Abstract
Streptococcus mutans is the primary causative agent of human dental caries, a ubiquitous infectious disease for which effective treatment strategies remain elusive. We investigated a 25-kDa SloR metalloregulatory protein in this oral pathogen, along with its target genes that contribute to cariogenesis. Previous studies have demonstrated manganese- and SloR-dependent repression of the sloABCR metal ion transport operon in S. mutans. In the present study, we demonstrate that S. mutans coordinates this repression with that of certain virulence attributes. Specifically, we noted virulence gene repression in a manganese-containing medium when SloR binds to promoter-proximal sequence palindromes on the S. mutans chromosome. We applied a genome-wide approach to elucidate the sequences to which SloR binds and to reveal additional “class I” genes that are subject to SloR- and manganese-dependent repression. These analyses identified 204 S. mutans genes that are preceded by one or more conserved palindromic SloR recognition elements (SREs). We cross-referenced these genes with those that we had identified previously as SloR and/or manganese modulated in microarray and real-time quantitative reverse transcription-PCR (qRT-PCR) experiments. From this analysis, we identified a number of S. mutans virulence genes that are subject to transcriptional upregulation by SloR and noted that such “class II”-type regulation is dependent on direct SloR binding to promoter-distal SREs. These observations are consistent with a bifunctional role for the SloR metalloregulator and implicate it as a target for the development of therapies aimed at alleviating S. mutans-induced caries formation.
Streptococcus mutans is a known colonizer of the tooth surface and primary etiologic agent of dental caries in humans (2), although it can also be associated with systemic infections that culminate in valvular endocarditis (33). Despite advances in water fluoridation, hygiene, and access to professional dental care in the United States, poor oral health prevails, particularly in communities of low socioeconomic status. Americans spend up to $102 billion on dental services each year, which is more than is spent on cancer, asthma, mental disorders, or trauma (5, 20).
The development of dental caries is dependent, in part, on the accumulation of plaque at the tooth surface, which creates a microenvironment suitable for fermentative metabolism. There are an estimated 600 different species of bacteria present in the human oral cavity (31), with S. mutans cited as the most cariogenic, owing to its acidogenicity (the ability to produce acid) and aciduricity (the ability to withstand acid) (13, 29). Specifically, S. mutans-induced cariogenesis is the result of lactic acid accumulation at the tooth surface, the product of fermentative metabolism of sugars consumed in the diet. Lactic acid, in turn, demineralizes the tooth enamel to initiate decay, which can progress into the underlying dentine and penetrate the tooth pulp (33, 36). The discomfort associated with active caries and the serious health risks posed by S. mutans once it accesses the bloodstream underscore the importance of continued research aimed at alleviating S. mutans-induced disease.
Divalent metal ions, such as iron and manganese, are required for the growth and survival of nearly all microorganisms, including S. mutans (25). Early reports in the literature reveal correlations between drinking water with elevated manganese content and high caries incidence (6). Consistent with this finding is the reported association of manganese in tooth enamel with increased caries frequency (7). More-recent reports propose a role for divalent metal ions in S. mutans sucrose-dependent adherence that is mediated by glucosyltransferases (14). In addition, manganese functions as a necessary cofactor for streptococcal enzymes during lactic acid fermentation (14) and for superoxide dismutase (SOD), which serves as a catalyst for the conversion of toxic superoxide radicals into less harmful by-products. High concentrations of free Mn2+ can also provide direct protection against radical oxygen species (ROS) (24) and so further promote aerotolerance in the oral streptococci (2, 4, 30).
Metal ion homeostasis in bacteria is maintained primarily via the regulation of membrane-associated transporters. Previous studies focused on mechanisms of iron homeostasis and the numerous problems posed by the accumulation of this transition metal in vivo. For instance, ferric iron (Fe3+) in the mammalian host can promote the production of ROS by Fenton chemistry (23, 34); hence, iron is tightly sequestered to host proteins, such as transferrin and lactoferrin, making the concentration of iron available to invading microbes (10−12 μM) too low to support microbial growth and persistence (40, 45). Consequently, bacterial pathogens have evolved specialized mechanisms to rob iron from host proteins, and so they maintain homeostasis by tightly regulating membrane transporters that bind iron for uptake.
Manganese uptake and utilization pose fewer challenges for invading microbes, since manganese is highly soluble and does not catalyze Fenton-type reactions (17). In fact, some bacterial pathogens preferentially accommodate manganese in lieu of iron, presumably to circumvent the toxicity effects associated with Fe utilization (4, 8, 22, 25, 30). Manganese functions as an essential cofactor for numerous bacterial metalloenzymes, including those that mediate photosynthesis, gluconeogenesis, glycolysis, sugar and amino acid metabolism, peptide cleavage, nucleic acid degradation, and signal transduction (25).
Like that of iron, manganese homeostasis is tightly controlled at the site of the bacterial membrane. Among the different families of manganese transport proteins that work to bring manganese into microbial cells are the ATP-binding cassette transporters (ABC transporters) that are controlled at the level of transcription and regulated in response to both intra- and extracellular concentrations of metal ions. Metal ion sensing in S. mutans is mediated by a SloR metalloregulatory protein that can bind Mn2+ or Fe2+ to modulate gene expression. SloR homologs in other microorganisms are similarly responsive to these metal cations and share a mechanism for regulating their permeases. Specifically, MntR in Bacillus subtilis, IdeR in Mycobacterium tuberculosis, ScaR in Streptococcus gordonii, and SloR (Dlg) in S. mutans bind to promoter sequences that precede the ABC transporter operon when either Mn2+ or Fe2+ is plentiful and so prevent transcription of the metal ion permease (26, 27, 28, 35). At lower metal ion concentrations, the metalloregulator dissociates from the promoter region so that expression of the permease is derepressed and metal ion scavenging can ensue. We hypothesize that such S. mutans gene regulation functions to coordinate the expression of essential Mn2+ and/or Fe2+ transporters with that of its virulence traits, particularly during extended periods of metal ion limitation that prevail in the plaque environment between meal times.
Metal ion accumulation in dental plaque lies at the very interface of the host pathogen interaction. In the present study, we propose an important role for SloR and its Mn2+ cofactor in S. mutans virulence gene expression. To address this, we pursued global genomic profiling to reveal S. mutans genes that are subject to SloR/Mn2+ control. Microarrays were used to examine the 1,960 genes present in the S. mutans genome and to reveal the Mn2+ and SloR regulons. An in silico analysis of the S. mutans UA159 genome was then conducted to identify the SloR recognition elements (SREs) that localize upstream of genes belonging to the manganese and/or SloR regulons. Taken together, our findings support manganese- and SloR-dependent, differential gene expression for a multitude of S. mutans virulence genes, many of which harbor a recognizable SRE in their promoter-proximal and/or -distal regions (see Table 2). The continued characterization of this “SloR metalloregulome” in S. mutans can improve our understanding of gene regulation in this important oral cariogen and elucidate potential new targets for anticaries therapy and/or prevention.
TABLE 2.
