Abstract
Bacillus anthracis spores are enclosed by an exosporium comprised of a basal layer and an external hair-like nap. The filaments of the nap are composed of trimers of the collagen-like glycoprotein BclA. The attachment of essentially all BclA trimers to the exosporium requires the basal layer protein BxpB, and both proteins are included in stable high-molecular-mass exosporium complexes. BclA contains a proteolytically processed 38-residue amino-terminal domain (NTD) that is essential for basal-layer attachment. In this report, we identify three NTD submotifs (SM1a, SM1b, and SM2, located within residues 21 to 33) that are important for BclA attachment and demonstrate that residue A20, the amino-terminal residue of processed BclA, is not required for attachment. We show that the shortest NTD of BclA—or of a recombinant protein—sufficient for high-level basal-layer attachment is a 10-residue motif consisting of an initiating methionine, an apparently arbitrary second residue, SM1a or SM1b, and SM2. We also demonstrate that cleavage of the BclA NTD is necessary for efficient attachment to the basal layer and that the site of cleavage is somewhat flexible, at least in certain mutant NTDs. Finally, we propose a mechanism for BclA attachment and discuss the possibility that analogous mechanisms are involved in the attachment of many different collagen-like proteins of B. anthracis and closely related Bacillus species.
Bacillus anthracis, a Gram-positive, rod-shaped, aerobic bacterium, is the causative agent of anthrax (17). When vegetative cells of B. anthracis are starved for certain essential nutrients, they form dormant spores that can survive in harsh soil environments for many years (12, 19). Spore formation starts with asymmetric septation that divides the starved vegetative cell into two genome-containing compartments, a mother cell compartment and a smaller forespore compartment. The mother cell then engulfs the forespore and surrounds it with three protective layers: a cortex composed of peptidoglycan, a closely apposed proteinaceous coat, and a loosely fitting exosporium (11). After a spore maturation stage, the mother cell lyses and releases the mature spore. When spores encounter an aqueous environment containing nutrients, they can germinate and grow as vegetative cells (18). Anthrax is typically caused by contact with spores (17).
The outermost layer of B. anthracis spores, the exosporium, has been studied intensively in recent years because it is both the first point of contact with the immune system of an infected host and the target of new detectors for agents of bioterrorism (21, 28, 32). The exosporium of B. anthracis and closely related pathogenic species, such as Bacillus cereus and Bacillus thuringiensis, is a prominent structure consisting of a paracrystalline basal layer and an external hair-like nap (1, 9). The filaments of the nap are formed by trimers of the collagen-like glycoprotein BclA (2, 29). Recent studies suggest that BclA plays a major role in pathogenesis by directing spores to professional phagocytic cells, a critical step in disease progression (4, 21). The basal layer is composed of approximately 20 different proteins (23, 25, 26), several of which have been shown to play key roles in exosporium assembly (3, 13, 27). One of these proteins is BxpB (also called ExsFA) (25, 30, 34), which is required for the attachment of approximately 98% of spore-bound BclA to the basal layer (26, 30). Residual BclA attachment requires the basal layer protein ExsFB, a paralog of BxpB (30).
BclA contains three distinct domains: a 38-residue amino-terminal domain (NTD), a central collagen-like region containing a strain-specific number of XXG (mostly PTG) repeats, and a 134-residue carboxyl-terminal domain (CTD) (25, 29, 31). The CTD apparently functions as the major nucleation site for trimerization of BclA (24), and CTD trimers form the globular distal ends of the filaments in the nap (2). The highly extended collagen-like region is extensively glycosylated (5), and its length determines the depth of the nap (2, 31). The NTD is the site of attachment of BclA to the basal layer, and deletion of the NTD prevents this attachment (2). The NTD is normally proteolytically processed to remove the first 19 amino acids, and it is this mature form of BclA that is attached to the basal layer (25, 29). In an earlier report, we suggested that NTD processing of BclA is required for basal-layer attachment, perhaps through a direct covalent linkage to BxpB (26).
Recently, Thompson and Stewart identified conserved 11-residue sequences in the NTDs of BclA and the minor B. anthracis collagen-like glycoprotein BclB and showed that these sequences are involved in the incorporation of BclA and BclB into the exosporium. These investigators used a truncated BclA NTD that lacked residues 2 through 19 but included the conserved 11-amino-acid sequence to target enhanced green fluorescent protein (EGFP) to the surface of the developing forespore (33). Thompson and Stewart also reported that cleavage of the BclA NTD occurred after its association with the forespore and suggested that this cleavage was involved indirectly in the attachment process. Actual cleavage sites were not determined in these studies, however. We have performed related studies of the attachment of BclA to the exosporium that provide a more detailed and somewhat different view of this process. In our studies, which are reported here, we identified short segments, or submotifs, of the BclA NTD that can be arranged in different combinations to produce 10-amino-acid motifs sufficient for tight attachment of BclA, and probably most proteins, to the exosporium basal layer. Additionally, we present direct evidence showing that BclA NTD cleavage is required for efficient attachment to the basal layer and that selection of the cleavage site can be somewhat flexible. Finally, we discuss a possible mechanism for BclA attachment and the likelihood that similar mechanisms are used for attachment of many different collagen-like proteins of B. anthracis and closely related Bacillus species.
MATERIALS AND METHODS
Bacterial strains.
The Sterne 34F2 strain of B. anthracis, which is not a human pathogen because it lacks the genes necessary to produce the capsule of the vegetative cell, was used as the wild-type strain (25). Spores produced by the Sterne strain are essentially identical to spores produced by virulent B. anthracis strains (23). A ΔbclA variant of the B. anthracis Sterne strain, designated CLT306, was used as the parent strain for transformations. Strain CLT306 was constructed by allelic exchange (5), which deleted the entire bclA gene and replaced it with a spectinomycin resistance cassette (6). The BclA polypeptide produced by the Sterne strain contains 400 amino acids, with 76 XXG repeats in the collagen-like region, and has a molecular mass of 36,836 Da.
Expression of plasmid-borne variants of the bclA operon.
The wild-type bclA operon (i.e., the bclA promoter, gene, and transcription terminator) was inserted into the cloning site of the multicopy plasmid pCLT1474 (6). Point mutations and deletions were introduced into the bclA operon of the recombinant plasmid by outward PCR (5), leading to the production of a set of plasmid variants. To reduce expression of plasmid-borne operons (i.e., BclA synthesis) to a level equivalent to that expressed from the chromosomal bclA operon, three T residues between the optimally positioned Shine-Dalgarno sequence and the initiation codon of the bclA gene were deleted in each plasmid construct (35). The appropriate size of this deletion was determined empirically. To construct gene fusions encoding NTD-EGFP fusion proteins, a segment of the wild-type bclA/pCLT1474 plasmid containing bclA codon 39 to the end of the bclA open reading frame was deleted and replaced with a DNA fragment containing the entire EGFP open reading frame, which was obtained from plasmid pEGFP-N1 (BD Biosciences Clontech). Deletions were introduced into the NTD-encoding region of the fusion gene as described above. Note that a wild-type bclA ribosome-binding site precedes NTD-EGFP fusion genes. All mutations and plasmid constructions were confirmed by DNA sequencing. Each recombinant plasmid was introduced into strain CLT306 (ΔbclA) by transformation (6), and the resulting transformants were used to assess the effects of the NTD mutations on the expression and exosporium attachment of BclA and NTD-EGFP.
