Abstract
The transition metal nickel plays a central role in the human gastric pathogen Helicobacter pylori because it is required for two enzymes indispensable for colonization, the nickel metalloenzyme urease and [NiFe] hydrogenase. To sustain nickel availability for these metalloenzymes while providing protection from the metal's harmful effects, H. pylori is equipped with several specific nickel-binding proteins. Among these, H. pylori possesses a particular chaperone, HspA, that is a homolog of the highly conserved and essential bacterial heat shock protein GroES. HspA contains a unique His-rich C-terminal extension and was demonstrated to bind nickel in vitro. To investigate the function of this extension in H. pylori, we constructed mutants carrying either a complete deletion or point mutations in critical residues of this domain. All mutants presented a decreased intracellular nickel content measured by inductively coupled plasma mass spectrometry (ICP-MS) and reduced nickel tolerance. While urease activity was unaffected in the mutants, [NiFe] hydrogenase activity was significantly diminished when the C-terminal extension of HspA was mutated. We conclude that H. pylori HspA is involved in intracellular nickel sequestration and detoxification and plays a novel role as a specialized nickel chaperone involved in nickel-dependent maturation of hydrogenase.
Helicobacter pylori is a Gram-negative, microaerophilic bacterium that is the only persistent inhabitant of the human stomach. Its presence in humans is associated with a variety of pathologies, ranging from gastric and duodenal peptic ulcers to the development of gastric adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma (1, 39). Indeed, H. pylori is the only formally recognized bacterial carcinogen for humans (17), infecting half of the world's population (19).
In H. pylori, metal ions play a central role, since the transition metal nickel is the cofactor of the urease enzyme and is also required for [NiFe] hydrogenase. Urease catalyzes the hydrolysis of urea into the buffering compounds bicarbonate and ammonia, enabling H. pylori to persist in the acidic environment of the stomach. This enzyme accounts for up to 6% of the soluble cellular proteins and requires 24 nickel ions per active enzymatic complex (16). The uptake-type hydrogenase of H. pylori is a nickel-dependent enzyme containing a binuclear [NiFe] active site. This [NiFe] hydrogenase catalyzes the oxidation of molecular hydrogen and permits the utilization of hydrogen as an energy source during respiration-based energy production in the mucosa (21). Both enzymes are important for host colonization, as shown with several animal models (9, 10, 28, 42, 43). To sustain nickel availability for urease and hydrogenase while providing protection from the metal's harmful effects, H. pylori possesses an elaborate and strictly controlled nickel metabolism.
The incorporation of nickel ions into apohydrogenase requires the participation of the HypAB (HP0869 and HP0900) accessory proteins; for apourease, both the UreEFGH (HP0070-0067) accessory proteins and HypAB are necessary (4, 29). Besides these widely distributed accessory proteins, H. pylori possesses several specific proteins that are present in all H. pylori strains, namely, the histidine-rich proteins Hpn (HP1427) and Hpn-like (HP1432). These cytoplasmic and abundant proteins (Hpn represents 2% of the total protein content) bind nickel ions (five Ni2+ ions per monomer; dissociation constant [Kd] for nickel of 7.1 μM) and protect H. pylori against metal overload (15). Furthermore, it has recently been proposed that Hpn and Hpn-like can compete for nickel ions with the urease enzyme and thus regulate its enzymatic activity. In vivo and in vitro experiments indicate that Hpn and Hpn-like sequester nickel ions at neutral pH but donate them for urease activation under acidic pH conditions (14, 35, 44). Hydrogenase activity was unchanged in the Δhpn and Δhpn-like mutants (35).
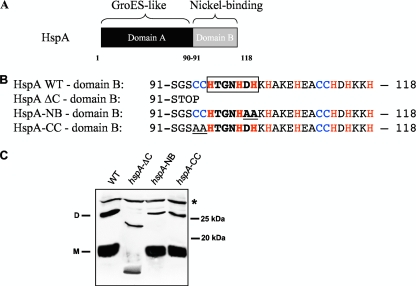
In addition to these proteins, H. pylori possesses a particular chaperone, HspA (HP0011), that is a homolog of the highly conserved and essential bacterial heat shock protein GroES (40). No other gene encoding a GroES homolog is found in the genome of H. pylori. GroES is the cochaperonin of the heptameric GroEL-GroES barrel complex, which mediates the correct folding of a variety of cellular proteins and which is conserved and essential in prokaryotes and eukaryotes (30). In addition to the conserved GroES chaperonin domain (domain A, amino acids 1 to 90) (Fig. 1A), HspA contains a C-terminal extension of 28 amino acids (domain B, amino acids 91 to 118) (Fig. 1A and B) that contains 8 His and 4 Cys residues. Based on this high number of His and Cys residues known to bind transition metal ions, the purified recombinant HspA protein specifically binds two nickel ions per molecule (Kd of 1.1 to 1.8 μM) (7, 18). This domain also contains an HX4DH motif (boxed in Fig. 1B) that is considered to be a nickel-binding signature sequence in the nickel-cobalt (NiCoT) transporter family (11). In addition, Loguercio et al. (20) observed that in vitro, the HspA C-terminal domain is folding into two vicinal disulfide bounds engaging two cysteine pairs that form a unique closed-loop structure. However, since HspA is a cytoplasmic protein, the in vivo relevance of this structure is uncertain.
