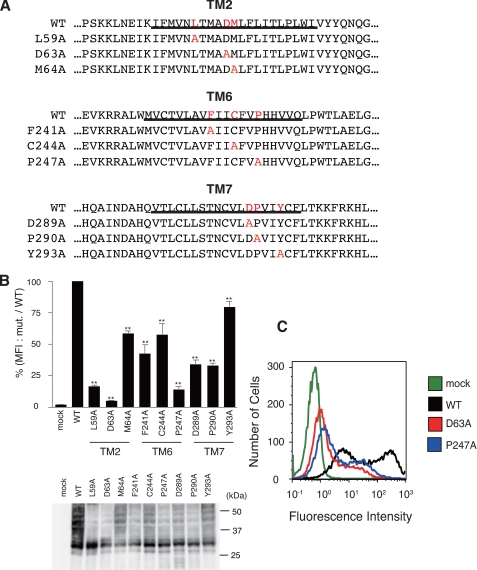
In the transmembrane domains of the rhodopsin-type GPCRs, several amino acid residues are highly conserved, e.g. aspartic acid and leucines in TM2; phenylalanine, cysteine, and proline in TM6; and asparagine/aspartic acid, proline, and tyrosine in TM7 (Fig. S1). To determine the importance of these residues for trafficking of GPCRs, we substituted alanine for these conserved residues in HA-tagged human PAFR (HA-hPAFR; Fig. 1A) and analyzed cell surface expression of these mutants semiquantitatively by flow cytometry in transiently transfected HeLa cells. Many of these mutant HA-hPAFRs, e.g. L59A and D63A in TM2 and P247A in TM6, showed a marked decrease in cell surface expression (Fig. 1, B and C). In particular, mutations of the aspartic acid in TM2 (Asp63) and the proline in TM6 (Pro247) resulted in drastic impairment of cell surface expression of PAFR; these residues are proposed to be structurally important in folding of the receptor based on structural analysis of GPCRs (7, 14, 15). These results suggest that these residues could play a pivotal role in cell surface expression of the rhodopsin-type GPCRs.
FIGURE 1.
Requirement of conserved residues in rhodopsin-type GPCRs for cell surface expression of HA-hPAFR. A, the 9 mutant HA-hPAFRs generated in this study are indicated. Conserved residues and substituted alanines in each mutant PAFR are indicated in red. Transmembrane domains are underlined. B, after transfection of WT and mutant HA-hPAFRs into HeLa cells, cell surface expression levels of each mutant HA-hPAFR were determined with flow cytometry. After the cells were stained with an anti-HA primary antibody and phycoerythrin-conjugated anti-rat IgG secondary antibody, the fluorescence intensity of each transfectant was measured. The mean fluorescence intensities (MFI), represented as a percentage of the MFI of WT HA-hPAFR, were used to evaluate the expression levels. Data are represented as mean ± S.E. (n = 3). Statistical significance was analyzed using analysis of variance with Dunnett post hoc pairwise comparisons. **, p < 0.01 (versus WT). Bottom, the protein levels of the tested receptors in HeLa cells were shown by Western blotting using anti-HA antibody. Approximate molecular sizes are examined in kDa at the right. C, a representative result from the flow cytometric analysis is shown. HeLa cells were transiently transfected with WT HA-hPAFR, D63A, P247A, or empty vector.
Accumulation of Mutant HA-hPAFRs in the ER
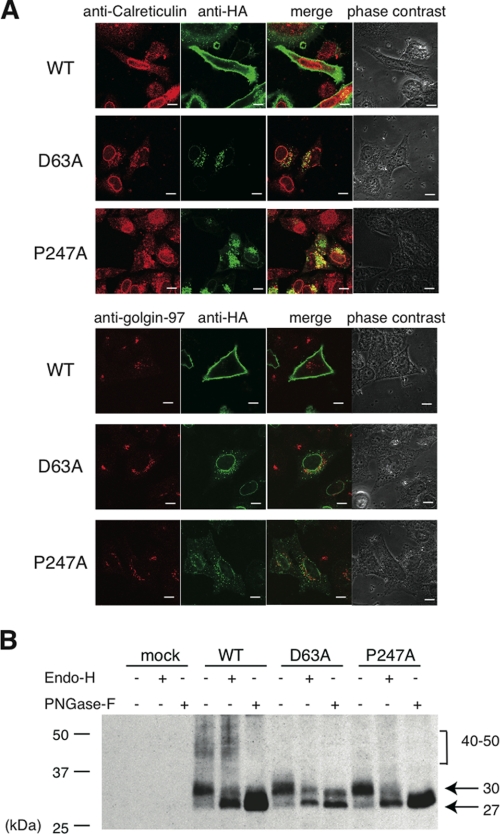
To investigate what caused the decrease in cell surface expression of the mutant HA-hPAFRs, we used confocal immunolocalization analyses to observe the subcellular localization of these receptors in HeLa cells. WT HA-hPAFR was expressed mainly on the plasma membrane 48 h after transfection of HeLa cells (Fig. 2A). In contrast, the D63A and P247A mutants were localized in the perinuclear regions of the cells, resulting in deficient cell surface expression. In merged images, mutant HA-hPAFRs overlapped with the ER marker calreticulin, but not with golgin-97, a Golgi apparatus marker. Accumulation of D63A and P247A mutants in the ER was further confirmed by examining glycosylation of the receptors, which are linked and further modified during trafficking to the cell surface (23). Western blot analyses of WT HA-hPAFR showed several versions of the protein with different molecular masses: a broad species (approximately 40–50 kDa), and 30 and 27 kDa (Fig. 2B). Because the broad species disappeared following treatment with PNGase-F, which cleaves all types of N-glycans, but not with Endo-H, which specifically digests high-mannose glycans, these proteins are likely to be fully glycosylated. The 30-kDa band was shifted to 27 kDa by treatment with PNGase-F and Endo-H, suggesting that the 30- and 27-kDa bands correspond to a core-conjugated glycochain and a nonglycosylated form of the protein, respectively, probably localized in the ER. As was previously observed (24), the detected proteins ran faster than their predicted molecular masses (∼39 kDa for nonglycosylated hPAFR). In contrast, the broad species was not detected in the D63A- or P247A-expressing cells, although similar shifts of the core chain band from the conjugated to the nonglycosylated form were observed after treatment with PNGase-F and Endo-H. It is likely that the 30-kDa band, especially in the D63A mutant, contains modification(s) resistant to glycosidases, because the band did not completely disappear upon treatment with these enzymes. Moreover, we confirmed similar accumulation of D63A and P247A in the ER in Chinese hamster ovary-K1 cells, suggesting that these deficiencies in ER export were observed in various cell types (supplemental Fig. S2). Taken together, these results imply that D63A and P247A mutants accumulate in the ER in HeLa cells.
