Abstract
The human oropharyngeal pathogen Aggregatibacter actinomycetemcomitans synthesizes multiple adhesins, including the nonfimbrial extracellular matrix protein adhesin A (EmaA). EmaA monomers trimerize to form antennae-like structures on the surface of the bacterium, which are required for collagen binding. Two forms of the protein have been identified, which are suggested to be linked with the type of O-polysaccharide (O-PS) of the lipopolysaccharide (LPS) synthesized (G. Tang et al., Microbiology 153:2447-2457, 2007). This association was investigated by generating individual mutants for a rhamnose sugar biosynthetic enzyme (rmlC; TDP-4-keto-6-deoxy-d-glucose 3,5-epimerase), the ATP binding cassette (ABC) sugar transport protein (wzt), and the O-antigen ligase (waaL). All three mutants produced reduced amounts of O-PS, and the EmaA monomers in these mutants displayed a change in their electrophoretic mobility and aggregation state, as observed in sodium dodecyl sulfate (SDS)-polyacrylamide gels. The modification of EmaA with O-PS sugars was suggested by lectin blots, using the fucose-specific Lens culinaris agglutinin (LCA). Fucose is one of the glycan components of serotype b O-PS. The rmlC mutant strain expressing the modified EmaA protein demonstrated reduced collagen adhesion using an in vitro rabbit heart valve model, suggesting a role for the glycoconjugant in collagen binding. These data provide experimental evidence for the glycosylation of an oligomeric, coiled-coil adhesin and for the dependence of the posttranslational modification of EmaA on the LPS biosynthetic machinery in A. actinomycetemcomitans.
The Gram-negative, nonmotile, microaerophilic, and oropharyngeal bacterium Aggregatibacter actinomycetemcomitans preferentially colonizes the subgingival region of the human oral cavity. This microorganism is implicated as the etiological agent of localized aggressive periodontitis (9, 13) and causes extraoral infections, including pneumonia, osteitis (30), and infective endocarditis (6). Recent studies also link this periodontal pathogen to cardiovascular diseases, such as atherosclerosis (20).
Typical of Gram-negative bacteria, the outer membrane of A. actinomycetemcomitans possesses an asymmetric lipid-protein bilayer. The inner leaflet of the outer membrane is mainly phospholipids, and the outer leaflet consists of lipopolysaccharide (LPS), phospholipids, and proteins (4). LPS molecules are ubiquitously distributed on the outer membrane and are essential for maintaining the membrane integrity (3). Intact LPS molecules are also required for the assembly of some large outer membrane proteins (3, 18, 41). A typical LPS molecule is composed of hydrophobic lipid A, a nonrepeat core oligosaccharide, and a repeating O-antigen or O-polysaccharide (O-PS). The distal O-PS is a major antigen, stimulating the host immune response, and the basis for serotyping Gram-negative bacteria (36), including A. actinomycetemcomitans (32, 50).
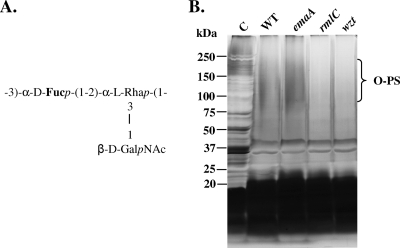
Six different serotypes (a to f) and the corresponding genetic loci have been identified for A. actinomycetemcomitans (19, 22, 27, 44, 50, 54, 55). Serotype b remains one of the common serotypes found in the human oral cavity (9, 13, 51). The serotype b O-PS of A. actinomycetemcomitans is encoded by an operon composed of 21 genes, which are responsible for the biosynthesis of the repeating trisaccharide unit of this particular serotype (53, 55). Each O-PS unit of serotype b contains a disaccharide backbone composed of d-fucose (d-Fuc) and l-rhamnose (l-Rha), linked by a nonreducing d-N-acetylgalactosamine (d-GalNAc) at the O-3 position of l-Rha (33) (Fig. 1A).
FIG. 1.
(A) O-PS structure of serotype b A. actinomycetemcomitans. (B) Silver-stained 5 to 15% polyacrylamide-SDS gel of serotype b LPS. A total of 1.0 ml of mid-logarithmic-phase cells were collected and lysed. Three lysates from each strain were combined and treated with proteinase K at 60°C for 60 min before electrophoresis, followed by silver staining. C, control: whole-cell lysate without proteinase K digestion; WT, wild type (VT1169); emaA, extracellular matrix protein adhesin A mutant; rmlC, rhamnose epimerase mutant; wzt, ATP-binding cassette sugar transport mutant. The dark brown staining of the high molecular weight (75,000 to 250,000) corresponds to polymerized O-PS.
The assembly of LPS molecules in Gram-negative bacteria involve diverse enzymes and pathways due to the variation of the O-PS structures among different bacteria (36). RmlC (previously RfbD), Wzt (previously AbcA or RfbB), and WaaL are three enzymes involved in different stages of the LPS synthesis of some Gram-negative bacteria (7, 36, 37). A homologue of RmlC, TDP-4-keto-6-deoxy-d-glucose 3,5-epimerase, which is required for l-Rha synthesis, has been identified in A. actinomycetemcomitans (53, 55). Wzt is an ATP binding cassette (ABC) transporter that exports saccharide polymers from the cytoplasm to the periplasmic space (7, 36). A homologue of wzt was originally identified from a serotype b strain of A. actinomycetemcomitans, based on protein sequence identity with Aeromonas salmonicida (55). Kaplan et al. (19) later showed that a serotype f wzt mutant strain of A. actinomycetemcomitans produces less O-PS. WaaL, an O-antigen ligase found in Escherichia coli and Pseudomonas aeruginosa, ligates an undecaprenol pyrophosphate-linked oligo- or polysaccharide onto the lipid A-core oligosaccharide in the periplasm (1, 36). A putative O-antigen ligase is located in the chromosome of a serotype b A. actinomycetemcomitans strain (HK1651), based on sequence homology (Oralgen, Los Alamos, NM).
Our earlier work suggested a correlation between the type of LPS molecule and the form of EmaA synthesized by A. actinomycetemcomitans (46). The EmaA of serotype b A. actinomycetemcomitans is a 202-kDa protein that forms the antennae-like appendages found on the surface of A. actinomycetemcomitans and is required for collagen binding (40). The appendages are composed of three EmaA monomers that oligomerize to form an ellipsoidal structure required for the collagen binding activity (56, 57). The ellipsoidal structure corresponds to the amino termini of the proteins and is located at the distal end of a long stalk domain that is attached to the outer membrane by the carboxyl termini (57). The carboxyl termini of the proteins assume β-barrel structures required for pore formation and translocation of the molecules through the outer membrane, similar to those of other type Vc autotransporter proteins (14). Recently, we have demonstrated that EmaA is important in the initiation of infective endocarditis in a rabbit model of infectious endocarditis (45).
Two transposon mutant strains (rmlC and wzt) and a waaL mutant strain generated by site-directed insertional mutagenesis have been developed and characterized in this study. The rmlC mutant did not synthesize l-Rha and did not produce detectable O-PS. The wzt and waaL mutant strains synthesized less O-PS than the wild-type strain. Complementation of the mutant strains restored the production of the serotype b O-PS to wild-type levels. An increase in the electrophoretic mobility of the EmaA monomer was observed in all three mutants, which suggests the presence of carbohydrate. The EmaA mobility reverted to wild-type mobility upon complementation. The presence of carbohydrate associated with EmaA was confirmed by lectin blotting, and in vitro collagen binding assessment demonstrated that the glycoconjugant is important for the full function of this adhesin. The experimental data suggest that EmaA contains carbohydrate similar to that present in O-PS and is a substrate for the O-antigen ligase of the LPS biosynthetic pathway of A. actinomycetemcomitans.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All A. actinomycetemcomitans mutant strains used in this study are based on the nonfimbriated strain VT1169, a spontaneous rifampin- and nalidixic acid-resistant mutant derived from the clinical strain SUNY465, and is referred to as the wild-type strain in this study (24) (Table 1). A. actinomycetemcomitans strains were grown statically in 3% Trypticase soy broth-0.6% yeast extract (TSBYE; Becton, Dickinson and Company) in a 37°C incubator with 10% humidified carbon dioxide. All mutants in this study retained growth characteristics similar to those of the wild-type strain. Escherichia coli strains were grown in 1% Bacto tryptone, 0.1% yeast extract, and 0.5% sodium chloride (Luria-Bertani [LB]) medium at 37°C under aerobic conditions with agitation.