FIG. 1.
(A) Representation of the HspA protein of H. pylori with the GroES-like domain A and the nickel-binding domain B. (B) Amino acid sequence of domain B of wild-type HspA and of three mutants: HspA-ΔC, with a complete deletion of this domain, and HspA-NB and -CC, each carrying two substitutions that are underlined. Cysteine and histidine residues are in blue and red, respectively. The HX4DH motif, which in the nickel-cobalt (NiCoT) transporter family is considered to be a nickel-binding signature sequence, is boxed. (C) Immunoblot experiment with whole-cell lysates from the H. pylori wild-type strain and from the three hspA mutants after denaturing SDS-PAGE and using the monoclonal antibody P1-1, which specifically recognizes a conserved epitope of HspA domain A. The predicted molecular mass of the wild-type HspA monomer is 13 kDa, and that of HspA-ΔC is 9.8 kDa. The monomeric (M) and dimeric (D) forms of the HspA wild type (WT) are indicated on the left side of the blot. A cross-reacting unspecific protein band is marked with a star (*) and served as a loading control. Molecular mass standards are indicated at right.
The domain B sequence is conserved in and restricted to H. pylori and the closely related Helicobacter acinonychis species but is absent from all other available sequenced Helicobacter species (see Fig. S1 in the supplemental material). When expressed in Escherichia coli, HspA protected bacteria from nickel overload (7) and increased urease activity 4-fold from the coexpressed H. pylori urease gene cluster (18). Therefore, HspA was hypothesized to function in nickel sequestration and as a specialized nickel donor protein for urease (18). However, no functional characterization of the C terminus was carried out for H. pylori due to the essential nature of HspA (40).
In this study, we investigated the role of the nickel-binding C terminus of HspA in H. pylori. We found that the unique C terminus of HspA is involved in nickel sequestration and protection against nickel overload. Contrary to previous data from heterologous studies of E. coli, HspA seemed not to provide nickel ions for urease activation. In contrast, we have found an unexpected and specific function of the HspA C-terminal region in the nickel-dependent maturation of the important colonization factor hydrogenase.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The H. pylori strains employed in this study were 26695 (41) and X47-2AL (12) and their respective mutants. H. pylori was grown on blood agar base 2 (Oxoid) plates supplemented with 10% defibrinated horse blood and an antibiotic/fungicide cocktail consisting of vancomycin (12.5 μg ml−1), polymyxin B (0.31 μg ml−1), trimethoprim (6.25 μg ml−1), and Fungizone (2.5 μg ml−1). Kanamycin (30 μg ml−1) or chloramphenicol (30 μg ml−1) was added for selection when needed. For growth in liquid medium, Brucella broth (Difco) supplemented with 0.2% β-cyclodextrin (Sigma) plus the antibiotic/fungicide cocktail was used; this medium (BBβ) was adjusted to either pH 7 or 5 (by adding HCl). Nickel sensitivity was monitored by following the growth of serial dilutions of bacterial suspensions during 4 to 5 days on Brucella broth agar (1%; Difco) supplemented with 10% fetal calf serum, the antibiotics/fungicide cocktail, and increasing concentrations of NiCl2 (Sigma). Plates and flasks were incubated under microaerobic conditions (Campygen gas pack; Oxoid) at 37°C. Escherichia coli strain MC1061 (6) grown at 37°C on solid or liquid Luria-Bertani medium (25) was used as a recipient for cloning and as a host for the preparation of the plasmids employed to transform H. pylori.
Molecular techniques and construction of H. pylori mutant strains.
DNA manipulations were carried out following standard procedures (32). Chromosomal DNA of H. pylori strain 26695 was used as a template for all PCR amplifications, and the primers are listed in Table S1 in the supplemental material. PCR was carried out according to the manufacturer's recommendations using the Expand High Fidelity DNA polymerase kit (Roche). DNA cassettes to target specific genomic loci of H. pylori were constructed following two strategies. For the hspA-ΔC mutant, the sequence encoding amino acids 1 to 90 of HspA (270 bp) was amplified by PCR with the primers KS1/KS2 and the product was cloned using EcoRI/BamHI into vector pUC18 (New England Biolabs). The antibiotic resistance cassette aphA-3 (37) was amplified with primers KS3/KS4 and inserted after this fragment with BamHI/XbaI. To ensure the native expression of the HspB protein, which is encoded in an operon with hspA, the 3′ sequence downstream of the hspA gene that contains the entire intergenic region sequence between hspA and hspB, as well as hspB sequence 1 to 106 (318 bp), was amplified with primers KS5/KS6 and cloned using XbaI/HindIII downstream of the aphA-3′ cassette. The absence of a polar effect on hspB was verified (see Fig. S2 in the supplemental material). The resulting plasmid was named pILL2201. For the hspA-NB and hspA-CC mutant strains, the hspA-ΔC sequence of pILL2201 was first replaced by the entire hspA wild-type sequence, which was amplified with primers KS1/KS228 and cloned using EcoRI/BamHI. This plasmid was used as a template for the introduction of point mutations using the QuikChange site-directed mutagenesis protocol (Stratagene, Inc.) and the Expand Long template polymerase (Roche). For the hspA-NB mutant, the mutagenic primers used were KS229/KS230, which changed DH (amino acids 101 and 102) into AA. For the hspA-CC mutant, the mutagenic primers used were KS231/KS232, which changed CC (amino acids 94 and 95) into AA. The Δhpn mutant was constructed using a three-step PCR procedure as described in reference 38. In the first step, the H. pylori genomic DNA was used to amplify the 500-bp fragments flanking the target sequence with primer pairs KS57/KS122 and KS123/KS60. In the second step, a chloramphenicol resistance gene (pCM4 Cartridge; Pharmacia) was introduced by PCR between these two fragments. H. pylori mutants were obtained by natural transformation as previously described (5) with approximately 2 μg of either plasmid DNA or the three-step PCR product. Midi Qiagen columns were employed for large-scale plasmid preparations. The hspA-ΔC/Δhpn double mutant strain was constructed by deletion of the hpn gene in the hspA-ΔC mutant strain. Clones that had undergone allelic exchange were selected after 4 to 5 days of growth on plates containing the appropriate antibiotic. Genomic DNA of the recombinant H. pylori mutants was isolated using a QIAamp DNA minikit (Qiagen). All insertions into the chromosome were verified by PCR and sequenced using an ABI 310 automated DNA sequencer (Perkin-Elmer).