FIGURE 2.
Accumulation of mutant HA-hPAFRs in the ER. A, subcellular localizations of WT and mutant HA-hPAFRs were analyzed by immunofluorescence confocal microscopy. HeLa cells were transfected with WT HA-hPAFR, D63A, or P247A and subjected to immunocytochemical analysis 48 h after transfection. Calreticulin, golgin-97, and HA-tagged hPAFRs were visualized using anti-calreticulin (upper, red), anti-golgin-97 (lower, red), and anti-HA (green) antibodies, respectively. White bars indicate 10 μm. The data are representative of three independent experiments with identical results. B, protein preparations from cells transfected with WT HA-hPAFR, D63A, P247A, or empty vector were treated with Endo-H (0.005 units) or PNGase-F (1 unit). The results were analyzed by Western blotting using an anti-HA antibody. Approximate molecular sizes are shown in kDa at the right. The data are representative of three independent experiments with identical results.
Deficiency in Cell Surface Expression of Other GPCRs by Mutating Crucial Residues
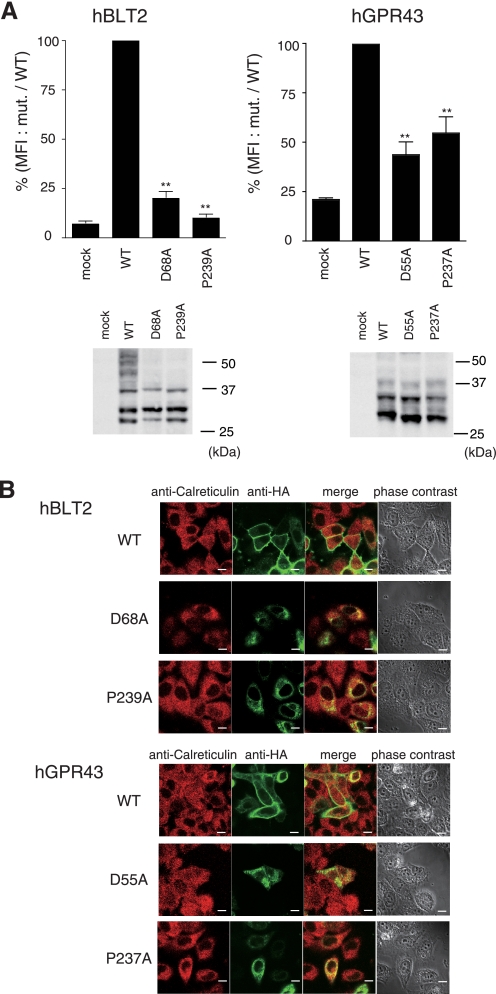
To gain further evidence for the importance of the conserved residues in TM2 and TM6 for GPCR trafficking, we constructed other mutant GPCRs using HA-tagged human leukotriene B4 type-2 receptor (HA-hBLT2) and human GPR43 (HA-hGPR43), which introduced mutations into positions corresponding to Asp63 and Pro247 in HA-hPAFR. Similar to the results obtained with HA-hPAFR, these mutants were impaired in cell surface expression and predominantly accumulated in the ER (Fig. 3, A and B), suggesting that these residues could play a pivotal role in cell surface expression of these GPCRs.
FIGURE 3.
