Abstract
The current biological studies have been advanced by the continuous development of robust, accurate and sensitive mass spectrometric technologies. The MALDI LTQ Orbitrap is a new addition to the orbitrap configurations, known for their high resolving power and accuracy. This configuration provides features inherent to the MALDI source, such as reduced spectra complexity, forgiveness to contaminants, and sample retention for follow-up analyses with targeted or hypothesis-driven questions. Here we investigate its performance for characterizing the composition of isolated protein complexes. To facilitate the assessment, we selected two well characterized complexes from Saccharomyces cerevisiae—Apl1 and Nup84. Manual and automatic MS and MS/MS analyses readily resolved their compositions, with increased confidence of protein identification when compared to our previous reports using MALDI QqTOF and MALDI IT. CID fragmentation of singly-charged peptides provided sufficient information for conclusive identification of the isolated proteins. We then assessed the resolution, accuracy and sensitivity provided by this instrument in the context of analyzing the isolated protein assemblies. Our analysis of complex mixtures of singly-charged ions up to m/z 4000 showed that 1) the resolving power, inversely proportional to the square root of m/z, had over 4 orders of magnitude dynamic range; 2) internal calibration led to improved accuracy, with an average absolute mass error of 0.5 ppm and a distribution centered at 0 ppm; and 3) subfemtomol sensitivity was achieved using both CHCA and DHB matrices. Additionally, our analyses of a synthetic phosphorylated peptide in mixtures showed subfemtomol level of detection using neutral loss scanning.
Introduction
In recent years, mass spectrometry has acquired a well deserved pivotal role in biological studies. Mass spectrometric analyses of complex biological samples have become an essential aspect in elucidating cellular function. Frequently utilized mass spectrometers include matrix-assisted laser desorption ionization (MALDI) configurations, shown to be robust and valuable tools for analyzing the composition of isolated protein complexes and larger assemblies (e.g. nuclear pore complex [1, 2] or postsynaptic densities [3]). These studies were made possible by the development of MALDI mass spectrometers as tandem hybrid or modular configurations that provided access to the combined strengths of various mass analyzers, such as Time-of-Flight (ToF) [4, 5], Quadrupole/Time-of-flight (Q-ToF) [6], QqToF [7], ToF-ToF [8] and ion traps (IT) [9]. For example, Krutchinsky and co-workers [10] have described MALDI- QqToF and MALDI-IT mass spectrometers as a modular instrument, allowing the analysis of complex protein mixtures by taking advantage of the levels of resolution, accuracy (QqToF), speed and sensitivity (IT). There are several features that make the use of MALDI configurations attractive, including ease of use and forgiveness to contaminants. The reduced sample complexity inherent to the MALDI-generated singly-charged ions provides the opportunity of carrying out analyses on the majority of the observed ions. While high-throughput automatic analyses are frequently employed using these configurations, sample retention provides the option for follow-up analyses, lowering the threshold of the initial level of detection. One well known limitation when using MALDI configurations is that the fragmentation of singly-charged ions is highly dependent on the amino acid sequence, occurring preferentially at the C-terminus of aspartyl and glutamyl residues and the N-terminus of prolyl residues [11–13], making their implementation less suitable for comprehensive sequencing. However, a MALDI-quadrupole ion trap was shown to provide the levels of detection necessary for observing the less prominent fragments and, in certain situations, enough information for de novo sequencing [14].
A new addition to the MALDI mass spectrometric configurations, the MALDI LTQ Orbitrap XL [15], couples a vacuum MALDI source to a hybrid LTQ Orbitrap [16]. Recent years have seen a rapid development and wide-spread implementation of the orbitrap mass analyzer and its configurations [17–20]. Several modifications to these ESI LTQ Orbitrap configurations, such as the integration of electron transfer dissociation [21], emerged to further extend performance [22] and reach of applications. The introduction of the MALDI LTQ Orbitrap XL [15] provides complementary merits to those offered by ESI. Karas and co-workers have recently reported its application, in conjunction with acid hydrolysis, to analyzing RNA oligonucleotide sequences [23]. Here we describe the performance of a MALDI LTQ Orbitrap configuration for elucidating the composition of protein complexes isolated via rapid immunoaffinity purifications on magnetic beads. To aid the assessment, we selected two protein complexes that we previously characterized using MALDI QqToF and MALDI-IT configurations, the yeast nuclear pore Nup84 subcomplex and the yeast clathrin associated protein complex AP-2 isolated via Apl1 [24]. We discuss the utility of this instrument for studying the composition of protein complexes and assess the resolving power, accuracy and sensitivity from the perspective of analyzing complex peptide mixtures.
Experimental
Cell line construction and Cell Culture
Saccharomyces cerevisiae yeast strains expressing Nup84 or Apl1 genomically tagged with protein A (PrA) or enhanced green fluorescent protein (EGFP), respectively, were grown to 2.0 × 107 cells/ml, harvested, and frozen as previously described [24, 25]. The Nup84-PrA strain was a kind gift from Michael P. Rout (The Rockefeller University) and the Apl1-EGFP strain was purchased from Invitrogen. Cells were harvested by centrifugation at 3,000 rpm, washed twice with PBS, weighed and suspended at 100 μl/g cell in freezing buffer (20 mM HEPES, pH 7.5, 1.2% w/v polyvinyl-pyrrolidone (Sigma), 1/100 v/v protease inhibitor cocktail (Sigma)). The resulting cell paste was frozen as small pellets in liquid nitrogen as previously described [24].
Cell Cryogenic Disruption and Lysis
2 g Nup84-PrA and 6 g Apl1-EGFP yeast cells were cryogenically disrupted using the Retsch MM301 Mixer Mill (Retch, Newtown, PA) by loading the cell pellets into 25 ml stainless steel jars (Retch), each with a 20 mm stainless steel grinding ball (McMaster, NJ). Approximately 90% efficiency of grinding, as assessed by light microscopy, was achieved using 10 steps of 2.5 min at 30 Hz with intermittent cooling in liquid nitrogen. The frozen cell powders were suspended in optimized lysis buffers (Supplementary Table S1) at 5 ml/g cell powder, homogenized for 10 s with a PT 10–35 Polytron (Kinematica), centrifuged for 10 min at 5,000 rpm at 4°C. The supernatant fractions were used for affinity purifications.
Conjugation of Magnetic Beads with Antibodies
A custom anti-GFP rabbit polyclonal antibody was prepared and affinity-purified [24]. Purified rabbit IgG (ICN Pharmaceutical) was used for isolations of the PrA tag. M-270 epoxy magnetic beads (Invitrogen Dynal) were conjugated as previously described [24] with 5 μg of anti-GFP antibodies or 10 μg IgG per mg of beads. 8 mg of conjugated beads were utilized for each immunoaffinity purification.
Immunoaffinity Purification
Conjugated magnetic beads were washed three times with corresponding lysis buffers and incubated with the cell lysates by gentle rotation at 4 °C for optimized periods of time—30 min for Nup84 and 1 hour for Apl1. After six washes with lysis buffer, the isolated protein complexes were eluted from the beads with 700 μl of a freshly-made 0.5 N NH4OH, 0.5 mM EDTA solution by shaking for 20 min at room temperature. The eluted fractions were frozen in liquid nitrogen and dried using vacuum centrifugation (Speed Vac Plus SC110A with gel pump GP110, Thermo Scientific). The dried samples were suspended in 30 μl LDS-PAGE buffer with reducing agent (Invitrogen), incubated at 70 °C for 10 min with agitation, alkylated with 100 mM iodoacetamide at room temperature in the dark for 30 min, and resolved by 1-D gel electrophoresis on 4–12% NuPAGE Novex Bis-Tris gel (Invitrogen). The gels were stained with SimplyBlue™ SafeStain (Invitrogen).