TABLE 1.
Strainsa
| Strain | Genotype/remarks | Source or reference |
|---|---|---|
| A. actinomycetemcomitans | ||
| VT1169 | Wild type, a Rifr Nalr derivative of SUNY465, serotype b | 24 |
| VT1169 (pKM2/emaA) | VT1169 transformed with plasmid pKM2/emaA (EmaA) | This study |
| emaA mutant (emaA::Sp) | Spectinomycin adenyltransferase gene (aad9) inserted into emaA | 24 |
| rmlC mutant (rmlC::pLOF/Sp) | Transposon pLOF/Sp inserted into rmlC (TDP-4-keto-6-deoxy-d-glucose 3,5-epimerase) | This study |
| rmlC mutant/rmlC | rmlC mutant complemented with plasmid pKM2/ltxP/rmlC | This study |
| wzt mutant (wzt::pLOF/Sp) | Transposon pLOF/Sp inserted into wzt (ATP binding cassette transporter) | This study |
| waaL mutant (waaL::pKM221) | Plasmid pKM221 inserted into waaL (O-antigen ligase) | This study |
| waaL mutant/waaL | waaL mutant complemented with plasmid pKM2/ltxP/waaL | This study |
| E. coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lazZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(are, leu)7697 galU galK λ−rpsL nupG tonA | Invitrogen, Carlsbad, CA |
| DH5α (λ pir) | endA1 hadR17(r− m+) supE44 thi-1 recA gyrA1(Nalr) relA1 Δ(lacIZYA-argF)U169 deoR (φ80dlacΔ(lacZ)M15) λ pir | 25 |
| SM10 (λ pir) | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km pir R6K | 25 |
Rif, rifampin; Nal, nalidixic acid; Sp, spectinomycin.
The rmlC and wzt mutants described in this study were isolated from a transposon mutant library, and the integration sites of the transposon within the A. actinomycetemcomitans chromosome were determined as described previously (24). The gene with the inserted transposon was identified based on the A. actinomycetemcomitans genomic DNA database (strain HK1651; Oralgen, Los Alamos, NM) (http://www.oralgen.lanl.gov/). The sequencing was performed at the University of Vermont Cancer Center DNA Analysis Facility.
The shuttle plasmid pKM2 was used for genetic complementation of the O-PS mutants (10) (Table 2). The 520-bp leukotoxin (ltx) promoter of A. actinomycetemcomitans (5) was used as the promoter for the expression of the rmlC and waaL genes in the complemented strains. The EmaA-overproducing strain was developed by transformation of the wild-type strain VT1169 with a plasmid containing the emaA sequence and 500 bp upstream of the start codon (pKM2/emaA).
TABLE 2.
Plasmidsa
| Plasmid | Description | Reference |
|---|---|---|
| pVT1461 | A derivative of pGP704, containing the spectinomycin adenyltransferase gene (aad9), Spr | 25 |
| pKM221 | pVT1461 containing 5-545 bp of waaL, Spr | This study |
| pLOF/Sp | Tn10-based transposon vector; Apr Spr | 24 |
| pKM2 | pPK1 containing chloramphenicol acetyltransferase, Cmr | 10 |
| pKM2/emaA | pKM2 containing ∼500-bp upstream sequence of emaA (the putative promoter region) + emaA sequence, Cmr | This study |
| pKM2/ltxP | pKM2 containing ∼500-bp ltx promoter, Cmr | This study |
| pKM2/ltxP/rmlC | pKM2 containing ∼500-bp ltx promoter + rmlC sequence, Cmr | This study |
| pKM2/ltxP/waaL | pKM2 containing ∼500-bp ltx promoter + waaL sequence, Cmr | This study |
Ap, ampicillin; Cm, chloramphenicol; Sp, spectinomycin.
Complementation of rmlC.
The complete rmlC (rfbD) sequence (GenBank accession no. DQ119107) was amplified using the following primers, 5′-CTCGAGATGAAAGTTATTG-3′ and 5′-GAATTCTTAAAATTTTACCG-3′, which were engineered with 5′-XhoI and 5′-EcoRI restriction endonuclease sites, respectively (italics indicate restriction sites). The 550-bp fragment amplified from the chromosomal DNA of VT1169 was sequenced and found to be identical to that of the rmlC gene of HK1651 (http://www.oralgen.lanl.gov/). The PCR product was cloned into a pCR2.1-TOPO cloning vector (Invitrogen), transformed into TOP10 One Shot E. coli competent cells, and selected on LB plates containing 100 μg/ml ampicillin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG). The 5′-XhoI-rmlC-EcoRI-3′ fragment was isolated and ligated with the complementary sites of pKM2/ltx, which was restricted with the same enzymes, followed by dephosphorylation with shrimp alkaline phosphatase (USB Corporation, Cleveland, OH). The ligation mixture was transformed into DH10B cells, and colonies were selected on LB agar containing 20 μg/ml chloramphenicol. The pKM2/ltx/rmlC plasmid was isolated and transformed into the A. actinomycetemcomitans rmlC mutant by electroporation (42). Colonies were selected on TSBYE agar containing 1 μg/ml of chloramphenicol and 50 μg/ml of spectinomycin.
Development of a waaL mutant strain by insertional mutagenesis.
A DNA fragment corresponding to base pairs 5 to 545 of the waaL gene was amplified from the wild-type strain VT1169 using the following primers: 5′-CGATGATAAAGTGCGGTCATT-3′ and 5′-GAAAACATGCCCAACGACAT-3′. The 541-bp sequence was cloned into pCR 2.1-TOPO, transformed into DH10B cells by electroporation, and selected on LB plates containing 50 μg/ml kanamycin, X-Gal, and IPTG. The plasmid was purified, restricted with EcoRI, and ligated with purified plasmid pVT1461 restricted with the same enzyme. The ligation mixture was transformed into competent DH5α λ pir E. coli cells, and colonies were selected on LB plates containing 50 μg/ml of spectinomycin. The resulting plasmid was transformed into SM10 λ pir cells for conjugation with the wild-type strain. The transconjugants were selected on a TSBYE plate, with 50 μg/ml of spectinomycin, 100 μg/ml of rifampin, and 50 μg/ml of nalidixic acid. The transconjugants were screened by colony PCR using the primers 5′-ATCGACACCGACACTCAATG-3′, corresponding to the sequence of the upstream gene fba (fructose bisphosphate aldolase, class II) of waaL, and 5′-CTCTTGCCAGTCACGTTACG-3′, corresponding to the sequence of the spectinomycin cassette aad9 (25), to confirm the integration site in the chromosome.
Complementation of waaL.
The waaL gene (1,311 bp) was amplified and engineered with XhoI and EcoRI restriction sites using the following primers: sense primer (5′-GCTCGAGATGCCGATGATA-3′) and antisense primer (5′-GAATTCTTATTCATGGCGGTT-3′) (italics indicate restriction sites). The amplified fragment (waaL) was found to be identical to the sequence of the HK1651 strain (gene ID AA01434) (http://www.oralgen.lanl.gov/). The fragment was treated with XhoI and EcoRI. The fragment 5′-XhoI-waaL-EcoRI-3′ was purified and ligated with the vector pKM2/ltxP, treated with the same enzyme, and dephosphorylated. The ligation mix was transformed into DH10B cells, and colonies were selected on LB agar plates with 20 μg/ml chloramphenicol. The new construct pKM2/ltxP/waaL was electrotransformed into the A. actinomycetemcomitans waaL mutant. The waaL complemented strain was selected on the TSBYE plate containing 1 μg/ml chloramphenicol and 50 μg/ml spectinomycin.
Isolation of LPS.