Deficiency of cell surface expression by mutation of the pivotal residues in HA-hBLT2 and HA-hGPR43. A, WT and two mutant receptors, with point mutations in the aspartic acid in TM2 or the proline in TM6, were prepared using HA-hBLT2 (left) and HA-hGPR43 (right). After transfection of these receptors into HeLa cells, cell surface expression levels of each GPCR were determined by flow cytometric analysis. Cells were stained with anti-HA and phycoerythrin-conjugated anti-rat IgG as primary and secondary antibodies, respectively, and the fluorescence intensity of each transfectant was measured. The expression levels were evaluated by mean fluorescence intensities (MFI), represented as a percentage of the WT receptor. Data are represented as mean ± S.E. (n = 3). Statistical significance was analyzed using analysis of variance with Dunnett post hoc pairwise comparisons, **, p < 0.01 (versus WT). Bottom, the protein levels of the tested receptors in HeLa cells were examined by Western blotting using anti-HA antibody. Approximate molecular sizes are shown in kDa at the right of each panel. B, subcellular localizations of WT and mutant HA-hBLT2 and hGPR43 were analyzed by immunofluorescence confocal microscopy. HeLa cells were transfected with WT-hBLT2, D68A-hBLT2, P239A-hBLT2, WT-hGPR43, D55A-hGPR43, or P237A-hGPR43 and subjected to immunocytochemical analysis 48 h after transfection. Calreticulin and HA-tagged receptors were visualized using anti-calreticulin (red) and anti-HA (green) antibodies, respectively. White bars indicate 10 μm. The data are representative of three independent experiments with identical results.
Action of Specific Ligands as Pharmacological Chaperones
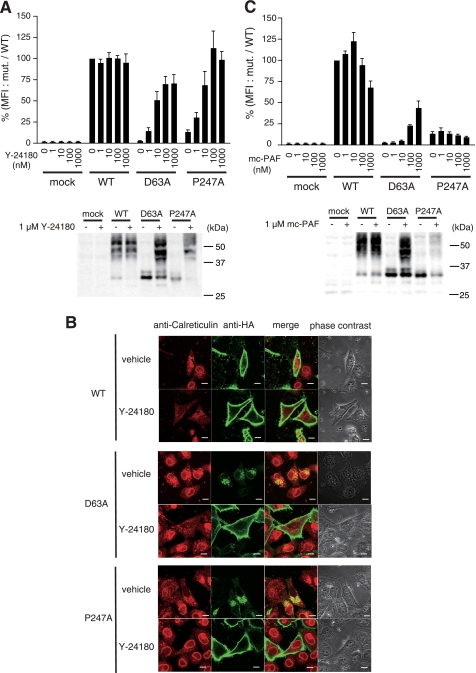
Recently, a class of compounds called pharmacological chaperones was shown to rescue the intracellular retention of several misfolded proteins by stabilizing their conformation and/or enhancing refolding (25). To test whether specific ligands for PAFR act as pharmacological chaperones, we used flow cytometry to examine the effects of the PAFR antagonist Y-24180 (26) on the export of mutant PAFR from the ER. Treatment with 1 μm Y-24180 facilitated cell surface expression of all mutant PAFRs previously retained in the ER (Figs. 1B and supplemental S3A). In particular, Y-24180 markedly increased the expression of D63A and P247A on the cell surface in a dose-dependent fashion (Fig. 4A). We further observed a similar effect of Y-24180 in PC12h cells. Namely, the deficiencies in surface expression of D63A and P247A in these cells were rescued by treatment with Y-24180 (supplemental Fig. S3B). To examine whether this specific ligand of PAFR promoted ER export of the mutant PAFRs, we used immunofluorescence confocal microscopy to examine subcellular localization of the mutant PAFRs after treatment with Y-24180. In these experiments, mutant receptors that had accumulated in the ER disappeared from the ER following treatment with Y-24180 and were detected mainly on the plasma membrane (Fig. 4B). In general, misfolded proteins that accumulate in the ER are ubiquitinated and then degraded by proteasomes. Therefore, we further examined the amount of ubiquitinated receptors with or without Y-24180 treatment using an anti-ubiquitin antibody. Without ligand treatment, the mutant receptors were highly ubiquitinated compared with WT HA-hPAFR, whereas Y-24180 treatment decreased ubiquitination (supplemental Fig. S4). Unexpectedly, even high doses of mc-PAF (27), an agonist of PAFR, did not enhance expression of P247A on the cell surface (Fig. 4C). Because mc-PAF substantially augmented the surface expression of D63A in a dose-dependent manner, we needed to analyze the P247A mutant behavior in more detail.
FIGURE 4.
Action of specific ligands as pharmacological chaperones. A, dose-dependent effects of Y-24180 on cell surface expression of the mutant HA-hPAFRs. These experiments were carried out using cell lines HeLa-WT, HeLa-D63A, HeLa-P247A, and HeLa-mock cells, which are induced to produce the receptors when Dox is added. Twenty-four hours after addition of Dox, the cells were treated with the indicated concentrations of Y-24180 for 24 h. The expression levels were evaluated by the mean fluorescence intensity (MFI), represented as a percentage of WT HA-hPAFR at 0 nm compound. Data are represented as mean ± S.E. (n = 3). Bottom, the protein levels of each receptor under the conditions of Y-24180 (1 μm) treatment were examined by Western blotting using anti-HA antibody. Approximate molecular sizes are shown in kDa at the right. B, effects of Y-24180 (1 μm) on subcellular localization of WT and mutant HA-hPAFRs were observed with immunofluorescence confocal microscopy. Twenty-four hours after transfection of HeLa cells with WT HA-hPAFR, D63A, or P247A, the cells were treated with vehicle (ethanol; upper) or Y-24180 (lower) for 24 h, then subjected to immunocytochemical analysis. Calreticulin and HA-tagged hPAFRs were visualized using anti-calreticulin (red) and anti-HA (green) antibodies, respectively. White bars indicate 10 μm. The data are representative of three independent experiments with identical results. C, dose-dependent effects of mc-PAF on cell surface expression of the mutant HA-hPAFRs. These experiments were carried out using the same cells and methods described in A. Bottom, the protein levels of each receptor under mc-PAF (1 μm) treatment conditions were examined by Western blotting using anti-HA antibody. Approximate molecular sizes are shown in kDa at the right.