Genes of interest that derive from genome-wide analyses of S. mutansa
| GenBank locus tag | Gene name | Description | SRE sequence (5′ to 3′) | Space (bp)b | Class | Fold change |
|---|---|---|---|---|---|---|
| SMU.91* | ropA | Trigger factor | GAAAGAAACTAACTTTAATTGC | 105 | II | 2 |
| SMU.113 | pfk | Fructose kinase | I | 4 | ||
| SMU.114 | Fructose PTS | I | 6 | |||
| SMU.115 | Fructose PTS | I | 6 | |||
| SMU.166 | Unknown | CAAAAATATTTTAGATATTTTG | 68 | I | 2 | |
| SMU.182* | sloA | Iron/Mn transport | AAAATTAACTTGACTTAATTTT | 35 | I | 11 |
| SMU.217c* | Unknown | AATAGAAATTAAAGATTTTTTG | 34 | I | 3 | |
| SMU.308 | srlD | Sorbitol dehydrogenase | I | 2 | ||
| SMU.309 | srlR | Sorbitol operon regulator | I | 2 | ||
| SMU.310 | srlM | Sorbitol operon activator | I | 2 | ||
| SMU.311 | srlA | Sorbitol PTS | I | 2 | ||
| SMU.312 | srlE | Sorbitol PTS | AAAAGTATCGTTGCTTATTTCA | 9 | I | 2 |
| SMU.313 | srlB | Sorbitol PTS EII | I | 3 | ||
| SMU.493 | pfl2 | Pyruvate formate lyase | I | 2 | ||
| SMU.494 | mipB | Fructose aldolase | I | 3 | ||
| SMU.495 | gldA | Glycerol dehydrogenase | I | 3 | ||
| SMU.610* | spaP | Surface antigen | GACAAAATCCTGACTTTTTTTG | 197 | II | 6 |
| SMU.629* | sod | Superoxide dismutase | CAAAGCAAGAATGTATATTTAG | 117 | II | 2 |
| SMU.670* | citB | Aconitate hydratase | AGAAAAAAGGAGACTGGTTATG | 0 | I | 3 |
| SMU.671* | citZ | Citrate synthase | I | 3 | ||
| SMU.672* | idh | Isocitrate dehydrog. | I | 3 | ||
| SMU.673 | Unknown | ATCAGAAAACTACCTTTTTTTG | II | 2 | ||
| SMU.770c* | hitA | Iron/Mn transport | AAAAGAAAAATACTTGAATTTA | 197 | I | 3 |
| SMU.872 | fruA | Fructose PTS | AAACGAAAATTATCTTACTTTA | I | 2 | |
| SMU.877 | agaL | α-Galactosidase | I | 2 | ||
| SMU.878 | msmE | Multiple sugar transport | I | 2 | ||
| SMU.879 | msmF | Multiple sugar transport | I | 2 | ||
| SMU.880 | msmG | Multiple sugar transport | I | 2 | ||
| SMU.881 | gtfA | Sucrose phosphorylase | I | 2 | ||
| SMU.882 | msmK | Multiple sugar transport | I | 2 | ||
| SMU.883 | dexB | Dextran glucosidase | I | 2 | ||
| SMU.924* | tpx | Thiol peroxidase | AAAATAAACATATTTTTATTGT | 154 | II | 2 |
| SMU.961 | AAAATTATGTAAAAATATTTTG | 150 | II | 2 | ||
| SMU.1012c | cpsY | Transcriptional regulation | AAAATAATAATGGATTAATTGG | 82 | I | 2 |
| SMU.1148 | lctF | ABC transport | TGAAAAATCCTTGCTTAATTGC | 92 | II | 2 |
| SMU.1165c | acrR | Transcriptional regulator | AAAAGAATGGTTCCTTGTTTTG | 133 | II | 2 |
| SMU.1390 | Unknown | TGAAGAATAATACTTTATTTGG | 284 | II | 2 | |
| SMU.1396c* | gbpC | Glucan binding | GAAAAAATGTCTTTTTATTGTT | 49 | I | 2 |
| SMU.1421* | pdhC | Acid dehydrogenase | I | 6 | ||
| SMU.1422* | pdhB | Pyruvate dehydrogenase | I | 8 | ||
| SMU.1423* | pdhA | Pyruvate dehydrogenase | I | 7 | ||
| SMU.1424* | Pyruvate dehydrogenase | I | 6 | |||
| SMU.1517* | vicR | Response regulator | II | 2 | ||
| SMU.1535* | phsG | Glycogen phosphorylation | GAAAGACAACTTTGAGTTTTCC | Coding sequence | I | 2 |
| SMU.1536* | glgA | Glycogen synthase | CAAATTCAGGAGTCATAATTTC | Coding sequence | I | 3 |
| SMU.1537* | glgD | Glycogen biosynthesis | CTAAATCAAAAGTGATATTGGC | Coding sequence | I | 3 |
| SMU.1538* | glgC | Glucose 1P transferase | I | 2 | ||
| SMU.1539* | glgB | Glycogen branching | I | 2 | ||
| SMU.1566c | malR | Transcriptional repressor | TAAAAAAACTTGACATTTTTCG | 163 | II | 2 |
| SMU.1596 | celD | Cellobiose PTSc | I | 4 | ||
| SMU.1597c | Hypothetical protein | I | 4 | |||
| SMU.1598 | celC | Cellobiose PTS | I | 5 | ||
| SMU.1599 | celR | Transcriptional regulator | I | 5 | ||
| SMU.1600 | celB | cellobiose PTS | I | 3 | ||
| SMU.1601 | celA | 6-P glucosidase | I | 2 | ||
| SMU.1746c | fabM | Acid tolerance | AAAAATAAAAATAGTTTTTTTA | 132 | II | 2 |
| SMU.1788c* | bta | Bacteriocin transport | AAAAGAAACCTTGCAAATTTTG | 167 | II | 2 |
| SMU.1917c* | comE | Genetic competence | AGAAGAAACTTTTCTTTTTTTG | 179 | II | 2 |
| SMU.1924c* | gcrR | Response regulator | AAAAAACTGATGGGTTATTTTG | 201 | II | 2 |
| SMU.1841 | scrA | Sucrose PTS | I | 2 | ||
| SMU.1843 | scrB | Sucrose 6P hydrolase | I | 2 | ||
| SMU.1844 | scrR | Sucrose operon repressor | I | 2 |
Shown is a select list of S. mutans genes that derive from the genome-wide analyses we describe in Materials and Methods. If a “recognizable” SRE in or near a promoter region could be drawn from our in silico analysis, its sequence and distance from the ATG initiation codon (space) is noted. Results for the microarray platform are presented as fold changes in expression for class I (SloR downregulated) and class II (SloR upregulated) genes. The asterisk denotes functional validation of expression in qRT-PCR experiments and/or confirmation of direct SloR binding in EMSA. Gene clusters that are organized in an operon-like arrangement on the UA159 chromosome have consecutive numbers in their GenBank locus tags.
Distance/spacing between the 3′ end of the SloR binding motif and the translation initiation codon.
PTS, phosphotransferase system.
MATERIALS AND METHODS
Bacterial strains and primers.
The bacterial strains used in this study (Table 1) include S. mutans UA159 (serotype c) and a UA159-derived SloR-deficient mutant, GMS584, that was constructed previously in our laboratory (35). All DNA primers were designed using MacVector 7.2 software and purchased from Sigma-Genosys (Table 1).
TABLE 1.