LPS was isolated using either proteinase K digestion of whole-cell lysates by following the method described by Hitchcock and Brown (15) or hot phenol-aqueous extraction described by Westphal and Jann (48). For the proteinase K digestion method, 1.0 ml of A. actinomycetemcomitans (optical density at 495 nm [OD495] = 0.4) culture was washed with phosphate-buffered saline (PBS; 10 mM sodium phosphate, 150 mM sodium chloride) at pH 7.4 and centrifuged at 10,000 × g for 2 min to pellet the cells. The pellet was solubilized in 50 μl lysis buffer (2% sodium dodecyl sulfate [SDS], 4% 14.3 M β-mercaptoethanol, 10% glycerol, 1 M Tris at pH 6.8, and bromophenol blue) and boiled for 5 min. Three cell lysates were incubated with 50 μg/μl of proteinase K (Sigma-Aldrich, Milwaukee, WI) at 60°C for 60 min. LPS samples were loaded into 5 to 15% polyacrylamide-SDS gels, separated by electrophoresis at 80 V for 16 h at 4°C, according to the method of Laemmli (21), and silver stained based on the protocol described by Hitchcock and Brown (15).
Alternatively, LPS isolated using the hot phenol-aqueous method (48) was used for carbohydrate analysis, enzyme-linked immunosorbent assay (ELISA), and immunoblotting. A total of 200 ml of stationary-phase cells were washed with 10 ml PBS, resuspended in 5 ml of water (65 to 68°C), and vigorously mixed with phenol for 10 to 15 min. After it cooled, the mixture was centrifuged, and the top aqueous phase containing the LPS was removed. The LPS solution was extensively dialyzed using a molecular porous dialysis membrane (molecular weight cutoff, 3,500; Spectra/Por) against deionized water at 4°C. The dialyzed samples were concentrated by lyophilization.
Gas chromatography/mass spectrometry (GC/MS) of LPS.
The glycosyl composition of the LPS extracted from the wild-type A. actinomycetemcomitans strain and the isogenic mutants was analyzed using combined GC/MS of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides, using acidic methanolysis at the Complex Carbohydrate Research Center, the University of Georgia. A total of 400 μg of each LPS sample was used for the analysis. Methyl glycosides were prepared by methanolysis of the samples in 1 M HCl in methanol at 80°C for 18 to 22 h, followed by re-N-acetylation with pyridine and acetic anhydride in methanol for detection of amino sugars. The samples were per-O-trimethylsilylated by treatment with Tri-Sil (Pierce Biotechnology, Rockford, IL) at 80°C for 30 min (52). GC/MS analyses of the TMS methyl glycosides were performed on an HP 6890 GC interfaced to a 5975B mass selective detector (MSD), using an Alltech EC-1-fused silica capillary column (inside diameter, 30 m by 0.25 mm).
Immunoreactivity of O-PS, determined by ELISA.
The hot phenol-water-extracted LPS preparations were dissolved in deionized water and incubated in the wells of a 96-well microtiter plate overnight at 4°C. The wells were rinsed with water and blocked for 30 min in PBS with 0.05% Tween 20, 1 mM EDTA, and 0.25% bovine serum albumin (BSA) (19). The wells were incubated with purified rabbit anti-A. actinomycetemcomitans immunoglobulins (24) for 1 h at room temperature. The nonbinding immunoglobulins were removed, and the wells were washed with buffer and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin (Jackson Laboratory, Bar Harbor, ME) in PBS for 1 h. Immunoglobulin complexes were detected using 100 μl of citrate-phosphate buffer (24.3 mM citric acid monohydrate, 51.4 mM dibasic sodium phosphate, pH 5.0) containing 0.04% o-phenylenediamine and 0.012% hydrogen peroxide. The reaction was stopped by addition of 50 μl of 4 M H2SO4, and absorbance was measured at 490 nm.
Immunoblot analysis of LPS.
A total of 0.5 μg of the isolated LPS sample from each strain was dissolved in loading buffer containing 10 mM HEPES, 2% SDS, 5% (vol/vol) 14.3 M β-mercaptoethanol, 10% (vol/vol) glycerol, and 0.05% (wt/vol) bromophenol blue, boiled for 5 min, and loaded into a 4 to 15% polyacrylamide Tris-HCl Ready Gel (Bio-Rad, Hercules, CA). The electrophoresis was performed at 60 V at 4°C for 120 min. Separated carbohydrate molecules were transferred to Optitran nitrocellulose membranes (Whatman Incorporated, Piscataway, NJ) at 70 V at 4°C for 90 min and probed with the purified rabbit anti-A. actinomycetemcomitans immunoglobulins mentioned above. Immune complexes were detected using HRP-conjugated goat anti-rabbit IgG and visualized using the chemiluminescent substrate (Pierce Biotechnology, Rockford, IL). Membranes were exposed to Kodak X-OMAT LS films (Carestream Health, Rochester, NY).
Isolation of total membrane proteins.
The membrane protein fraction of A. actinomycetemcomitans was prepared as described previously (24, 46). Briefly, 200 ml late logarithmic phase cells were harvested and resuspended in 2.5 ml of 10 mM HEPES (pH 7.4) with 1 mM phenylmethylsulfonyl fluoride (PMSF; USB Corporation, Cleveland, OH) and 1× complete protease inhibitor cocktail (Roche Diagnostic Corporation, Atlanta, GA). Cells were lysed by three cycles of 9,000 lb/in2 (62,100 kPa) at 4°C using a French pressure minicell. Whole-cell lysates were centrifuged at 7,650 × g to remove cell debris, followed by centrifugation at 100,000 × g for 40 min to pellet the membrane fraction. The protein concentration was estimated using absorbance at 280 nm (43).
Immunoblot analysis of EmaA.
Equivalent amounts of membrane protein, determined by absorbance at 280 nm, were prepared in the loading buffer as described by the immunoblotting of LPS and loaded into 4 to 15% polyacrylamide Tris-HCl gels. Electrophoresis was performed at 40 V and 4°C for 15 h. The separated proteins were transferred to an Optitran nitrocellulose membrane at 70 V and 4°C for 105 min and probed with an anti-EmaA monoclonal antibody (46). The immune complex was detected using HRP-conjugated goat anti-mouse IgG (Jackson Laboratory, Bar Harbor, ME) and visualized as described by the immunoblotting of LPS.
Lectin blot analysis.
Equivalent amounts of membrane protein were prepared as described above and transferred to nitrocellulose membrane filters as described for the immunoblotting. The filter was blocked in PBS (pH 7.4) with 0.5% Tween 20 (PBS-T), and probed with biotinylated fucose-specific Lens culinaris agglutinin (LCA; Vector Laboratories, Burlingame, CA) for 1 h. The membrane was washed in PBS-T for 5 min with 6 changes and then incubated with HRP-conjugated avidin D (Vector Laboratories, Burlingame, CA) for 1 h. After being washed six times, the lectin-avidin complex was visualized as described by the immunoblotting of LPS.
Liquid chromatography/mass spectrometry (LC/MS) analysis.
Equivalent amounts of membrane proteins from the parent and emaA mutant strains were dissolved in electrophoresis loading buffer as described above, boiled for 5 min, and loaded onto a 5 to 15% gradient polyacrylamide-SDS gel, with a 3% stacking gel (24). Electrophoresis was performed at 60 V and 4°C for 24 h. The gel was fixed with 50% methanol-10% acetic acid for 10 min, incubated in colloidal blue stain (Invitrogen, Carlsbad, CA) for 12 h, and destained in deionized water for 8 h. EmaA from the wild-type strain and the corresponding area of the emaA mutant strain were excised for LC/MS (10). The LC/MS was performed at the Vermont Genetics Network Proteomics Facility located at the University of Vermont.
In vitro competition binding assay to rabbit mitral valves.