Protein Digestion
For digesting isolated proteins, the entire gel lanes were cut into approximately 30 slices, diced and destained in 200 μl 50% (v/v) acetonitrile in 50 mM ammonium bicarbonate with 10 min agitation at 4 °C. The gel pieces were then dehydrated in 100% acetonitrile and digested with 12.5 ng/μl sequencing grade modified trypsin (Promega). The resulting peptides were extracted on reverse phase resin (POROS 20 R2, Applied Biosystems), packed on ZipTip with C18 resin (Millipore), washed twice with 20 μl 0.1% (v/v) trifluoroacetic acid (TFA), and eluted with 25 mg/ml 2,5-dihydroxybenzoic acid (DHB) or 2 mg/ml α-cyano-4-hydroxycinnamic acid (CHCA) in 70% (v/v) acetonitrile and 0.1% (v/v) TFA directly onto a MALDI target. Both metal MALDI targets (Thermo), as well as electrically-conductive magnetic MALDI targets [10] were utilized. For in-solution digestion of bovine serum albumin (BSA, 50 pmol, New England Biolabs) was performed with 0.2 μg trypsin in 50 mM ammonium bicarbonate at 37 °C for 10 h with gentle agitation.
Mass Spectrometry
Mass spectrometric analyses were performed on a MALDI LTQ Orbitrap XL mass spectrometer (Thermo Electron, Bremen, Germany). The MALDI Duo source configuration on the LTQ Orbitrap has been modified from the previous model on the vMALDI LTQ instrument, which incorporated a nitrogen laser with 20 Hz firing rate, coupled to the LTQ mass spectrometer via a fiber optic cable. The MALDI Duo source on this configuration is equipped with a Nitrogen Laser MNL 100 (LTB, Berlin, Germany), operating at 337.1 nm wavelength, with up to 60 Hz repetition rate, 75 μJ maximum energy per pulse, and 3 ns pulse duration [15]. The laser beam is focused using two adjustable flat mirrors and a single plano-convex lens. This direct beam configuration was designed to improve the consistency and efficiency of the laser energy transmission of the vMALDI interface. The instrument was operated in positive mode. MALDI laser energy was set to 7–10 or 15–20 μJ for samples embedded in CHCA or DHB matrices, respectively. As linear traps have limited ion capacity, which leads to readily occurring space charge effects and reduced mass accuracy, we performed the MS analyses of digested proteins in the orbitrap. MS spectra were acquired for a mass range of up to m/z 4000 with the resolution setting 60,000 at m/z 400. For manual mode acquisition, parameters were: automated spectrum filter (ASF) off, 200 scans/step, automated gain control (AGC) on, allowing storage of 5e5 ions. For acquisition in automated mode, the parameters were: crystal positioning system (CPS), ASF off, 3 scans/step and AGC on. Tandem mass spectrometric (MS/MS) analyses were carried out by MALDI IT collision-induced dissociation (CID) using parameters previously described for analyses on MALDI-IT (LCQ Deca XP Plus, Thermo Electron) [24, 26] and vMALDI-LTQ (Thermo Electron) [10] mass spectrometers. Utilized parameters included: precursor ion isolation in the linear ion trap, 4 mass units isolation width, normalized collision energy 33%, activation q 0.25, activation time 30–300 ms. For automatic MS/MS acquisition, the mass spectrometer was operated in a data-dependent mode to switch between orbitrap MS and LTQ MS/MS analyses. The 50 most prominent ions were sequentially isolated for CID fragmentation in the linear ion trap and subsequently excluded for 900 sec. Mass spectra were visualized and processed in Qual Browser (version 2.0.7, Thermo Fisher Scientific). Protein candidates were identified by database searching against the National Center for Biotechnology Information nonredundant protein database, version 06/10/16, using the computer algorithms XProteo (http://www.xproteo.com) or ProFound [27]. Search parameters for MS data were: species: Saccharomyces cerevisiae (11105 sequences); Protein Mass: 0–500 kDa; Protein pI: 1–14; Mixture search: Auto, Number of candidates displayed: 50; Enzyme: trypsin; Miscleavage: 1; Mass type: monoisotopic; Charge state: MH+; Mass tolerance: 0.02 Da; Fixed modification: carbamidomethylation of Cys; Variable modifications: oxidation of Met and phosphorylations on Ser, Thr or Tyr. Candidate peptides were confirmed by manual or automatic CID MS/MS analyses by assessment of the b and y ion fragments and the preferential cleavages C-terminus of Asp and Glu [11] or N-terminus of Pro residues [12, 13]. Additional parameters used for searching MS/MS data were: Precursor tolerance: 0.02 Da; Fragment tolerance: 0.5 Da; Instrument: MALDI_I_TRAP. All XProteo assignments made based on MS/MS data acquired in automated mode were checked manually. The XProteo probability scores, based on an improved version of the ProFound Bayesian algorithm [27], indicate d′ (discriminability) values for each candidate protein as the normalized distance between the score distribution (of the candidate protein) and the distribution of randomly matched proteins (in units of standard deviation). A score of d′=4 correspond to a true positive rate of 0.99 and a false positive rate of 0.05. While proteins with d′>4 were readily verified by our MS/MS analyses, several proteins with lower scores could also be confirmed (Supplementary Tables S3, S4 and S5).
Calibration
External calibration was performed using standard peptide mixtures (ProteoMass™ calibration kit, Sigma) for either the normal (m/z 150–2000) or high mass range (m/z 200–4000). Internal calibration was performed by enabling the lock mass option in MS mode. The trypsin peptide LGENHIDVLEGNEQFINAAK (C96H152N27O33; m/z 2211.104586) was used for internal calibration.
Accuracy determination
109 peptides confirmed by peptide fragmentation from proteins immunopurified with Nup84 were selected for determining accuracy. Immediately following external calibration, MS were acquired as described above, with or without the lock mass. The m/z values of the confirmed peptides, recorded to 6 digits after the decimal points, were used to calculate the mass error (Supplementary Table S2).
Sensitivity Assay
Angiotensin II (ProteoMass™ Sensitivity Standard, Sigma) was spotted onto a MALDI target with either recrystallized CHCA or DHB as a serial dilution with final concentration ranging from 100 fmol to 0.05 fmol. The laser energy was set to 10 μJ for CHCA or 20 μJ for DHB.
Resolution Assessment
Duplicate isolations of Nup84 were processed into 28 samples per gel lane and spotted using either DHB or CHCA. MALDI Orbitrap MS of Nup84 samples were acquired using resolution settings of 60,000 and 30,000 at m/z 400. The resolution values, as indicated in Qual Browser (Thermo Scientific), were obtained from approximately 26,000 peaks for each resolution setting and plotted against m/z using MatLab, where exponential regression was calculated.
Neutral Loss Scanning
Synthetic phosphorylated peptide RRLIEDAE-pY-AARG (SRC peptide, ANA SPEC, San Jose, CA) was spotted onto a MALDI target as a serial dilution with final concentrations ranging from 100 fmol/μl to 0.25 fmol/μl, as a mixture with: 1) 50 fmol/μl three-peptide mixture (des-Arg-bradykinin, neurotensin and glucagon), 2) 50 fmol/μl BSA digested with trypsin and 3) trysin-digested 100 kDa synthetic weight-marker (Invitrogen). Neutral loss scanning was performed using stepwise acquisition of parent ion scanning, precursor ion isolation window of 4 Da over a m/z range of 1100–1900, product ion width of 1 Da, 35% collision energy, and neutral loss set at −98 Da for the loss of phosphoric acid.
