Abstract
We studied the two mreB genes, encoding actinlike cytoskeletal elements, in the predatory bacterium Bdellovibrio bacteriovorus. This bacterium enters and replicates within other Gram-negative bacteria by attack-phase Bdellovibrio squeezing through prey outer membrane, residing and growing filamentously in the prey periplasm forming an infective “bdelloplast,” and septating after 4 h, once the prey contents are consumed. This lifestyle brings challenges to the Bdellovibrio cytoskeleton. Both mreB genes were essential for viable predatory growth, but C-terminal green fluorescent protein tagging each separately with monomeric teal-fluorescent protein (mTFP) gave two strains with phenotypic changes at different stages in predatory growth and development. MreB1-mTFP cells arrested growth early in bdelloplast formation, despite successful degradation of prey nucleoid. A large population of stalled bdelloplasts formed in predatory cultures and predation proceeded very slowly. A small proportion of bdelloplasts lysed after several days, liberating MreB1-mTFP attack-phase cells of wild-type morphology; this process was aided by subinhibitory concentrations of an MreB-specific inhibitor, A22. MreB2-mTFP, in contrast, was predatory at an almost wild-type rate but yielded attack-phase cells with diverse morphologies, including spherical, elongated, and branched, the first time such phenotypes have been described. Wild-type predatory rates were seen for all but spherical morphotypes, and septation of elongated morphotypes was achieved by the addition of A22.
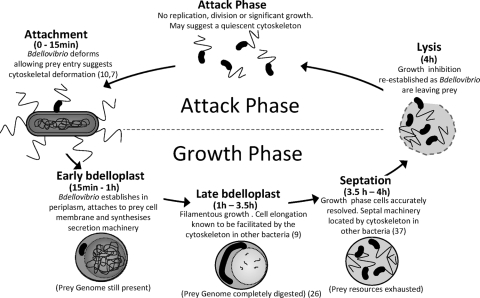
The predatory bacterium Bdellovibrio bacteriovorus shows novel filamentous growth within the periplasm of the Gram-negative prey bacterium on which it feeds. This study focuses on the cytoskeletal protein MreB and the role that two homologues of it play in B. bacteriovorus predatory or host-dependent (HD) growth. The HD B. bacteriovorus life cycle can be split into two phases: an attack phase and a growth phase (Fig. 1). The attack-phase B. bacteriovorus is a small free-swimming, highly motile cell within which replication has been arrested and which does not take up organic nutrients from the environment or grow extensively (24, 26). Once an attack-phase cell has collided with a suitable prey bacterium, B. bacteriovorus opens and squeezes through a small hole, formed in the outer membrane, using type IV pili to pull itself inside (7, 10). The B. bacteriovorus reseals the hole upon entering the periplasm. Once inside, the prey is killed rapidly within 15 min, and the prey cell wall is partially digested (35), forming a rounded structure called the bdelloplast (see Fig. 1 and 4Ac and d). The HD B. bacteriovorus cell then enters the second, growth phase, part of the life cycle, (Fig. 1), whereby it grows filamentously while simultaneously coordinating the digestion and transportation of monomers from the prey cytoplasm. The mature growth-phase cell is multiploid and elongates typically 3 to 10 times the length of an attack-phase cell, its length being a reflection of the nutritional resources available in the prey (20). Once resources within the bdelloplast are depleted, the mature filament septates sequentially from one pole to form multiple progeny. These lyse the exhausted bdelloplast, mature into attack-phase cells, and repress growth once again (Fig. 1). B. bacteriovorus can be cultured slowly, without prey, as host-independent (HI) cells growing upon peptone-rich medium (30). In these conditions they grow pleiomorphically as mainly long filamentous or serpentine cells, from which some small attack-phase cells septate (30).
FIG. 1.