Immunoblotting.
Immunoblotting was performed on crude extracts (sonication followed by centrifugation to eliminate cell debris) according to standard protocols. The protein amounts of the crude extracts were calibrated using the Bradford assay (Bio-Rad) with bovine serum albumin (BSA) as a standard. Extracts were migrated in denaturing SDS-PAGE. The H. pylori HspA protein was specifically detected with the monoclonal P1 antibody (I. Kansau and A. Labigne, unpublished data), used at a dilution of 1:100. Horseradish peroxidase (HRP)-conjugated antimouse antibodies were used as secondary antibodies, and detection was achieved using the ECL reagent (Pierce).
Nickel content measurements by inductively coupled plasma mass spectrometry (ICP-MS).
Overnight cultures of H. pylori bacteria were diluted to an optical density at 600 nm (OD600) of 0.5 in fresh BBβ medium without added nickel or with 1 or 10 μM NiCl2, each adjusted to pH 5. Bacteria were incubated for 6 h, and then 6 ml of culture was centrifuged at 400 × g at 4°C for 5 min through 0.3 ml of a 1:2 mixture of the silicone oils AR20/AR200 (SPCI) in order to separate the cells from the medium. Cells were lysed with 400 μl 0.2 M NaOH-1% SDS for 60 min at 90°C. Protein contents were calibrated with standard series of cell cultures using the Bradford assay for protein determination (Bio-Rad). The samples were acidified with ultrapure 65% nitric acid (Normatom quality grade; Prolabo) and diluted in ultrapure water to 2%. Nickel concentrations were measured by ICP-MS using an X7 series quadrupole instrument (Thermo Electron Corporation, Cergy-Pontoise, France). Calibration curves were obtained by analysis of a range of Spex certiPrep nickel standards (Metuchen), and yttrium was used as an internal standard (1 μg liter−1). Measurement of each strain under every condition was performed in triplicates.
Measurement of urease activity in H. pylori strains.
Urease activities were measured as previously described (8, 36) on crude bacterial extracts prepared by lysis with cell lysis buffer (Sigma). One unit (U) of urease activity was defined as the amount of enzyme required to hydrolyze 1 μmol of urea (producing 2 μmol of ammonia) per min per mg of total protein. The amount of ammonia released was determined from a standard curve. The protein concentration was determined using a commercial version of the Bradford assay (Sigma Chemicals) using BSA as a standard.
Measurement of [NiFe] hydrogenase activity in H. pylori strains.
Hydrogen uptake activity was determined spectrophotometrically at 604 nm by following the color change of methyl viologen (MV) from a colorless oxidized form to a dark-violet reduced form. All enzyme assays were performed at 30°C under anaerobic conditions in a glove box to preserve the hydrogenase enzyme from oxygen inhibition. The reaction mixture, containing gas-free 50 mM Tris (pH 8.5) and 1 mM MV in a final volume of 1 ml, was first saturated with gaseous hydrogen (H2) in an assay cuvette. Fifteen ml of H. pylori cultures were harvested by centrifugation at an OD600 of 1.5 and resuspended in 1 ml of 50 mM Tris (pH 8.5). After two cycles of freezing/thawing, 100 μl of cell suspensions were injected into the assay cuvette and the absorbance at 604 nm was followed. Rates of MV reduction were calculated using an absorption coefficient of 13.6 mM−1·cm−1. Protein content in each sample was calibrated using the Bradford assay for protein determination (Bio-Rad). One unit (U) of hydrogenase activity was defined as the amount of enzyme that catalyzes the oxidation of 1 nmol H2 per min per mg of total proteins.
Mouse infection with H. pylori strain X47-2AL.
H. pylori bacteria were grown on blood agar plates for 24 h and suspended in peptone broth at a concentration of 109 bacteria/ml. Female NMRI mice (4 weeks old; Iffa-Credo) were inoculated orogastrically with 108 bacteria (100 μl) as described previously (13). Six mice were inoculated with peptone broth as a negative control. The precise inoculated dose was determined by enumeration of CFU on blood agar plates. Mice were sacrificed after 4 weeks of infection, and the gastric tissue was homogenized in peptone broth. Bacterial infection efficacies were determined by enumeration of CFU on blood agar plates containing the antibiotic/antifungal cocktail, 200 μg/ml bacitracin, and 10 μg/ml nalidixic acid.