Results and Discussion
Characterizing protein complexes using rapid isolations and MALDI mass spectrometry
In previous work, we observed that MALDI mass spectrometric configurations provide valuable tools for analyzing the composition of isolated protein complexes or larger assemblies [1–3, 24, 28]. To assess the performance of the MALDI LTQ Orbitrap for characterizing protein complexes, we selected as case studies two protein assemblies that have been previously analyzed using modular MALDI QqTOF and MALDI IT mass spectrometers—the yeast clathrin associated protein complex AP-2R, isolated via Apl1, and the yeast nuclear pore Nup84 subcomplex [24]. We utilized Saccharomyces cerevisiae yeast strains expressing endogenous levels of Nup84 or Apl1 tagged with protein A (PrA) or enhanced green fluorescent protein (EGFP), respectively. Following harvesting and cryogenic cell lysis, Apl1 and Nup84 complexes were isolated by one-step immunoaffinity purifications on magnetic beads coated with antibodies against the tag, as previously reported (Supplementary Fig S1A) [24]. Custom high-affinity anti-GFP antibodies were utilized for isolating Apl1-GFP and IgG for Nup84-PrA. Co-isolated proteins were resolved by 1-D gel electrophoresis, subjected to trypsin digestion and deposited onto MALDI targets. We utilized both commercially-available metal targets and electrically-conductive magnetic targets [10], which can be made in-house using polyethylene film coated with indium tin oxide. Due to the elimination of the washing and reusing steps required for the common metal targets, the magnetic targets were amenable for sample preservation, and, therefore, valuable for targeted follow-up experiments. The use of magnetic targets was an economical alternative to the metal targets, especially when considering the numerous users and ongoing parallel experiments. While readily incorporated for manual and automatic MS and MS/MS analyses, these magnetic targets were not found to be optimal for performing instrument calibrations. One possible explanation for this observation is the requirement of a stable flow of ions during the calibration process. Factors such as differences in the crystallization process on the two different surfaces, spot-to-spot variations, and, occasionally, possible matrix depletion leading to the production of polymer ions from the plastic surface of the target, can affect the stability of ion production. Therefore, we utilized metal targets for the calibration and sensitivity assays, and magnetic targets for the analyses of samples derived from isolating protein complexes.
Resulting peptides were analyzed by MALDI MS and MS/MS analyses using a MALDI LTQ Orbitrap (Supplementary Fig. S1B). The FTMS peptide fingerprint analyses, executed in the orbitrap, benefited from high accuracy and resolving power, while the LTQ offered speed and sensitivity of detection for MALDI LTQ CID MS/MS analyses (Figures 2, 3, and Supplementary Fig. S2). Examples of MALDI CID MS/MS spectra with detection at the linear trap or orbitrap levels are shown in Supplementary Fig. S2. For the orbitrap detection, an increased number of scans (compared to the linear trap detection) were necessary to generate a product ion spectrum that could confidently confirm peptide identity. However, the accuracy and resolution provided by the orbitrap proved critical for resolving and detecting peptides in complex mixtures (as described in the following sections). The peptides of interest can also be investigated using two additional methods for ion fragmentation available on this configuration—PQD (pulsed Q dissociation) [29–31] and HCD (higher-energy C-trap dissociation) [32]. While not the focus of this manuscript, these fragmentations were reported to be suitable for quantitative studies on the ES LTQ orbitrap configuration [30, 31].
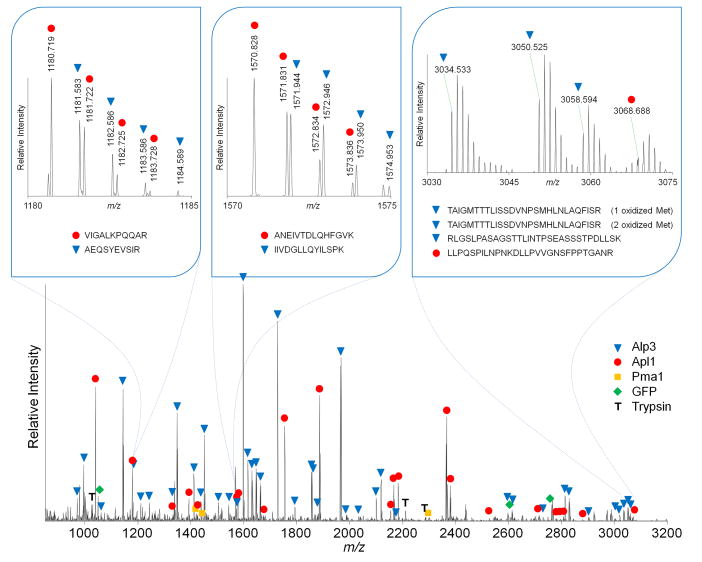
Fig. 2. Representative MALDI LTQ Orbitrap MS of selected proteins co-isolated with Apl1.
The MS shows a mixture of peptides corresponding to Apl1, Apl3, Pma1 and GFP, co-migrating at ~110 kDa. Examples of peptides from each protein are labeled in the spectrum. The panel insets show expanded MS regions containing Apl1 and Apl3 peptides, illustrating the ability of the orbitrap to resolve adjacent peaks.
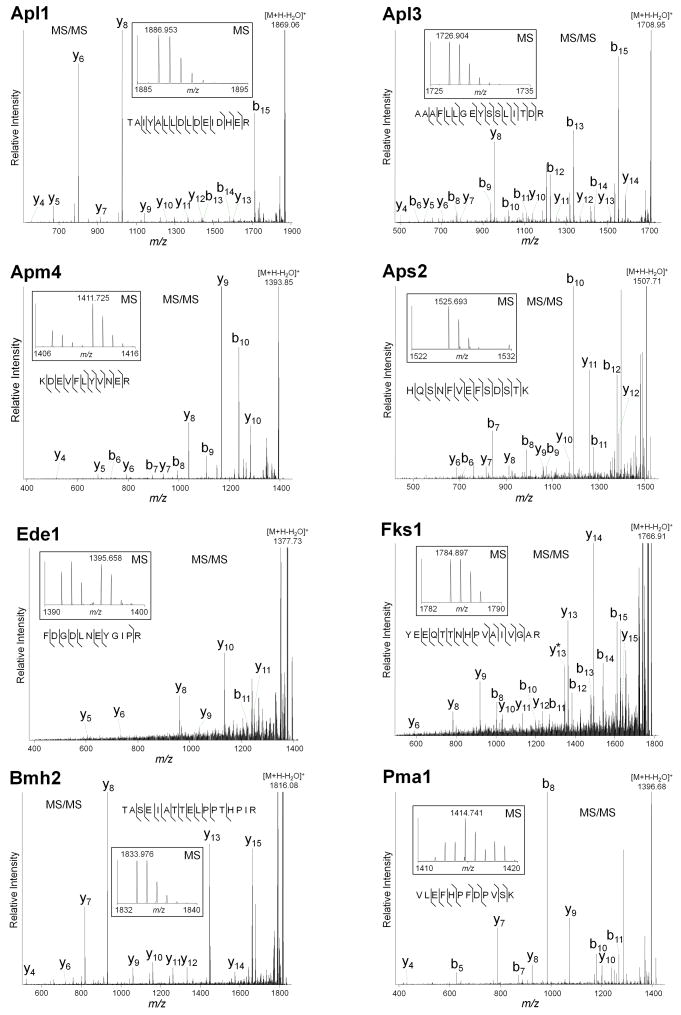
Fig. 3. Representative MALDI LTQ Orbitrap CID spectra of [M + H]+ ions of peptides from proteins co-isolated with Apl1.
The small panel insets illustrate the MS regions containing the peptides selected for fragmentation. The corresponding monoisotopic masses are indicated. Following CID, the generated fragment ions were detected at the linear trap level.
Apl1 and Nup84 candidate interacting partners were identified by database searching using the XProteo computer algorithm and verified manually for: precursor mass errors less than 4 ppm (according to our accuracy assessment, Figure 5), correct assignment of b and y ions, preferential cleavages at Asp, Glu and Pro residues, and MS/MS confirmation of a minimum of 2 peptides per protein candidate. Complete lists of identified proteins from the three isolations are given as supplementary material (Supplementary Tables S3 and S4).
Fig. 5. Characterization of the mass accuracy of the MALDI LTQ Orbitrap.
Mass errors of 109 peptides were calculated from externally (dark blue points) or internally (light pink triangles) calibrated MS spectra of proteins co-isolated with Nup84. A) The absolute values of mass errors are plotted against the m/z of the peptides (left panel). The distribution of the real mass errors is shown (right panel). B) Summary of mass error statistics.
