Abstract
Glycine sarcosine methyltransferase (Gsm) and sarcosine dimethylglycine methyltransferase (Sdm) catalyze glycine betaine synthesis from glycine. Disruption of the M. xanthus gsmA (MXAN 7068) or sdmA (MXAN 3190) gene, encoding Gsm or Sdm homologue proteins, respectively, generated mutants that exhibited a longer lag period of growth and delayed spore germination under osmostress.
Myxococcus xanthus is a Gram-negative bacterium that exhibits a complex multicellular developmental cycle (6, 7). These bacteria live in soil, where they prey on other microbes for food. In response to nutritional stress, hundreds of thousands of vegetative cells aggregate to form multicellular fruiting bodies containing differentiated myxospores. Once conditions become favorable for growth, the desiccation- and heat-resistant spores can germinate and initiate vegetative growth.
It was reported previously that the receptor-type adenylyl cyclases CyaA and CyaB of M. xanthus act as osmosensors during spore germination and growth, respectively (8, 9). Glycine betaine is a very efficient osmolyte found in a wide range of prokaryotic and eukaryotic organisms, where it is accumulated at high cytoplasmic concentrations in response to osmotic stress (4, 16). In this study, it is reported that in M. xanthus glycine betaine can be biosynthesized from glycine and mainly functions as an osmoprotectant for cell growth and spore germination under osmotic stress conditions.
Genes encoding enzymes involved in glycine betaine synthesis in M. xanthus.
In many bacteria, plants, and animals, glycine betaine is synthesized from choline in two steps: (i) choline to betaine aldehyde and (ii) betaine aldehyde to glycine betaine. Choline dehydrogenase (Cdh) or choline oxidase (Cox) catalyzes the first step in bacteria and animals, and the second step is catalyzed by NAD+-dependent betaine aldehyde dehydrogenase (Badh) (10, 18). The M. xanthus genome database at The Institute for Genomic Research (TIGR) was searched for these enzymes using the BlastP program. The M. xanthus proteins MXAN 0138, 1504, 3822, 3925, and 5165 belong to a family of glucose-methanol-choline (Gmc) oxidoreductases that showed approximately 25 to 27% identity with Cdh or Cox from bacteria, and various aldehyde dehydrogenases (e.g., MXAN 0121, 0607, 0921, 2326, 5040, and 6986) of M. xanthus showed approximately 30 to 40% identity with Badh from bacteria. Because many homologues involved in the synthesis of glycine betaine from choline were identified by BlastP homology searches, it was difficult to determine which enzymes catalyze the two-step reactions.
On the other hand, several halophilic bacteria synthesize glycine betaine from simple carbon sources (13, 14). In these bacteria, glycine betaine is synthesized from glycine through three steps of methylation reactions with S-adenosylmethionine as a methyl donor; sarcosine and dimethylglycine serve as the intermediates. The three-step methylation reactions are catalyzed by glycine sarcosine methyltransferase (Gsm) and sarcosine dimethylglycine methyltransferase (Sdm). Gsm catalyzes the methylation of glycine or sarcosine to form sarcosine or dimethylglycine, and Sdm catalyzes the methylation of sarcosine or dimethylglycine to dimethylglycine or glycine betaine, respectively. The M. xanthus genome sequence database was also searched with the BlastP program for genes that encode homologues of glycine sarcosine dimethylglycine methyltransferase (Gsdm) from Actinopolyspora halophila and Gsm or Sdm from Halorhodospira halochloris (13), and at least two homologues (MXAN 7068, designated GsmA, and MXAN 3190, designated SdmA) of Gsm or Sdm were found. GsmA and SdmA consisted of 268 and 256 amino acids with calculated molecular masses of 29.5 and 27.9 kDa, respectively. GsmA shared 27% and 25% identity with Gsm from H. halochloris and Gsdm from A. halophila, respectively, and SdmA shared 25% and 26% identity with Sdm from H. halochloris and Gsdm from A. halophila, respectively (Fig. 1). Putative S-adenosylmethionine binding motifs existed in the N-terminal regions of M. xanthus GsmA and SdmA.
FIG. 1.