Bacterial strains and oligonucleotide primers used in this study
| Strain, assay, or primer name | Genotype or phenotype description or nucleotide sequence (5′ to 3′) | Annealing temp (°C) | Amplicon length (bp) |
|---|---|---|---|
| Strain | |||
| UA159a | Wild type (serotype c) | ||
| GMS584b | SloR insertion-deletion mutant (UA159-derived); Emr | ||
| Assay and primer name | |||
| qRT-PCR | |||
| bta.RT.kpo.F | GCTCCTAAGTTGAGTCAGGTTGCTG | 53.8 | 191 |
| bta.RT.kpo.R | GCCTCTTGTGAAAGCGATGAATC | ||
| citZ.F | AGTGCCATCAAGGATAATAAACTCTCG | 50.7 | 109 |
| citZ.R | AATGAAGATTCCAAAGGAGGTAAATGAC | ||
| hk11.RT.F | GCTGGCTAATAATGTCATCAAGC | 50.8 | 88 |
| hk11.RT.R | CTCAACAGTTACTTCAATCTCCTCC | ||
| pdh.RT.kpo.F | GTGGCTTCCTTTGGACTGAGTG | 52.8 | 117 |
| pdh.RT.kpo.R | TCCTTGCGTCTCTGTTGATGC | ||
| sloR.RT.F | CATCTCTTTATCGCAAGCATCG | 50.7 | 127 |
| sloR.RT.R | CGTTCAACAAACACATCAGAAACAG | ||
| Smu.217c.RT.kpo.F | ACATTATGGAAGAGGTTGTTC | 45.3 | 105 |
| Smu.217c.RT.kpo.R | AGTTATCGCTTTGTTTTCTATC | ||
| tpx.RT.kpo.F | TGGAAATACGGTGACACTTG | 49.6 | 144 |
| tpx.RT.kpo.R | ATAGATGGCACAACGCTAATC | ||
| EMSA | |||
| pre.bta.F | CGCTTCTGTAAAATGTGACA | 48.3 | 317 |
| pre.bta.R1 | TGACACTTGAAGGGGAAG | ||
| pre.bta.R2 | GAACATAGGTCATTAGTCG | 45.2 | 155 |
| pre.sloABC.For | ATCGGTGAATCGCACTGTCG | 51.1 | 310 |
| pre.sloABC.Rev | TAAGGTTGACTTGCCCGCAC | ||
| recA.Gel.LN.F | CGGTTATCCAAAAGGGCGTATC | 55.3 | 212 |
| recA.Gel.LN.R | CCTGTTCTCCTGAATCTGGTTGTG | ||
| pre.smu.217c.F | CAATCCATAATTGAATATC | 44.7 | 320 |
| pre.smu.217c.R | GGTAAGCTAGTCTATAACG |
ATCC 700610.
See reference 35.
In silico analysis.
Nucleotide sequences preceding the S. mutans sloABC, comD and -E, spaP, sod, ropA, and gcrR genes, shown previously to bind SloR in gel mobility shift experiments (19, 35), were submitted to MEME (12) in search of a conserved motif for SloR binding. Previous work performed by Kitten and coworkers revealed a 22-bp interrupted palindrome in the promoter region of the S. mutans sloABCR operon that shares significant nucleotide sequence identity with the DtxR binding sequence in Corynebacterium diphtheria (27). We therefore set the MEME algorithm to identify S. mutans motifs of 22 bp in length with “sequence is a palindrome” mandated and used this as an input to search the first 300 bp of sequence preceding the sloABC, comD and -E, spaP, sod, ropA, and gcrR genes. The resulting motif that derived from this analysis was used to generate a 4-by-22 position-specific scoring matrix (PSSM). This matrix was then used to search all intergenic regions (IGRs) in the S. mutans UA159 genome for significantly similar motifs with the PROMSCAN (42) and MAST (11) software packages. To calculate nucleotide position frequencies within the SRE motif, we applied the outputs of the PROMSCAN and MAST SRE search to a “make_matrix” PERL script (http://www.promscan.uklinux.net/software.html). Each significant hit was manually located using the Oralgen servers (available at http://oralgen.lanl.gov/), and its distance from the ATG initiation codon of any downstream gene was recorded.
S. mutans high-density microarrays.
The complete sequence of the S. mutans UA159 genome is composed of 2,030,936 bp and contains 1,963 open reading frames (2). Custom high-density microarrays for S. mutans UA159 were designed by NimbleGen and purchased from Affymetrix (Santa Clara, CA) as a NimbleExpress array. The S. mutans arrays contained 190,000 single-stranded probes (25-mers) for interrogation of 1,960 genes and 920 intergenic regions. The controls included a B2 oligonucleotide used for grid alignment and lys, phe, thr, and dap genes as RNA spike-in controls to provide quality control data on synthesis and hybridization efficiencies as well as to estimate assay sensitivity.
Target preparation, hybridization, and GeneChip processing.
Target preparation, hybridization, washing, staining, and array scanning were performed according to the prokaryotic protocols in the Affymetrix GeneChip expression analysis technical manual by the University of Vermont's Microarray Core Facility. Ten micrograms of total RNA was isolated from S. mutans UA159 cells grown to mid-logarithmic phase (optical density at 600 nm [OD600], ∼0.5) in a semidefined medium (SDM) containing 0.1 μM or 10.0 μM Mn2+ and from UA159 and GMS584 cells grown as described above in a conventional manganese-containing SDM. The final metal ion content of these media was confirmed by inductively coupled argon plasma (ICP) analysis. Intact RNAs deriving from two independent experiments were assessed for quality with an Agilent 2100 Bioanalyzer and reverse transcribed into single-stranded cDNA by using random primers (Invitrogen Life Technologies), followed by controlled cDNA fragmentation with DNase I. The resulting fragments were 3′ end labeled with biotin by using terminal transferase and GeneChip DNA labeling reagent (Affymetrix). After confirmation of >80% biotin labeling in a gel shift assay, the labeled product was prepared as a hybridization cocktail according to the Affymetrix technical manual and hybridized to the S. mutans GeneChip for 16 h at 45°C with rotation at 60 rpm in an Affymetrix model 320 hybridization system. Immediately following hybridization, the arrays were washed and stained with streptavidin-phycoerythrin (SAPE) with an automated Affymetrix GeneChip fluidics 450 station, followed by scanning with an Affymetrix GeneChip GS3000 scanner.
Microarray data analysis.
Analysis was performed using tools made available through the Bioconductor Project (http://www.biocondutor.org/). Probe-set expression statistics, referred to here as RMA, were calculated from probe intensities (CEL files) by using the Affymetrix package and the background correction method of Speed (41), the qspline normalization method of Workman et al. (44), and the median polish summary statistic. Paired differences in RMA expression statistics were averaged prior to calculation of the log2 fold change. Differential expression levels of operons between paired samples were evaluated using a t test with paired-probe-set-expression statistics. The assignment of probe sets to operons was guided by the correlation coefficients of adjacent probe set expression patterns across 17 arrays.
Validating the microarray platform with quantitative reverse transcription-PCR (qRT-PCR).
Total RNA was isolated using a modified version of the protocol of Chen et al. (16). Briefly, 10 ml of S. mutans UA159 and GMS584 cells grown to mid-log phase (OD600, 0.5 ± 0.05) was harvested and then treated with RNAprotect (Qiagen) according to the instructions provided by the supplier. The cell pellet was resuspended in 250 μl of a 50 mM Tris-10 mM EDTA buffer before addition of 5 μl of 20% SDS, 300 μl of acid phenol, and 250 μl of glass beads. The solution was then subjected to two rounds of mechanical disruption (30 s each) in a Bio 101 FastPrep machine (MP Biomedicals, OH) and in the presence of zirconium beads prior to centrifugation at 18,000 × g in a Micromax refrigerated microcentrifuge (Thermo Electron Corp., Milford, MA) for 10 min. Two hundred microliters of the supernatant was purified and DNase treated on an RNeasy column (Qiagen) according to the manufacturer's instructions. The resulting intact RNA was reverse transcribed using a first-strand cDNA synthesis kit (MBI Fermentas, Ontario, CA) according to the instructions included with the kit. Reaction mixtures containing no RNA template (NTC) or no reverse transcriptase (RT−) served as controls. The cDNA products (diluted 1.5:100) were used as a template for PCR amplification, along with 150 nM primers (Table 1). Amplification in real time proceeded in a Cepheid SmartCycler II (Sunnyvale, CA), with SYBR green I as the intercalating dye (Invitrogen, Carlsbad, CA). RT− controls were run in parallel with the same primer sets to confirm the absence of contaminating chromosomal DNA in the samples. PCR cycling was set as follows: 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, primer annealing at the optimal temperature for 30 s (Table 1), and primer extension at 72°C for 30 s. Gene expression was standardized relative to the expression of an hk11 endogenous control gene, which we determined does not change in either UA159 or GMS584 when grown under the experimental test conditions. Standard curves were generated for each test gene with a UA159 chromosomal DNA dilution series as a template and gene-specific primer sets, so that DNA copy number could be calculated. Threshold cycle (CT) values were assigned a cycle number at which the second derivative of the fluorescence curve was at its maximum.