The clathrin adaptor complex AP-2R, which links clathrin to receptors on the membrane surface of a coated vesicle, is a heterotetramer composed of two large (Apl3 and Apl1), one medium (Apm4) and one small (Aps2) subunits. Our isolation of Apl1 resulted in the identification of all its previously reported interacting partners, Apl3, Apm4, Aps2 [33, 34] and Ede1 [24] (Figure 1–3 and Supplementary Table S3). The expression levels of the isolated proteins were estimated in the S. cerevisiae GFP fusion protein localization database at approximately 1670 (Apl1), 1590 (Apl3), 1390 (Aps2), 3410 (Apm4) and 1380 (Ede1) molecules per haploid cell [35]. Several additional proteins were co-isolated with Apl1, such as Fks1 (expression levels undetermined), which is known to be involved in cell wall remodeling [36] and may indicate new avenues to pursue in further elucidating the molecular functions of Apl1. The specificity of the new binding partners, including Bmh2, a 14-3-3 protein reported to be involved in exocytosis and vesicle transport [37], and Pma1, a Plasma membrane H+-ATPase [38], which are present at higher copy number per cell (47,600 and 1260,000, respectively), is still to be determined.
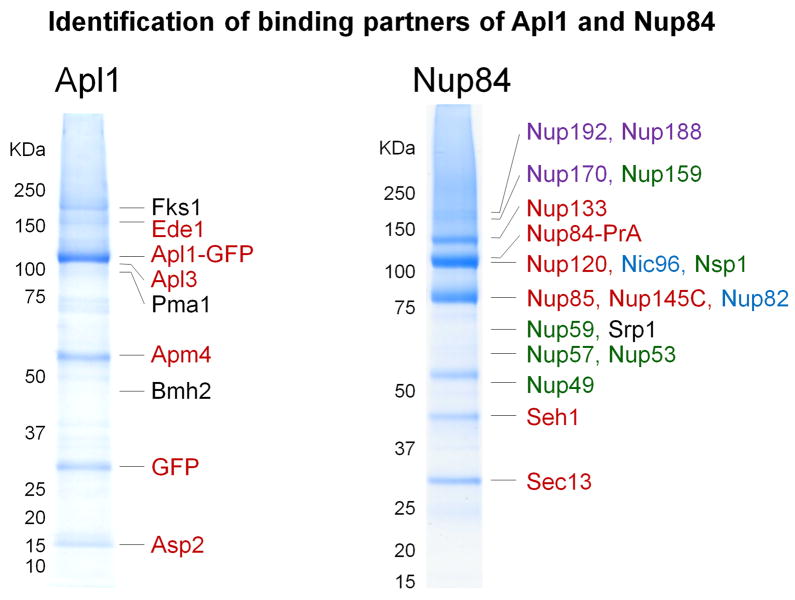
Fig. 1. Characterization of protein complexes using the MALDI LTQ Orbitrap.
Isolation of endogenous levels of genomically-tagged Apl1-EGFP and Nup84-PrA from yeast. The main observed interacting partners are indicated and color coded according to known (red) and novel (black) associations. The colors in the Nup84 isolation illustrate the presence of multiple subcomplexes of the yeast nuclear pore.
Representative MALDI LTQ Orbitrap MS and MALDI IT CID fragmentations of [M + H]+ ions of peptides from proteins co-isolated with Apl1 are presented in Figures 2 and 3, illustrating the complementary merits of the orbitrap and linear trap. The detection of adjacent peaks was greatly assisted by the resolving power of the orbitrap, as we could readily assign peptides corresponding to Apl1, Apl3, Pma1 and GFP in a mixture (Figure 2). As shown in Figure 3, the MALDI CID fragmentation of singly-charged ions provided sufficient information for the conclusive identification of peptides corresponding to Apl1 interacting partners. We compared these results to our previous observations from equivalent Apl1 isolations (same sample amounts, magnetic beads and lysis buffer), using as a reference point the data reported from a MALDI-QqToF instrument [24]. To keep the data interpretation consistent, we reanalyzed our results in a similar manner with our previous study, by using the ProFound search engine [27] (Table 1). For the proteins identified in both isolations, this comparison indicated equivalent results in terms of protein sequence coverage. Several factors can contribute to this observation. First, the possible presence of false peptide assignments, as a consequence of the higher mass errors following Q-ToF analyses (allowed for in the database search to match the instrument performance), could erroneously inflate the sequence coverage. Second, given the equivalent sample amounts utilized for these isolations, the resulting peptides were expected to be present at similar levels. Therefore, unless peptide recognition was obstructed by the noise level or neighboring peaks in the Q-ToF experiments and aided by the resolving power and accuracy of the orbitrap (e.g. Figure 2), similar protein sequence coverage would be expected. Third, although these isolations were carried out under identical conditions (amount of cells and beads, lysis buffer), some experimental variations may still occur (e.g. during protein digestion). These factors may also in part account for the identification of the three proteins not observed in the Q-ToF experiment, Fsk1, Pma1 and Bmh2. Another observation from this comparison was that, as expected, the average absolute mass errors of the observed peptides were significantly improved in the MALDI LTQ Orbitrap MS analyses, which led to improved expectation values (Table 1) and therefore, conclusive protein identifications.
Table 1.
Equivalent Apl1-EGFP immunoisolations analyzed using MALDI LTQ Orbitrap or MALDI QqToF.
| MALDI LTQ Orbitrap MS | MALDI QqTOF MS | |||||
|---|---|---|---|---|---|---|
| Protein | Sequence Coverage | Mass Error (Da) | Expectation | Sequence Coverage | Mass Error (Da) | Expectation |
| Apl1 | 53% | 0.003 (±0.002) | 1.6×10−9 | 47% | 0.017 (±0.012) | 1.6×10−6 |
| Apl3 | 53% | 0.003 (±0.002) | 9.0×10−15 | 46% | 0.016 (±0.016) | 2.4×10−9 |
| Ede1 | 19% | 0.005 (±0.004) | 8.5×10−7 | 23% | 0.022 (±0.016) | 6.8×10−10 |
| Apm4 | 58% | 0.002 (±0.001) | 2.3×10−19 | 51% | 0.011 (±0.013) | 1.6×10−14 |
| Aps2 | 48% | 0.003 (±0.003) | 6.9×10−5 | 45% | 0.014 (±0.011) | 9.2×10−5 |
| Pmd1 | 6% | 0.006 (±0.003) | 2.4×10−2 | 9% | 0.031 (±0.015) | 9.4×10−3 |
| Fks1 | 11% | 0.003 (±0.001) | 1.6×10−7 | n/o | ||
| Pma1 | 19% | 0.002 (±0.002) | 5.0×10−7 | n/o | ||
| Bmh2 | 45% | 0.005 (±0.001) | 9.7×10−6 | n/o | ||
Protein sequence coverages (%), average absolute mass errors (Da), and expectation values (ProFound) are shown for interacting partners detected using a MALDI LTQ Orbitrap as compared with our previous reports using a MALDI QqToF [24]; n/o – not observed.
As a second test, we selected another well characterized protein assembly and isolated PrA-tagged Nup84, present at endogenous level in yeast cells. The nuclear pore Nup84 subcomplex is an assembly of seven proteins [39–42], Nup133, Nup120, Nup145C, Nup85, Nup84, Seh1, and Sec13. These proteins constitute the outer rings of nuclear pore complexes [1, 2], which are the gatekeepers of nucleo-cytoplasmic transport. The isolation of Nup84 yielded all the seven members of the Nup84 subcomplex [1, 2] and several nuclear pore proteins that were previously identified in isolations of Nup84, such as Nup170, Nic96 [43] and Nup188, Nup192, Nup59, Nup53, Nup57 and Nsp1 [24]. We previously showed that this inter-subcomplex association with members of the Nup170 subcomplex, which is weaker than the intra-subcomplex interactions between the 7 members of the Nup84 subcomplex, is preserved by using short times of incubation with magnetic beads (e.g. 30 minutes or less) [24]. Additionally, we found other members of the nuclear pore complex, including Nup49, Nup82 and Nup159. As demonstrated in a recent detailed study of the architecture of the nuclear pore complex [1, 2], these proteins are present in subcomplexes neighboring the Nup84 subcomplex. Figure 1 illustrates the identified proteins isolated with Nup84-PrA, color coded according to their affiliated subcomplexes (Figure 1 and Supplementary Table S4) [1, 2]. Supplementary Table S4 presents a complete list of the identified proteins, classified according to their affiliation to structural subunits of the nuclear pore complex, or proteins that we and others [25, 44] have observed as common contaminants in isolations from yeast cell extracts. A direct comparison between results obtains from the MS analyses of the same samples using manual and automatic modes of acquisition demonstrated that these experiments can be carried out in a high-throughput manner (Table S4). Similarly, MS/MS analyses performed in automated mode can benefit from the accurate selection of the peaks via a pre-scan of the ion population in the orbitrap. While these experiments provided promising results, we asked how these findings translate in terms of basic instrument performance and seek to characterize the resolving power and mass accuracy in the context of our routine analyses of isolated protein complexes. The ability to resolve and detect peptides within mixtures is critical when using mass fingerprinting for identifying or quantifying peptides and posttranslational modifications. This is equally true when studying isolated protein assemblies, where the complexity and stoichiometry of protein components can present challenges for their detection. Therefore, we asked what the distribution of the orbitrap resolving power over the full mass range is for analyzing mixtures of peptides present at various levels.