(A) Alignment of the deduced amino acid sequences of M. xanthus GsmA (Mxa) with H. halochloris Gsm (Hha) and A. halophila Gsdm (Aha). (B) Alignment of the deduced amino acid sequences of M. xanthus SdmA (Mxa) with H. halochloris Sdm (Hha) and A. halophila Gsdm (Aha). Identical amino acid residues are indicated by asterisks. Putative S-adenosylmethionine binding motifs are overlined.
Phenotypes of gsmA and sdmA mutants. (i) Construction of mutants.
To examine the role of glycine betaine synthesized from glycine in M. xanthus, the chromosomal gsmA or sdmA genes were disrupted by insertion of a 1.2-kb fragment containing a kanamycin resistance gene. The oligonucleotide primers gsmA1 (5′-CCGCCATGCTCTGGTCCGACACCG-3′) and gsmA2 (5′-ATGAACAGCTCGCTGCAGGGGGGC-3′) plus sdmA1 (5′-ACCACGAACTGCTGGACCTGGGGC-3′) and sdmA2 (5′-TCATGCAGTTGCGACTGGGGCAGG-3′) were used to amplify the gsmA and sdmA genes, respectively, from the M. xanthus genome. The PCR products were ligated into a pT7BlueT vector (Takara Bio) to construct pT7gsmA and pT7sdmA, respectively. A 1.2-kb DNA fragment containing a kanamycin resistance (Kmr) gene was amplified by PCR with TnV (3) as a template and a pair of primers, 5′-GTGCTGACCCCGGGTGAATGTCAG-3′ and 5′-ATCGAGCCCGGGGTGGGCGAAGAA-3′. The resulting DNA fragment was inserted into the EheI-PmaCI sites of pT7gsmA and the SmaI-EheI sites of pT7sdmA, respectively. The disrupted genes constructed as described above were amplified by PCR using the above-mentioned oligonucleotides. The PCR products thus obtained were introduced into M. xanthus cells by electroporation, which was performed as described by Plamann et al. (15). M. xanthus kanamycin-resistant colonies were grown in Casitone-yeast extract (CYE) medium containing 100 μg of kanamycin/ml, and chromosomal DNAs were prepared from the mutants. Using PCR and restriction enzyme analyses, we confirmed that the kanamycin resistance gene was inserted into the gsmA or sdmA gene on the chromosomes of M. xanthus mutants in the same orientation. From the M. xanthus chromosome map, the gsmA or sdmA gene seems to form an operon with one (MXAN 7069) or two (MXAN 3188 and 3189) genes, respectively, and the gsmA or sdmA gene is the last gene in the operon, suggesting that mutations by insertion of the kanamycin resistance gene into the gsmA or sdmA gene would not affect the transcription of downstream genes.
(ii) Cell growth and fruiting body formation under osmotic stress.
gsmA and sdmA mutants showed normal growth in CYE medium at 30°C. The phenotypic differences between the wild-type and mutant strains under osmotic stress were investigated. When inoculated at 2 × 106 cells/ml and grown in CYE medium containing 0.2 M NaCl or 0.2 M sucrose at 30°C, wild-type cells initiated growth after a lag time of 85 h or 66 h, respectively. gsmA and sdmA mutants under osmotic stress conditions exhibited longer lag periods of about 5 to 12 h compared to the wild type (data not shown). gsmA and sdmA mutants cultured in CYE medium containing 0.2 M NaCl or 0.2 M sucrose had lag times of 92 h and 97 h, and 71 h and 73 h, respectively. No significant differences were found between the final cell densities and maximum growth rates of the wild-type and mutant strains under osmotic stress.
gsmA and sdmA gene expression was determined by reverse transcriptase PCR (RT-PCR). M. xanthus cells grown in CYE medium with or without 0.2 M NaCl were harvested, and total RNA was extracted from these cells with a urea buffer and purified as previously described (11). Contaminating DNA was removed by digestion of DNase (Promega), and cDNA was synthesized with BcaBEST polymerase (Takara Bio). PCR was performed with PrimeStar GXL DNA polymerase (Takara Bio), gsmA primers (CCATGCCACCGATTTGAGC and CGTCAGCAGATGCGTCACC) or sdmA primers (TTCAACGGCATCCTCATCC and GAGGCCTTGGGAGCAAAGC), and the synthesized cDNA. As shown in Fig. 2, the expected 172-bp and 140-bp RT-PCR products for the gsmA and sdmA genes, respectively, were amplified from RNA of M. xanthus cells under osmotic and nonosmotic stress conditions. The expected product was not amplified without a reverse transcriptase (data not shown).