EMSA.
Primers were designed to generate amplicons ranging in size from 155 to 317 bp for use in electrophoretic mobility shift assays (EMSA) (Table 1). The PCR products were purified using a QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions and end labeled with 10 U T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and 10 μCi (370 kBq) [γ-32P]ATP (PerkinElmer, Waltham, MA). The end-labeling reaction mixture was incubated at 37°C for 30 min and then at 70°C for 20 min before the final volume was adjusted to 50 μl with sterile distilled H2O. The samples were passed through a TE Select-D G-25 spin column (Roche Applied Science, Indianapolis, IN) to remove unincorporated radiolabel and stored at −80°C. Binding reactions were prepared as follows in a final reaction volume of 16 μl: 1 μl of end-labeled DNA (∼38 picograms), 0.8 μM or 4.4 μM SloR-maltose binding protein (SloR-MBP) fusion prepared previously (19), and 3.2 μl of a freshly made 5× binding buffer (42 mM NaH2PO4, 58 mM Na2HPO4, 250 mM NaCl, 25 mM MgCl2, 50 μg/ml of bovine serum albumin, 1 mg sonicated salmon sperm DNA, 50% [vol/vol] 100% glycerol, and 625 μM MnCl2). EDTA was added at a final concentration of 1.5 mM to select samples to determine the metal ion dependency of SloR binding. To rescue a band shift that was compromised by EDTA, additional MnCl2 (0.32 μl of a fresh 100 mM stock) was added, bringing the final concentration of MnCl2 to 2.1 mM. Reaction mixtures were mixed gently by vortexing and allowed to stand at room temperature for 20 min before they were loaded onto a 6% nondenaturing polyacrylamide gel (3 ml 20× bis-Tris borate (pH 7.4), 74 μl 100 mM MnCl2, 1.5 ml 100% glycerol, 12 ml 30% acrylamide (37.5:1 acrylamide-bis), 300 μl 15% ammonium persulfate (APS), and 90 μl TEMED [N,N,N′,N′-tetramethylethylenediamine]), which was prerun for 1 h at 30 V. Each gel was run at 300 V for 2 to 3 h and then exposed to Kodak BioMax film for 10 to 20 h at −80°C in the presence of an intensifying screen.
Microarray data accession number.
The complete UA159 and GMS584 microarray data sets have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE10215 (19).
RESULTS
Microarray studies.
We used a microarray platform to reveal the S. mutans transcriptome and identify gene expression patterns that are manganese and SloR responsive. In light of reports in the literature that describe heightened transcription of the S. mutans vicR, ftf, and gtfB and -C genes in exponentially growing cells (37), we proceeded to isolate RNA from S. mutans mid-logarithmic-phase cells (OD600, 0.5 ± 0.05) so that we might reveal a role for SloR in the regulation of these and possibly other virulence attributes in expression profiling experiments. From these studies, we noted a total of 131 genes (6.6% of the UA159 genome) that demonstrated at least 2-fold-differential expression when grown in the presence of 0.1 μM versus 10.0 μM manganese; in contrast, 686 genes (35% of the genome) were at least 2-fold differentially expressed in the UA159 versus GMS584 microarray platform (Table 2).
The functional classification for S. mutans manganese- and SloR-responsive genes is represented in Fig. 1, with the majority of the genes being hypothetical. Also prevalent, however, are genes whose products mediate cellular processes, such as oxidative stress tolerance, signal transduction, amino acid biosynthesis, and energy metabolism. We focused our investigation on a subset of genes belonging to these diverse functional classes, all of which encode products that are important to S. mutans-induced caries formation, are SloR modulated in the presence of manganese, and/or are preceded by a predicted SloR recognition element (SRE).
FIG. 1.
Genes belonging to the S. mutans SloR metalloregulome organized by functional class. Shown are the numbers of S. mutans genes that are Mn2+ responsive, SloR regulated, and/or preceded by a predicted SRE, categorized by functional class. Most highly represented across all three of these genome-wide datasets are hypothetical genes, although genes that are involved in cellular processes, protein biosynthesis, transport, and energy metabolism are also prevalent. SloR seems to affect the most S. mutans genes across all 17 functional classes. AA, amino acid; PPNN, purines, pyrimidines, nucleosides, and nucleotides.
SRE identification throughout the S. mutans genome.
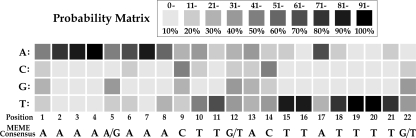
MEME analysis (12) revealed a 22-bp palindromic motif in the promoter regions that precede a plethora of S. mutans virulence genes, including sloABCR, gcrR, sod, spaP, ropA, and comD and -E, all of which we showed to bind SloR directly (19, 35). The motif that the algorithm recognized in the sloABCR promoter region is identical to that reported previously by Kitten et al. (27) and conserved across sloABC homologues in other streptococcal species. All six conserved motifs were used to build a 4-by-22 position-specific scoring matrix (PSSM) which was subsequently used to scan the intergenic regions in the S. mutans UA159 genome for additional interrupted palindromes by using PROMSCAN (42) and MAST (11). The collective results of both searches revealed a total of 273 SREs that reside within the IGRs of S. mutans and that precede a total of 204 unique genes. Herein, we present a probability matrix for the 22-bp SRE motif that derived from these analyses to reflect the likelihood that a given nucleotide will occur at each position within the predicted SloR binding sequence (Fig. 2). The analysis identified all 6 input motifs, thus validating the application of PROMSCAN and MAST for SRE prediction. The algorithms did not reveal any SREs within the promoter region of the S. mutans recA gene, consistent with the results of EMSA that support the absence of a SloR binding motif at this locus and qRT-PCR experiments that reveal recA expression that is independent of SloR control (data not shown).
FIG. 2.
Identification of SloR recognition elements (SREs) in S. mutans. Shown is a probability matrix that was derived from the results of PROMSCAN (42) and MAST (11) searches of the S. mutans UA159 genome. The searches revealed 273 SloR binding palindromes that share significant nucleotide sequence similarity or identity, with a 4-by-22 position-specific scoring matrix (PSSM) generated as described in Materials and Methods. The degree of shading in each square corresponds to the probability that a given nucleotide occurs at that position in the SRE motif. The highest nucleotide probability at each position was used to generate the consensus SRE sequence that is shown below the matrix.
Validation of SloR-modulated gene expression.