The competition assay was performed as described previously (45). Briefly, 1 ml of mid-logarithmic-phase cells from the wild type and the rmlC mutant, equivalent to 2.5 × 108 CFU in sterile PBS, was used for each assay. One leaflet of the mitral valve treated with trypsin was incubated with the inoculum at 37°C for 1 h. Serial dilutions of the original inoculum and the homogenized, infected valve samples were enumerated on both TSBYE agar and TSBYE containing 50 μg/ml spectinomycin. Plates with spectinomycin were used to determine the concentration of the rmlC mutant. The number of wild-type cells was obtained by subtracting this value (obtained from plates with spectinomycin) from the value obtained from plates without antibiotics. The in vitro competition assay was performed in triplicate, using mitral valves from three rabbits. The competitive index (CI) was calculated as the ratio of the number of mutant to wild-type CFU in the cardiac valve samples divided by the ratio of the number of mutant to wild-type CFU in the inoculum. The CI values from each valve leaflet were compared to a value of 1 using the paired t test with GraphPad Prism software (version 5.02). P values of <0.05 were set as statistically significant.
RESULTS
Characterization of A. actinomycetemcomitans serotype b LPS.
Visualization of LPS isolated from the wild-type strain using the proteinase K method (15), by silver staining the polyacrylamide-SDS gels, revealed the presence of a dark brown stain located in the high-molecular-weight region of the gel (75,000 to 250,000) (Fig. 1B), which represents polymerized O-PS of serotype b strains (Fig. 1A). A similar LPS profile was demonstrated for the emaA mutant strain. These two profiles differed from the LPS profiles of both the rmlC and wzt mutants, due to the absence or reduction of O-PS (Fig. 1B). However, the LPS isolated from the rmlC and wzt mutant strains contained core oligosaccharides profiles similar to those of the wild-type or emaA mutant strains (Fig. 1B).
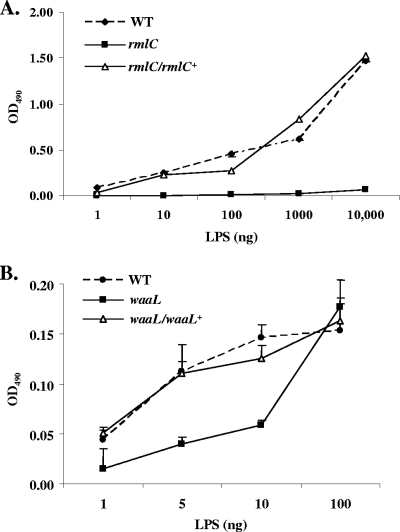
Hot phenol-water-extracted LPS (48) was used both in ELISA and in immunoblot assays, in which polyclonal antiserum raised against the whole bacteria was used to determine the relative amount of O-PS associated with the LPS. Antibody binding in wells adsorbed with as little as 10 ng of LPS isolated from the wild-type strain was observed. The signal increased with increasing amounts of LPS. In contrast, a weak signal in wells adsorbed with a 1,000-fold increase (10 μg) of LPS isolated from the rmlC mutant was detected. A binding pattern similar to that of the wild-type strain was observed, using an LPS sample isolated from the mutant strain complemented with rmlC, driven by the ltx promoter in trans (Fig. 2A).
FIG. 2.
Analyses of purified LPS from O-PS mutants by ELISA. The phenol-water-extracted LPS samples were adsorbed to wells of 96-well microtiter plates and detected using purified rabbit anti-A. actinomycetemcomitans antibodies. (A) WT, wild type (VT1169); rmlC, rhamnose epimerase mutant; rmlC/rlmC+, rhamnose epimerase complemented. (B) waaL, O-antigen ligase mutant; waaL/waaL+, O-antigen ligase complemented.
The potential that other LPS biosynthetic enzymes are involved in EmaA modification was determined by characterizing a waaL mutant strain. The putative A. actinomycetemcomitans waaL gene is predicted to encode the lipid A-core O-antigen ligase. The waaL gene of A. actinomycetemcomitans was identified based on the homology of the translated protein sequence with Haemophilus influenzae (42% amino acid identity) (GenBank accession no. ZP 00202015). A single open reading frame was identified in the A. actinomycetemcomitans genome (gene ID AA01434) (http://www.oralgen.lanl.gov/), which is homologous to known O-antigen ligases. The waaL gene is located outside of the 21-gene serotype b O-PS operon, which includes rmlC and wzt.
Insertional inactivation of the waaL gene resulted in a strain that synthesized a reduced level of LPS. The reduction corresponded to a decrease in the binding of anti-A. actinomycetemcomitans antibodies, determined by ELISA (Fig. 2B), compared with that of the wild-type strain. However, the amount of immunoreactivity was greater than that found with the LPS preparation of the rmlC mutant strain (Fig. 2A). The amount of antibody binding to the LPS preparation of the waaL complemented strain was similar to the wild-type strain. Collectively, the reduction in the level of O-PS isolated from this mutant and the high protein homology with other O-antigen ligases suggest a similar role of the WaaL protein in A. actinomycetemcomitans LPS biosynthesis.
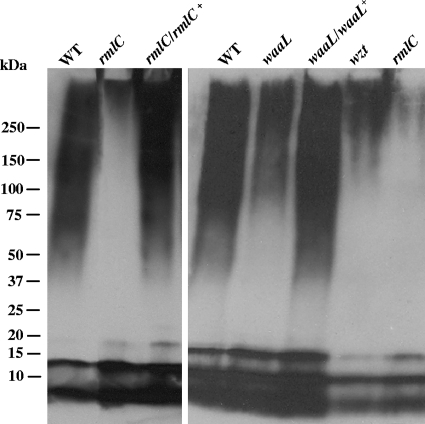
The difference in antibody binding between the O-PS mutants and the wild-type LPS preparations was also visualized by immunoblotting. A larger amount of immunoreactive material was present in the LPS samples isolated from the wild-type and the complemented strains than in those isolated from the mutant strains. The presence of a small amount of O-PS staining in the rmlC mutant may be attributed to some non-O-PS polymers that also reacted with the antibodies or to another rmlC-like gene that partially complemented the mutation. The data clearly demonstrate a gradient in the amount of sugars associated with the LPS isolated from the waaL, wzt, and waaL mutant strains (Fig. 3).
FIG. 3.
LPS immunoblot using an anti-A. actinomycetemcomitans antibody. A total of 500 ng of phenol-water-purified LPS was resolved on 4 to 15% polyacrylamide Tris-HCl Ready Gels. The separated carbohydrate molecules were transferred to a nitrocellulose membrane and probed with purified rabbit anti-A. actinomycetemcomitans immunoglobulins. WT, wild type (VT1169); rmlC, rhamnose epimerase mutant; rmlC/rlmC+, rhamnose epimerase complemented; waaL, O-antigen ligase mutant; waaL/waaL+, O-antigen ligase complemented; wzt, ATP-binding cassette sugar transport mutant.
The quantification of the carbohydrates associated with the LPS isolated from different mutants and the wild-type strain was determined using GC/MS. Carbohydrate analysis revealed that the LPS sample from the three mutants contained less carbohydrate on a mass basis than the wild-type LPS (Table 3). This difference can be attributable to the absence or reduction in the saccharides associated with the serotype b O-PS of A. actinomycetemcomitans, l-Rha, d-Fuc, and GalNAc. Rha and GalNAc were not detected, and Fuc was greatly diminished in the rmlC mutant compared with that in the wild-type strain. Different from the rmlC mutant, Rha and GalNAc were present in the wzt mutant but at reduced levels compared to those in the wild-type LPS sample. The level of Fuc in the wzt mutant was comparable to the level found in the rmlC mutant. Similar to the other two mutants, the waaL mutant LPS showed a reduction in the amounts of both Rha and Fuc in comparison with those in the wild-type strain. In contrast to the LPS isolated from the rmlC and wzt mutants, an increase in the amount of GlcNAc was observed in the waaL mutant LPS.
TABLE 3.