Interaction of P247A with mc-PAF
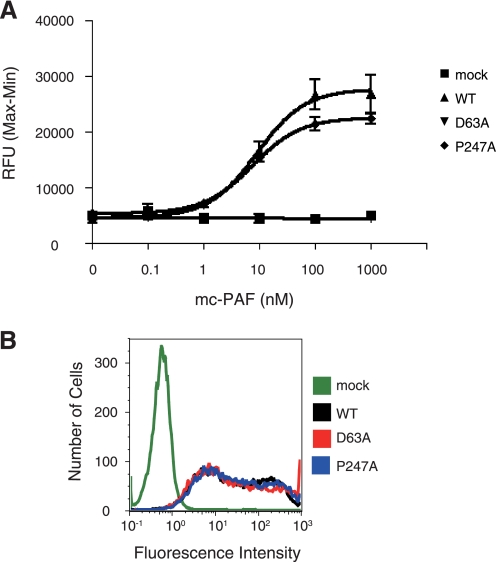
To clarify whether P247A physically interacts with mc-PAF, we examined whether mc-PAF activates the P247A trafficked to the cell surface. The P247A expressing cells were pre-treated with Y-24180 (antagonist) to enhance surface expression of this mutant. Because P247A was not activated by treatment with this compound, the trafficked P247A was retained on the surface. After P247A was sorted to the cell surface by treatment with Y-24180, we washed the cells to remove the ligand and then tested intracellular Ca2+ mobilization by stimulation with mc-PAF. mc-PAF-induced Ca2+ mobilization with similar dose dependence in WT and P247A expressing cells (Fig. 5A). These results suggest that P247A is activated by mc-PAF. However, we did not detect any mc-PAF-elicited Ca2+ mobilization in D63A expressing cells, even after cell surface expression of the receptor (Fig. 5B). These results also demonstrate that Asp63 is crucial for activation of PAFR. Thus, we hypothesized that following treatment with mc-PAF, P247A could have been trafficked to the cell surface, but was internalized immediately thereafter. In contrast, D63A is deficient in agonist-elicited activation of G-proteins (Fig. 5). Hence, mc-PAF surface-trafficked D63A was retained on the surface, resulting in detection by flow cytometric analysis (Fig. 4C).
FIGURE 5.
mc-PAF elicits an increase in intracellular Ca2+ through the mutant HA-PAFRs sorted to the cell surface by Y-24180. A, the mc-PAF-elicited intracellular Ca2+ increase in HeLa cells transfected with WT HA-hPAFR, D63A, P247A, or empty vector with Y-24180 (1 μm) treatment was measured with a FlexStation fluorometer. Relative fluorescence units (RFU, Max − Min) are indicated. Data are represented as mean ± S.E. (n = 3). B, a representative result from the flow cytometric analysis is shown. HeLa cells were transiently transfected with WT HA-hPAFR, D63A, P247A, or empty vector, then treated with Y-24180 (1 μm).
Signaling and Internalization of P247A after Treatment with mc-PAF
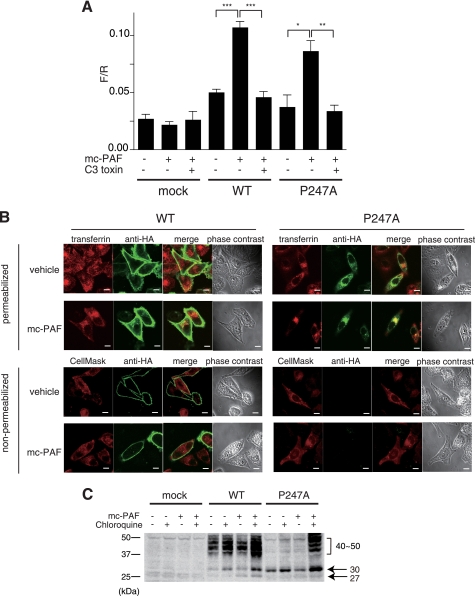
When a reporter gene system (22) was used to evaluate intracellular signaling, mc-PAF treatment increased the F/R ratio, which represents signal transduction, in P247A expressing cells as well as in WT expressing cells (Fig. 6A). Moreover, coexpression with C3 toxin diminished elevation of the F/R ratio, indicating that P247A might be transported to the cell surface by mc-PAF, after which its signals are transduced via Gq and/or G12/13. Furthermore, a confocal immunolocalization analysis revealed that WT HA-hPAFR was present not only in the early endosomes but also on the plasma membrane after treatment with mc-PAF, whereas P247A localized predominantly in the early endosomes (Fig. 6B). Thus, it is possible that P247A trafficked to the surface by mc-PAF is immediately internalized into the early endosomes. We further hypothesized that P247A is not properly trafficked, e.g. enhanced internalization, deficiency in recycling, and/or accelerated degradation, which was supported by a dramatic increase in fully glycosylated P247A in mc-PAF-exposed cells following treatment with chloroquine, a lysosomal inhibitor (28) (Fig. 6C).
FIGURE 6.