We performed qRT-PCR experiments to validate the SloR and/or manganese responsiveness of genes deriving from the microarray platform and falling within the intersects of the Venn diagram shown in Fig. 3. The results of these experiments corroborate the microarray data shown in Table 2 and reveal expression profiles for the S. mutans pdh, tpx, citZ, bta, glgBCDA, and smu.217c genes that differ more than 2-fold in the wild-type UA159 strain and the isogenic SloR-deficient GMS584 mutant (Table 3). Specifically, the pdh, citZ, glgBCDA, and smu.217c genes are subject to classical repression by SloR since their expression was derepressed in the SloR-deficient GMS584 mutant. In contrast, expression of the tpx and bta genes was decreased in GMS584, consistent with a gene-“activating” role for SloR in the wild-type UA159 strain. These findings implicate SloR as a bifunctional regulator of S. mutans gene expression with so-called class I genes, which are subject to SloR repression, and class II genes, which are upregulated by the metalloregulator.
FIG. 3.
Venn diagram of S. mutans genes that are subject to metalloregulation. The Venn diagram reveals the numbers of S. mutans genes that were identified in one or more of our genome-wide analyses. Only genes that were significantly manganese and/or SloR regulated (P < 0.05) and/or preceded by a predicted SRE are represented. We selected the S. mutans genes that reside at the intersects of all three analyses (shaded region) for further investigation because (i) many of them encode S. mutans virulence attributes (e.g., pdh, tpx, citZ, bta, and glgBCDA), (ii) they are manganese responsive, (iii) they are expressed differently in the wild-type UA159 strain and the GMS584 SloR-deficient mutant and hence are SloR-regulated, and (iv) they are preceded by a predicted SRE. For the PROMSCAN and MAST analyses of the SRE data set, we applied a PERL script to remove duplications.
TABLE 3.
Expression levels of S. mutans genes derived from qRT-PCR experiments support the results for the microarray platforma
| Gene name | Gene product description | Expression level in GMS584 | Fold change | Class for regulation |
|---|---|---|---|---|
| pdhC | Pyruvate dehydrogenase | Increased | 4.0 | I |
| citZ | Citrate synthase | Increased | 2.3 | I |
| glgBCDA | Glycogen biosynthesis protein | Increased | 6.0 | I |
| SMU.217c | Hypothetical protein | Increased | 4.2 | I |
| bta | Bacteriocin transport accessory protein | Decreased | 2.4 | II |
| tpx | Thiol peroxidase | Decreased | 2.2 | II |
Shown are the results for qRT-PCR experiments performed with RNA isolated from S. mutans UA159 cells and its SloR-deficient derivative, GMS584, grown in a manganese-replete SDM. These expression patterns support those revealed by the microarray platform. A gene whose expression is decreased in GMS584 implicates SloR as an activator or derepressor of transcription in the UA159 wild-type strain (class II-type regulation). In contrast, increased expression in GMS584 supports repression by SloR in the wild type (class I-type regulation).
SRE positioning is predictive of class I and class II gene regulation.
We noted an interesting correlation between SloR regulation of S. mutans class I and class II genes and SRE positioning relative to the translational start site. Namely, the class I genes sloA, citZ, srlE, gbpC, and smu.217c all harbor SREs that localize within 50 bp of the ATG initiation codon and hence share overlap with the promoter regions for these genes (Table 2). Other class I genes, including those that comprise the glgBCDA operon, harbor a predicted SRE within the 5′ end of their coding sequence. In contrast, the class II genes bta, tpx, gcrR, sod, spaP, ropA, and comE all harbor predicted SREs that localize ∼100 to 300 nucleotides upstream of the translational start site (Table 2). Taken together, these observations suggest a possible mechanism for bifunctional virulence gene regulation by the S. mutans SloR metalloregulator.
SloR binding to class I and class II promoter regions is direct.
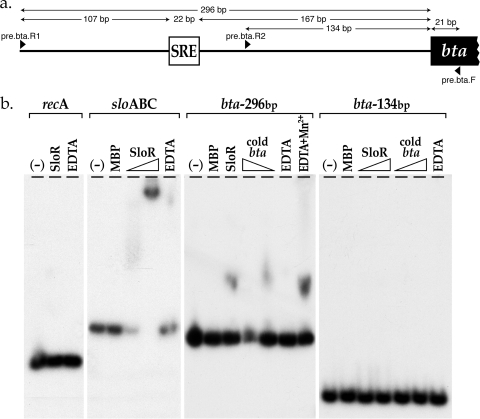
The S. mutans bta gene joins a list of other virulence gene loci previously characterized in our laboratory that are upregulated by SloR (19, 35). To confirm direct SloR binding to the promoter-distal SRE that is located 167 bp upstream of the bta translation start site, and to substantiate a putative relationship between SRE positioning and class II gene regulation in this oral pathogen, we generated a 317-bp amplicon that harbors the bta SRE and a 155-bp deletion derivative that lacks the predicted SRE for subsequent EMSA experiments. Migration of the 317-bp amplicon was hindered in these experiments when the SloR-MBP fusion protein was present in the reaction mixture. This band shift was abrogated, however, when as little as 1.5-fold and as much as 3-fold cold bta competitor DNA was added. The band shift was also abrogated when an EDTA metal ion chelator was included in the reaction, consistent with the metal ion dependence of the SloR-SRE interaction. Migration of the truncated amplicon (designated bta-134) was not retarded by the SloR-MBP fusion protein however (Fig. 4), consistent with SloR binding to the predicted SRE that is resident on the bta-296 amplicon and positioned promoter distal to the bta start site on the S. mutans chromosome.
FIG. 4.
SloR binds directly to the bta promoter region. The results of EMSA confirm a direct and specific interaction between the S. mutans bta promoter region and a SloR-MBP fusion protein. Specifically, a 317-bp amplicon that harbors 296 bp of upstream sequence relative to the bta start codon and the predicted promoter-distal SRE (a) was end labeled and mixed with up to 4.4 μM SloR-MBP before the protein-DNA mixture was resolved on a nondenaturing polyacrylamide gel (b). Run in parallel was the naked 317-bp amplicon in the absence of the fusion protein. A 212-bp amplicon on which the recA promoter region is resident and a 310-bp amplicon that harbors the sloABC promoter region served as negative and positive controls, respectively. A band shift was evident in the sloABC and bta-296 reaction mixtures, indicating direct SloR binding to these SRE-containing amplicons. There was no observable shift of the recA negative control, which lacks a predicted SRE. There was also no band shift when the bta promoter region was incubated with MBP alone, supporting the interaction of the SloR portion of the fusion protein with the DNA binding site. Importantly, a bta amplicon of 155 bp that lacks the predicted SRE (a) was also not shifted by SloR-MBP, supporting a specific interaction between the SloR fusion protein and its target SRE, which is resident on the bta-296 amplicon. The addition of cold bta competitor DNA or EDTA (1.5 mM) to the sloABC and bta-296 reaction mixtures abrogated the band shifts, whereas the addition of manganese to the EDTA-containing mixture rescued the shift. These findings support both the specificity and the metal ion dependence for binding of SloR to an SRE that is located promoter distal to the class II gene bta.