Dynamic range of resolution up to m/z 4000–While the MALDI LTQ Orbitrap XL allows spectra acquisition in the orbitrap at a maximum resolving power of 100,000 at m/z 400, we routinely employ a resolution setting of 60,000 at m/z 400 for peptide analysis as a compromise between scanning speed (0.9 sec per scan cycle) [20] and resolving power. For high-throughput analyses, where scanning speed is critical, a lower resolution setting of 30,000 (0.5 sec per scan cycle) may be preferred. As the majority of the MALDI-generated singly-charged peptide ions that we select for protein characterization usually range between m/z 600 and 4000, we performed the assessment of the resolution and its dynamic range over the full mass range (up to m/z 4000). To render this information accurate for the analysis of isolated protein assemblies, we performed these measurements using samples from immunopurified Nup84 protein complex (Figure 1). The measured resolution values were plotted as a function of m/z and dynamic range, the colored heatmap indicating the relative peak intensities (Figure 4 and S3). The obtained data was divided into three categories according to peak intensity, and illustrated separately (Figure 4). Several observations were made from these plots. First, in agreement with the prediction from the theoretical model [16, 45], the experimental resolution values for these complex peptide mixtures were inversely proportional to the square root of m/z (Figure 4C). Second, over the full m/z 200–4,000 range, the vast majority of peaks had resolutions above 20,000 or 10,000 for resolution settings of 60,000 or 30,000 at m/z 400, respectively. For example, at the setting of 60,000 at m/z 400, peaks had resolutions above 40,000 at m/z 1,000, above 30,000 at m/z 2,000, and above 20,000 at m/z 4,000. This enabled a clear determination of peptide isotopic distributions and separation of neighboring peaks in these peptide mixtures. Third, peaks of various intensities, ranging over 4 to 5 orders of magnitude, had resolutions that obey a similar trend (Figure 4A and B). This is consistent with the previously reported 3 orders of magnitude of dynamic range for accuracy for ions below m/z 2000 [46]. An alignment of the resolution pattern for the different peak intensities and examples of spectra are given as supplementary material (Supplemental Fig. S3). Fourth, a few data points deviated from the curve. These seem to partially derive from spectra complexity (overlapping peaks) or background chemical noise.
Fig. 4. The resolving power of MALDI-LTQ Orbitrap XL as a function of m/z and relative peak intensity.
Resolution values measured from MS spectra of the isolated Nup84 samples were scattered against m/z. Results are shown for spectra acquired at resolution settings of 60,000 (panel A) and 30,000 (panel B). The colors of the data points indicate relative peak intensity. Each data set, containing approximately 26,000 measurements, was split according to the peak intensity into —Top 1/3, —Middle 1/3 and —Low 1/3 categories and plotted separately. The peak intensities were drawn to the same scale for all plots. The color bars illustrate the corresponding intensity ranges, while the histograms represent the distribution of the observed peaks. The parameters of the correlations between resolving power and m/z are summarized in Panel C.
Effects of external and internal calibrations on mass accuracy over the full m/z range
Internal calibrations (e.g. polycyclodimethylsiloxane and MRFA peptide) have been reported to improve the accuracy and reduce the calibration drift on the ES LTQ Orbitrap configuration [19, 20]. MALDI mass spectrometers provide an innate ease for implementing internal calibrations due to the constant presence ions corresponding to matrix clusters and trypsin autolysis fragments [43]. We tested external and internal calibrations for improved mass accuracy of measurements of peptides from the isolated Nup84 subcomplex. External calibrations were carried out using two peptide mixtures for the normal (less than m/z 2000) and high (up to m/z 4000) mass ranges. For internal calibration, well characterized trypsin autolysis peptides [47] were used as lock masses. The use of a single lock mass proved sufficient for improved accuracy, as we illustrated for the m/z 2211.104586 trypsin autolysis peak (Figure 5). The mass errors of 109 MS/MS-confirmed peptides from proteins co-isolated with Nup84 (Table S2), with m/z values ranging from approximately 800 to 3700, were used to evaluate the resulting accuracy following external or internal calibrations. The absolute mass error was plotted against m/z (Figure 5A). External calibration resulted in an average mass error and standard deviation of 1.02 ± 0.73 ppm, with nearly 90% (97/109) of measurements having <2 ppm mass errors and 10% falling within the range of 2 – 3.5 ppm. As previously reported for the ES LTQ Orbitrap [20], we observed a long-term stability of mass measurements with external mass calibration, which remains stable within 3 ppm for 3 days (data not shown). Internal calibration using the trypsin autolysis peptide further improved accuracy, leading to a smaller average mass error and a tighter error distribution of 0.50 ± 0.54 ppm (p values < 0.001 and 0.01, respectively). Moreover, the distribution of mass error with external calibration was centered at 1 ppm (Figure 5B), implicating a small systematic error that may be caused by other factors, such as temperature instability [19] and space charge effects [48]. The ability to prevent space charge effects is challenging for MALDI experiments, where the shot-to-shot variation can drastically affect the ion yield [49]. The automatic gain control (AGC), used for controlling ion flux, can reduce space charge effects and was reported to achieve the target range of number of charges for the majority (~ 95%) of data acquisitions [15]. Our results indicated that the internal calibration can compensate for the systematic shift that may be caused by such factors, leading to a distribution of mass accuracy centered at 0 ppm. Although we haven’t tested other internal calibrants, matrix cluster ions may also be a suitable option [47].
Sensitivity using DHB and CHCA matrices
The ability to detect and identify peptides present at various levels within complex mixtures relies on the sensitivity of the instrument. While the sensitivity provided by a ES LTQ Orbitrap configuration was reported at femtomol levels [20, 50], the concentration of the utilized matrix sensitivity will be critical for achieving optimal sensitivity when using a MALDI mass spectrometer [51, 52]. As the use of different matrices for MALDI MS and MS/MS analyses can be advantageous in accommodating various samples, we assessed the optimal concentrations of two matrices that we most commonly use for peptide analysis—CHCA and DHB. Table 2 illustrates the reproducibility of the mass spectrometric signal detected from triplicate analyses of a serial dilution of 0.05 to 25 fmol Angiotensin II in different concentrations of CHCA or DHB matrices. Both matrices provided subfemtomol sensitivity when analyzing this single peptide, with a higher sensitivity of detection for decreased matrix concentrations. These matrix concentrations were optimized for the detection of a single peptide. The comprehensive characterization of complex peptide mixtures, such as those resulting from the isolation of protein complexes, requires longer acquisition times. For such studies, 2 mg/ml for CHCA and 20 mg/ml for DHB provide a balance between sensitivity and slower sample depletion.
Table 2.