RESULTS
Construction of H. pylori hspA mutant strains.
In order to elucidate the specific function of the nickel-binding domain and because HspA is essential (40), we constructed H. pylori mutants that either lacked the C-terminal extension (hspA-ΔC) or contained modifications in the His or Cys residues known to bind nickel. We targeted the following: (i) the predicted His-containing nickel-binding motif (boxed in Fig. 1B) that is considered to be a nickel-binding signature sequence in the nickel-cobalt (NiCoT) transporter family (mutant hspA-NB) or (ii) the first Cys pair (mutant hspA-CC) of the C-terminal domain (Fig. 1B). In the hspA-NB mutant (NB for nickel binding), HX4DH was changed into HX4AA, and in the hspA-CC mutant, the first CC pair was altered into AA (Fig. 1B). With use of a nonpolar kanamycin (Km) resistance cassette (24), the C-terminal extension could be successfully deleted in the sequenced H. pylori strain 26695 (41). This demonstrated that the C-terminal extension of HspA was not essential for growth of H. pylori. The hspA-NB and hspA-CC mutants were obtained by replacing the wild-type 3′ end of hspA with site-specific modified sequences and selection with the above-mentioned nonpolar Km resistance cassette.
To assess the protein synthesis level of HspA in all constructed mutant strains, immunoblot experiments were performed. We used the monoclonal antibody P1-1 (Kansau and Labigne, unpublished), which recognized a conserved GroES epitope of domain A present in both wild-type HspA and the mutant HspA versions. In addition to the monomeric and dimeric HspA forms that were visible on the immunoblot (at 13 kDa and 26 kDa, respectively), the P1-1 antibody recognized an unspecific protein (*) that served as a loading control. We observed that complete deletion of the C-terminal extension led to a 10-fold-decreased amount of the monomeric and dimeric forms of the HspA-ΔC protein, which both run at a lower molecular weight than the wild-type protein (Fig. 1C; see also Fig. S2-A in the supplemental material). This decrease in the protein amount is probably due to a partial loss of protein stability in the absence of the C-terminal domain. Despite the diminished amount of the truncated HspA protein, the growth rate of the hspA-ΔC mutant strain in liquid medium was not affected (see Fig. S3).
In contrast, the expression levels of the mutated HspA-NB and HspA-CC proteins in the respective H. pylori strains did not differ from that of the wild-type strain, indicating that their stability was not affected by the introduced mutations (Fig. 1C).
Intracellular nickel concentration of HspA mutants.
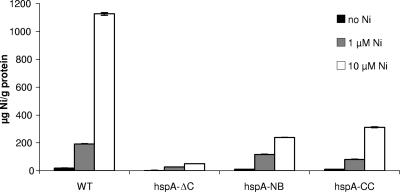
Because HspA was shown to bind nickel ions in vitro and is a highly abundant protein in H. pylori, its role in global nickel homeostasis was first investigated. We analyzed the nickel content of lysed H. pylori wild-type and mutant bacteria that were grown in acidic Brucella broth either without added nickel or with 1 μM or 10 μM NiCl2 by inductively coupled plasma mass spectrometry (ICP-MS) (Fig. 2). Under this condition, H. pylori has been shown to actively acquire nickel ions (33). When grown in the presence of increased concentrations of nickel, wild-type bacteria accumulated this metal up to 1,126.4 (±9.3) μg nickel/g total protein at 10 μM NiCl2.
FIG. 2.
Nickel content of wild-type H. pylori strain 26695 and of the isogenic mutants hspA-ΔC, hspA-NB, and hspA-CC without nickel (black bars) or with increasing NiCl2 concentrations, 1 μM (gray bars) and 10 μM (white bars). The nickel content was measured in cell lysates using ICP-MS and is expressed in μg of nickel per g of proteins. Measurements were performed in triplicate.
In contrast, the three HspA mutant strains presented a strongly diminished nickel content, even in the presence of increasing concentrations of this metal in the growth medium. The hspA-ΔC mutant almost completely lacked the ability to accumulate nickel, whereas the hspA-NB and hspA-CC mutants showed a highly significant decrease in nickel accumulation compared to the wild-type strain. Indeed, the nickel contents of the hspA-NB and hspA-CC mutant strains were 21% and 28% of that of the wild-type strain, respectively. This result indicated that the C terminus of HspA binds nickel and plays an important role in nickel homeostasis in vivo.
Tolerance of the HspA mutants toward nickel.
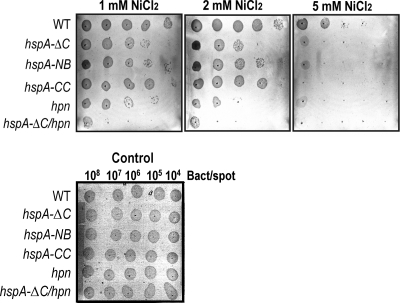
To analyze whether the nickel-binding capacity of HspA may protect H. pylori from nickel toxicity, growth assays on Brucella agar in the presence of high nickel concentrations (1, 2, and 5 mM NiCl2) were performed (Fig. 3). Growth of wild-type bacteria started to be affected at 2 mM NiCl2 supplementation (Fig. 3, middle panel), while growth of the three hspA mutants was already inhibited at 1 mM NiCl2, with a stronger sensitivity of the hspA-ΔC strain than of the hspA-NB and hspA-CC mutants (Fig. 3, left and middle panels). This result was consistent with the nickel content analysis, demonstrating that the C-terminal domain of HspA is important in vivo for resisting nickel overload.