Preparation of spores and deglycosylated exosporia.
Spores were prepared by growing B. anthracis strains at 37°C on LB agar plates until sporulation was complete, typically 3 to 4 days. The spores were washed from the plates with cold (4°C) sterile water, collected by centrifugation, purified by sedimentation through a two-step gradient of 20% and 50% Renografin, and washed extensively with cold sterile water (10). The spores were stored at 4°C in sterile water and quantitated spectrophotometrically at 580 nm as previously described (20). Exosporia were purified from the spores as previously described (25). To prepare deglycosylated exosporia, approximately 2 mg of a dried sample of purified exosporia was treated with trifluoromethanesulfonic acid according to the instructions in a GlycoProfile IV chemical deglycosylation kit (Sigma).
Gel electrophoresis, immunoblotting, and amino-terminal protein sequencing.
Spores (107) or deglycosylated exosporium samples were boiled for 8 min in 20 μl of sample buffer containing 125 mM Tris-HCl (pH 6.8), 4% sodium dodecyl sulfate (SDS), 100 mM dithiothreitol, 0.024% bromophenol blue, and 10% (vol/vol) glycerol. Solubilized proteins were separated by SDS-polyacrylamide gradient gel electrophoresis (PAGE) in a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen). For immunoblotting, spore proteins were transferred from a polyacrylamide gel to a nitrocellulose membrane and detected by staining them as previously described (25). The intensity of staining was measured by densitometry. The purified mouse monoclonal antibody (MAb) probes EF12 (anti-BclA), 10-44-1 (anti-BxpB), and G9-3 (anti-ExsY/CotY) were prepared as described previously (3, 25, 28). For amino-terminal protein sequencing, deglycosylated protein bands were transferred from a gel to a polyvinylidene difluoride membrane, detected by staining with Coomassie brilliant blue, excised, and subjected to six cycles of automated Edman degradation at the Molecular Structure Facility, University of California, Davis.
Flow cytometry and phase-contrast and fluorescence microscopy.
Flow cytometry was used to detect binding to spores of fluorescently (Alexa Fluor 488) labeled anti-BclA MAb EF12 or of an equivalently labeled isotype control MAb, which was unable to bind to spores (28). Briefly, 2 × 107 spores were mixed with 10 μg/ml of fluorescently labeled MAb in 20 μl of phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA), and the mixtures were incubated with shaking (150 rpm) at 4°C for 2 h. The spores were washed four times with 200 μl of PBS containing 1% BSA, suspended in 200 μl of PBS containing 1% BSA, and analyzed using a FACSCalibur fluorescence-activated cell sorter with CellQuest Pro software (Becton Dickinson Biosciences). The spores treated with Alexa-labeled MAb EF12 were further analyzed by phase-contrast and fluorescence microscopy. A 10-μl sample of these spores was placed on a poly-l-lysine-coated microscope slide and allowed to dry in air. A drop of Fluoromount-G (Southern Biotech) was applied to the sample to reduce fluorescence fading. The spores were examined using a Nikon Eclipse E600 microscope equipped with a Y-FL epifluorescence attachment. Phase-contrast and fluorescence images were captured with a Spot charge-coupled-device digital camera (Diagnostic Instruments, Inc.), and these images were displayed and merged by using Spot (v4.0) software.
To detect BclA NTD-EGFP fusion proteins, B. anthracis strains producing these proteins were grown on LB agar plates at 37°C for approximately 40 h. An isolated colony was picked and suspended in a drop of water on a poly-l-lysine-coated microscope slide. Sporangia and spores were examined by phase-contrast and fluorescence microscopy as described above.
RESULTS
Exosporium protein complexes containing chromosome-encoded or plasmid-encoded BclA.
The B. anthracis exosporium contains stable high-molecular-mass complexes that include BclA, BxpB, ExsY, and perhaps the ExsY paralog CotY and other exosporium proteins (23, 26). To inspect these complexes in more detail, exosporium proteins were extracted by boiling purified B. anthracis spores in sample buffer containing 4% SDS and a high level of reducing agent. Solubilized proteins and protein complexes were separated by SDS-PAGE and analyzed by immunoblotting them with anti-BclA, anti-BxpB, and anti-ExsY/CotY MAbs. The anti-BclA MAb reacts with the CTD of BclA (2), the anti-BxpB MAb does not react with the BxpB paralog ExsFB (26), and the anti-ExsY/CotY MAb reacts similarly with ExsY and CotY (3). Three major bands with apparent molecular masses of approximately 180 kDa, 440 kDa, and 660 kDa (the last a rough extrapolation) reacted with the anti-BclA MAb (Fig. 1A, chr.). The 440-kDa and 660-kDa bands also reacted with the anti-BxpB MAb (Fig. 1B, chr.), while only the 660-kDa band reacted convincingly with the ExsY/CotY MAb (data not shown). These results indicated that the 440-kDa and 660-kDa bands contained highly stable exosporium complexes, with the 440-kDa complex containing BclA and BxpB and the 660-kDa complex containing BclA, BxpB, and ExsY and/or CotY. It is also possible that these complexes contain other exosporium proteins not examined here. The 180-kDa band, which appeared to contain approximately 8% of the total extractable BclA (Fig. 1A, chr.), reacted only with the anti-BclA MAb. Preliminary characterization of this band suggested that it contained only fully glycosylated monomeric BclA (data not shown). Hereafter, we refer to the BclA in the 180-kDa band as “free” BclA. A band containing free monomeric BxpB can also be seen in the immunoblot with anti-BxpB MAb (Fig. 1B, chr.).
FIG. 1.
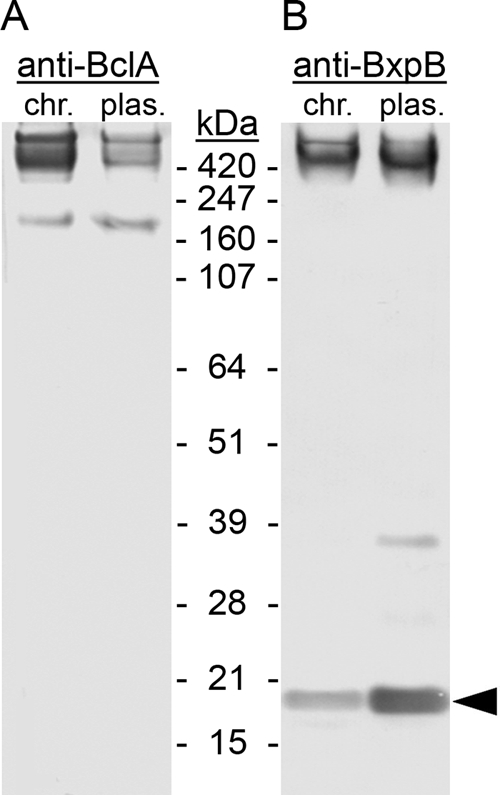
Exosporium protein complexes containing BclA and BxpB. Exosporium complexes were extracted under denaturing and reducing conditions from spores of the Sterne strain, which carries a chromosomal (chr.) copy of the wild-type bclA operon, and from spores of a ΔbclA mutant strain expressing a plasmid (plas.)-borne wild-type bclA gene from the bclA promoter. Complexes and proteins were separated by SDS-PAGE and visualized by immunoblotting them with anti-BclA (A) and anti-BxpB (B) MAbs. Prestained protein standards were included in the analysis, and their gel locations and molecular masses are indicated. The arrowhead points to the band containing monomeric BxpB.