Signaling and internalization of P247A after treatment with mc-PAF. A, PC12h cells were transiently transfected with reporter plasmids (zif 268-firefly luciferase and cytomegalovirus-Renilla luciferase as an internal control) and an HA-hPAFR expression plasmid with or without C3 exoenzyme (pEF-C3). The cells were cultured for 36 h after transfection and subsequently stimulated with 1 μm mc-PAF for 12 h. The ratios of firefly luciferase activity to Renilla luciferase are shown. Data are mean ± S.E. (n = 3). The data shown are representative of three independent experiments with similar results. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (unpaired t test) compared with the corresponding mc-PAF-stimulated cells without pEF-C3. B, subcellular localizations of WT HA-hPAFR and P247A with or without exposure to mc-PAF were observed by immunofluorescence confocal microscopy. Twenty-four hours after transfection of HeLa cells with WT HA-hPAFR or P247A, the cells were treated with vehicle (ethanol) or mc-PAF (1 μm) for 24 h, then subjected to immunocytochemical analysis. Top, the early endosomes and HA-tagged hPAFRs were visualized with Alexa Fluor 546-conjugated transferrin (upper, red) and an anti-HA antibody (green), respectively, under permeabilized conditions. Bottom, localization of the receptors on the plasma membrane was confirmed under non-permeabilized conditions by merging with CellMaskTM Orange Plasma Membrane Marker (red). Under these conditions, P247A was not detected with or without mc-PAF stimulation. White bars indicate 10 μm. The data are representative of three independent experiments with identical results. C, Western blot analysis was performed using HeLa-WT, HeLa-P247A, and HeLa-mock cells. After incubation in the presence of Dox, these cells were treated with or without mc-PAF (1 μm) and chloroquine (30 μm) for 24 h, followed by preparation of cell lysates. Molecular standards are indicated at the left of the gel. The data are representative of three independent experiments with identical results.
Aberrant Trafficking of P247A after Stimulation with Agonist
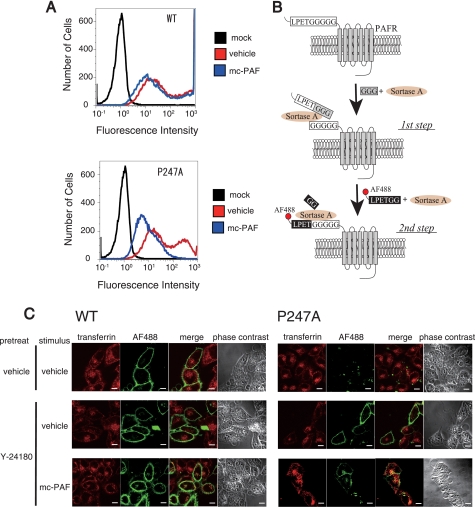
Next, to examine trafficking of P247A, we tested whether the cell surface-trafficked P247A accumulated in the early endosomes after stimulation with mc-PAF. After HeLa-WT and HeLa-P247A were treated with Dox and Y-24180 to facilitate biosynthesis and cell surface trafficking of the receptors, respectively, these cells were stimulated with mc-PAF, and internalization of the receptors was evaluated with flow cytometry. The amount of WT-PAFR on the cell surface was slightly reduced (Fig. 7A), probably due to the continuous internalization and recycling of the receptors from the early endosomes (29), whereas the amount of cell surface-trafficked P247A markedly decreased after stimulation with mc-PAF, suggesting aberrant trafficking of this mutant. A site-specific method for labeling the N termini of cell surface proteins by sortase-A was recently developed (Fig. 7B) (16, 17). By using this technique, we specifically labeled cell surface PAFRs in HeLa cells with Alexa Fluor 488 and compared translocation of the labeled P247A induced by mc-PAF with that of the WT by using confocal microscopy. The Alexa Fluor -488-labeled receptors were not detected in P247A expressing cells without treatment of Y-24180. Although a small portion of Alexa Fluor 488-labeled WT was transported into the early endosomes, a considerable amount of the receptor was detected on the plasma membrane due to their effective recycling (Fig. 7C). In contrast, Alexa Fluor 488-labeled P247A accumulated substantially in early endosomes after stimulation with mc-PAF.
FIGURE 7.
Aberrant trafficking of P247A after stimulation with mc-PAF. A, flow cytometric analysis was performed using HeLa-WT, HeLa-P247A, and HeLa-mock cells to evaluate the internalization of WT- and P247A-hPAFRs. After incubation in the presence of Dox, these cells were treated with Y-24180 (100 nm) for 12 h, followed by washes with PBS and incubation in DMEM (0.1% BSA) for 1 h to remove Y-24180. Thereafter, these cells were or were not stimulated with mc-PAF (3 μm) for 1 h. The data are representative of three independent experiments with identical results. B, N-terminal labeling of LPETGGGGG-tagged cell surface PAFRs with LPETGG-tethered Alexa Fluor 488 by sortase-A. The first step, preincubation of the cells expressing LPETGGGGG-tagged PAFRs with sortase-A and triglycine, allowed cleavage between the threonine and glycine of the LPETGGGGG tag, resulting in presentation of a pentaglycine tag at the N termini of the target PAFRs. In the following step, labeling of the target receptors with chemical probes was achieved by incubation of the cells with sortase-A and LPETGG-tethered Alexa Fluor 488. C, subcellular localizations of WT-hPAFR and P247A with or without stimulation of mc-PAF were observed by immunofluorescence confocal microscopy. After HeLa-WT and HeLa-P247A were cultured in the presence of Dox, these cells were pre-treated with or without Y-24180 (100 nm) for 12 h, followed by washes with PBS. The cell surface-trafficked receptors were specifically labeled with Alexa Fluor 488 by sortase-A as described in B. Thereafter, these cells were stimulated with or without mc-PAF (3 μm) for 1 h in the presence of Alexa Fluor 546-conjugated transferrin (50 μg/ml), and then subjected to immunocytochemical analysis. The early endosomes were visualized by Alexa Fluor 546-conjugated transferrin (red), and Alexa Fluor 488-labeled PAFRs are shown in green. White bars indicate 10 μm. The data are representative of three independent experiments with identical results.