FIG. 3.
Sensitivity to increasing amounts of nickel chloride for the wild-type H. pylori strain 26695 and for different isogenic mutants, hspA-ΔC, hspA-NB, hspA-CC, the Δhpn mutant, and the hspA-ΔC/Δhpn double mutant. Growth of serial dilutions of exponentially growing bacteria (108, 107, 106, 105, or 104 bacteria per spot) was tested on Brucella broth agar plates with 10% fetal calf serum that contained increasing toxic nickel concentrations (1, 2, and 5 mM NiCl2) or no added metal as a control. Growth was followed for 4 days; the data are from an individual assay but are representative of three independent experiments.
Because the Hpn protein was reported to protect from nickel overload, we additionally analyzed nickel tolerance in H. pylori strains deficient in hpn or in an hspA-ΔC/Δhpn double mutant. We constructed a Δhpn mutant by replacement of the gene with a nonpolar chloramphenicol (Cm) resistance cassette. The double hspA-ΔC/Δhpn mutant was constructed by the interruption of the hpn gene in the hspA-ΔC mutant strain. In the Δhpn mutant, growth inhibition by nickel was more important than that of the hspA-ΔC mutant. The most pronounced phenotype of nickel sensitivity was observed with the hspA-ΔC/Δhpn double mutant (Fig. 3, left panel). Thus, HspA and Hpn are both required for nickel detoxification in H. pylori, and their functions are overlapping and partially redundant.
Role of HspA in urease and hydrogenase enzymatic activities.
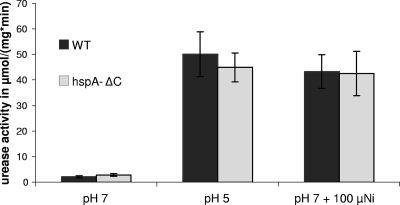
The C-terminal extension of HspA was shown to increase urease activity in E. coli strains expressing both H. pylori hspA and urease genes (18). Therefore, we assayed urease activity in the H. pylori hspA-ΔC mutant under various conditions. Unexpectedly, urease activity of the hspA-ΔC mutant did not differ significantly from that of the wild-type strain at neutral pH or under nickel-replete (100 μM NiCl2) and acidic conditions (pH 5), under which urease is activated (Fig. 4). This result indicated that at least under these conditions, HspA is not involved in nickel incorporation into urease.
FIG. 4.
Urease activity of wild-type 26695 H. pylori strain (black bars) and its isogenic hspA-ΔC mutant (gray bars) grown in liquid Brucella broth at pH 7 or pH 5 in unsupplemented medium or medium with added NiCl2 (100 μM). Error bars represent the standard deviations for three to four independent cultures.
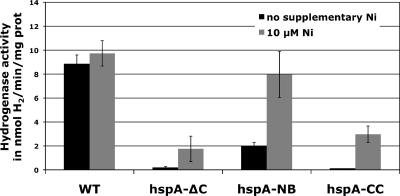
We measured [NiFe] hydrogenase uptake activity in H. pylori bacteria grown at pH 7 (Fig. 5). The wild-type strain presented hydrogenase activity of 8.8 (±0.8) U/mg of protein, which was comparable to previous results (28). Very interestingly, all hspA mutants showed strongly diminished hydrogenase activity. While the hspA-ΔC and hpsA-CC mutants presented marginal activities at the detection limit of the assay of 0.1 U/mg protein, hydrogenase activity of the hspA-NB mutant amounted to 22% of the wild-type value. Thus, lack of or mutations in the C terminus of HspA strongly affected the activity of H. pylori hydrogenase.
FIG. 5.
Hydrogenase uptake activity of wild-type H. pylori strain 26695 and of isogenic mutants hspA-ΔC, hspA-NB, and hspA-CC without nickel (black bars) or with 10 μM nickel (gray bars). One unit (U) of hydrogenase activity was defined as the amount of enzyme that catalyzes the oxidation of 1 nmol H2 per min per mg of total proteins. Error bars represent the standard deviations for at least three independent cultures.
To analyze whether the effect of mutations in the C-terminal extension of HspA can be compensated by the addition of nickel, we grew the wild-type strain and HspA mutants in medium supplemented with 10 μM NiCl2. It was previously shown that mutations in hydrogenase accessory genes are suppressed by high nickel concentrations in the growth medium. Hydrogenase activity of the wild-type H. pylori strain did not significantly increase in the presence of 10 μM NiCl2 (Fig. 5), as reported previously (28). Interestingly, when hspA mutant strains were grown in the presence of 10 μM nickel, hydrogenase activity of the hspA-NB mutant increased to wild-type levels whereas the hydrogenase activity of the hspA-ΔC and hspA-CC strains showed only partial restoration. Together these results demonstrated that HspA was required for hydrogenase activity in H. pylori. Furthermore, the nickel-binding motif is required to sustain full hydrogenase activity under nickel-restricted conditions but could be compensated by the addition of nickel.