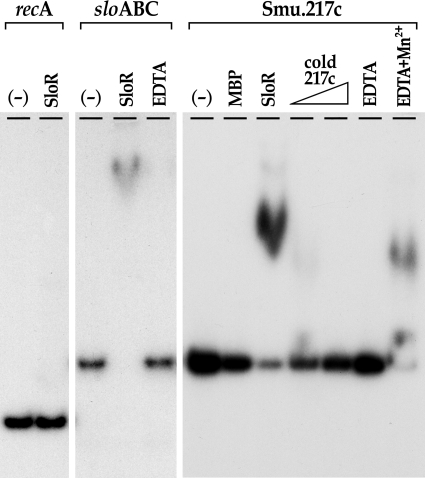
Additional gel mobility shift experiments were performed with the promoter region of the class I gene smu.217c, which is subject to SloR repression. The smu.217c gene harbors a putative promoter-proximal SRE some 34 bp upstream of the ATG initiation codon. As little as 0.8 μM and as much as 4.4 μM SloR-MBP was sufficient to shift a 320-bp amplicon on which the smu.217c promoter region and its predicted SRE are resident (Fig. 5). The addition of up to 3-fold cold competitor DNA to the reaction mixture abrogated the band shift. MnCl2, when added to an EDTA-containing reaction mixture at a molar ratio of 1.4:1, rescued the band shift that was otherwise abrogated upon addition of EDTA alone. These results suggest direct binding of SloR to a promoter-proximal SRE at the smu.217c locus that is both direct and metal ion dependent. Taken collectively, the EMSA results support direct SloR binding to both class I- and class II-type promoter regions, lending further support to a bifunctional role for the SloR metalloregulator in S. mutans.
FIG. 5.
SloR binds directly to the promoter region of the Smu.217 gene cluster. To determine whether the SloR fusion protein binds directly to the Smu.217c promoter region, EMSA experiments were performed with a 320-bp end-labeled amplicon on which the proposed promoter-proximal SRE is resident. The mobility of this amplicon was retarded by the SloR-MBP fusion protein when resolved on a nondenaturing polyacrylamide gel. The band shift was abrogated when up to 3-fold cold competitor DNA or EDTA (1.5 mM) was added to the reaction mixture. The addition of excess Mn2+ (2.1 mM) to the EDTA-containing mixture rescued the band shift. These findings confirm direct SloR binding to the Smu.217c promoter region that is manganese dependent and uphold a correlation between class I gene regulation in S. mutans and a SloR-SRE interaction that is promoter proximal.
DISCUSSION
This report validates a significant role for manganese in the expression patterns of genes in the Streptococcus mutans genome, including genes that are required for virulence (1, 9, 35). In addition, expression profiling of a GMS584 SloR-deficient mutant supports Mn-activated SloR as a global regulator of virulence gene control in this important dental pathogen. We recognize that some of the genes identified as both SloR and manganese responsive in the microarray platform may not be subject to direct regulation by SloR-Mn2+ complexes and that there can be a high rate of false positives associated with these experiments. In addition, we acknowledge that other signaling molecules, such as iron, can bind and activate SloR, which could explain why there are fewer genes at the intersects of the microarray datasets than one might predict if manganese served as the only SloR cofactor. We performed real-time qRT-PCR to validate the microarray expression profiling data. In addition, we undertook an in silico analysis of the S. mutans UA159 genome to identify those SloR-modulated genes that are also associated with a SloR binding recognition element (SRE).
Among the virulence genes that derive from at least two of our three genome-wide analyses and so reside at the intersects of the Venn diagram shown in Fig. 3 are those that comprise the pdhABC locus. The pdhABC gene products are known to function as intermediaries of S. mutans sugar and amino acid metabolism, and more recently, they have been implicated as playing a role in S. mutans aciduricity (29). Specifically, these investigators describe an S. mutans pdhA mutant that is significantly less aciduric than its wild-type UA159 progenitor, and the results of qRT-PCR experiments support pdhA expression that is upregulated under conditions of acid stress. The S. mutans pdhABC genes are organized as an operon on the UA159 chromosome, and we present evidence to support class I-type repression of these genes by SloR. In fact, the impact of SloR on pdhA expression is among the most robust that we observed in our expression profiling experiments, with 4-fold derepression noted for the GMS584 SloR-deficient mutant. Although the class I repressible effect of SloR on pdhABC expression is consistent with our analysis that took place at the neutral pH, we were puzzled by the apparent absence of a recognizable SRE binding palindrome at this locus. We can attribute this (and other SloR-modulated genes listed in Table 2 that lack a recognizable SRE) to the low G+C content of the S. mutans genome, which may preclude the identification of the SRE that is itself A/T rich. Alternatively, it is also possible that SloR binds to nonconsensus SREs or that some genes are targets of SloR-modulated small RNAs (sRNAs). In fact, we did identify several unique sRNAs in silico that reside in the intergenic regions (IGRs) of the UA159 genome and that harbor predicted SREs in their 5′ untranslated regions (UTRs). We went on to cross-reference these sRNAs with the results of our expression profiling experiments to elucidate those IGRs that are actively transcribed and dependent on SloR and/or manganese for expression. We are presently investigating the role of three candidate sRNAs that we identified with TargetRNA analyses (43).
The S. mutans citZ gene is also subject to class I-type repressible regulation by SloR (Table 2). Its gene product is a citrate synthetase which participates in glutamate synthesis as part of the partial tricarboxylic acid (TCA) cycle in S. mutans. Glutamate biosynthesis has been implicated in the S. mutans stringent response to facilitate bacterial survival in the lower layers of the plaque biofilm where nutrients become limiting (18). In this study, we report on the results of qRT-PCR experiments that reveal heightened citZ expression in the SloR mutant (Table 3), consistent with class I-type repression at this locus in the wild-type strain. We also noted that citZ transcription, along with that of the adjacent citB and citC genes, was increased up to 7.5-fold in a manganese-limiting (0.1 μM MnCl2) SDM relative to its expression in the same medium supplemented with 10 μM MnCl2 (data not shown). These observations corroborate the results of the microarray platform and support class I-type repressible regulation of the S. mutans stringent response by a SloR-Mn2+ complex. The SRE that is associated with the citBZC locus is positioned immediately upstream of the S. mutans citB gene and shares overlap with the citB start codon (Table 2).
In previous reports, we describe S. mutans intracellular polysaccharide (IPS) accumulation encoded by the glgBCDA operon as a significant contributor to caries formation (38, 39), given the ability of S. mutans to metabolize these glycogen-like storage polymers when exogenous dietary carbohydrates become limiting in the oral cavity, such as during periods of famine. Herein, we confirm S. mutans glg gene expression that is SloR dependent and subject to class I-type repression. In fact, from the more than 3,000 genes that we analyzed in the UA159 and GMS584 microarray platforms, the glgBCDA genes are among the most robust for differential expression. The results of qRT-PCR experiments corroborate these findings, with glgBCDA transcription that is upregulated 6-fold in the GMS584 SloR-deficient mutant relative to the wild-type level (Table 3). The identification of SloR as a class I regulator of S. mutans IPS accumulation is significant because the mobilization of these intracellular stores during periods of famine essentially extends the period of exposure of host tissues to organic acids well beyond mealtimes, thereby exacerbating tooth decay. SloR repression of the IPS-encoding glg genes makes sense during mealtimes when manganese is available as a corepressor and exogenous dietary carbohydrates are readily available for metabolism. In contrast, when Mn is depleted shortly after a mealtime but excess carbon prevails, the S. mutans glg genes become derepressed and the synthesis of carbon storage reserves is facilitated. Interestingly, several SREs are predicted at the glg locus (Table 2), each localized within the coding sequence of genes that comprise the glgBCDA operon.
Among the SloR- and manganese-responsive virulence genes that are subject to class II-type upregulation in S. mutans is tpx, which encodes a thiol peroxidase that acts as an antioxidant to convert toxic oxygen radicals into harmless by-products. Hydroxyl and peroxide radicals are major stressors for S. mutans in the plaque environment, and as a consequence, evolution has selected for an adaptive response that depends on antioxidants for S. mutans survival and pathogenesis (20). In the present study, we confirm S. mutans tpx expression that is upregulated more than 2-fold in the SloR-proficient UA159 strain and downregulated in the GMS584 SloR-deficient mutant (Table 3). We also noted increased tpx expression in UA159 cells grown in a manganese-replete (10 μM MnCl2) as opposed to a manganese-deplete (0.1 μM MnCl2) SDM (data not shown). The S. mutans tpx gene harbors a promoter-distal SRE ∼154 bp upstream of the ATG initiation codon, consistent with class II-type upregulation by the SloR metalloregulator.