Taken together, these results demonstrate that the P247A mutant is transported to the cell surface by chaperones and activated by mc-PAF. However, the mutant has abnormality in trafficking, e.g. enhanced internalization, deficiency in recycling, and/or accelerated degradation.
DISCUSSION
Although comparison of the amino acid sequences reveals little identity among the rhodopsin-type family of GPCRs, many GPCRs of this family share several common residues in the seven transmembrane helices (supplemental Fig. S1) (1, 2). In this report, to elucidate the importance of conserved residues in quality control and intracellular trafficking of GPCRs in living cells we generated various mutated PAFRs in which these conserved residues were replaced with alanines (Fig. 1A). We successfully identified several residues, including Lle59, Asp63, Pro247, Asp289, and Pro290 that could be crucial for correct folding by evaluating cell surface expression of PAFRs containing mutations in these residues (Fig. 1, B and C). In particular, mutations of an aspartic acid in TM2 or proline in TM6 resulted in a drastic decrease in cell surface expression of the receptors and their accumulation in the ER (Figs. 2 and supplemental S2). Helices in a peptide chain tend to be bent at proline residues, and defects in the GPCR ligand binding ability due to mutations of proline residues in TMs have been previously reported (14, 15, 30), suggesting that proline residues in TMs are crucial for correct folding of GPCRs in the ER. Therefore, it is likely that mutating Pro247 caused a marked conformational change of the receptor. Likewise, the crystal structure of rhodopsin (7) suggests that the aspartic acid in TM2 makes a hydrogen bond with asparagine in TM1 and alanine in TM7; therefore, it too might contribute to the correct folding of GPCRs. Interestingly, single nucleotide polymorphisms of prolines in TM6 of rhodopsin (31) and the V2 vasopressin receptor (32, 33) and aspartic acids in TM2 of the P2Y5/LPA6 receptor (34, 35) and melanocortin 1 receptor (36) have been reported. Although precise analyses of these mutants have not been completed, these mutant receptors might be eliminated by the ER-associated degradation system because of their aberrant structures. In this study, Y-24180 was able to augment cell surface expression and reduce ubiquitination of the mutant PAFRs, implying that specific ligands for PAFR act as pharmacological chaperones to enhance the ER export of these mutants (Figs. 4, A and B, and supplemental S3 and S4).
We observed that most of the mutant PAFRs had impaired cell surface expression, probably due to lack of ER export. In general, accumulation of aberrant GPCRs in the ER can lead to two different phenotypes in cells: loss of function because of the deficiency in the cell surface expression (8, 9) and ER stress by the accumulation of aberrant proteins (37). Pharmacological chaperones can rescue both phenotypes by promoting protein trafficking from the ER to the cell surface (38–43).
Previously, several groups have demonstrated that the conserved aspartic acid in TM2 is structurally required for G-protein coupling to the receptor in PAFR and other GPCRs (44). For example, Parent et al. (45) reported that substitution of Asp63 to asparagine in PAFR resulted in impairment of the activation of coupled G-proteins without significant alteration of the binding affinity to PAF. In Fig. 5, we obtained supporting results that PAF-elicited Ca2+ mobilization was not observed in D63A expressing cells, although a significant amount of the receptors was trafficked to the cell surface after Y-24180 treatment. It supports the idea that Asp63 is pivotal for activation of coupled G-proteins. More structural studies will clarify the importance of this residue for the function of GPCRs.
In this study, we further found that mc-PAF does not increase the expression of P247A on the cell surface. We concluded that following treatment with mc-PAF, P247A is trafficked to the cell surface but is then immediately internalized after ligand stimulation, because 1) mc-PAF activated P247A and the WT receptor similarly (Figs. 5A and 6A); 2) P247A accumulated in the early endosomes after treatment with mc-PAF (Fig. 6B); and 3) the fully glycosylated form of P247A appeared after treatment with chloroquine, a lysosomal inhibitor (Fig. 6C). Moreover, we suggest that, unlike the WT receptor, P247A has an abnormality in trafficking, such as enhanced internalization, deficiency in recycling, and/or accelerated degradation after stimulation with agonist (Fig. 7A). The aberrance of P247A trafficking is also supported by the results using a novel labeling method for cell surface receptors (Fig. 7, B and C). Several groups have recently reported that carrier proteins, e.g. Rab4 and Rab11, are required for recycling of GPCRs (46, 47). The structural aberrance of P247A might result in both deficient ER export and a lack of interaction with various proteins required for recycling from the early endosomes.
In summary, our data demonstrate that some of the conserved residues of GPCRs have important roles in correct folding and trafficking. However, what defines the behavior of GPCRs is not fully understood, and further studies are needed to address this issue. Although many studies have focused on the roles of conserved amino acids in GPCR functions, new methods, such as forced trafficking by pharmacological chaperones and sortase-A-mediated labeling for cell surface proteins, could provide additional information.