Glycosyl mass comparison of isolated LPS
| Glycosyl residue | Mass (μg) of LPS inb: |
|||
|---|---|---|---|---|
| Wild type | rmlC::pLOF/Sp | wzt::Plof/Sp | waaL::pKM221 | |
| Tetradecanoic acid | + | + | + | + |
| Ribose (Rib) | 9.1 | 5.1 | 3.4 | 5.9 |
| Rhamnose (Rha) | 31.0* | ND | 3.8 | 5.9 |
| Fucose (Fuc) | 24.8 | 2.5 | 3.1 | 6.1 |
| 3-OH tetradecanoic acid | + | + | + | + |
| Glucuronic acid (GlcUA) | ND | ND | ND | + |
| Galacturonic acid (GalUA) | ND | ND | ND | + |
| Mannose (Man) | ND | ND | ND | 0.2 |
| Galactose (Gal) | 5.9 | 3.4 | 2.6 | 2.6 |
| Glucose (Glc) | 29.8 | 25.6* | 9.8 | 10.0 |
| N-acetyl galactosamine (GalNAc) | 10.1 | ND | 2.1 | 30.0* |
| N-acetyl glucosamine (GlcNAc) | 26.3 | 14.3 | 17.9* | 22.6 |
| Heptose (Hep) | 29.0 | 24.8 | 11.7 | 12.2 |
| 3-Deoxy-2-manno-2-octulosonic acid | ND | ND | ND | ND |
| Sum | 166 | 76 | 54 | 96 |
| Total loading amt | 400 | 400 | 400 | 400 |
| % Carbohydratea | 41.5 | 19.0 | 13.6 | 23.9 |
The total percentage of carbohydrate of each LPS sample (400 μg per sample).
ND, not detectable; +, detected; *, the most predominant glycosyl residues.
Characterization of EmaA in the wild-type and O-PS mutant strains.
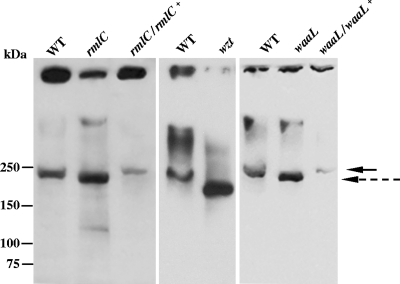
The EmaA monomer of serotype b strains is identified as a 202-kDa molecule, based on the predicted amino acid sequence and immunoblots using an EmaA-specific monoclonal antibody (24, 46). EmaA-immunoreactive material has been also found associated with the stacking gel, which may represent protein aggregation (57). Similar observations were associated with the wild-type and mutant strains in this study (Fig. 4). However, the EmaA monomer of the three O-PS mutants displayed an increase in electrophoretic mobility, which corresponded to a lower molecular mass than that of the wild-type EmaA monomer. In addition to the change in the electrophoretic mobility, the amount of the EmaA monomer in the three O-PS mutant strains was greater than that in the wild-type strain. The increase in the amount of the monomer correlated with a decrease in the amount of immunoreactive material found associated with the stacking gel in the mutant strains. The aggregation state and molecular weight change of EmaA in the O-PS mutant strains can be reverted to the wild-type phenotype after the complementation of the mutants in trans. Transmission electron microscopy images indicated that the EmaA appendages were present on the cell surface of the mutants (data not shown).
FIG. 4.
Characterization of EmaA in the O-PS mutants. Equivalent amounts of membrane protein from each strain was prepared and separated by electrophoresis using 4 to 15% gradient polyacrylamide Tris-HCl gels. The proteins were transferred to nitrocellulose and probed with a monoclonal antibody specific for EmaA. The solid arrow indicates the electrophoretic mobility of the EmaA monomers associated the wild type and complemented strains in the separating gel. The dashed arrow corresponds to the mobility of the EmaA monomers associated with the rmlC mutant strain in the separating gel. The immunoreactive material at the top of the immunoblot corresponds to EmaA aggregates associated with the stacking gel. WT, wild type (VT1169); rmlC, rhamnose epimerase mutant; rmlC/rlmC+, rhamnose epimerase complemented; wzt, ABC sugar transport mutant; waaL, O-antigen ligase mutant; waaL/waaL+, O-antigen ligase complemented.
O-PS sugar is associated with EmaA.
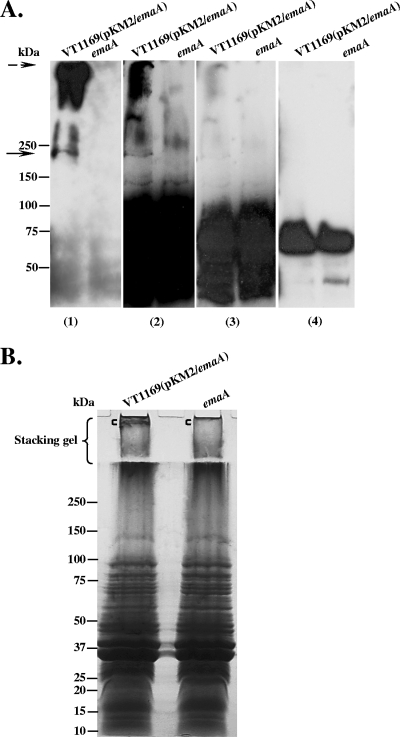
The change in the electrophoretic mobility of the EmaA monomer in the three O-PS mutant strains suggested that the protein is associated with carbohydrate, and the carbohydrate is composed of sugars associated with the serotype b O-PS. We tested this hypothesis by employing a biotinylated, fucose-specific lectin, isolated from Lens culinaris (23) (LCA; Vector Laboratories, Burlingame, CA), in blots of membrane fractions isolated from an EmaA overexpression strain and the emaA mutant strain. The lectin interacted with an ∼200-kDa protein in the lane corresponding to membrane proteins of the EmaA-overproducing strain VT1169 (pKM2/emaA) (Fig. 5A, panel 2). The lectin binding at this molecular weight was absent in the membrane protein lane corresponding to the emaA mutant (Fig. 5A, panel 2). Intense lectin binding material was also observed in the stacking gel of the EmaA overexpression strain, which was negligible in the corresponding gel region of the emaA mutant strain (Fig. 5A, panel 2). The lectin binding material at ∼200 kDa and in the stacking gel matched the immunoreactive-staining pattern of EmaA using a monoclonal antibody specific for EmaA (Fig. 5A, panel 1). The intensely stained region of the lectin blot (below the 100-kDa marker) (Fig. 5A, panel 2) is attributed to the binding of avidin to a 70-kDa membrane protein, as described previously by Mintz and Fives-Taylor (26). This is depicted in a shorter exposure time of the lectin blot (Fig. 5A, panel 3) than that of an identical blot with similar exposure time using HRP-avidin alone (Fig. 5A, panel 4).
FIG. 5.
Fucose-specific Lens culinaris agglutinin (LCA) blots of membrane proteins from EmaA-producing and emaA mutant strains. Equivalent amounts of membrane proteins from the EmaA-overproducing strain VT1169 (pKM2/emaA) and the emaA mutant (emaA) were prepared, loaded into the 4 to 15% polyacrylamide Tris-HCl gel, and transferred to nitrocellulose membranes. (A) The same protein-transferred membrane was probed with anti-EmaA monoclonal antibody (panel 1); biotinylated LCA, with different exposure times of the film (panels 2 and 3); and avidin alone (nonlectin control) (panel 4). Antibody binding was detected using goat anti-mouse antibodies, and lectin binding was detected using avidin. The solid arrow at ∼200 kDa corresponds to the EmaA monomer. The dashed arrow corresponds to EmaA aggregates associated with the stacking gel. (B) Colloidal blue stain of membrane proteins. Equivalent amounts of membrane proteins from the EmaA-overproducing strain VT1169(pKM2/emaA) and the emaA mutant strain were separated in a 5 to 15% polyacrylamide-SDS gel with a 3% stacking gel. Following electrophoresis, the gel was stained with colloidal blue. The region of the gel corresponding to the EmaA aggregates, shown with the square bracket ([) in the stacking gel, and a similar region of the gel in the emaA mutant ([) were excised and analyzed using LC/MS (Table 4).
The above-described data suggest that the lectin binding activity is related to the presence of EmaA. LC/MS analysis was performed to determine if the lack of lectin binding activity in the mutant strain was due to the absence of EmaA but not due to a change in the protein composition. Equivalent amounts of membrane protein from each strain were separated by gel electrophoresis and stained (Fig. 5B). In the lane corresponding to the EmaA-overproducing strain, stained material was present in the stacking gel, which was absent in the emaA mutant strain, shown with the square bracket in Fig. 5B. The band in the stacking gel and the corresponding gel region of the emaA mutant strain were excised for analysis by LC/MS. The LC/MS results indicated that EmaA was the most abundant protein present in the EmaA-overproducing strain but was absent in the mutant strain (Table 4). The protein composition of the mutant strain was similar to that of the EmaA-overproducing strain, except for the absence of EmaA. However, some variation in the concentration of the individual proteins was observed. The electrophoretic mobility change of the EmaA monomer in the O-PS mutants, the lectin blot, and the LC/MS data support the hypothesis that EmaA contains a sugar associated with serotype b O-PS.