Schematic host-dependent (HD) predatory cycle for B. bacteriovorus on E. coli prey, showing the different phases of growth and inferred demands on the B. bacteriovorus cell cytoskeleton. References where the roles of the cytoskeleton in cell development have been proven for other bacteria are provided in parentheses. The status of the prey genome is both drawn from an earlier study (26) and confirmed by our work in the present study (see Fig. 4).
The predatory lifestyle of B. bacteriovorus presents a number of novel developmental challenges to the B. bacteriovorus cell and its cytoskeleton. It is not known how the attack-phase cells deform, allowing the B. bacteriovorus to squeeze through a pore it makes in the prey outer membrane that is narrower than the width of an attack-phase cell, as was imaged by Burnham et al. in the 1960s and more recently by Evans et al. (7, 10). It is also not known how the growth-phase filamentous cell within the bdelloplast is generated and remains resistant to division until terminal sequential septation begins, despite having multiple potential sites for septation along its length while elongating.
The processes of cell elongation in rod-shaped bacteria are coordinated by an internal MreB cell cytoskeleton (9). MreB is a eukaryotic actin homologue and has been well studied in Escherichia coli, Bacillus subtilis, and Caulobacter crescentus (38). MreB monomers polymerize on ATP binding, forming helical structures in vivo that appear to associate with the cytoplasmic side of the bacterial cytoplasmic membrane (11, 18, 31). Bacterial two-hybrid experiments in Escherichia coli suggest that MreB forms a transmembrane complex with the two proteins MreC and MreD, each of which have been shown to form helical structures in vivo (21). A complex of MreBCD, together with the RodA protein, influences the shape of the peptidoglycan cell wall and thus the shape of the cell by positioning the peptidoglycan biosynthetic machinery so that its action is directionally specific (9, 19, 37). The MreB filament has also been shown to have roles in chromosome segregation, septation, and cell polarity (13, 14, 22, 37).
Depletion of the MreB protein levels in E. coli and B. subtilis led to cells taking on a spherical morphology and eventual loss of viability since new peptidoglycan is not synthesized evenly along the cell wall (9, 36). Uneven incorporation of new peptidoglycan is potentially driven by the tubulin homologue FtsZ (4, 36). A similar phenotype is achieved by addition of the MreB inhibitor A22 that causes the reversible loss of MreB filament localization in vivo (14, 17). A22 was discovered in a chemical library being screened for the ability to generate anucleate minicells from E. coli (16, 17). It has been used extensively by others to examine MreB function in bacteria of many different genera. The addition of A22 to E. coli cells at 3.13 μg/ml leads to the breakdown of MreB filaments, spheroplasting and the generation of minicells (16). In B. subtilis, A22 at concentrations in excess of 100 μg/ml is required to generate spheroplasts; in C. crescentus 6-h incubations of A22 at 10 μg/ml are needed before any change to the cell shape can be observed—in this case cells take on a characteristic “lemon shape” (14, 16). Isolation and sequencing of A22-resistant mutants of C. crescentus, as well as biochemical evidence from purified MreB from Thermotoga maritima, revealed that A22 binds in the nucleotide binding pocket of MreB (3, 14). In vitro light scattering assays of MreB filamentation showed that A22 acted as a competitive inhibitor of ATP binding and was able in inhibit the formation of MreB filaments, presumably by sequestering and inactivating MreB monomers preventing their recycling (3). This study also demonstrated that in vitro A22 can have a role in stabilizing ADP-bound MreB (3).
In the present study we investigated the functions of the two MreB homologues found in B. bacteriovorus, testing the role each has in predation and cell morphology by a combination of genetic approaches and A22 treatment. A reduction in function in both MreB proteins, achieved by C-terminal teal fluorescent protein (TFP) tagging, suggests that the MreB1 (Bd0211) protein was required in the growth-phase cell in bdelloplasts, whereas the MreB2 (Bd1737) protein is required later in the predatory process, for maintaining and allowing successful resolution of the growth-phase filament into short attack-phase vibroid cells.