The consensus SRE palindrome that directs binding of SloR to these and other virulence gene loci on the S. mutans chromosome is presented in Fig. 2. On the basis of the 204 predicted SREs that were derived from our genome-wide search of the S. mutans UA159 chromosome and the probability determination for nucleotide positioning within this consensus palindrome, we predict that the adenines at positions 2, 3, 4, and 7 and the thymines at positions 15, 16, and 18 to 21 are likely paramount for SloR-SRE binding. We are currently generating nucleotide substitutions at these positions to reveal their putative vulnerability to DNase I digestion in DNA footprinting experiments using a C-terminal His-tagged SloR fusion protein.
As noted above and in Table 2, many of the SloR-repressible genes (class I) in S. mutans harbor predicted SREs that localize within 50 bp of the ATG initiation codon. In contrast, SloR upregulated genes (class II) have predicted SREs that localize ∼100 to 300 bp upstream of the initiation codon. Based on these observations, we set out to investigate these SloR-modulated genes in EMSA to reveal whether binding of SloR to their promoter-proximal and -distal SREs is direct and extrapolative to class I/II gene regulation. We focused our analyses on the S. mutans smu.217c and bta genes that are SloR-modulated, preceded by promoter-proximal and -distal SREs, respectively, and subject to respective class I- and class II-type control.
The S. mutans bta gene is annotated as a putative bacteriocin transport accessory protein (Smu.1788c) (http://oralgen.lanl.gov) that shares more than 40% nucleotide sequence identity with bta genes from pneumococcus and group B streptococci. Bacteriocins are bacterial toxins with a narrow-spectrum killing capacity, and those that are elaborated by S. mutans can promote colonization of the tooth surface by inhibiting the growth of closely related bacteria in the plaque environment (32). Bacteriocin transport can be mediated by type II secretion machinery, but in S. mutans, it is mediated by ABC-type transport systems (21). The results of EMSA experiments presented in Fig. 4 support a manganese requirement for binding of SloR to a 317-bp bta promoter fragment, which harbors a predicted SRE that resides 167 bp upstream of the bta translation start site on the S. mutans UA159 chromosome. We observed no such binding to a truncated 155-bp bta promoter fragment, which does not harbor the predicted SRE. These findings support a correlation between class II-type transcriptional control and a promoter-distal SloR-SRE interaction that likely allows promoter access to RNA polymerase and subsequent transcription.
The Smu.217 locus harbors a gene cluster that occupies a 16.7-kb region on the S. mutans chromosome. In the present study, we noted that expression of smu.217c, the first gene in the cluster that encodes a hypothetical protein, is derepressed more than 4-fold in a GMS584 SloR-deficient mutant relative to the level for its UA159 wild-type progenitor. Hence, the result of SloR binding to the promoter-proximal SRE is class I-type repressible regulation of the smu.217 gene cluster. The promoter-proximal positioning of this SRE, some 34 bp upstream of the smu.217c start codon, likely overlaps regulatory sequences that direct the expression of this gene cluster, thereby precluding transcription. Additional gel mobility shift experiments support SloR binding to a promoter-proximal sequence at this locus that is both specific and metal ion dependent (Fig. 5). These results also implicate SloR binding at promoter-proximal SREs as foretelling of class I-type repressible regulation.
Taken together, this work demonstrates that both transcriptional up- and downregulation can derive from the direct binding of SloR to promoter-associated SREs. This led us to propose a role for SloR as a bifunctional regulator of S. mutans gene expression, with SloR acting as a repressor when it binds to promoter-proximal SREs and as an activator when it binds to SREs at promoter-distal sites. Specifically, we propose that class I-type repressible regulation occurs when SloR binds to promoter-proximal SREs with high affinity and as a homodimer when manganese is available and that such binding leads to transcriptional repression upon blocking of promoter access by RNA polymerase. The high-affinity, homodimeric binding of SloR to SREs that localize downstream of the transcription start site could also lead to gene repression by impeding transcription elongation. This model is consistent with SRE positioning within the coding sequences of class I repressed genes that comprise the S. mutans glgBCD operon. For class II-type regulation, we predict that SloR binding to promoter-distal SREs occurs with lower affinity to stabilize the downstream RNA polymerase-promoter interaction and facilitate transcription. There is precedence in the literature to support these regulatory models in S. mutans. Namely, the DtxR metalloregulator in Corynebacterium diphtheriae, a homolog of SloR, is similarly bifunctional, with DtxR-mediated repression versus activation defined, in part, by the location of the DtxR binding site relative to the transcription start site of the DtxR-regulated gene (15). Transcription factors in Escherichia coli also function as repressors when the DNA binding protein binds to a promoter-proximal sequence and as activators upon binding to sequences that localize further upstream of the transcription start site (10, 30). The work of Babu and Teichmann (10) extends the model for class I-type regulation to include binding sites that localize well downstream of the transcription start site. By and large, the mechanistic details that underlie bifunctional regulation in these and other microbial pathogens remain largely unknown, however, and warrant further investigation. To confirm a role for SRE positioning in the bifunctionality of SloR regulation, we are currently monitoring S. mutans transcriptional fusions in a manganese-replete medium to reveal the putative impact of ectopic SREs on the promoter activity of class I and class II genes.
In sum, this report underscores the importance of metalloregulation in S. mutans virulence gene expression and suggests potential mechanisms for bifunctional regulation by the SloR metalloregulator. In previous reports, we and others describe manganese as a necessary cofactor for SloR-DNA binding, which, in turn, can affect the expression of downstream genes (40). This study presents the results of a genome-wide analysis of the SloR metalloregulome and the repertoire of genes that are subject to SloR and manganese control and preceded by a predicted SRE. Indeed, the expression of genes belonging to a diverse array of functional classes is affected by SloR, thereby supporting the SloR protein as a global regulator in S. mutans. Future work should more fully elucidate the details of the SloR-SRE interaction in this important oral pathogen, including the identification of critical amino acids that affect SloR binding specificity and affinity. Whether increasing concentrations of manganese bring on conformational changes in the SloR protein that can influence SloR binding to promoter-proximal and/or -distal SREs should also be explored. Accumulating evidence could reveal the SloR-SRE interaction as an appropriate target for rational drug design so that virulence gene expression in S. mutans may be constitutively repressed and the caries-forming process alleviated.
Acknowledgments
This research was supported by funds from the National Institutes of Health (NIH), grant R01 DE014711, to G.A.S.; from the Vermont Genetics Network to K.P.O. through the Institutional Development Award Network of Biomedical Research Excellence (INBRE) Program of the National Center for Research Resources, a component of the NIH; and from the Middlebury College Department of Biology.
We thank Scott Tighe at the University of Vermont Cancer Center DNA Analysis Facility for performing the microarray hybridizations and Gary Xie at the Los Alamos National Laboratories for his mentorship and guidance with the in silico analyses of the S. mutans genome.
Footnotes
Published ahead of print on 13 November 2009.