Acknowledgments
We thank Dr. Jun-ichi Miyazaki (Osaka University, Japan) for supplying pCXN2, Dr. Shuh Narumiya (Kyoto University) for pEF-C3, and Japan Tobacco Co. for zif 268-firefly luciferase-pGL2. We also thank Chizu Kanokoda and laboratory members for technical advice and useful discussions.
This work was supported in part by a grant-in-aid for Challenging Exploratory Research (to M. N.) and a grant-in-aid for Specially Promoted Research (to T. S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and grants from the Japan Society for the Promotion of Science (Global COE program) and the Center for NanoBio Integration, the University of Tokyo.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and primer sequences.
- GPCR
- G-protein coupled receptor
- Dox
- doxycycline
- Endo-H
- endoglycosidase H
- PAF
- platelet-activating factor
- mc-PAF
- methylcarbamyl-PAF
- PAFR
- platelet-activating factor receptor
- PNGase-F
- N-glycosidase F
- TM
- transmembrane
- WT
- wild-type
- ER
- endoplasmic reticulum
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- DMEM
- Dulbecco's modified Eagle's medium
- GTPγS
- guanosine 5′-O-(thiotriphosphate).
REFERENCES
- 1.Bockaert J., Pin J. P. (1999) EMBO J. 18, 1723–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagerström M. C., Schiöth H. B. (2008) Nat. Rev. Drug. Discov. 7, 339–357 [DOI] [PubMed] [Google Scholar]
- 3.Nakamura M., Honda Z., Izumi T., Sakanaka C., Mutoh H., Minami M., Bito H., Seyama Y., Matsumoto T., Noma M., et al. (1991) J. Biol. Chem. 266, 20400–20405 [PubMed] [Google Scholar]
- 4.Parent J. L., Gouill C. L., Escher E., Rola-Pleszczynski M., Staková J. (1996) J. Biol. Chem. 271, 23298–23303 [DOI] [PubMed] [Google Scholar]
- 5.Ishii I., Izumi T., Tsukamoto H., Umeyama H., Ui M., Shimizu T. (1997) J. Biol. Chem. 272, 7846–7854 [DOI] [PubMed] [Google Scholar]
- 6.Honda Z., Nakamura M., Miki I., Minami M., Watanabe T., Seyama Y., Okado H., Toh H., Ito K., Miyamoto T., Shimizu T. (1991) Nature 349, 342–346 [DOI] [PubMed] [Google Scholar]
- 7.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 8.Bernier V., Lagacé M., Bichet D. G., Bouvier M. (2004) Trends Endocrinol. Metab. 15, 222–228 [DOI] [PubMed] [Google Scholar]
- 9.Conn P. M., Ulloa-Aguirre A., Ito J., Janovick J. A. (2007) Pharmacol. Rev. 59, 225–250 [DOI] [PubMed] [Google Scholar]
- 10.Yasuda D., Okuno T., Yokomizo T., Hori T., Hirota N., Hashidate T., Miyano M., Shimizu T., Nakamura M. (2009) FASEB J. 23, 1470–1481 [DOI] [PubMed] [Google Scholar]
- 11.Strader C. D., Sigal I. S., Register R. B., Candelore M. R., Rands E., Dixon R. A. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neve K. A., Cox B. A., Henningsen R. A., Spanoyannis A., Neve R. L. (1991) Mol. Pharmacol. 39, 733–739 [PubMed] [Google Scholar]
- 13.Chanda P. K., Minchin M. C., Davis A. R., Greenberg L., Reilly Y., McGregor W. H., Bhat R., Lubeck M. D., Mizutani S., Hung P. P. (1993) Mol. Pharmacol. 43, 516–520 [PubMed] [Google Scholar]
- 14.Xu W., Li J., Chen C., Huang P., Weinstein H., Javitch J. A., Shi L., de Riel J. K., Liu-Chen L. Y. (2001) Biochemistry 40, 8018–8029 [DOI] [PubMed] [Google Scholar]
- 15.Conner A. C., Hay D. L., Simms J., Howitt S. G., Schindler M., Smith D. M., Wheatley M., Poyner D. R. (2005) Mol. Pharmacol. 67, 20–31 [PubMed] [Google Scholar]
- 16.Yamamoto T., Nagamune T. (2009) Chem. Commun. 9, 1022–1024 [DOI] [PubMed] [Google Scholar]
- 17.Tsukiji S., Nagamune T. (2009) ChemBioChem 10, 787–798 [DOI] [PubMed] [Google Scholar]
- 18.Zhang H. M., Yuan J., Cheung P., Luo H., Yanagawa B., Chau D., Stephan-Tozy N., Wong B. W., Zhang J., Wilson J. E., McManus B. M., Yang D. (2003) J. Biol. Chem. 278, 33011–33019 [DOI] [PubMed] [Google Scholar]
- 19.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 20.Xia S., Kjaer S., Zheng K., Hu P. S., Bai L., Jia J. Y., Rigler R., Pramanik A., Xu T., Hökfelt T., Xu Z. Q. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15207–15212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peshenko I. V., Olshevskaya E. V., Dizhoor A. M. (2008) J. Biol. Chem. 283, 21747–21757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami N., Yokomizo T., Okuno T., Shimizu T. (2004) J. Biol. Chem. 279, 42484–42491 [DOI] [PubMed] [Google Scholar]