FIG. 2.
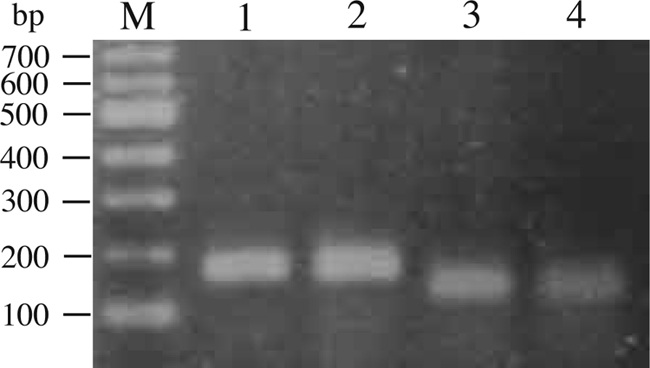
RT-PCR analysis of gsmA (lanes 1 and 2) and sdmA (lanes 3 and 4) gene expression in M. xanthus. RT-PCR analysis was performed on RNA prepared from cultures grown in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 0.2 M NaCl for 4 h. The molecular sizes of DNA fragments are given in base pairs (lane M).
Glycine betaine also acts as a thermoprotectant for Escherichia coli and Bacillus subtilis cells (2, 5). Incubation of gsmA or sdmA mutant cells at high temperature (37°C) showed growth similar to that of wild-type cells (data not shown). When developed on clone fruiting (CF) agar, both mutants showed normal developmental processes. There were no significant differences in numbers of fruiting bodies on CF containing 0.15 M NaCl or 0.15 M sucrose between the wild type and mutants.
For the determination of intracellular glycine betaine concentrations, cells were harvested by centrifugation, washed twice with distilled water, and disrupted by boiling them for 5 min. The extracts were analyzed for glycine betaine by high-performance liquid chromatography (HPLC) using an ODS-80Ts column (Tosoh) as described previously (1). Glycine betaine was synthesized and accumulated in wild-type vegetative cells in response to osmotic stress. As shown in Fig. 3A, the glycine betaine levels of wild-type cells under nonosmotic stress conditions was 19 nmol/mg protein, and the glycine betaine levels of wild-type cells increased 5- to 7-fold with 5.5 h of incubation in the presence of 0.2 M NaCl or 0.2 M sucrose at 30°C. In gsmA or sdmA mutant cells, glycine betaine accumulation increased during the first 1.2 h of incubation, but a remarkable increase in the glycine betaine concentration was not seen after that. After 5.5 h of incubation under osmotic stress, the levels of glycine betaine in mutant cells were about 4- to 10-fold lower than in wild-type cells. The results indicated that glycine betaine is synthesized from glycine in response to osmotic stress and functions as an osmoprotectant in M. xanthus vegetative cells. M. xanthus also possesses other Gsm homologues (e.g., MXAN 6164 and 3033) and Sdm homologues (e.g., MXAN 0096 and 2601). These proteins or Cdh, Cox, or Badh homologuess may be involved in glycine betaine synthesis because glycine betaine accumulation was detected in gsmA or sdmA mutants. Trehalose is also known as a general osmoprotectant in many organisms. In M. xanthus, trehalose is synthesized and accumulated in vegetative cells in response to osmotic stress (12, 17). In addition, myxospores accumulate large amounts of trehalose (3.2 μmol/mg of protein) during fruiting body formation under normal conditions (12). gsmA and sdmA mutants under osmotic stress may be able to grow after a slightly prolonged lag period relative to the wild type and form fruiting bodies and spores like the wild type because trehalose is synthesized in both mutants.
FIG. 3.