REFERENCES
- 1.Adkins, B. L., and F. L. Losee. 1970. A study of the covariation of dental caries prevalence and multiple trace element content of water supplies. N. Y. State Dent. J. 36:618-622. [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Anjem, A., S. Varghese, and J. A. Imlay. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72:844-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anonymous. 2009. Oral health: preventing cavities, gum disease and tooth loss. Centers for Disease Control and Prevention National Center for Chronic Disease Prevention and Health Promotion, Atlanta, GA.
- 6.Aranha, H., R. C. Strachan, J. E. Arceneaux, and B. R. Byers. 1982. Effect of trace metals on growth of Streptococcus mutans in a Teflon chemostat. Infect. Immun. 35:456-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archibald, F. 1986. Manganese: its acquisition by and function in the lactic acid bacteria. Crit. Rev. Microbiol. 13:63-109. [DOI] [PubMed] [Google Scholar]
- 8.Archibald, F. S., and I. Fridovich. 1981. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J. Bacteriol. 145:442-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arirachakaran, P., E. Benjavongkulchai, S. Luengpailin, D. Ajdic, and J. A. Banas. 2007. Manganese affects Streptococcus mutans virulence gene expression. Caries Res. 41:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babu, M. M., and S. A. Teichmann. 2003. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 19:75-79. [DOI] [PubMed] [Google Scholar]
- 11.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. [DOI] [PubMed] [Google Scholar]
- 12.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 13.Banas, J. A. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 9:1267-1277. [DOI] [PubMed] [Google Scholar]
- 14.Bauer, P. D., C. Trapp, D. Drake, K. G. Taylor, and R. J. Doyle. 1993. Acquisition of manganous ions by mutans group streptococci. J. Bacteriol. 175:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brune, I., H. Werner, A. T. Huser, J. Kalinowski, A. Puhler, and A. Tauch. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, Y. Y., C. A. Weaver, D. R. Mendelsohn, and R. A. Burne. 1998. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180:5769-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheton, P. L., and F. S. Archibald. 1988. Manganese complexes and the generation and scavenging of hydroxyl free radicals. Free Radic. Biol. Med. 5:325-333. [DOI] [PubMed] [Google Scholar]
- 18.Cvitkovitch, D. G., J. A. Gutierrez, and A. S. Bleiweis. 1997. Role of the citrate pathway in glutamate biosynthesis by Streptococcus mutans. J. Bacteriol. 179:650-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunning, D. W., L. W. McCall, W. F. Powell, Jr., W. T. Arscott, E. M. McConocha, C. J. McClurg, S. D. Goodman, and G. A. Spatafora. 2008. SloR modulation of the Streptococcus mutans acid tolerance response involves the GcrR response regulator as an essential intermediary. Microbiology 154:1132-1143. [DOI] [PubMed] [Google Scholar]
- 20.Fox, C. 2009. Tooth decay: oral, dental, and craniofacial research can lead to improvements in oral health for all Americans. American Association for Dental Research, Alexandria, VA.
- 21.Hale, J. D. F., N. C. K. Heng, R. W. Jack, and J. R. Tagg. 2005. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. J. Bacteriol. 187:5036-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horsburgh, M. J., S. J. Wharton, M. Karavolos, and S. J. Foster. 2002. Manganese: elemental defence for a life with oxygen. Trends Microbiol. 10:496-501. [DOI] [PubMed] [Google Scholar]
- 23.Horsburgh, M. J., S. J. Wharton, A. G. Cox, E. Ingham, S. Peacock, and S. J. Foster. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44:1269-1286. [DOI] [PubMed] [Google Scholar]
- 24.Inaoka, T., Y. Matsumura, and T. Tsuchido. 1999. SodA and manganese are essential for resistance to oxidative stress in growing and sporulating cells of Bacillus subtilis. J. Bacteriol. 181:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakubovics, N. S., and H. F. Jenkinson. 2001. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology 147:1709-1718. [DOI] [PubMed] [Google Scholar]
- 26.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 27.Kitten, T., C. L. Munro, S. M. Michalek, and F. L. Macrina. 2000. Genetic characterization of a Streptococcus mutans LraI family operon and role in virulence. Infect. Immun. 68:4441-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolenbrander, P. E., R. N. Andersen, R. A. Baker, and H. F. Jenkinson. 1998. The adhesion-associated sca operon in Streptococcus gordonii encodes an inducible high-affinity ABC transporter for Mn2+ uptake. J. Bacteriol. 180:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korithoski, B., C. M. Levesque, and D. G. Cvitkovitch. 2008. The involvement of the pyruvate dehydrogenase E1alpha subunit, in Streptococcus mutans acid tolerance. FEMS Microbiol. Lett. 289:13-19. [DOI] [PubMed] [Google Scholar]
- 30.McEwan, A. G. 2009. New insights into the protective effect of manganese against oxidative stress. Mol. Microbiol. 72:812-814. [DOI] [PubMed] [Google Scholar]
- 31.Nasidze, I., J. Li, D. Quinque, K. Tang, and M. Stoneking. 2009. Global diversity in the human salivary microbiome. Genome Res. 19:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen, T., Z. Zhang, I. H. Huang, C. Wu, J. Merritt, W. Shi, and F. Qi. 2009. Genes involved in the repression of mutacin I production in Streptococcus mutans. Microbiology 155:551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paik, S., A. Brown, C. L. Munro, C. N. Cornelissen, and T. Kitten. 2003. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J. Bacteriol. 185:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierre, J. L., and M. Fontecave. 1999. Iron and activated oxygen species in biology: the basic chemistry. Biometals 12:195-199. [DOI] [PubMed] [Google Scholar]
- 35.Rolerson, E., A. Swick, L. Newlon, C. Palmer, Y. Pan, B. Keeshan, and G. Spatafora. 2006. The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J. Bacteriol. 188:5033-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell, R. 2000. Pathogenesis of oral streptococci, p. 272. In V. Fischetti, R. Novick, J. Ferretti, D. Portnoy, and J. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, DC.
- 37.Shemesh, M., A. Tam, M. Feldman, and D. Steinberg. 2006. Differential expression profiles of Streptococcus mutans ftf, gtf, and vicR genes in the presence of dietary carbohydrates at early and late exponential growth phases. Carbohydr. Res. 341:2090-2097. [DOI] [PubMed] [Google Scholar]
- 38.Spatafora-Harris, G., S. M. Michalek, and R. Curtiss III. 1992. Cloning of a locus involved in Streptococcus mutans intracellular polysaccharide accumulation and virulence testing of an intracellular polysaccharide-deficient mutant. Infect. Immun. 60:3175-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spatafora, G., K. Rohrer, D. Barnard, and S. Michalek. 1995. A Streptococcus mutans mutant that synthesizes elevated levels of intracellular polysaccharide is hypercariogenic in vivo. Infect. Immun. 63:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spatafora, G., M. Moore, S. Landgren, E. Stonehouse, and S. Michalek. 2001. Expression of Streptococcus mutans fimA is iron-responsive and regulated by a DtxR homologue. Microbiology 147:1599-1610. [DOI] [PubMed] [Google Scholar]
- 41.Speed, T. P. (ed.). 2003. Interdisciplinary statistics: statistical analysis of gene expression microarray data. Chapman & Hall/CRC Press, Boca Raton, FL.
- 42.Studholme, D. J., and R. Dixon. 2003. Domain architectures of sigma54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tjaden, B., S. S. Goodwin, J. A. Opdyke, M. Guillier, D. C. Fu, S. Gottesman, and G. Storz. 2006. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 34:2791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Workman, C., L. J. Jensen, H. Jarmer, R. Berka, L. Gautier, H. B. Nielser, H. H. Saxild, C. Nielsen, S. Brunak, and S. Knudsen. 2002. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3(9):research0048.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]