TABLE 4.
Protein composition of the EmaA-overproducing strain and the emaA mutant
| VT1169 (pKM2/emaA) proteina | emaA mutant abundanceb |
|---|---|
| 1. Extracellular matrix adhesin A (EmaA) | Not detected |
| 2. NAD(P) transhydrogenase subunit alpha (PntA) | 2 |
| 3. Protein-export membrane protein (SecD) | 5 |
| 4. Leukotoxin A (LtxA) | 3 |
| 5. Fumarate reductase flavoprotein subunit A | |
| (FrdA) | 9 |
| 6. Conserved hypothetical protein | 6 |
| 7. Signal peptide peptidase (SppA) | 23 |
| 8. Acriflavine resistance protein (AcrB) | 14 |
| 9. Glycerol-3-phosphatase transporter (GlpT) | 12 |
| 10. Conserved hypothetical protein | 15 |
The proteins are listed based on their relative amount, as detected by LC/MS. Only the 10 most abundant proteins in the EmaA-producing strain are listed.
The numbers represent the relative abundance of the corresponding proteins of the EmaA-overproducing strain found in the emaA mutant strain, as determined by LC/MS (Fig. 5B).
Assessment of collagen binding activity of the O-PS mutant using an in vitro tissue model.
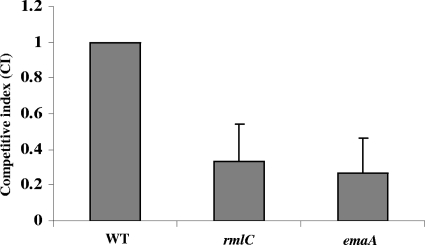
Equivalent amounts of bacteria were incubated with trypsin-treated rabbit mitral valves to assess the role of the modification of EmaA in collagen binding activity (Fig. 6). The competitive index (CI) between the rmlC mutant and the wild type was 0.33 (paired t test; P = 0.008), which was equivalent to that of the emaA mutant strain (CI = 0.27). A CI value of 1 indicates no difference in competitiveness between the mutant and wild-type strains. These data suggest that the rmlC mutant strain, as well as the emaA mutant strain, colonized the heart valve approximately 3-fold less effectively than the wild-type strain. These data suggest that the modification of the adhesin is important for the interaction with collagen.
FIG. 6.
Assessment of collagen binding activities using trypsin-treated rabbit heart valves. Rabbit heart valves were surgically removed and treated with trypsin to remove the endothelia. Equal CFU numbers of the wild-type (WT) and the rmlC (rhamnose epimerase mutant) bacteria were added to the treated valves and incubated. The competitive index (CI) was calculated as the ratio of the numbers of mutant to wild-type CFU in the cardiac valve samples divided by the ratio of the numbers of mutant to wild-type CFU in the inoculum (CI, 0.33; Paired t test; P = 0.008), which was similar to that of the emaA mutant (CI, 0.27).
DISCUSSION
The A. actinomycetemcomitans serotype b O-PS is a trisaccharide-repeating unit composed of d-Fuc, l-Rha, and d-GalNAc residues (2, 29, 33, 50). Rha and Fuc were the main sugars identified in the analysis of the LPS isolated from the serotype b strain used in this study. The isolated LPS gave a typical silver stain profile for serotype b A. actinomycetemcomitans following electrophoresis (29, 50). The O-PS of serotype b appeared as a broad smear in the high-molecular-weight region of the polyacrylamide-SDS gel, and the sugars and fatty acids that are typical of the LPS core oligosaccharides and lipid A were observed in the lower-molecular-weight region of the gel (29, 50). The smear is most likely due to different numbers of repeating saccharide units present in the O-PS. O-PS is the immunodominant material when probed with anti-A. actinomycetemcomitans antibodies, which is similar to the observation obtained with serum samples from patients with periodontal disease (29, 50).
Rha and Fuc, the predominant sugars of the serotype b O-PS, are synthesized following a shared biosynthetic pathway involving d-glucose-1-phosphate and dTTP (53). Following the generation of the intermediate dTDP-4-keto-6-deoxy-d-glucose, the intermediate is converted to dTDP-l-rhamnose via an epimerase (RmlC) and a reductase (RmlD) or to dTDP-d-fucose using a reductase (Fcd) (53). The integral nature of RmlC in Rha synthesis was demonstrated by the absence of Rha in the LPS isolated from the rmlC mutant, as analyzed by GC/MS. d-GalNAc was also not detected in the LPS isolated from this mutant. Both fcd and the genes associated with the synthesis of d-GalNAc are downstream of rmlC (53). Restoration of O-PS following complementation of the mutant with the full-length rmlC gene in trans indicates that disruption of the gene does not have a polar effect on the transcription of the downstream genes in the operon. There was, however, a decrease in the molar percentage of Fuc associated with the LPS isolated from the rmlC mutant. The presence of fucose may represent undecaprenyl phosphate (Und-PP)-linked fucose that copurifies with the modified LPS. The absence of Rha and GalNAc, the loss of high-molecular-weight staining material in the silver-stained SDS-PAGE gels, and the apparent decreased immunoreactivity in the ELISA and LPS immunoblots with the rmlC mutant LPS are consistent with those of a strain of A. actinomycetemcomitans lacking O-PS.
Wzt and WaaL are two enzymes that function in the latter stages of the LPS biosynthetic pathway. Wzt is an ATP binding cassette (ABC) transporter that exports saccharide polymers from the cytoplasm to the periplasmic space (36). The carbohydrate analysis and the antibody reactivity studies collectively indicate a substantial decrease in the amount of O-PS sugars associated with the LPS in the wzt mutant. The presence of O-PS sugar residues associated with the mutant LPS suggests that the mutant bacterium may be able to express either an active truncated protein or another enzyme, albeit with a lower affinity, for transport of the oligosaccharides across the cytoplasmic membrane. Mutation of wzt in a serotype f strain of A. actinomycetemcomitans has also been shown to cause defects in O-PS synthesis (19).
The O-antigen ligase (WaaL) of Enterobacteriaceae has been shown to be responsible for attachment of polysaccharides to the lipid A core (49). The enzyme ligates an undecaprenol pyrophosphate-linked oligo- or polysaccharide onto the lipid A-core oligosaccharide in the periplasm of the bacterium (1, 34). Mutants with interrupted waaL genes of both Escherichia coli and Salmonella enterica serovar Typhimurium are unable to attach the O antigen to the lipid A core (36). Inactivation of the A. actinomycetemcomitans waaL ortholog resulted in the LPS containing reduced amounts of Rha and Fuc and caused a decrease in immunoreactivity to the anti-A. actinomycetemcomitans antibodies compared with that of the wild-type strain. However, we did not observe a complete loss of the O-PS moiety in this mutant. The presence of O-PS in the mutant-extracted LPS may be contributed to contamination of Und-PP-linked O-PS precursors or other carbohydrate containing impurities that copurify during the LPS extraction. The data, however, do suggest that waaL is associated with LPS biosynthesis in A. actinomycetemcomitans. Biological and biochemical characterization of the protein is required to address the specific activity of this enzyme.
In this study, the disruption in O-PS biosynthesis was associated with a change in the properties of EmaA. Changes in the electrophoretic mobility and aggregation of the EmaA molecules were observed for the three mutants representing individual steps in the biosynthesis and assembly of O-PS molecules. The change in the apparent molecular weight of EmaA is consistent with the loss of a posttranslational modification of the protein. The electrophoretic mobilities of other autotransporter proteins of A. actinomycetemcomitans, including the buccal epithelial adhesin Aae (39, 46) and the trimeric adhesin/invasin Omp100 (58), were not affected in the same O-PS mutant background (data not shown). The fucose-specific lectin, binding to the region of the gels corresponding to EmaA, lends additional support to the hypothesis that EmaA is a glycoprotein.