To examine the roles of certain BclA NTD amino acids in BclA attachment to the exosporium, we constructed a set of B. anthracis strains lacking the chromosomal copy of bclA (ΔbclA) and carrying a plasmid from which wild-type and mutant bclA genes could be expressed from the bclA promoter. We used these strains in the experiments described below. The plasmid vector was designed to allow a level of BclA synthesis comparable to that directed by the wild-type chromosomal bclA operon (see Materials and Methods). The exosporium protein complexes formed with the plasmid-borne wild-type bclA gene also were similar to those formed with the chromosomal bclA gene, as revealed by immunoblots using anti-BclA (Fig. 1A, plas.) and anti-BxpB (Fig. 1B, plas.) MAbs. One minor difference was that the levels of free BclA (20% of the total) and free BxpB compared to the total amount of each protein were slightly higher with the plasmid-borne bclA gene. Additionally, the total amount of BclA detected with the plasmid-borne bclA gene (Fig. 1A, plas.) was somewhat less than that detected with the chromosomal bclA gene (Fig. 1A, chr.), but this difference is irrelevant when only plasmid-borne bclA operons are compared. All comparisons were performed using equal numbers of spores.
Residue A20 is not required for BclA attachment.
The form of BclA attached to spores lacks residues 1 through 19, and the amino-terminal residue of this truncated BclA is A20 (Fig. 2A). Because the cleavage event that exposes residue A20 is associated with BclA attachment to the basal layer, it seems reasonable to suspect that the free amino group of A20 is directly involved in the attachment process. This possibility appears even more likely because there are few amino acids in the NTD of cleaved BclA that possess side chains with functional groups (Fig. 2A). To determine if residue A20 plays a critical role in BclA attachment, we examined the effects of mutations that either deleted residue A20 (Δ20) or changed this residue to proline (A20P). Spores were produced using strains expressing the two mutant bclA genes and the wild-type bclA gene as a control, and exosporium protein complexes extracted from these spores were analyzed by immunoblotting them as described above. Our results showed that neither mutation decreased the levels of BclA-containing high-molecular-mass complexes; instead, the mutations resulted in a 2-fold to 3-fold increase in the levels of these complexes, as well as the level of free BclA (Fig. 2B). Additionally, an approximately 330-kDa complex containing both BclA and BxpB was readily observed with Δ20 and A20P spores; this complex could also be seen with wild-type spores, but it was a minor band in the immunoblots (Fig. 2B).
FIG. 2.

Effects of mutations altering BclA residue A20 on the attachment of BclA to the exosporium. (A) Sequence of the NTD of BclA; the arrowhead indicates the normal cleavage site between residues S19 and A20. (B) Immunoblots with anti-BclA and anti-BxpB MAbs examining exosporium protein complexes and free BclA extracted from spores produced by strains expressing the wild-type (WT), Δ20, and A20P versions of BclA. Only the relevant parts of the immunoblots are shown, and the locations and molecular masses (in kDa) of protein standards are indicated. (C) Analysis of WT, Δ20, and A20P spores by flow cytometry following treatment of the spores with a fluorescently (Alexa Fluor 488) labeled anti-BclA MAb (histograms with bold lines); the filled histograms were obtained with a comparably labeled control MAb that does not bind B. anthracis spores.
To further examine the effects of the Δ20 and A20P mutations on BclA attachment to the exosporium, spores produced by the strains expressing the mutant and wild-type bclA genes were treated with a fluorescently labeled anti-BclA MAb and analyzed by flow cytometry. This assay specifically examines BclA exposed on the spore surface. The results showed that all spore types were extensively labeled by the anti-BclA MAb, with the mutant spores labeled more uniformly and to a slightly higher level (Fig. 2C). These results were entirely consistent with the immunoblotting data. We also examined the fluorescently labeled spores by fluorescence microscopy. All spore types were brightly and uniformly fluorescent, but it was not possible to visually discern differences between them (data not shown).
Deletion analysis of the NTD of BclA to locate sequences required for attachment.
To locate sequences required for BclA attachment, we examined mutations that introduced a set of nested deletions in the NTD. All deletions began at residue S2, and the end points of the deletions analyzed in detail were residues S19 (Δ2-19), A20 (Δ2-20), N24 (Δ2-24), L25 (Δ2-25), and L30 (Δ2-30) (Fig. 3A). The effects of these mutations on the attachment of BclA were determined by immunoblotting (Fig. 3B) as described above for the A20 point mutations. In the case of the Δ2-19 mutation, the patterns of BclA- and BxpB-containing bands were essentially the same as the patterns observed with the A20 point mutations, indicating that amino acids 2 to 19 were not required for efficient attachment of BclA. A minor difference was that the level of free BclA extracted from the Δ2-19 spores was slightly higher, representing approximately 40% of the total BclA (Fig. 3B). In sharp contrast, the Δ2-20 mutation reduced the level of extractable BclA-containing material by a factor of 25 compared to the level from Δ2-19 spores, and most of this material was free BclA. A similar reduction was observed with BxpB-containing complexes. This result indicated that extending the Δ2-19 deletion by a single amino acid nearly eliminated the NTD signal for BclA attachment.
FIG. 3.
Analysis of a nested set of BclA NTD deletions to locate sequences required for attachment to the exosporium. (A) Sequence of the NTD of BclA; the arrowhead indicates the normal NTD cleavage site. The amino acids indicated by numbers correspond to the end points of a set of deletions that begin at residue S2. Spores produced by strains expressing the wild-type and the indicated deleted versions of BclA were analyzed by immunoblotting with anti-BclA and anti-BxpB MAbs (B) and by flow cytometry following treatment with a fluorescently labeled anti-BclA MAb (C) as described in the legend to Fig. 2.