Colonization capacity of the hspA-ΔC mutant.
Finally, we tested whether the C-terminal extension of HspA might play a critical role in vivo by assessing the mouse colonization capacity. Because strain 26695 does not colonize mice, we have constructed an hspA-ΔC mutant in the mouse adapted strain X47-2AL. We did not find any difference between the phenotypes of 26695 hspA-ΔC and X47-2AL hspA-ΔC (data not shown). Eight NMRI female mice were infected with 108 bacteria per mouse. The colonization rate was analyzed 4 weeks postinfection. Colonization by the X47-2AL hspA-ΔC mutant was similar to that of the parental strain (see Fig. S4 in the supplemental material). This showed that the lack of the C-terminal extension of HspA, as well as a lower protein expression level, did not prevent in vivo colonization of the mouse model.
DISCUSSION
In this study, we have analyzed the function of the unique C-terminal nickel-binding domain of HspA in H. pylori. We found that the C terminus played a major role in nickel sequestration and nickel detoxification and was required for hydrogenase activity.
In vivo nickel-binding activity of HspA.
We found that the nickel-binding domain of HspA binds nickel ions in vivo and provides H. pylori with a protective mechanism against nickel overload. A good correlation between the decrease in the intracellular nickel content of the three hspA mutants and their nickel sensitivity was observed. The hspA-ΔC mutant strain sequestered less nickel and was most sensitive to nickel ions (Fig. 3), whereas the hspA-CC and hspA-NB mutants showed intermediate phenotypes. The stronger sensitivity of the hspA-ΔC mutant likely resulted from diminished amounts of the HspA protein (Fig. 1C). Together, these results indicated that we have identified residues that are crucial for nickel binding in the HspA C-terminal extension that allow H. pylori HspA to sequester nickel ions upon nickel overload.
Estimations from 63Ni2+ uptake experiments suggest that H. pylori accumulates about 50 times more nickel than E. coli (2) due to the unique TonB-dependent nickel uptake system through the outer membrane protein FrpB4 (33, 34) and the high-affinity nickel transporter NixA (26). Thus, the presence of a nickel-binding tail on the highly abundant and constantly expressed GroES proteins could enable H. pylori to balance efficient nickel uptake and to deal with the constant intracellular presence of nickel ions. We have found that nickel binding by HspA has a role similar to the nickel sequestration function of the Hpn protein (14, 15, 27, 35). However, the hpn deletion strain showed a greater sensitivity to nickel overload, in agreement with the fact that Hpn binds 2 to 3 more nickel ions per monomer than HspA, having-binding affinities of the same range. Remarkably, the functions of HspA and Hpn in protection from nickel overload are overlapping and partly redundant, since the lack of these two sinks for nickel ions strongly reduced nickel tolerance.
Nickel incorporation into urease and [NiFe] hydrogenase.
Initially, it was proposed that HspA is a specialized nickel-binding protein that is needed to saturate the huge amounts of the urease enzyme with its cofactor nickel (18). However, the H. pylori hspA-ΔC mutant did not exhibit decreased urease activity under various conditions tested, rendering this function of HspA unlikely. We think that the enhanced urease activity observed in E. coli (18) could result from a total increase in cellular nickel content and a lack of proteins competing for nickel in this heterologous system. In E. coli, urease activity was found to increase linearly with the nickel concentration (26). In contrast, the nickel-binding residues of HspA were required for [NiFe] hydrogenase activity. Previously it was reported that the GroES/GroEL complex homolog of E. coli was involved in the nickel-dependent processing of [NiFe] hydrogenases of E. coli (31). Total hydrogenase activity was found to be reduced to 60% in groES and groEL thermosensitive E. coli mutant strains, indicating that the GroES/GroEL chaperonin assisted the precursor of hydrogenase in reaching a conformation competent for nickel insertion (31). In H. pylori, HspA could have a similar role as a hydrogenase chaperone; however, our data point to an additional specific function of the nickel-binding C-terminal domain in nickel delivery to hydrogenase. Indeed, strongly diminished hydrogenase activity was measured in H. pylori strains expressing the mutant HspA-CC and HspA-NB versions that did not reveal protein instability. Moreover, nickel supplementation increased hydrogenase activity of the hspA-ΔC and hspA-CC mutants and restored hydrogenase activity to wild-type levels in the mutant hspA-NB. Thus, we propose that the C-terminal nickel-binding domain of HspA is involved in nickel-dependent maturation of hydrogenase, probably playing a role in delivery of nickel to this enzyme. The fact that the hspA-CC mutant has a behavior similar to that of the hspA-ΔC strain suggests that the first cysteine pair is in addition critical for the HspA chaperone function.