Glycosylation of Gram-negative bacterial adhesins has been identified. These include fimbriae (pili), found in E. coli (28, 47), Neisseria meningitidis (34), and Pseudomonas aeruginosa (35), as well as the nonfimbrial adhesin HMW1 found in Haemophilus influenzae. N. meningitides pilin glycosylation proceeds through a homologue of O-antigen ligase (PglL), which is not associated with LPS assembly in this bacterium (34). The pilin glycosylation of P. aeruginosa requires an oligosaccharyltransferase (PilO), which is also independent from the LPS biosynthetic pathway, although the conjugated oligosaccharide is identical to the O-antigen-repeating unit of this organism (16, 35). Different from the above-mentioned O-linked pilin glycosylation pathways, the N-linked glycosylation of the nonfimbrial adhesin HMW1 found in H. influenzae uses an independent glycosylation machinery, requiring HMW1C and phosphoglucomutase (11, 12). The glycosylation of HMW1 occurs in the cytoplasm (11) instead of in the periplasmic space associated with the fimbrial adhesins (34, 35).
Secreted glycosylated proteins have been found in the periodontal pathogen Porphyromonas gingivalis (31). The extracellular cysteine proteinases Arg-gingipains (RgpsA and RgpsB) contain sugar moieties similar to those of the anionic polysaccharides (APS) of the cell surface associated with this organism (31). APS is identified as an essential surface structure different from either the LPS or the capsule polysaccharide (31). Recently, the O-antigen ligase (WaaL) of P. gingivalis has been shown to ligate the O antigen to the lipid A core and assemble the APS sugar repeat units (38). However, the role of WaaL in the modification of the gingipains is unknown.
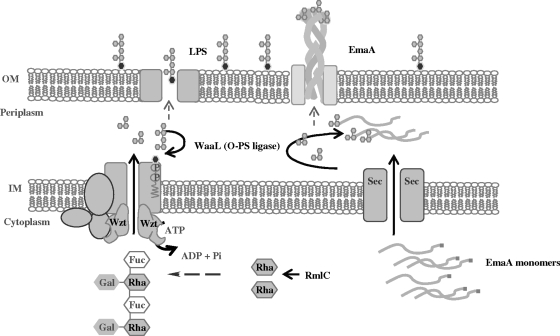
In this study, we present both genetic and biochemical evidence that A. actinomycetemcomitans strain VT1169 synthesizes a trimeric autotransporter adhesin that contains carbohydrate. These data also suggest that the enzymes involved in the modification of EmaA overlap with the enzymes of the LPS biosynthetic machinery. The genetic studies imply that saccharide assembly for EmaA is mediated by the O-antigen ligase (WaaL), which is also required for ligation of the O-polysaccharide to the lipid A-core oligosaccharide of LPS (Fig. 7). It is possible that the loss of the intact LPS molecules in the waaL mutant has destabilizing effects on another protein responsible for EmaA glycosylation. At this time, we cannot exclude this possibility. However, the data presented here support the least complicated hypothesis that EmaA modification is dependent on the LPS biosynthetic pathway. This dependency makes EmaA unique among the Gram-negative glycosylated proteins, which are independent of LPS biosynthetic enzymes.
FIG. 7.
Hypothetical pathway for LPS and EmaA biosynthesis in A. actinomycetemcomitans. The proposed LPS biosynthetic pathway is based on enzymatic reactions that are known or proposed, based on protein homology, for other Gram-negative bacteria. We propose that EmaA monomers are translocated across the inner membrane (IM) using the Sec translocon mediated by the signal sequence (data not shown). Once in the periplasmic space, the O-PS sugars are transferred to the individual EmaA monomers by the O-antigen ligase (WaaL) before translocation through the outer membrane (OM) and presentation on the bacterial surface.
EmaA is one of two reported glycosylated proteins associated with A. actinomycetemcomitans. The fimbriae of this organism are also suggested to be glycosylated (17); however, the mechanism of fimbrillin glycosylation is unknown. In this study, we present the first evidence for the posttranslational modification of a trimeric autotransporter protein adhesin. The evidence presented in this study suggests that the nonfimbrial collagen adhesin EmaA of A. actinomycetemcomitans is posttranslationally modified by enzymes associated with the LPS biosynthetic pathway. Furthermore, this modification may be important for full biological activity of this adhesin. Biochemical experiments are under way to confirm the nature and the site(s) of glycosylation of EmaA.
Acknowledgments
We gratefully acknowledge the contributions of Marni Slavik and Travis Bellville for their help in the isolation of the transposon mutants used in this study. We also thank Teresa Ruiz, Claude Gallant, Chunxiao Yu, and Erin Chicoine for their helpful discussions.
We also acknowledge the Vermont Genetics Network and grant P20 RR16462 from the INBRE program of the National Center for Research Resources (NCRR). This research was supported by NIH-NIDCR grants RO1-DE13824 and RO1-DE09760.
Footnotes
Published ahead of print on 8 January 2010.
REFERENCES
- 1.Abeyrathne, P., C. Daniels, K. Poon, M. Matewish, and J. Lam. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187:3002-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano, K., T. Nishihara, N. Shibuya, T. Noguchi, and T. Koga. 1989. Immunochemical and structural characterization of a serotype-specific polysaccharide antigen from Actinobacillus actinomycetemcomitans Y4 (serotype b). Infect. Immun. 57:2942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bengoechea, J., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol. Microbiol. 52:451-469. [DOI] [PubMed] [Google Scholar]
- 4.Beveridge, T. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brogan, J., E. Lally, K. Poulsen, M. Kilian, and D. Demuth. 1994. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect. Immun. 62:501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouqui, P., and D. Raoult. 2001. Endocarditis due to rare and fastidious bacteria. Clin. Microbiol. Rev. 14:177-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu, S., and T. Trust. 1993. An Aeromonas salmonicida gene which influences a-protein expression in Escherichia coli encodes a protein containing an ATP-binding cassette and maps beside the surface array protein gene. J. Bacteriol. 175:3105-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Fine, D., K. Markowitz, D. Furgang, K. Fairlie, J. Ferrandiz, C. Nasri, M. McKiernan, and J. Gunsolley. 2007. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 45:3859-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallant, C., M. Sedic, E. Chicoine, T. Ruiz, and K. Mintz. 2008. Membrane morphology and leukotoxin secretion are associated with a novel membrane protein of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190:5972-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St. Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 12.Gross, J., S. Grass, A. Davis, P. Gilmore-Erdmann, R. R. Townsend, and J. W. St. Geme III. 2008. The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 283:26010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haubek, D., O. Ennibi, K. Poulsen, N. Benzarti, and V. Baelum. 2004. The highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans and progression of periodontal attachment loss. J. Dent. Res. 83:767-770. [DOI] [PubMed] [Google Scholar]
- 14.Henderson, I., F. Navarro-Garcia, M. Desvaux, R. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hitchcock, P., and T. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horzempa, J., C. Dean, J. Goldberg, and P. Castric. 2006. Pseudomonas aeruginosa 1244 pilin glycosylation: glycan substrate recognition. J. Bacteriol. 188:4244-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue, T., H. Ohta, I. Tanimoto, R. Shingaki, and K. Fukui. 2000. Heterogeneous post-translational modification of Actinobacillus actinomycetemcomitans fimbrillin. Microbiol. Immunol. 44:715-718. [DOI] [PubMed] [Google Scholar]
- 18.Jain, S., P. van Ulsen, I. Benz, M. Schmidt, R. Fernandez, J. Tommassen, and M. Goldberg. 2006. Polar localization of the autotransporter family of large bacterial virulence proteins. J. Bacteriol. 188:4841-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, J., M. Perry, L. MacLean, D. Furgang, M. Wilson, and D. Fine. 2001. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 69:5375-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozarov, E., B. Dorn, C. Shelburne, W. Dunn, and A. Progulske-Fox. 2005. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Thromb. Vasc. Biol. 25:e17-e18. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lakio, L., S. Paju, G. Alfthan, T. Tiirola, S. Asikainen, and P. Pussinen. 2003. Actinobacillus actinomycetemcomitans serotype d-specific antigen contains the O antigen of lipopolysaccharide. Infect. Immun. 72:5005-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumura, K., K. Higashida, H. Ishida, Y. Hata, K. Yamamoto, M. Shigeta, Y. Mizuno-Horikawa, X. Wang, E. Miyoshi, J. Gu, and N. Taniguchi. 2007. Carbohydrate binding specificity of a fucose-specific lectin from Aspergillus oryzae: a novel probe for core fucose. J. Biol. Chem. 282:15700-15708. [DOI] [PubMed] [Google Scholar]