Surprisingly, extending the Δ2-19 deletion by five (but not fewer) amino acids, creating the Δ2-24 mutation, partially restored the signal for BclA attachment (Fig. 3B). In this case, the level of extractable BclA-containing material was approximately 30% of that obtained from Δ2-19 spores; however, 80% of this material was free BclA. This result indicated that although the Δ2-24 mutation permitted significant BclA association with spores, only a small fraction of this BclA was included in stable high-molecular-mass complexes. As observed with the Δ2-19 mutation, extending the Δ2-24 deletion by a single amino acid, creating the Δ2-25 mutation, reduced the level of extractable BclA-containing material by a factor of 25. Extraction of Δ2-25 spores produced barely detectable levels of BclA-containing material and BxpB-containing complexes. The significance of the parallel effects of extending Δ2-19 and Δ2-24 by a single amino acid is discussed below. Deletions longer than the Δ2-25 mutation (e.g., a Δ2-30 mutation) reduced extractable BclA-containing material and BxpB complexes to undetectable levels. As a control, we demonstrated that each strain examined above (and below) produced comparable levels of BclA during sporulation (data not shown).
Our set of nested NTD deletions was also examined by flow cytometry using spores that had been treated with a fluorescently labeled anti-BclA MAb as described for the A20 point mutations (Fig. 3C). As expected, the histogram for Δ2-19 spores was essentially identical to those for Δ20 and A20P spores, indicating extensive and uniform labeling of surface-exposed BclA. In contrast, the histogram for Δ2-20 spores indicated that they were only weakly and nonuniformly labeled. The histogram for Δ2-24 spores also indicated extensive labeling of surface-exposed BclA, similar to that observed with wild-type spores. The labeling was more heterogeneous and somewhat less intense than that observed with Δ2-19 spores, however. The histogram for Δ2-25 spores was similar to that of Δ2-20 spores, indicating relatively weak labeling. As expected, the histogram for Δ2-30 spores indicated essentially no BclA exposed on the spore surface.
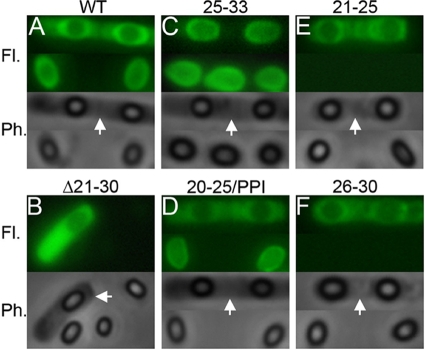
To verify the conclusions drawn from flow cytometry, we examined the same fluorescently labeled spores by fluorescence microscopy (Fig. 4). As suggested, Δ2-19 and Δ2-24 spores were uniformly and highly fluorescent. In contrast, Δ2-20 and Δ2-25 spores exhibited random and patchy fluorescence, with most spores only weakly fluorescent. The Δ2-30 spores were not fluorescently labeled. Thus, immunoblotting, flow cytometry, and fluorescence microscopy provided a complementary and generally consistent view of BclA attachment to the exosporia of the mutant spores.
FIG. 4.

Effects of the nested set of NTD deletions on BclA attachment examined by fluorescence microscopy. Spores produced by strains expressing the wild-type and the indicated deleted versions of BclA were treated with a fluorescently (Alexa Fluor 488) labeled anti-BclA MAb prior to analysis. Merged fluorescence and phase-contrast images are shown in each panel.
Identification of NTD submotifs important for BclA attachment.
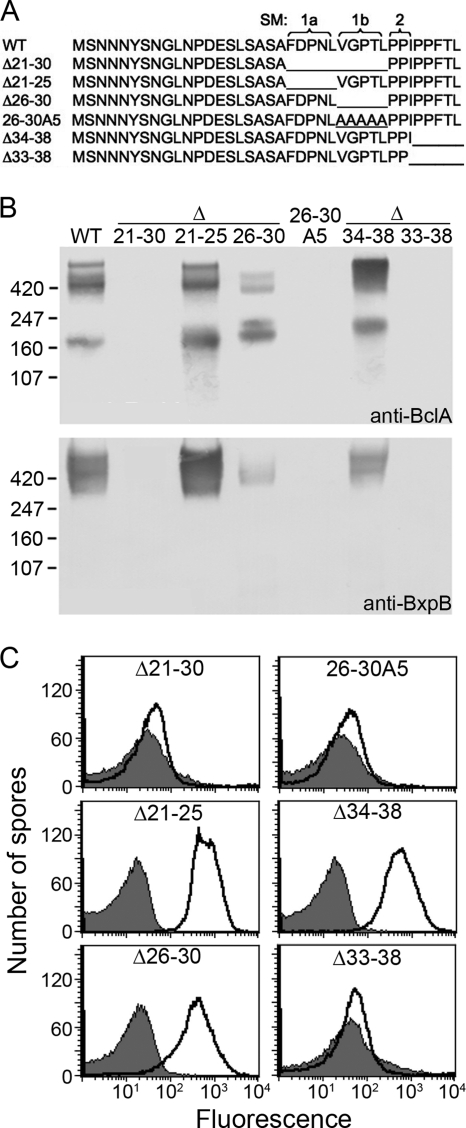
Based on the BclA deletion analysis described above and the fact that residues 21 to 25 (FDPNL) and 26 to 30 (VGPTL) possessed similarities in their sequences, we constructed a set of internal deletions and a multiple-base-substitution mutation (Fig. 5A) to more precisely map NTD sequences required for BclA attachment to the exosporium. Again, the effects of the mutations on BclA attachment were analyzed initially by immunoblotting (Fig. 5B). As expected, when residues 21 to 30 were deleted (Δ21-30), high-molecular-mass complexes containing BclA and BxpB, as well as free BclA, were not detected. On the other hand, when residues 21 to 25 (Δ21-25) were deleted, we detected levels of the high-molecular-mass complexes and free BclA that were comparable to (actually somewhat higher than) the levels observed with wild-type spores. When residues 26 to 30 (Δ26-30) were deleted, we again detected significant levels of the high-molecular-mass complexes and free BclA; however, the level of total BclA was clearly lower than that of Δ21-25 (and even wild-type) spores. Additionally, the Δ26-30 mutation resulted in a redistribution of BclA toward atypical lower-molecular-mass complexes and free BclA, perhaps indicating a deficiency in complex formation. Taken together, though, these results suggested that residues 21 to 25 and residues 26 to 30 were partially interchangeable sequence motifs required for BclA attachment. Surprisingly, when residues 26 to 30 were changed to five A residues (26-30A5), no high-molecular-mass complexes or free BclA was detected. This result suggested that BclA attachment required proper spacing between distinct NTD sequence motifs; in this case, one motif preceded residue 26 and another followed residue 30. Finally, deletion of residues 34 to 38 (Δ34-38) had a slight stimulatory effect on BclA attachment, while deletion of residues 33 to 38 (Δ33-38) eliminated BclA attachment. This result indicated that residue 33, or perhaps residues 31 to 33 (PPI), played a critical role in the attachment of BclA. Residues 21 to 25, residues 26 to 30, and residues 31 to 33 were designated submotifs SM1a, SM1b, and SM2, respectively (Fig. 5A).
FIG. 5.