The complex maturation process of hydrogenase is poorly studied for H. pylori. Only the HypA (HP0869) and HypB (HP0900) accessory proteins that are involved in nickel incorporation have been characterized for H. pylori (22). H. pylori possesses a gene encoding an ortholog of SlyD, the nickel-binding peptidyl-prolyl cis-trans isomerase, involved in hydrogenase maturation in E. coli; however, its function in H. pylori has not been addressed so far. Our finding that the C-terminal extension of the GroES homologue HspA is required for hydrogenase activity identifies a novel hydrogenase accessory protein that is specific to H. pylori species. In Bradyrhizobium japonicum and Rhizobium leguminosarum, HypB contains a histidine-rich tail domain that is thought to function in nickel delivery. However, H. pylori HypB does not contain this extension and does not bind nickel ions. We propose that HspA might compensate for the absence of such a nickel-binding domain in the H. pylori HypB protein. Thus, the HspA protein of H. pylori could be a very nice example of how evolution has selected a highly specialized protein that possesses both chaperone and nickel delivery functions, allowing an optimization of hydrogenase maturation.
A unique feature of H. pylori is the interconnectivity of the maturation process of hydrogenase and urease. H. pylori mutants that are deficient in HypA or HypB lacked hydrogenase activity and presented diminished urease activity (23, 29). In contrast, hydrogenase activity is not modified by urease accessory proteins, since deletion of ureE and ureG has no effect on hydrogenase activity (3, 23). HypA and UreE were found to interact in vitro in a biochemical protein cross-link assay (4). Recently we found that the urease accessory protein UreG physically interacts with the hydrogenase accessory protein HypB in vivo (38). However, despite this interconnectivity, it seems that different nickel-binding proteins of H. pylori specifically sustain nickel incorporation into either urease or hydrogenase. Hpn and Hpn-like proteins have been shown to act as nickel reservoirs that under certain conditions supply this metal to urease but not to hydrogenase (3, 23, 35). In the present study, we demonstrated that the nickel-binding domain of HspA is required for hydrogenase activation and not for urease activity. This indicates that the nickel homeostasis in H. pylori is complex and precisely controlled.
Colonization experiments using the mouse model demonstrate that the hspA-ΔC mutant has the same colonization ability as a wild-type strain. Deletion of the hydrogenase structural genes led to decreased colonization ability; however, several H. pylori bacteria were recovered from stomachs of infected mice (28). Thus, the partial hydrogenase activity measured for the hspA mutant strains seems to be sufficient for the colonization of the mouse model. Alternatively, the known strain variability of H. pylori (X47-AL in this study and SS1 in reference 28) might account for the observed differences.
Conclusion.
We conclude that the C-terminal domain of HspA is involved in intracellular nickel sequestration and detoxification. In addition, HspA plays a critical role as a specialized nickel chaperone in achieving hydrogenase maturation. Because nickel ions play a crucial role during H. pylori colonization but seem not to be utilized by humans, the nickel metabolism of H. pylori is an attractive therapeutic target, particularly in the age of increasing microbial antibiotic resistance. Our study thus provides key insights for the development of alternative antimicrobials.
Supplementary Material
Acknowledgments
We are grateful to Barbara Gouget, Clarisse Mariet, and Francine Carrot for their help with the ICP-MS analyses. We thank Jean-Michel Thiberge for excellent technical assistance with the mouse experiment. Kerstin Stingl is gratefully acknowledged for many useful comments and critical reading of the manuscript and Laurent Terradot for constructive suggestions.
K.S. received financial support from the German Academic Exchange Service (DAAD) and the European Network of Excellence PathoGenomics.
Footnotes
Published ahead of print on 8 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Atherton, J. C. 2006. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. Mech. Dis. 1:63-96. [DOI] [PubMed] [Google Scholar]
- 2.Bauerfeind, P., R. M. Garner, and L. T. Mobley. 1996. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect. Immun. 64:2877-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit, S., and R. J. Maier. 2003. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J. Bacteriol. 185:4787-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benoit, S. L., N. Mehta, M. V. Weinberg, C. Maier, and R. J. Maier. 2007. Interaction between the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation. Microbiology 153:1474-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bury-Moné, S., S. Skouloubris, C. Dauga, J. M. Thiberge, D. Dailidiene, D. E. Berg, A. Labigne, and H. De Reuse. 2003. Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect. Immun. 71:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadaban, M., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusions and cloning in E. coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 7.Cun, S., H. Li, R. Ge, M. C. Lin, and H. Sun. 2008. A histidine-rich and cysteine-rich metal-binding domain at the C terminus of heat shock protein A from Helicobacter pylori: implication for nickel homeostasis and bismuth susceptibility. J. Biol. Chem. 283:15142-15151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cussac, V., R. L. Ferrero, and A. Labigne. 1992. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 174:2466-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton, K. A., J. V. Gilbert, E. A. Joyce, A. E. Wanken, T. Thevenot, P. Baker, A. Plaut, and A. Wright. 2002. In vivo complementation of ureB restores the ability of Helicobacter pylori to colonize. Infect. Immun. 70:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton, K. A., and S. Krakowka. 1994. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 62:3604-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eitinger, T., J. Suhr, L. Moore, and A. C. Smith. 2005. Secondary transporters for nickel and cobalt ions: themes and variations. BioMetals 18:399-405. [DOI] [PubMed] [Google Scholar]