Effects of matrix type and concentration on sensitivity of orbitrap detection.
| A. CHCA Concentration vs. Sensitivity | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| CHCA Concentration | Angiotensin II (fmol) | 0.05 | 0.1 | 0.25 | 0.5 | 1.0 | 2.5 | 10 | 25 |
| 3 mg/mL | + | +++ | +++ | +++ | +++ | ||||
| 2 mg/mL | + | +++ | +++ | +++ | +++ | ||||
| 1 mg/mL | + | ++ | ++ | +++ | +++ | +++ | +++ | +++ | |
| 0.5 mg/mL | ++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | |
| B. DHB Concentration vs. Sensitivity | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| DHB Concentration | Angiotensin II (fmol) | 0.05 | 0.1 | 0.25 | 0.5 | 1.0 | 2.5 | 10 | 25 |
| 40 mg/mL | + | + | ++ | ++ | +++ | +++ | +++ | ||
| 20 mg/mL | + | + | ++ | ++ | +++ | +++ | +++ | ||
| 10 mg/mL | ++ | ++ | ++ | ++ | ++ | +++ | +++ | +++ | |
| 5 mg/mL | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | |
Detection levels of Angiotensin II peptide (DRVYIHPF, 1046.5423 m/z) are illustrated for single scan MALDI LTQ Orbitrap FTMS acquisitions using various concentrations of A) CHCA and B) DHB matrices. +, ++ and +++ indicate the reproducibility of the detection from a triplicate experiment.
We next inquired about the sensitivity of detecting phosphorylation sites, an important aspect of characterizing protein complexes and understanding biological processes. To render this information relevant for analyzing peptide mixtures, we selected a synthetic phosphorylated peptide and analyzed it using neutral loss scanning within mixtures of increased complexity. We analyzed a series dilution of the tyrosine phosphorylated SRC peptide RRLIEDAEpYAARG. Following analyses of the pure sample, the peptide was mixed with standard peptides (calibrants), tryptic peptides from BSA or the 100 kDa molecular weight marker to increase spectra complexity. MS2 and MS3 analyses were used to confirm the identity of the phosphorylated peptide (Figure S4). It is noteworthy that the collision-induced dissociation of the phosphorylated SRC peptide, in contrast to the usual loss of the metaphosphoric acid (HPO3, 80 Da) for pTyr-peptides, led to a more abundant [M + H − 98]+ ion (Figure S4). A neutral loss of phosphoric acid (H3PO4, 98 Da) was previously reported for the same phosphotyrosine-containing SRC peptide and shown to be dependent on the presence of the arginine and aspartic acid residues [53]. The proposed mechanism involved a transfer of the phosphate group from tyrosine to aspartic acid, with following cleavage of H3PO4 via succinimide formation [53]. A recent study [54] has investigated this interesting loss of H3PO4 from singly deprotonated phosphotyrosine-peptides during Q-ToF and IT CID, demonstrating that this elimination occurs from an intramolecular relocation of the pTyr phosphate, and not the consecutive losses of HPO3 and water. Our MALDI CID MS2 of the pTyr-peptide was consistent with these previous reports, as the loss of 98 Da was the most prominent fragment detected, the 80 Da loss being close to noise level. Therefore, we selected the ion corresponding to the neutral loss of 98 Da for assessing the sensitivity achieved using neutral loss scanning. Our results showed that this ion was detected following CID fragmentation of as low as 0.5 fmol of SRC peptide (Table 3). A limitation in implementing neutral ion scanning is the lengthy duration of analysis and, consequently, consumption of a large amount of sample. Therefore, as a general practice, when implementing neutral ion scanning in studies of other isolated protein complexes (where phosphorylation played important regulatory roles), we performed these analyses after the initial identification of the proteins present within those samples (data not shown).
Table 3.
Sensitivity of detection of the phosphorylated RRLIEDAEpYAARG (pSRC) peptide using neutral loss scanning.
| Concentration of pSRC peptide (fmol/μL) | 0.25 | 0.5 | 1.0 | 2.5 | 5.0 | 10 | 25 | 50 | 100 |
|---|---|---|---|---|---|---|---|---|---|
| pSRC + 50 fmol/μL calibrants | +++ | ++++ | ++++ | +++ | +++ | ++++ | ++++ | ++++ | |
| pSRC + 50 fmol/μL BSA | ++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | |
| pSRC + 100 kDa marker | +++ | ++++ | +++ | ++++ | ++++ | ++++ | +++ | ++++ |
Detection levels are shown for pSRC peptides as a mixture with 50 fmol calibrant (ProteoMass vMALDI calibrant, Sigma), 50 fmol in-solution digested BSA, and 50 fmol in-gel trypsin digested 100 kDa marker; + symbol indicate the reproducibility from four experiments.
Conclusions
Resolution, accuracy and sensitivity are key features of the performance of a mass spectrometric instrumentation, defining the achievable depth in characterizing complex biological samples. Here, we determined the performance of the MALDI LTQ Orbitrap configuration for analyzing the composition of isolated protein complexes. As illustrated for the isolations of Apl1 and Nup84, the combination of resolving power, accuracy, and sensitivity greatly improved the detection of co-isolated proteins present at various levels, inherent of the complex stoichiometry, or as part of neighboring subcomplexes. Our assessment of the orbitrap resolving power by measuring 20,000 peptides from these isolated protein complexes demonstrated, to our knowledge for the first time using large-scale experimental data up to m/z 4000, that the resolution is a function of the square root of m/z as previously predicted [16, 45], and has a dynamic range of over four orders of magnitude. This dynamic range is valuable for detecting peptides with reduced ionization efficiencies or present at low levels. As the MALDI mass spectrometers are easily amenable to the implementation of internal calibrations [43], we utilized a tryptic autolysis peak as mass lock and showed an improved accuracy to 0.5 ppm absolute mass error with centered mass error distribution. In addition, subfemtomol sensitivity was achieved using both CHCA and DHB matrices in a matrix concentration-dependent manner. These results indicate that the MALDI LTQ Orbitrap is well suited for studies of protein complex composition, reemphasizing the value of MALDI configurations for such analyses.
In terms of analyzing isolated protein complexes, the enhanced sensitivity of the current mass spectrometric methods has led to a lower threshold of detection and the identification of an increased proportion of co-isolated proteins. While this is desired and necessary for comprehensive studies of protein assemblies or cellular compartments, these analyses also highlighted the increasing need for improved methodologies for clean and robust isolations of protein complexes. The isolation of protein complexes is usually accompanied by non-specific associations present at various levels, as well as indirect associations of neighboring complexes, which are increasingly detectable with the current sensitivity (e.g. the immunoaffinity purification of Nup84). The choice of antibodies, resin used for affinity purifications, and time of incubation of the cell lysate with resin can critically affect the levels of contaminations observed. Non-specific associations may not only derive from the direct binding to the resin or antibodies utilized, but also from binding to the isolated protein complex. Purified proteins can act as new interacting sites for numerous proteins present in the cell lysate (e.g. abundant cytoskeletal and ribosomal proteins) during the isolation process. This can interfere with the interpretation of the results in terms of pursuing specific protein function. Incorporation of strategies that result in clean isolations, such as fast (minutes) immunoaffinity purifications [28] and two-step isolations (e.g. [55, 56]), or that allow distinguishing specific and non-specific interactions (e.g. differential labeling with stable isotopes, I-DIRT [44]) are a necessity.
Supplementary Material
Supplementary Fig. S1. Isolating and identifying proteins and their interacting partners. A) Strategy for visualizing and immunoaffinity purifying tagged proteins, together with interacting partners, from yeast or mammalian cells (adapted from Cristea et al. [24]). Confocal fluorescence microscopy is illustrated for Nup84-EGFP in S. cerevisiae. B) Configuration of the MALDI LTQ Orbitrap XL.
Supplementary Fig. S2. MALDI IT CID fragmentation of singly-charged ions using linear trap or orbitrap detection. The CID fragmentation of the Nup84 peptide FWHLVELLLVFR (m/z 1571.910230), derived from the isolation of the yeast Nup84 complex, is illustrated following 1 or 9 scan acquisitions. The results indicate the speed and sensitivity of the linear trap detection versus the accuracy and resolving power obtained following Orbitrap detection.
Supplementary Fig. S3. Summarized distribution of the resolving power as a function of m/z. Data is given for A) 60,000 and B) 30,000 resolution settings (at m/z 400). The heat maps indicate relative peak intensities. Insert panels illustrate examples of observed peptides of various intensity levels.