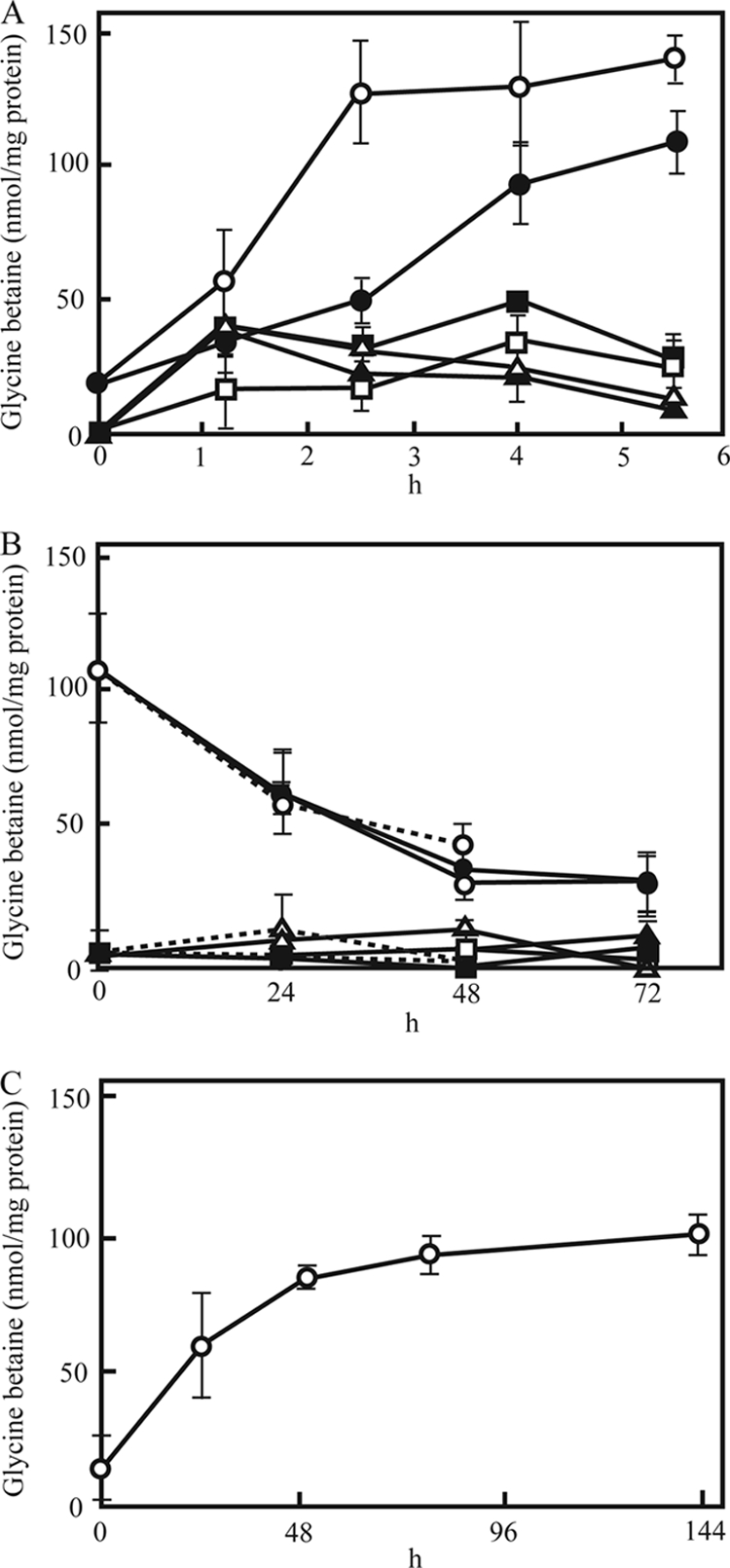
(A) Changes in intracellular levels of glycine betaine in wild-type vegetative cells and gsmA and sdmA mutant vegetative cells during osmotic stress. Wild-type cells (circles) and gsmA (squares) and sdmA (triangles) mutant cells were incubated in CYE medium with 0.2 M NaCl (open symbols) or 0.2 M sucrose (closed symbols). (B) Changes in intracellular levels of glycine betaine in wild-type spores and gsmA and sdmA mutant spores under osmotic stress. Wild-type spores (circles) and gsmA (squares) and sdmA (triangles) mutant spores harvested from 6- to 8-day-old fruiting bodies on CF plates were incubated in CYE medium with 0.2 M NaCl (open symbols) or 0.2 M sucrose (closed symbols) for 72 h or without either (dashed lines) for 48 h. The spores were harvested by centrifugation, washed twice with distilled water, and sonicated with 0.2 g of 0.4-mm glass beads for 5 min. (C) Changes in intracellular levels of glycine betaine in wild-type cells during development under nonosmotic stress conditions. Cells were harvested from CF agar at various times during development, and glycine betaine was measured in the cell extracts. Experiments were repeated three times. The standard deviations are shown by error bars.