Identification of NTD submotifs necessary for BclA attachment. (A) Sequences of the wild-type NTD and of mutant NTDs containing internal deletions and a 5-base substitution are shown. Submotifs (SM) are enclosed by brackets and labeled in the wild-type sequence; lines replace deleted amino acids in the mutant sequences. Spores produced by strains expressing the wild-type and the indicated mutant versions of BclA were analyzed by immunoblotting with anti-BclA and anti-BxpB MAbs (B) and by flow cytometry following treatment with a fluorescently labeled anti-BclA MAb (C) as described in the legend to Fig. 2.
We further examined the internal deletions and 26-30A5 mutation by flow cytometry using spores that had been treated with a fluorescently labeled anti-BclA MAb as described above (Fig. 5C). Consistent with the immunoblotting data, the histograms for Δ21-30, 26-30A5, and Δ33-38 spores were essentially identical to those for spores treated with a control MAb, indicating no detectable BclA on the surfaces of these spores. On the other hand, the histograms for Δ21-25, Δ26-30, and Δ34-38 spores indicated extensive and uniform fluorescent labeling, with fluorescence intensities proportional to the levels of total BclA extracted from these spores. Inspection of the fluorescently labeled spores by fluorescence microscopy was consistent with these conclusions (data not shown).
Sequence motifs sufficient for BclA attachment.
To identify the shortest BclA NTD sequence sufficient for attachment to the exosporium, we constructed mutations that resulted in the production of BclA proteins with condensed NTDs. Important goals in these constructions were to include the following: an initiating methionine residue; in some cases, an arbitrary residue 2, which provides a possible cleavage site between residues 1 and 2; either SM1a or SM1b; and a complete or partial SM2 (Fig. 6A). The levels of high-molecular-mass complexes containing BclA and BxpB and the level of free BclA were determined again by immunoblotting (Fig. 6B). When the NTD contained the 10-residue sequence MAFDPNLPPI (20-25/PPI), which was assembled from wild-type residues 1, 20 to 25 (A plus SM1a), and 31 to 33 (SM2), a low level of 440-kDa complexes and a large amount of BclA-containing material with apparent masses between approximately 70 kDa and 330 kDa were detected. The 440-kDa complexes contained both BclA and BxpB, but most of the heterogeneous BclA-containing material apparently did not include BxpB. The heterogeneity in the 70- to 330-kDa material might reflect, at least in part, variable glycosylation of BclA, which is suggested by the fact that unglycosylated free BclA monomers migrate with an apparent mass of 70 kDa during SDS-PAGE (25). When the A residue preceding SM1a (21-25/PPI) or the last residue of SM2 (20-25/PP) was omitted from the 20-25/PPI NTD, high-molecular-mass complexes and free BclA were barely detectable. In contrast, an NTD with the 10-residue sequence MLVGPTLPPI (25-33), which was assembled from wild-type residues 1 and 25 to 33 (L plus SM1b through SM2), directed a level of BclA attachment comparable to that observed with wild-type BclA. Unlike the wild-type situation, however, most of the BclA-containing material was free BclA in the case of the 25-33 NTD. When the L residue preceding SM1b (26-33) or the last residue of SM2 (25-32) was deleted from the 25-33 NTD, attachment of BclA was severely reduced or effectively eliminated, respectively. These results were strikingly similar to those obtained with the modified versions of the 20-25/PPI NTD, and together they suggested a critical role in BclA attachment for either the residue or spacing between the initiating methionine and SM1(a or b, hereafter referred to as SM1a/b) and for the terminal residue of an adjacent SM2. Additionally, these results indicated that SM1b, which is normally adjacent to SM2, was more efficient than SM1a in the incorporation of BclA into high-molecular-mass complexes.
FIG. 6.
Identification of sequence motifs sufficient for BclA attachment. (A) Sequences of condensed versions of the NTD of BclA; deleted amino acids are replaced by lines, and submotifs are enclosed by brackets and labeled. Spores produced by strains expressing the wild-type and the indicated mutant versions of BclA were analyzed by immunoblotting with anti-BclA and anti-BxpB MAbs (B) and by flow cytometry following treatment with a fluorescently labeled anti-BclA MAb (C) as described in the legend to Fig. 2.
Flow cytometry was used to demonstrate that the attachment of BclA containing either the 20-25/PPI NTD or 25-33 NTD to the spore surface was equivalent to that observed with wild-type BclA (Fig. 6C). Analysis of the terminally truncated versions of the 20-25/PPI and 25-33 NTDs by flow cytometry indicated only low or marginal levels of BclA attachment (data not shown).
Sequence motifs sufficient for attachment of an NTD-EGFP fusion protein.
To determine whether the minimized NTDs that directed attachment of BclA to the exosporium were capable of directing similar attachment of a foreign protein, we constructed a plasmid designed to express a recombinant operon consisting of the bclA promoter-leader region, selected BclA NTD codons fused in frame to the EGFP gene, and the bclA transcription terminator. The plasmids were transformed individually into our ΔbclA parent strain of B. anthracis, and sporulating cells (sporangia) and free spores produced by these strains were examined by fluorescence microscopy. All but two of the NTDs analyzed were identical to NTD sequences shown in Fig. 5A and 6A. The two new NTDs contained a methionine residue followed by BclA amino acids 21 to 25 (SM1a) or 26 to 30 (SM1b); these NTDs were termed 21-25 and 26-30, respectively.
As positive and negative controls, we examined the attachment of fusion proteins containing the wild-type and Δ21-30 NTDs, respectively. In the case of the wild-type NTD, free spores were highly fluorescent, indicating that the fusion protein was efficiently attached to the exosporium (Fig. 7A). Additionally, we observed that during sporulation, wild-type fusion protein was produced in the mother cell compartment and localized to the forespore (Fig. 7A). As expected, the Δ21-30 spores were not fluorescent (Fig. 7B), again indicating that residues 21 to 30 were critical for attachment to the exosporium. Although not attached to spores, the Δ21-30 fusion protein was produced in sporangia at a level comparable to that observed with the wild-type fusion protein. Unlike the wild-type protein, the Δ21-30 fusion protein was not localized to the forespore during spore development (Fig. 7B). Similar results were described in the recent report by Thompson and Stewart (33).
FIG. 7.
Spore attachment and forespore localization of recombinant BclA NTD-EGFP fusion proteins. Spore formation by strains producing fusion proteins in which wild-type and mutant versions of the BclA NTD were fused to the amino terminus of EGFP was monitored by fluorescence microscopy. The NTD of the fusion protein examined is indicated at the top of each panel. Each panel includes a fluorescence (Fl.) and phase-contrast (Ph.) image of sporangia containing nearly fully developed forespores and mature spores shortly after their release from mother cells. The white arrows in the phase-contrast images indicate the sporangia.