Supplementary Fig. S4. MALDI LTQ Orbitrap MS, MS2 and MS3 analyses of the pSRC peptide. The [M + H − 98]+ ion was selected for the CID MS3 fragmentation. Consistent with previous reports of the pSRC peptide, the MS2 spectrum illustrates a prominent loss of 98 Da. Although only the y and b ions corresponding to the location of the +80 Da (MS2) and −18 Da (MS3) on the tyrosine residue are shown here, the CID MS2 and MS3 analyses also indicated (at lower levels) the presence of equivalent ions on the aspartic acid residue. The underlying mechanisms require further investigation.
Supplementary Table S1. Experimental conditions used for isolating Apl1 and Nup84 from yeast cells. The optimized parameters, including the amount of cells, time of incubation with magnetic beads and amount of beads used, are indicated.
Supplementary Table S2. List of peptides utilized for assessment of accuracy following external and internal calibrations. The first section (green) summarizes the information regarding the 109 used peptides–protein names, peptide sequences, theoretical masses and the position (sample number) on the gel where the peptides were found. Peptides shown in red have oxidized methionine residues; star * indicates one oxidized and one unmodified methionine; cysteine residues are carbamidomethylated. The second and third sections (blue and red) summarize the observed masses and errors following external or internal calibrations, respectively.
Supplementary Table S3. List of proteins identified in the immunoisolates of EGFP-tagged Apl1 from yeast cells. The detection and confirmation of the co-isolated proteins is indicated, together with information regarding the gi protein number, sequence coverage, number of peptides observed, scores (d′ values from the database search using the XProteo software) obtained following the analysis of the MS data and confirmed peptides sequences following MS/MS analyses. For the proteins that we categorized as likely contaminants, corresponding to proteins that we routinely encounter in isolates from yeast cells, only a limited number of peptides was selected for MS/MS analyses.
Supplementary Table S4. List of proteins identified in the immunoisolates of Protein A-tagged Nup84. Results are shown of two replicate experiments from 2.1 g yeast cells each. The manual and automatic acquisition of MS data was compared and the similarity between the observed number of peptides, sequence coverage and scores (d′ values from the database search using the XProteo software) is illustrated. MALDI LTQ CID MS/MS analyses were performed to confirm the protein identities. The peptide sequences selected and confirmed by MS/MS analyses are indicated; (n/o) - not observed.
Supplementary Table S5. MALDI LTQ Orbitrap MS analysis results for Apl1 and associated proteins using the ProFound search engine.
Acknowledgments
This work was supported by Award Number DP1DA026192 from the National Institute On Drug Abuse and Princeton University Start-up funding to IMC. The authors of the article have no competing financial interests. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute On Drug Abuse or the National Institutes of Health. We are grateful to Rosa Viner (Thermo Fisher Scientific) for software training.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A. Determining the architectures of macromolecular assemblies. Nature. 2007;450(7170):683–94. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 2.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450(7170):695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 3.Selimi F, Cristea IM, Heller E, Chait BT, Heintz N. Proteomic studies of a single CNS synapse type: the parallel fiber/purkinje cell synapse. PLoS Biol. 2009;7(4):e83. doi: 10.1371/journal.pbio.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60(20):2299–301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 5.Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem. 1991;63(24):1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 6.Loboda AV, Krutchinsky AN, Bromirski M, Ens W, Standing KG. A tandem quadrupole/time-of-flight mass spectrometer with a matrix-assisted laser desorption/ionization source: design and performance. Rapid Communications in Mass Spectrometry. 2000;14(12):1047–1057. doi: 10.1002/1097-0231(20000630)14:12<1047::AID-RCM990>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 7.Krutchinsky AN, Zhang W, Chait BT. Rapidly switchable matrix-assisted laser desorption/ionization and electrospray quadrupole-time-of-flight mass spectrometry for protein identification. J Am Soc Mass Spectrom. 2000;11(6):493–504. doi: 10.1016/S1044-0305(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 8.Medzihradszky KF, Campbell JM, Baldwin MA, Falick AM, Juhasz P, Vestal ML, Burlingame AL. The characteristics of peptide collision-induced dissociation using a high-performance MALDI-TOF/TOF tandem mass spectrometer. Anal Chem. 2000;72(3):552–8. doi: 10.1021/ac990809y. [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Ruud J, Chait BT. A practical ion trap mass spectrometer for the analysis of peptides by matrix-assisted laser desorption/ionization. Anal Chem. 1996;68(10):1784–91. doi: 10.1021/ac9511612. [DOI] [PubMed] [Google Scholar]
- 10.Blethrow JD, Tang C, Deng C, Krutchinsky AN. Modular mass spectrometric tool for analysis of composition and phosphorylation of protein complexes. PLoS ONE. 2007;2(4):e358. doi: 10.1371/journal.pone.0000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsaprailis G, Nair H, Somogyi A, Wysocki VH, Zhong WQ, Futrell JH, Summerfield SG, Gaskell SJ. Influence of secondary structure on the fragmentation of protonated peptides. Journal of the American Chemical Society. 1999;121(22):5142–5154. [Google Scholar]
- 12.Schilling B, Wang W, McMurray JS, Medzihradszky KF. Fragmentation and sequencing of cyclic peptides by matrix-assisted laser desorption/ionization post-source decay mass spectrometry. Rapid Commun Mass Spectrom. 1999;13(21):2174–9. doi: 10.1002/(SICI)1097-0231(19991115)13:21<2174::AID-RCM771>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 13.Breci LA, Tabb DL, Yates JR, 3rd, Wysocki VH. Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Anal Chem. 2003;75(9):1963–71. doi: 10.1021/ac026359i. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Krutchinsky AN, Chait BT. “De novo” peptide sequencing by MALDI-quadrupole-ion trap mass spectrometry: a preliminary study. J Am Soc Mass Spectrom. 2003;14(9):1012–21. doi: 10.1016/S1044-0305(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 15.Strupat K, Kovtoun V, Bui H, Viner R, Stafford G, Horning S. MALDI Produced Ions Inspected with a Linear Ion Trap-Orbitrap Hybrid Mass Analyzer. J Am Soc Mass Spectrom. 2009 [Google Scholar]
- 16.Makarov A. Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal Chem. 2000;72(6):1156–62. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- 17.Hardman M, Makarov AA. Interfacing the orbitrap mass analyzer to an electrospray ion source. Anal Chem. 2003;75(7):1699–705. doi: 10.1021/ac0258047. [DOI] [PubMed] [Google Scholar]
- 18.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40(4):430–43. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 19.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4(12):2010–21. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78(7):2113–20. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 21.McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Anal Chem. 2007;79(10):3525–34. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Q, Cooks RG, Noll RJ. Phase-enhanced selective ion ejection in an orbitrap mass spectrometer. J Am Soc Mass Spectrom. 2007;18(6):980–3. doi: 10.1016/j.jasms.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Bahr U, Aygun H, Karas M. Sequencing of single and double stranded RNA oligonucleotides by acid hydrolysis and MALDI mass spectrometry. Anal Chem. 2009;81(8):3173–9. doi: 10.1021/ac900100x. [DOI] [PubMed] [Google Scholar]
- 24.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4(12):1933–41. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Archambault V, Li CX, Tackett AJ, Wasch R, Chait BT, Rout MP, Cross FR. Genetic and biochemical evaluation of the importance of Cdc6 in regulating mitotic exit. Mol Biol Cell. 2003;14(11):4592–604. doi: 10.1091/mbc.E03-06-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krutchinsky AN, Kalkum M, Chait BT. Automatic identification of proteins with a MALDI-quadrupole ion trap mass spectrometer. Anal Chem. 2001;73(21):5066–77. doi: 10.1021/ac010682o. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Chait BT. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal Chem. 2000;72(11):2482–9. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 28.Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem. 2006;281(40):30269–78. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz JC, Syka JP, Quarmby ST. Improving the Fundamentals of MSn on 2D Linear Ion Traps: New Ion Activation and Isolation Techniques. 53rd ASMS Conference on Mass Spectrometry; 2005; San Antonio, Texas. [Google Scholar]