(iii) Spore germination.
Under nonstress conditions, fruiting body spores of both mutants germinated normally in CYE medium. Approximately 50% of the spores of the wild type and mutants germinated after about 55 to 65 h of incubation and elongated into rod-shaped cells (Table 1). When cultured in CYE medium containing 0.2 M NaCl or 0.2 M sucrose, about 50% of wild-type spores germinated by 87 h or 83 h, respectively. However, gsmA and sdmA mutant spores incubated in CYE medium containing 0.2 M NaCl or 0.2 M sucrose germinated about 21 to 33 h or 27 to 38 h later than the wild-type spores, respectively. On the other hand, there was no difference in germination times between wild-type glycerol-induced spores and gsmA and sdmA mutant glycerol-induced spores when these spores were incubated in CYE medium containing 0.2 M NaCl or 0.2 M sucrose (data not shown). Wild-type spores harvested after 6 days of development contained 106.5 nmol of glycine betaine/mg of protein (Fig. 3B). The levels of glycine betaine in wild-type spores decreased gradually during incubation in CYE medium and were not increased by the addition of 0.2 M NaCl or sucrose. Glycine betaine levels in gsmA or sdmA mutant spores were much lower than the levels in wild-type spores. We next determined glycine betaine levels in wild-type cells during development under nonosmotic conditions, because wild-type spores exhibited about 5- to 6-fold higher levels of glycine betaine than wild-type vegetative cells. When wild-type cells were incubated on CF agar, the level of glycine betaine in the wild-type cells was increased about 6-fold after 6 days of development (Fig. 3C). These data suggest that the difference in glycine betaine accumulation between wild-type and mutant spores may cause the difference in germination times under osmotic stress, although glycine betaine was not synthesized in wild-type spores in response to osmotic stress. McBride and Zusman reported that trehalose is also synthesized in developing M. xanthus cells, and the trehalose content of spores decreased rapidly during the incubation of spores in nutrient-rich media (12).
TABLE 1.
Germination times of gsmA and sdmA mutant spores under osmotic stress
| Spore type | Germination time± SD (h)a |
||
|---|---|---|---|
| No addition | 0.2 M NaCl | 0.2 M sucrose | |
| Wild type | 56.3 ± 7.5 | 86.6 ± 8.1 | 83.3 ± 9.6 |
| gsmA | 54.6 ± 12.5 | 107.6 ± 7.4 | 109.9 ± 18.1 |
| sdmA | 64.6 ± 1.9 | 119.6 ± 14.0 | 121.6 ± 13.6 |
Germination time is the time at which about 50% of spores had germinated. Spores were harvested from 6- to 8-day-old fruiting bodies on CF plates, sonicated for 2 min, and treated with heat (60°C for 15 min). The spores were inoculated to 2 × 107 spores/ml in CYE medium containing 0.2 M NaCl or sucrose and incubated at 30°C with continuous shaking. The ungerminated spores in each culture were counted using a hemocytometer. Experiments were repeated three times.