We next examined the attachment of the fusion protein containing the 25-33 NTD, the 10-residue NTD most efficient at attaching BclA to the exosporium. Spores produced by the 25-33 strain were as fluorescent as spores with attached wild-type fusion protein, indicating that the 25-33 fusion protein was efficiently attached to the spore (Fig. 7C and flow cytometry not shown). Furthermore, the 25-33 fusion protein was produced at a high level in the mother cell and was localized efficiently to the forespore (Fig. 7C). We obtained similar results with a fusion protein containing the 20-25/PPI NTD (Fig. 7D), the 10-residue NTD that directed BclA attachment with moderate efficiency. In apparent contradiction of these results, Thompson and Stewart reported that they did not detect the attachment of a 25-35 fusion protein to spores (33). Possible reasons for their failure to detect attachment are discussed below.
Finally, we examined fusion proteins with NTDs containing either SM1a (21-25) or SM1b (26-30) alone. Both fusion proteins yielded essentially the same results; each fusion protein was produced at a high level in the mother cell and was localized efficiently around the developing forespore (Fig. 7E and F). However, spores produced by the 21-25 and 26-30 strains were not fluorescent (Fig. 7E and F). These results indicated that SM1a and SM1b function as independent forespore localization signals and that additional NTD sequences are required for efficient attachment to the spore.
NTD cleavage is required for efficient attachment of BclA.
To examine the role of NTD cleavage in BclA attachment, we determined the apparent cleavage site, or absence of such a site, in selected NTD mutants (Table 1). Cleavage sites were established by determining the amino-terminal residue of mature BclA purified from trifluoromethanesulfonic acid-treated exosporia isolated from mutant spores. We showed previously that treatment with trifluoromethanesulfonic acid produced deglycosylated BclA that migrated as a single band during SDS-PAGE (25). Initially, we confirmed that the amino-terminal residue of mature wild-type BclA was residue A20. We also found that cleavage of mutant NTDs lacking only SM1a (Δ21-25) or SM1b (Δ26-30) was the same as that observed with the wild-type NTD (Table 1). In each mutant construct, an SM1a/b submotif immediately followed residue A20, as observed in the wild-type NTD. Evidently, SM1a and SM1b were equivalent in terms of targeting cleavage to the S19-A20 peptide bond.
TABLE 1.
Cleavage sites in mutant variants of the NTD of BclA
| NTD type | Cleavage site in NTD sequencea |
|---|---|
| Wild type | MSNNNYSNGLNPDESLSAS▾AFDPNLVGPTLPPIPPFTL |
| Δ21-25 | MSNNNYSNGLNPDESLSAS▾A-----VGPTLPPIPPFTL |
| Δ26-30 | MSNNNYSNGLNPDESLSAS▾AFDPNL-----PPIPPFTL |
| Δ20 | MSNNNYSNGLNPDESLSAS-▾FDPNLVGPTLPPIPPFTL |
| A20P | MSNNNYSNGLNPDESLSASP▾FDPNLVGPTLPPIPPFTL |
| Δ2-19 | M------------------A▾FDPNLVGPTLPPIPPFTL |
| Δ2-20b | M-------------------FDPNLVGPTLPPIPPFTL |
| Δ2-24 | M-----------------------L▾VGPTLPPIPPFTL |
| Δ2-25b | M------------------------VGPTLPPIPPFTL |
The arrowheads indicate the sites of NTD cleavage. The dashes represent deleted amino acids.
No cleavage occurred in this NTD.
We next determined the effects of mutations that altered residue A20, namely, Δ20 and A20P. In each case, cleavage occurred at the peptide bond immediately preceding SM1a (Table 1). In the case of Δ20, the apparent shift could be simply a consequence of the deletion; however, the A20P mutation clearly altered the position of cleavage. This alteration might reflect a key role for A20 in targeting cleavage or a uniquely disruptive effect of the A20P mutation. Interestingly, NTD cleavage of the Δ2-19 mutant also occurred immediately preceding SM1a. Furthermore, significantly elevated levels of exosporium-attached BclA (and BxpB) were detected only with the Δ20, A20P, and Δ2-19 NTDs (Fig. 2 and 3). This observation suggested that the site of NTD cleavage has a major effect on the efficiency of BclA attachment to the exosporium.
The Δ2-19 and Δ2-24 NTDs are similar in that 2 amino acids precede an SM1a/b submotif. Cleavage of each mutant NTD was also similar, occurring immediately before SM1a/b (Table 1). As observed with the Δ2-19 NTD, the level of Δ2-24 NTD attachment was substantial (Fig. 3). Extension of the Δ2-19 and Δ2-24 deletions created the Δ2-20 and Δ2-25 NTDs, respectively. In the Δ2-20 and Δ2-25 constructs, only the initiating methionine preceded SM1a/b. Unlike all other NTDs examined, the Δ2-20 and Δ2-25 NTDs were not cleaved. Presumably, this failure was due to excessive truncation of the NTD, specifically, the sequence preceding SM1a/b. More importantly, the level of attachment of the Δ2-20 and Δ2-25 NTDs was about 4% of that observed with the Δ2-19 and Δ2-24 NTDs, respectively (Fig. 3B). This result indicated that efficient attachment of BclA required cleavage of the NTD.
DISCUSSION
Many different proteins with critical roles in metabolism, structure, and pathogenesis are attached to the surfaces of bacteria. This attachment can be noncovalent or covalent and can occur by a variety of mechanisms. Two mechanisms have been described for covalent attachment, each involving enzyme-catalyzed transpeptidation (7, 16). Sortases, which are found primarily in Gram-positive bacteria, catalyze the attachment of surface proteins to the cell wall. Sortases cleave an LPXTG (or equivalent) motif located within the C-terminal region of the protein substrate to produce an acyl-sortase intermediate, which is resolved by nucleophilic attack of an amino group provided by a cell wall cross bridge of a peptidoglycan precursor. In Gram-negative bacteria, an l,d-transpeptidase catalyzes the attachment of lipoproteins, via the C-terminal residue, to cell wall peptidoglycan. This reaction is mechanistically similar to that described for sortases. In this study, we investigated another example of surface protein attachment, namely, attachment of BclA to the B. anthracis exosporium. The mechanism of BclA attachment shares key features with the established mechanisms for covalent attachment. The similarities include a conserved amino acid sequence motif and the involvement of proteolytic cleavage. In the case of BclA, the critical attachment motif is a set of previously unrecognized short sequences termed submotifs that follow the cleavage site within a 38-residue NTD.
The submotifs include residues 21 to 25 (SM1a), 26 to 30 (SM1b), and 31 to 33 (SM2), with SM1a following the normal site of NTD cleavage between residues S19 and A20. Characterization of the submotifs suggested that SM1a and SM1b perform similar functions; both are sufficient to localize noncleaved BclA to the developing forespore, and either can support attachment of BclA to the exosporium. In fact, we showed that 10-residue NTD variants containing only an initiating methionine, an apparently arbitrary residue 2, SM1a or SM1b, and SM2 direct efficient attachment of BclA, with SM1b providing a somewhat stronger attachment platform. The same 10-residue NTDs also supported attachment of an NTD-EGFP fusion protein, indicating that these short NTDs provide a general signal for spore attachment. In apparent contradiction, Thompson and Stewart reported that a 12-residue NTD variant, which contained our more efficient 10-residue NTD plus two additional amino acids, did not direct spore attachment of a fluorescent reporter protein (33). One reason for the different results might be that the fluorescent reporter protein attached to spores produced by Thompson and Stewart was degraded prior to examination. We observed that NTD-EGFP fusion proteins attached to spores were rapidly degraded, especially in liquid medium. Producing spores on solid medium and immediately purifying released spores minimized fusion protein degradation. To avoid this problem and to keep the attachment process as natural as possible, we focused on the attachment of BclA. Another factor that might contribute to the apparent contradiction is that Thompson and Stewart used a B. anthracis strain that produced normal levels of wild-type BclA in addition to an NTD-reporter protein fusion. Competition between these two proteins might have significantly lowered the level of reporter protein attachment, at least in some cases.