MATERIALS AND METHODS
Strains.
A full list of all strains and plasmids used in the present study can be found in Table 1. The B. bacteriovorus genome-sequenced strain HD100 (20) was used throughout as a parental strain. All B. bacteriovorus strains were grown by predation on E. coli S17-1 in CaHEPES buffer (25 mM HEPES, 2 mM CaC12 [pH 7.6]) using standard culturing and maintenance methods described in reference (24). The prey strains E. coli S17-1 (32) and a kanamycin-resistant S17-1 strain carrying the pZMR100 plasmid (28) were used as needed. All molecular cloning was carried out using the E. coli strain DH5α (15). Conjugation of plasmids into B. bacteriovorus was achieved by using E. coli S17-1 as the donor strain.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1 | thi pro hsdR−hsdM+recA; integrated plasmid RP4-Tc::Mu-Kn::Tn7 used as donor for conjugation of plasmids into Bdellovibrio | 32 |
| DH5α | F′ endA1 hsdR17 (rK− mK−) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF)U169 deoR [80dlacΔ(lacZ)M15]; used as a cloning host strain | 15 |
| S17-1:pZMR100 | S17-1 strain containing pZMR100 plasmid to confer Kmr; used as Kmr prey for Bdellovibrio | 28 |
| B. bacteriovorus | ||
| HD100 | Type strain; genome sequenced | 27 |
| HID13 | Host-independent derivative of HD100 | This study |
| HD100mreB1-mtfp | HD100 strain carrying integrated plasmid pAKF41a at the mreB1 (Bd0211) locus | This study |
| HD100mreB2-mtfp | HD100 strain carrying integrated plasmid pAKF40a at the mreB2 (Bd1737) locus | This study |
| HD100Bd2345::Kmr | HD100 strain with a kanamycin-interrupted ABC gene Bd2345; Kmr cassette placed in the equivalent genome position of the Bdellovibrio sp. strain 109JK, which has no predatory phenotype | 24; L. Hobley, unpublished data |
| HD100filC1/filC1::Kmr | Merodiploid HD100 with both wild-type and kanamycin-interrupted fliC1 | 10 |
| Plasmids | ||
| pUC19 | Ampr; high-copy-number cloning vector | 39 |
| pUC4K | High-copy-number vector used as a source of Kmr cassette in Bdellovibrio kanamycin interruption knockouts | 39 |
| pSET151 | Suicide vector used for conjugation and recombination into Bdellovibrio genome | 5 |
| pK18mobsacB | Kmr suicide vector used for conjugation and recombination into Bdellovibrio genome | 29 |
| pAKF10 | Derivative of pSET151 containing kanamycin-interrupted HD100 mreB1 ORF with 1-kb 5′ and 3′ flanking genomic DNA | This study |
| pAKF20 | Derivitive of pSET151 containing kanamycin-interrupted HD100 mreB2 ORF with 1-kb 5′ and 3′ flanking genomic DNA | This study |
| pAKF40a | Derivative of pK18mobsacB containing 1,010-bp 3′ fragment of mreB2 ORF lacking the stop codon fused, with mtfp | This study |
| pAKF41a | Derivative of pK18mobsacB containing 948-bp 3′ fragment of mreB1 ORF lacking the stop codon fused, with mtfp | This study |
Nalr, nalidixic acid resistance; Kmr, kanamycin resistance; Ampr, ampicillin resistance.
mreBCD RT-PCR detection of transcripts.
B. bacteriovorus total RNA preparations and reverse transcription-PCR (RT-PCR) reactions (30 amplification cycles) were carried out as described previously (10). In the present study primers were designed to specifically amplify internal regions of the mreB1, mreB2, mreC, and mreD genes giving expected product sizes of: 95, 90, 110, and 105 bp, respectively.
mreB gene inactivation attempts.