- 30.Griffin TJ, Xie H, Bandhakavi S, Popko J, Mohan A, Carlis JV, Higgins L. iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. J Proteome Res. 2007;6(11):4200–9. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bantscheff M, Boesche M, Eberhard D, Matthieson T, Sweetman G, Kuster B. Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol Cell Proteomics. 2008;7(9):1702–13. doi: 10.1074/mcp.M800029-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4(9):709–12. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 33.Boehm M, Bonifacino JS. Adaptins: the final recount. Mol Biol Cell. 2001;12(10):2907–20. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boehm M, Bonifacino JS. Genetic analyses of adaptin function from yeast to mammals. Gene. 2002;286(2):175–86. doi: 10.1016/s0378-1119(02)00422-5. [DOI] [PubMed] [Google Scholar]
- 35.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 36.Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, Mazur P, Baginsky W, Li W, el-Sherbeini M, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc Natl Acad Sci U S A. 1994;91(26):12907–11. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gelperin D, Weigle J, Nelson K, Roseboom P, Irie K, Matsumoto K, Lemmon S. 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1995;92(25):11539–43. doi: 10.1073/pnas.92.25.11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serrano R, Kielland-Brandt MC, Fink GR. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature. 1986;319(6055):689–93. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- 39.Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84(2):265–75. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 40.Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149(1):41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21(3):387–97. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148(4):635–51. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutzmann M, Kunze R, Stangl K, Stelter P, Toth KF, Bottcher B, Hurt E. Reconstitution of Nup157 and Nup145N into the Nup84 complex. J Biol Chem. 2005;280(18):18442–51. doi: 10.1074/jbc.M412787200. [DOI] [PubMed] [Google Scholar]
- 44.Tackett AJ, DeGrasse JA, Sekedat MD, Oeffinger M, Rout MP, Chait BT. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4(5):1752–6. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- 45.Scigelova M, Makarov A. Orbitrap mass analyzer--overview and applications in proteomics. Proteomics. 2006;6(Suppl 2):16–21. doi: 10.1002/pmic.200600528. [DOI] [PubMed] [Google Scholar]
- 46.Makarov A, Denisov E, Lange O, Horning S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J Am Soc Mass Spectrom. 2006;17(7):977–82. doi: 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Harris WA, Janecki DJ, Reilly JP. Use of matrix clusters and trypsin autolysis fragments as mass calibrants in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2002;16(18):1714–22. doi: 10.1002/rcm.775. [DOI] [PubMed] [Google Scholar]
- 48.Jeffries JB, Barlow SE, Dunn GH. Theory of space-charge shift of ion cyclotron resonance frequencies. International Journal of Mass Spectrometry and Ion Processes. 1983;54(1–2):169–187. [Google Scholar]
- 49.Easterling ML, Mize TH, Amster IJ. Routine Part-per-Million Mass Accuracy for High- Mass Ions: Space-Charge Effects in MALDI FT-ICR. Analytical Chemistry. 1998;71(3):624–632. doi: 10.1021/ac980690d. [DOI] [PubMed] [Google Scholar]
- 50.Macek B, Waanders LF, Olsen JV, Mann M. Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol Cell Proteomics. 2006;5(5):949–58. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Cohen SL, Chait BT. Influence of matrix solution conditions on the MALDI-MS analysis of peptides and proteins. Anal Chem. 1996;68(1):31–7. doi: 10.1021/ac9507956. [DOI] [PubMed] [Google Scholar]
- 52.Laugesen S, Roepstorff P. Combination of two matrices results in improved performance of MALDI MS for peptide mass mapping and protein analysis. J Am Soc Mass Spectrom. 2003;14(9):992–1002. doi: 10.1016/S1044-0305(03)00262-9. [DOI] [PubMed] [Google Scholar]
- 53.Metzger S, Hoffmann R. Studies on the dephosphorylation of phosphotyrosine-containing peptides during post-source decay in matrix-assisted laser desorption/ionization. J Mass Spectrom. 2000;35(10):1165–77. doi: 10.1002/1096-9888(200010)35:10<1165::AID-JMS44>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 54.Edelson-Averbukh M, Shevchenko A, Pipkorn R, Lehmann WD. Gas-phase intramolecular phosphate shift in phosphotyrosine-containing peptide monoanions. Anal Chem. 2009;81(11):4369–81. doi: 10.1021/ac900244e. [DOI] [PubMed] [Google Scholar]
- 55.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17(10):1030–2. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 56.Deng C, Xiong X, Krutchinsky AN. Unifying fluorescence microscopy and mass spectrometry for studying protein complexes in cells. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800397-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Isolating and identifying proteins and their interacting partners. A) Strategy for visualizing and immunoaffinity purifying tagged proteins, together with interacting partners, from yeast or mammalian cells (adapted from Cristea et al. [24]). Confocal fluorescence microscopy is illustrated for Nup84-EGFP in S. cerevisiae. B) Configuration of the MALDI LTQ Orbitrap XL.
Supplementary Fig. S2. MALDI IT CID fragmentation of singly-charged ions using linear trap or orbitrap detection. The CID fragmentation of the Nup84 peptide FWHLVELLLVFR (m/z 1571.910230), derived from the isolation of the yeast Nup84 complex, is illustrated following 1 or 9 scan acquisitions. The results indicate the speed and sensitivity of the linear trap detection versus the accuracy and resolving power obtained following Orbitrap detection.
Supplementary Fig. S3. Summarized distribution of the resolving power as a function of m/z. Data is given for A) 60,000 and B) 30,000 resolution settings (at m/z 400). The heat maps indicate relative peak intensities. Insert panels illustrate examples of observed peptides of various intensity levels.
Supplementary Fig. S4. MALDI LTQ Orbitrap MS, MS2 and MS3 analyses of the pSRC peptide. The [M + H − 98]+ ion was selected for the CID MS3 fragmentation. Consistent with previous reports of the pSRC peptide, the MS2 spectrum illustrates a prominent loss of 98 Da. Although only the y and b ions corresponding to the location of the +80 Da (MS2) and −18 Da (MS3) on the tyrosine residue are shown here, the CID MS2 and MS3 analyses also indicated (at lower levels) the presence of equivalent ions on the aspartic acid residue. The underlying mechanisms require further investigation.
Supplementary Table S1. Experimental conditions used for isolating Apl1 and Nup84 from yeast cells. The optimized parameters, including the amount of cells, time of incubation with magnetic beads and amount of beads used, are indicated.
Supplementary Table S2. List of peptides utilized for assessment of accuracy following external and internal calibrations. The first section (green) summarizes the information regarding the 109 used peptides–protein names, peptide sequences, theoretical masses and the position (sample number) on the gel where the peptides were found. Peptides shown in red have oxidized methionine residues; star * indicates one oxidized and one unmodified methionine; cysteine residues are carbamidomethylated. The second and third sections (blue and red) summarize the observed masses and errors following external or internal calibrations, respectively.
Supplementary Table S3. List of proteins identified in the immunoisolates of EGFP-tagged Apl1 from yeast cells. The detection and confirmation of the co-isolated proteins is indicated, together with information regarding the gi protein number, sequence coverage, number of peptides observed, scores (d′ values from the database search using the XProteo software) obtained following the analysis of the MS data and confirmed peptides sequences following MS/MS analyses. For the proteins that we categorized as likely contaminants, corresponding to proteins that we routinely encounter in isolates from yeast cells, only a limited number of peptides was selected for MS/MS analyses.
Supplementary Table S4. List of proteins identified in the immunoisolates of Protein A-tagged Nup84. Results are shown of two replicate experiments from 2.1 g yeast cells each. The manual and automatic acquisition of MS data was compared and the similarity between the observed number of peptides, sequence coverage and scores (d′ values from the database search using the XProteo software) is illustrated. MALDI LTQ CID MS/MS analyses were performed to confirm the protein identities. The peptide sequences selected and confirmed by MS/MS analyses are indicated; (n/o) - not observed.
Supplementary Table S5. MALDI LTQ Orbitrap MS analysis results for Apl1 and associated proteins using the ProFound search engine.