We also investigated the role of BclA NTD cleavage in attachment. Our results indicated that cleavage was required for efficient attachment of the NTD to the exosporium. Furthermore, these results showed that cleavage was flexible but not random. When the normal cleavage site was altered, cleavage was directed to the peptide bond preceding SM1a or, when SM1a was deleted, to the peptide bond preceding SM1b. Interestingly, the site of cleavage appeared to influence the efficiency of BclA attachment, perhaps reflecting different interactions between the NTD and other exosporium proteins. Cleavage also was restricted to NTDs containing at least 2 amino acids preceding SM1a/b. In all cases, cleavage required the SM2 submotif positioned immediately after SM1a/b. Taken together, these results suggest that cleavage is required to generate an amino-terminal residue that is positioned, relative to other critical protein elements, to optimize BclA attachment. Presumably, the amino-terminal residue participates directly in attachment.
Although several of the mutant NTD constructs examined in this study (e.g., Δ2-24, 20-25/PPI, and 25-33) supported a high level of BclA attachment to the B. anthracis exosporium, the process appeared to differ from that with wild-type BclA in an important way. The mutant NTD constructs were typically less efficiently incorporated into high-molecular-mass complexes containing both BclA and BxpB, as indicated by the extraction of much higher than wild-type levels of free BclA (Fig. 3 and 6). This difference suggests that the mutant NTD constructs formed fewer or less stable high-molecular-mass complexes. Stated another way, BclA can be attached to the exosporium in different ways, and the wild-type NTD is better than the mutant NTD constructs in forming the most stable type of these attachments. Although the proposed different attachments have not been defined, the stability of BclA/BxpB-containing high-molecular-mass complexes suggests the most stable attachment is covalent. This covalent linkage cannot be a standard peptide bond, however, because this bond—unlike the stable BclA linkage—is resistant to trifluoromethanesulfonic acid. Accordingly, the less stable complexes would be formed by noncovalent interactions. Free BclA monomers could be included along with covalently attached BclA monomers as part of collagen-like BclA trimers. In addition, BclA could be bound to the exosporium through noncovalent NTD interactions with exosporium proteins.
The foregoing observations, as well as previously published results, are consistent with the following model for attachment of wild-type BclA. Following BclA synthesis and trimerization in the mother cell, the SM1a and SM1b submotifs interact with a receptor protein, which could be BxpB in the basal layer or a currently unrecognized adapter protein. SM1a and SM1b could be redundant or unique targeting signals that bind to the same or separate sites on the receptor protein, respectively. If an adapter protein is involved, it facilitates specific interactions between BclA and its binding partner that are necessary for NTD cleavage. Cleavage could be catalyzed by sequence elements within BclA and/or its binding partner or by a separate protease. Regardless of the participating proteins, the BclA submotif SM2 plays an essential role in this process. After cleavage, the new amino-terminal residue of BclA is appropriately positioned to facilitate a covalent linkage to a specific exosporium protein, with BxpB a likely candidate. Although all BclA monomers are cleaved, a small fraction is not attached covalently. Monomers in this fraction could be bound to the exosporium through trimerization interactions with covalently attached BclA monomers, through submotif interactions with binding-partner proteins, or via complex associations with other basal-layer proteins.
Finally, a search of all available genome sequences identified 18 proteins possessing sequences in their NTDs that closely resemble BclA sequences required for attachment to the B. anthracis exosporium (Table 2). Each protein contains a collagen-like region and is produced by a strain within the B. cereus group, which includes B. anthracis and four closely related species (22). Fifteen of the proteins contain sequences resembling submotifs SM1a, SM1b, and SM2, with the SM1a-like sequence typically preceded by an alanine residue. The other three proteins possess NTDs that are identical in length and very similar in sequence, and each NTD contains sequences resembling submotifs SM1b and SM2. The latter group of proteins includes the B. anthracis exosporium glycoprotein BclB. The functions and cellular or spore locations of the 17 uncharacterized proteins in Table 2 are unknown. However, the genes encoding these proteins are generally preceded by sequences resembling promoters that are transcribed in the mother cell during sporulation (8), suggesting that the proteins are incorporated into spore integuments. These observations suggest that the mechanism for BclA attachment might be a general mechanism for stable attachment of many different collagen-like proteins to spore surfaces and other spore structures.
TABLE 2.
Potential NTD attachment motifs in collagen-like proteins of the B. cereus group
| Protein/gene bank name | NTD sequencea | Bacillus sp. |
|---|---|---|
| BclA | MSNNNYSNGLNPDESLSASA FDPNL VGPTL PPI PPFTL | anthracis |
| AAT55598b | MSEKYIILHGTA LEPNL IGPTL PPI PPFTF | anthracis |
| AAT56919b,c | MEGNGGKSKIKSPLNSNFK ILSDL VGPTF PPV PTGMT | anthracis |
| AAP09378 | MSRKDRFNSPKIKSEIS ISPDL VGPTF PPI PSFTL | cereus |
| AAP09528 | MDELLSSTL INPDL LGPTL PAI PPFTL | cereus |
| AAP09594 | MFDKNKILQANA FNSNL IGPTL PPI PPFTL | cereus |
| AAY60601c | MENKKGSKHNEFLSAKA FNPNL VGPTL PPV PSFTL | cereus |
| ACK93566 | MNEEYNTLHGPA LEPNL IGPTL PSI PPFTF | cereus |
| ABK85529 | MDEFLSSAA INPNL VGPTL PPV PPFTL | thuringiensis |
| ABK86394 | MNEKYIILYGTA LEPNL IGPTL PPI PPFTF | thuringiensis |
| ABY43499 | MFDKNEIQKINGILQANA LNPNL IGPTL PPI PPFTL | weihenstephanensis |
| ABY43578 | MDEFLSFAA LNPGS IGPTL PPV PPFQF | weihenstephanensis |
| ABY44375 | MSKENIILHGPA LEPNL IGPTL PPI PPFTF | weihenstephanensis |
| ABY44744 | MFDKNEMKKTNEVLQANA LDPNI IGPTL PPI PPFTL | weihenstephanensis |
| ABY46500c | MERKNKWYGLNSNVNLSASS FDPNL VGPTL PPI SPISV | weihenstephanensis |
| ABY46741 | MSDKHQMKKISEVLQAHA LDPNL IGPPL PPI TPFTF | weihenstephanensis |
| BclB | MKQNDKLWLDKGIIGPENIGP TFPVL PPI HIPTG | anthracis |
| AAP09344 | MKRNDNLSLNKGMIGPENIGP TFPIL PPI YIPTG | cereus |
| AAS41401 | MNSNEKLSLNKGMVRPENIGP TFPVL PPI YIPTG | cereus |
Sequences resembling BclA submotifs SM1a (FDPNL), SM1b (VGPTL), and SM2 (PPI) are in boldface.