To the best of our knowledge, this is the first example of the production of glycine betaine from glycine in a nonhalophilic aerobic heterotroph. Because M. xanthus has a very large genome (total size, 9.14 Mb; total number of genes, 7,457), the species may have the necessary biosynthetic enzymes for the production of glycine betaine from simple carbon sources. In this study, we suggested that glycine betaine biosynthesized from glycine mainly functions as an osmoprotectant for cell growth and spore germination of M. xanthus under osmotic stress conditions. The glycine-glycine betaine synthesis pathway may be an important facet of the process of cellular adaptation of M. xanthus to high-osmolarity stress, because glycine betaine can be synthesized from simple carbon. However, glycine betaine synthesis from simple carbon sources is very costly to the cell. The 3-fold methylation of 1 mol of glycine, with S-adenosylmethionine acting as the methyl group donor, consumes 36 mol of ATP. Since trehalose is a major compatible solute of nonhalophilic bacteria, we are currently investigating the role of trehalose as a compatible solute of M. xanthus cells.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Bessieres, M. A., Y. Gibon, J. C. Lefeuvre, and F. Larher. 1999. A single-step purification for glycine betaine determination in plant extracts by isocratic HPLC. J. Agric. Food Chem. 47:3718-3722. [DOI] [PubMed] [Google Scholar]
- 2.Caldas, T., N. Demont-Caulet, A. Ghazi, and G. Richarme. 1999. Thermoprotection by glycine betaine and choline. Microbiology 145:2543-2548. [DOI] [PubMed] [Google Scholar]
- 3.Furuichi, T., M. Inouye, and S. Inouye. 1985. Novel one-step cloning vector with a transposable element: application to the Myxococcus xanthus genome. J. Bacteriol. 164:270-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galinski, E. A., and H. G. Truper. 1994. Microbial behavior in salt-stressed ecosystems. FEMS Microbiol. Rev. 15:95-108. [Google Scholar]
- 5.Holtmann, G., and E. Bremer. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186:1683-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jelsbak, L., and L. Søgaard-Andersen. 2003. Cell behavior and cell-cell communication during fruiting body morphogenesis in Myxococcus xanthus. J. Microbiol. Methods 55:829-839. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 58:75-98. [DOI] [PubMed] [Google Scholar]
- 8.Kimura, Y., Y. Mishima, H. Nakano, and K. Takegawa. 2002. An adenylyl cyclase, CyaA, of Myxococcus xanthus functions in signal transduction during osmotic stress. J. Bacteriol. 184:3578-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura, Y., M. Ohtani, and K. Takegawa. 2005. An adenylyl cyclase, CyaB, acts as an osmosensor in Myxococcus xanthus. J. Bacteriol. 187:3593-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamark, T., I. Kaasen, M. W. Eshoo, P. Falkenberg, J. McDougall, and A. R. Strom. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049-1064. [DOI] [PubMed] [Google Scholar]
- 11.Lee, M., and L. Shimkets. 1994. Cloning and characterization of the socA locus which restores development to Myxococcus xanthus C-signaling mutants. J. Bacteriol. 176:2200-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride, M. J., and D. R. Zusman. 1989. Trehalose accumulation in vegetative cells and spores of Myxococcus xanthus. J. Bacteriol. 171:6383-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyyssölä, A., J. Kerovuo, P. Kaukinen, N. von Weymarn, and T. Reinikainen. 2000. Extreme halophiles synthesize betaine from glycine by methylation. J. Biol. Chem. 275:22196-22201. [DOI] [PubMed] [Google Scholar]
- 14.Nyyssölä, A., T. Reinikainen, and M. Leisola. 2001. Characterization of glycine sarcosine N-methyltransferase and sarcosine dimethylglycine N-methyltransferase. Appl. Environ. Microbiol. 67:2044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plamann, L., J. M. Davis, B. Cantwell, and J. Mayor. 1994. Evidence that asgB encodes a DNA-binding protein essential for growth and development of Myxococcus xanthus. J. Bacteriol. 176:2013-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes, D., and A. D. Hanson. 1993. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44:357-384. [Google Scholar]
- 17.Ueki, T., and S. Inouye. 1998. A new sigma factor, SigD, essential for stationary phase is also required for multicellular differentiation in Myxococcus xanthus. Genes Cells 3:371-385. [DOI] [PubMed] [Google Scholar]
- 18.Weretilnyk, E. A., and A. D. Hanson. 1989. Betaine aldehyde dehydrogenase from spinach leaves: purification, in vitro translation of the mRNA, and regulation by salinity. Arch. Biochem. Biophys. 271:56-63. [DOI] [PubMed] [Google Scholar]