- 12.Ermak, T. H., P. J. Giannasca, R. Nichols, G. A. Myers, J. Nedrud, R. Weltzin, C. K. Lee, H. Kleanthous, and T. P. Monath. 1998. Immunization of mice with urease vaccine affords protection against Helicobacter pylori infection in the absence of antibodies and is mediated by MHC class II-restricted response. J. Exp. Med. 188:2277-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrero, R. L., J.-M. Thiberge, M. Huerre, and A. Labigne. 1998. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect. Immun. 66:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge, R., R. M. Watt, X. Sun, J. A. Tanner, Q.-Y. He, J.-D. Huang, and H. Sun. 2006. Expression and characterization of a histidine-rich protein, Hpn: potential for Ni2+ storage in Helicobacter pylori. Biochem. J. 393:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, J., J. Ramakrishna, F. Sunderman, Jr., A. Wright, and A. Plaut. 1995. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect. Immun. 63:2682-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha, N.-C., S.-T. Oh, J. Y. Sung, K. A. Cha, M. H. Lee, and B.-H. Oh. 2001. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat. Struct. Biol. 8:505-509. [DOI] [PubMed] [Google Scholar]
- 17.International Agency for Research on Cancer. 1994. Working Group on the Evaluation of Carcinogenic Risks to Humans. Helicobacter pylori. IARC Monogr. Eval. Carcinog. Risks Hum. 61:177-240. [Google Scholar]
- 18.Kansau, I., F. Guillain, J.-M. Thiberge, and A. Labigne. 1996. Nickel binding and immunological properties of the C-terminal domain of the Helicobacter pylori GroES homologue (HspA). Mol. Microbiol. 22:1013-1023. [DOI] [PubMed] [Google Scholar]
- 19.Kusters, J. G., A. H. van Vliet, and E. J. Kuipers. 2006. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19:449-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loguercio, S., C. Dian, A. Flagiello, A. Scannella, P. Pucci, L. Terradot, and A. Zagari. 2008. In HspA from Helicobacter pylori vicinal disulfide bridges are a key determinant of domain B structure. FEBS Lett. 582:3537-3541. [DOI] [PubMed] [Google Scholar]
- 21.Maier, R. J., C. Fu, J. Gilbert, F. Moshiri, J. Olson, and A. G. Plaut. 1996. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 141:71-76. [DOI] [PubMed] [Google Scholar]
- 22.Mehta, N., S. Benoit, and R. J. Maier. 2003. Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb. Pathog. 35:229-234. [DOI] [PubMed] [Google Scholar]
- 23.Mehta, N., J. W. Olson, and R. J. Maier. 2003. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 185:726-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Mobley, H. L., R. M. Garner, and P. Bauerfeind. 1995. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol. Microbiol. 16:97-109. [DOI] [PubMed] [Google Scholar]
- 27.Mobley, H. L., R. M. Garner, G. R. Chippendale, J. V. Gilbert, A. V. Kane, and A. G. Plaut. 1999. Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter 4:162-169. [DOI] [PubMed] [Google Scholar]
- 28.Olson, J. W., and R. J. Maier. 2002. Molecular hydrogen as an energy source for Helicobacter pylori. Science 298:1788-1790. [DOI] [PubMed] [Google Scholar]
- 29.Olson, J. W., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 30.Radford, S. E. 2006. GroEL: more than Just a folding cage. Cell 125:831-833. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigue, A., N. Batia, M. Muller, O. Fayet, R. Bohm, M. A. Mandrand-Berthelot, and L. F. Wu. 1996. Involvement of the GroE chaperonins in the nickel-dependent anaerobic biosynthesis of NiFe-hydrogenases of Escherichia coli. J. Bacteriol. 178:4453-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Schauer, K., B. Gouget, M. Carriere, A. Labigne, and H. de Reuse. 2007. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63:1054-1068. [DOI] [PubMed] [Google Scholar]
- 34.Schauer, K., D. A. Rodionov, and H. de Reuse. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33:330-338. [DOI] [PubMed] [Google Scholar]
- 35.Seshadri, S., S. L. Benoit, and R. J. Maier. 2007. Roles of His-rich Hpn and Hpn-like proteins in Helicobacter pylori nickel physiology. J. Bacteriol. 189:4120-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skouloubris, S., A. Labigne, and H. De Reuse. 1997. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol. Microbiol. 25:989-998. [DOI] [PubMed] [Google Scholar]
- 37.Skouloubris, S., J.-M. Thiberge, A. Labigne, and H. De Reuse. 1998. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 66:4517-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stingl, K., K. Schauer, C. Ecobichon, A. Labigne, P. Lenormand, J. C. Rousselle, A. Namane, and H. de Reuse. 2008. In vivo interactome of Helicobacter pylori urease revealed by tandem affinity purification. Mol. Cell Proteomics 7:2429-2441. [DOI] [PubMed] [Google Scholar]
- 39.Suerbaum, S., and C. Josenhans. 2007. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 40.Suerbaum, S., J. M. Thiberge, I. Kansau, R. L. Ferrero, and A. Labigne. 1994. Helicobacter pylori hspA-hspB heat-shock gene cluster: nucleotide sequence, expression, putative function and immunogenicity. Mol. Microbiol. 14:959-974. [DOI] [PubMed] [Google Scholar]
- 41.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 42.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wirth, H. P., M. H. Beins, M. Yang, K. T. Tham, and M. J. Blaser. 1998. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 66:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng, Y. B., D. M. Zhang, H. Li, and H. Sun. 2008. Binding of Ni(2+) to a histidine- and glutamine-rich protein, Hpn-like. J. Biol. Inorg. Chem. 13:1121-1131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.