- 24.Mintz, K. 2004. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology 150:2677-2688. [DOI] [PubMed] [Google Scholar]
- 25.Mintz, K., C. Brissette, and P. Fives-Taylor. 2002. A recombinase A-deficient strain of Actinobacillus actinomycetemcomitans constructed by insertional mutagenesis using a mobilizable plasmid. FEMS Microbiol. Lett. 206:87-92. [DOI] [PubMed] [Google Scholar]
- 26.Mintz, K., and P. M. Fives-Taylor. 1994. Identification of an immunoglobulin Fc receptor of Actinobacillus actinomycetemcomitans. Infect. Immun. 62:4500-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano, Y., Y. Yoshida, N. Suzuki, Y. Yamashita, and T. Koga. 2000. A gene cluster for the synthesis of serotype d-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim. Biophys. Acta 1493:259-263. [DOI] [PubMed] [Google Scholar]
- 28.Okuda, S., and G. Weinbaum. 1968. An envelope-specific glycoprotein from Escherichia coli B. Biochemistry 7:2819-2825. [DOI] [PubMed] [Google Scholar]
- 29.Page, R., T. Sims, L. Engel, B. Moncla, B. Bainbridge, J. Stray, and R. Darveau. 1991. The immunodominant outer membrane antigen of Actinobacillus actinomycetemcomitans is located in the serotype-specific high-molecular-mass carbohydrate moiety of lipopolysaccharide. Infect. Immun. 59:3451-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paju, S., P. Carlson, H. Jousimies-Somer, and S. Asikainen. 2000. Heterogeneity of Actinobacillus actinomycetemcomitans strains in various human infections and relationships between serotype, genotype, and antimicrobial susceptibility. J. Clin. Microbiol. 38:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paramonov, N., M. Rangarajan, A. Hashim, A. Gallagher, J. Aduse-Opoku, J. Slaney, E. Hounsell, and M. Curtis. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 58:847-863. [DOI] [PubMed] [Google Scholar]
- 32.Perry, M., L. MacLean, J. Brisson, and M. Wilson. 1996. Structures of the antigenic O-polysaccharides of lipopolysaccharides produced by Actinobacillus actinomycetemcomitans serotypes a, c, d and e. Eur. J. Biochem. 242:682-688. [DOI] [PubMed] [Google Scholar]
- 33.Perry, M., L. MacLean, R. Gmur, and M. Wilson. 1996. Characterization of the O-polysaccharide structure of lipopolysaccharide from Actinobacillus actinomycetemcomitans serotype b. Infect. Immun. 64:1215-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Power, P., K. Seib, and M. Jennings. 2006. Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem. Biophys. Res. Commun. 347:904-908. [DOI] [PubMed] [Google Scholar]
- 35.Qutyan, M., M. Paliotti, and P. Castric. 2007. PilO of Pseudomonas aeruginosa 1244: subcellular location and domain assignment. Mol. Microbiol. 66:1444-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raetz, C., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajakumar, K., B. Jost, C. Sasakawa, N. Okada, M. Yoshikawa, and B. Adler. 1994. Nucleotide sequence of the rhamnose biosynthetic operon of Shigella flexneri 2a and role of lipopolysaccharide in virulence. J. Bacteriol. 176:2362-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangarajan, M., J. Aduse-Opoku, N. Paramonov, A. Hashim, N. Bostanci, O. Fraser, E. Tarelli, and M. Curtis. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 190:2920-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose, J., D. Meyer, and P. Fives-Taylor. 2003. Aae, an autotransporter involved in adhesion of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 71:2384-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz, T., C. Lenox, M. Radermacher, and K. Mintz. 2006. Novel surface structures are associated with the adhesion of Actinobacillus actinomycetemcomitans to collagen. Infect. Immun. 74:6163-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen, K., and H. Nikaido. 1991. Lipopolysaccharide structure required for in vitro trimerization of Escherichia coli OmpF porin. J. Bacteriol. 173:926-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sreenivasan, P., D. LeBlanc, L. Lee, and P. Fives-Taylor. 1991. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect. Immun. 59:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoscheck, C. 1990. Quantitation of protein. Methods Enzymol. 182:50-68. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, N., Y. Nakano, Y. Yoshida, H. Nakao, and Y. Yamashita. 2000. Genetic analysis of the gene cluster for the synthesis of serotype a-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim. Biophys. Acta 1517:135-138. [DOI] [PubMed] [Google Scholar]
- 45.Tang, G., T. Kitten, C. Munro, G. Wellman, and K. Mintz. 2008. EmaA, a potential virulence determinant of Aggregatibacter (Actinobacillus) actinomycetemcomitans in infective endocarditis. Infect. Immun. 76:2316-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, G., T. Ruiz, R. Barrantes-Reynolds, and K. Mintz. 2007. Molecular heterogeneity of EmaA, an oligomeric autotransporter adhesin of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Microbiology 153:2447-2457. [DOI] [PubMed] [Google Scholar]
- 47.Tomoeda, M., M. Inuzuka, and T. Date. 1975. Bacterial sex pili. Prog. Biophys. Mol. Biol. 30:23-56. [DOI] [PubMed] [Google Scholar]
- 48.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedures. In R. C. Whistler (ed.), Methods in carbohydrate chemistry, vol. 5. Academic Press, New York, NY.
- 49.Whitfield, C., P. Amor, and R. Köplin. 1997. Modulation of the surface architecture of gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol. Microbiol. 23:629-638. [DOI] [PubMed] [Google Scholar]
- 50.Wilson, M., and R. Schifferle. 1991. Evidence that the serotype b antigenic determinant of Actinobacillus actinomycetemcomitans Y4 resides in the polysaccharide moiety of lipopolysaccharide. Infect. Immun. 59:1544-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, H., S. Asikainen, B. Dogan, R. Suda, and C. Lai. 2004. Relationship of Actinobacillus actinomycetemcomitans serotype b to aggressive periodontitis: frequency in pure cultured isolates. J. Periodontol. 75:592-599. [DOI] [PubMed] [Google Scholar]
- 52.York, W., A. Darvill, M. McNeil, T. Stevenson, and P. Albersheim. 1986. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118:3-40. [Google Scholar]
- 53.Yoshida, Y., Y. Nakano, T. Nezu, Y. Yamashita, and T. Koga. 1999. A novel NDP-6-deoxyhexosyl-4-ulose reductase in the pathway for the synthesis of thymidine diphosphate-D-fucose. J. Biol. Chem. 274:16933-16939. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida, Y., Y. Nakano, N. Suzuki, H. Nakao, Y. Yamashita, and T. Koga. 1999. Genetic analysis of the gene cluster responsible for synthesis of serotype e-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim. Biophys. Acta 1489:457-461. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida, Y., Y. Nakano, Y. Yamashita, and T. Koga. 1998. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycetemcomitans. Infect. Immun. 66:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, C., K. P. Mintz, and T. Ruiz. 2009. Investigation of the three-dimensional architecture of the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans by electron tomography. J. Bacteriol. 191:6253-6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, C., T. Ruiz, C. Lenox, and K. Mintz. 2008. Functional mapping of an oligomeric autotransporter adhesin of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190:3098-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yue, G., J. Kaplan, D. Furgang, K. Mansfield, and D. Fine. 2007. A second Aggregatibacter actinomycetemcomitans autotransporter adhesin exhibits specificity for buccal epithelial cells in humans and old world primates. Infect. Immun. 75:4440-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]