- 23.Tan C. M., Nickols H. H., Limbird L. E. (2003) J. Biol. Chem. 278, 35678–35686 [DOI] [PubMed] [Google Scholar]
- 24.Ishii S., Kihara Y., Shimizu T. (2005) J. Biol. Chem. 280, 9083–9087 [DOI] [PubMed] [Google Scholar]
- 25.Petäjä-Repo U. E., Hogue M., Bhalla S., Laperrière A., Morello J. P., Bouvier M. (2002) EMBO J. 21, 1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terasawa M., Aratani H., Setoguchi M., Tahara T. (1990) Prostaglandins 40, 553–569 [DOI] [PubMed] [Google Scholar]
- 27.O'Flaherty J. T., Redman J. F., Jr., Schmitt J. D., Ellis J. M., Surles J. R., Marx M. H., Piantadosi C., Wykle R. L. (1987) Biochem. Biophys. Res. Commun. 147, 18–24 [DOI] [PubMed] [Google Scholar]
- 28.Misinzo G., Delputte P. L., Nauwynck H. J. (2008) J. Virol. 82, 1128–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii I., Saito E., Izumi T., Ui M., Shimizu T. (1998) J. Biol. Chem. 273, 9878–9885 [DOI] [PubMed] [Google Scholar]
- 30.Eilers M., Hornak V., Smith S. O., Konopka J. B. (2005) Biochemistry 44, 8959–8975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souied E., Gerber S., Rozet J. M., Bonneau D., Dufier J. L., Ghazi I., Philip N., Soubrane G., Coscas G., Munnich A., et al. (1994) Hum. Mol. Genet. 3, 1433–1434 [DOI] [PubMed] [Google Scholar]
- 32.Pan Y., Metzenberg A., Das S., Jing B., Gitschier J. (1992) Nat. Genet. 2, 103–106 [DOI] [PubMed] [Google Scholar]
- 33.Pan Y., Wilson P., Gitschier J. (1994) J. Biol. Chem. 269, 31933–31937 [PubMed] [Google Scholar]
- 34.Azeem Z., Jelani M., Naz G., Tariq M., Wasif N., Kamran-Ul-Hassan Naqvi S., Ayub M., Yasinzai M., Amin-Ud-Din M., Wali A., Ali G., Chishti M. S., Ahmad W. (2008) Hum. Genet. 123, 515–519 [DOI] [PubMed] [Google Scholar]
- 35.Shimomura Y., Wajid M., Ishii Y., Shapiro L., Petukhova L., Gordon D., Christiano A. M. (2008) Nat. Genet. 40, 335–339 [DOI] [PubMed] [Google Scholar]
- 36.Valverde P., Healy E., Sikkink S., Haldane F., Thody A. J., Carothers A., Jackson I. J., Rees J. L. (1996) Hum. Mol. Genet. 5, 1663–1666 [DOI] [PubMed] [Google Scholar]
- 37.Kitao Y., Imai Y., Ozawa K., Kataoka A., Ikeda T., Soda M., Nakimawa K., Kiyama H., Stern D. M., Hori O., Wakamatsu K., Ito S., Itohara S., Takahashi R., Ogawa S. (2007) Hum. Mol. Genet. 16, 50–60 [DOI] [PubMed] [Google Scholar]
- 38.Morello J. P., Salahpour A., Laperrière A., Bernier V., Arthus M. F., Lonergan M., Petäjä-Repo U., Angers S., Morin D., Bichet D. G., Bouvier M. (2000) J. Clin. Invest. 105, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janovick J. A., Maya-Nunez G., Conn P. M. (2002) J. Clin. Endocrinol. Metab. 87, 3255–3262 [DOI] [PubMed] [Google Scholar]
- 40.Noorwez S. M., Kuksa V., Imanishi Y., Zhu L., Filipek S., Palczewski K., Kaushal S. (2003) J. Biol. Chem. 278, 14442–14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan J. Q., Ishii S., Asano N., Suzuki Y. (1999) Nat. Med 5, 112–115 [DOI] [PubMed] [Google Scholar]
- 42.Frustaci A., Chimenti C., Ricci R., Natale L., Russo M. A., Pieroni M., Eng C. M., Desnick R. J. (2001) N. Engl. J. Med. 345, 25–32 [DOI] [PubMed] [Google Scholar]
- 43.Matsuda J., Suzuki O., Oshima A., Yamamoto Y., Noguchi A., Takimoto K., Itoh M., Matsuzaki Y., Yasuda Y., Ogawa S., Sakata Y., Nanba E., Higaki K., Ogawa Y., Tominaga L., Ohno K., Iwasaki H., Watanabe H., Brady R. O., Suzuki Y. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15912–15917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose P. M., Krystek S. R., Jr., Patel P. S., Liu E. C., Lynch J. S., Lach D. A., Fisher S. M., Webb M. L. (1995) FEBS Lett. 361, 243–249 [DOI] [PubMed] [Google Scholar]
- 45.Parent J. L., Le Gouill C., Rola-Pleszczynski M., Stanková J. (1996) Biochem. Biophys. Res. Commun. 219, 968–975 [DOI] [PubMed] [Google Scholar]
- 46.Moore R. H., Millman E. E., Alpizar-Foster E., Dai W., Knoll B. J. (2004) J. Cell Sci. 117, 3107–3117 [DOI] [PubMed] [Google Scholar]
- 47.Wang F., Chen X., Zhang X., Ma L. (2008) Mol. Endocrinol. 22, 1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]