The predicted amino terminus shown is slightly different from (but more likely than) the terminus indicated in the current genome database annotation (14).
Acknowledgments
This work was supported by NIH grants AI057699 and AI081775.
We thank Sylvia McPherson and Mei Li for helpful suggestions, John Kearney for providing MAbs, and Evvie Allison for editorial assistance.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Ball, D. A., R. Taylor, S. J. Todd, C. Redmond, E. Couture-Tosi, P. Sylvestre, A. Moir, and P. A. Bullough. 2008. Structure of the exosporium and sublayers of spores of the Bacillus cereus family revealed by electron crystallography. Mol. Microbiol. 68:947-958. [DOI] [PubMed] [Google Scholar]
- 2.Boydston, J. A., P. Chen, C. T. Steichen, and C. L. Turnbough, Jr. 2005. Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis. J. Bacteriol. 187:5310-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boydston, J. A., L. Yue, J. F. Kearney, and C. L. Turnbough, Jr. 2006. The ExsY protein is required for complete formation of the exosporium of Bacillus anthracis. J. Bacteriol. 188:7440-7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bozue, J., K. L. Moody, C. K. Cote, B. G. Stiles, A. M. Friedlander, S. L. Welkos, and M. L. Hale. 2007. Bacillus anthracis spores of the bclA mutant exhibit increased adherence to epithelial cells, fibroblasts, and endothelial cells but not to macrophages. Infect. Immun. 75:4498-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daubenspeck, J. M., H. Zeng, P. Chen, S. Dong, C. T. Steichen, N. R. Krishna, D. G. Pritchard, and C. L. Turnbough, Jr. 2004. Novel oligosaccharide side-chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J. Biol. Chem. 279:30945-30953. [DOI] [PubMed] [Google Scholar]
- 6.Dong, S., S. A. McPherson, L. Tan, O. N. Chesnokova, C. L. Turnbough, Jr., and D. G. Pritchard. 2008. Anthrose biosynthetic operon of Bacillus anthracis. J. Bacteriol. 190:2350-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dramsi, S., S. Magnet, S. Davison, and M. Arthur. 2008. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol. Rev. 32:307-320. [DOI] [PubMed] [Google Scholar]
- 8.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerhardt, P., and E. Ribi. 1964. Ultrastructure of the exosporium enveloping spores of Bacillus cereus. J. Bacteriol. 88:1774-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 11.Henriques, A. O., and C. P. Moran, Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555-588. [DOI] [PubMed] [Google Scholar]
- 12.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, M. J., S. J. Todd, D. A. Ball, A. M. Shepherd, P. Sylvestre, and A. Moir. 2006. ExsY and CotY are required for the correct assembly of the exosporium and spore coat of Bacillus cereus. J. Bacteriol. 188:7905-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa, M., S. Goto, M. Hattori, K. F. Aoki-Kinoshita, M. Itoh, S. Kawashima, T. Katayama, M. Araki, and M. Hirakawa. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leski, T. A., C. C. Caswell, M. Pawlowski, D. J. Klinke, J. M. Bujnicki, S. J. Hart, and S. Lukomski. 2009. Identification and classification of bcl genes and proteins of Bacillus cereus group organisms and their application in Bacillus anthracis detection and fingerprinting. Appl. Environ. Microbiol. 75:7163-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 18.Moir, A. 2006. How do spores germinate? J. Appl. Microbiol. 101:526-530. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., West Sussex, United Kingdom.
- 21.Oliva, C. R., M. K. Swiecki, C. E. Griguer, M. W. Lisanby, D. C. Bullard, C. L. Turnbough, Jr., and J. F. Kearney. 2008. The integrin Mac-1 (CR3) mediates internalization and directs Bacillus anthracis spores into professional phagocytes. Proc. Natl. Acad. Sci. U. S. A. 105:1261-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priest, F. G., M. Barker, L. W. Baillie, E. C. Holmes, and M. C. Maiden. 2004. Population structure and evolution of the Bacillus cereus group. J. Bacteriol. 186:7959-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redmond, C., L. W. Baillie, S. Hibbs, A. J. Moir, and A. Moir. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 150:355-363. [DOI] [PubMed] [Google Scholar]
- 24.Réty, S., S. Salamitou, I. Garcia-Verdugo, D. J. Hulmes, F. Le Hégarat, R. Chaby, and A. Lewit-Bentley. 2005. The crystal structure of the Bacillus anthracis spore surface protein BclA shows remarkable similarity to mammalian proteins. J. Biol. Chem. 280:43073-43078. [DOI] [PubMed] [Google Scholar]
- 25.Steichen, C., P. Chen, J. F. Kearney, and C. L. Turnbough, Jr. 2003. Identification of the immunodominant and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steichen, C. T., J. F. Kearney, and C. L. Turnbough, Jr. 2005. Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J. Bacteriol. 187:5868-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steichen, C. T., J. F. Kearney, and C. L. Turnbough, Jr. 2007. Non-uniform assembly of the Bacillus anthracis exosporium and a bottle cap model for spore germination and outgrowth. Mol. Microbiol. 64:359-367. [DOI] [PubMed] [Google Scholar]
- 28.Swiecki, M. K., M. W. Lisanby, C. L. Turnbough, Jr., and J. F. Kearney. 2006. Monoclonal antibodies for Bacillus anthracis spore detection and functional analyses of spore germination and outgrowth. J. Immunol. 176:6076-6084. [DOI] [PubMed] [Google Scholar]
- 29.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 30.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2005. Contribution of ExsFA and ExsFB proteins to the localization of BclA on the spore surface and to the stability of the Bacillus anthracis exosporium. J. Bacteriol. 187:5122-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2003. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J. Bacteriol. 185:1555-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamborrini, M., D. B. Werz, J. Frey, G. Pluschke, and P. H. Seeberger. 2006. Anti-carbohydrate antibodies for the detection of anthrax spores. Angew. Chem. Int. Ed. Engl. 45:6581-6582. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, B. M., and G. C. Stewart. 2008. Targeting of the BclA and BclB proteins to the Bacillus anthracis spore surface. Mol. Microbiol. 70:421-434. [DOI] [PubMed] [Google Scholar]
- 34.Todd, S. J., A. J. Moir, M. J. Johnson, and A. Moir. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J. Bacteriol. 185:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vellanoweth, R. L., and J. C. Rabinowitz. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6:1105-1114. [DOI] [PubMed] [Google Scholar]