PCR-amplified fragments from the HD100 genome of both mreB genes plus 1 kb of 5′ and 3′ flanking genomic sequence were inserted into pUC19 by using XbaI sites introduced by the primers. Inactivation of mreB genes was by insertion of a blunt-ended HincII kanamycin cassette fragment from pUC4K (39) into the unique restriction sites in each mreB open reading frame (ORF): SnaBI for mreB1 and BglII for mreB2. Constructs were transferred to the mobilizable pSET151 vector by using XbaI sites generating constructs with kanamycin interruption of each mreB gene for conjugation into B. bacteriovorus: pAKF10 (mreB1::Km) and pAKF20 (mreB2::Km) (5). Conjugation of pAKF10 and pAKF20 plasmids using the S17-1 strain of E. coli is described in detail elsewhere (10, 24). Multiple candidates from independent conjugations and independently generated HI strains of merodiploids were screened in efforts to knock out these genes. A total of 62 HD and 266 HI knockout candidates were screened for an mreB1 knockout strain, and 136 HD and 436 HI knockout candidates were screened for an mreB2 knockout. This number has been successfully used to isolate multiple other knockout mutants in B. bacteriovorus (10, 23). The addition of 20 mM MgCl2 to B. bacteriovorus prey lysates to support growth of potential mreB knockouts was also attempted, as described previously (12, 19).
MreB C-terminal GFP fusions.
PCR amplification of 958- and 1,020-bp regions of the HD100 genome gave 3′ ORF fragments from the B. bacteriovorus mreB1 and mreB2 genes, respectively. These fragments, if translated, would run from glycine 11 to the C-terminal glutamic acid residue 347 and represent 97% of the MreB1 protein and between glycine 48 to the final valine residue 347 and represent 86% of MreB2. In each case, primers removed the stop codon and introduced EcoRI and KpnI sites, which were used for insertion, in frame, with the bright, monomeric GFP ORF (from Allelebiotech) named mTFP1, hereafter called mTFP (2). Insertion of each mreB ORF fragment generated short linker coding regions between the last codon of the mreB ORF fragment and the ATG start codon of the mtfp gene; this linker codes for the amino acids VQRSS in both cases. mreB-mTFP fusion constructs were removed intact using EcoRI and BamHI sites and inserted into pK18mobsacB, creating the pAKF41a and pAKF40a containing the mreB1-mtfp and mreB2-mtfp constructs, respectively (Table 1). The pK18mobsacB plasmid backbone encoded both a kanamycin resistance cassette and mobilization apparatus, which is acted upon by donor strain E. coli S17-1 (29). Plasmids were conjugated into B. bacteriovorus HD100 as described previously (24). Candidate strains containing a genome integrated plasmid were selected on medium containing kanamycin. Strains with recombination of the mreB-mtfp fusion construct into the genome at the correct mreB locus were confirmed by amplification using primers that bound to the genomic DNA both upstream of cloned ORF fragments and pK18mobsacB, followed by DNA sequencing of those PCR products That a single insertion of the plasmid in each genome had occurred was verified by Southern blotting.
Electron microscopy.
For electron microscopy, cells were prepared and imaged as described previously (10).
Fluorescence microscopy.
The mTFP activity in B. bacteriovorus cells was detected by using a Nikon Eclipse E600 epifluorescence microscope and a CFP filter block (excitation, 462 nm; emission, 429 nm). Images were captured by using a Hamamatsu Orca ER camera and analyzed by using IPLab, version 3.64. Fluorescence images were background corrected by using the 3D filter tool and normalized within the IPLab software. All GFP merged images were generated within the IPLab software. Due to the lack of color options within IPLab, DAPI merged images were achieved by using the duplicate channels tool in Photoshop version 5.5; 100% opacity was used in all cases.
E. coli predatory killing curves.
Fifty milliliters of B. bacteriovorus cultures that had fully lysed E. coli prey was filtered through two 0.45-μm-pore-size filters to remove any residual prey cells, and B. bacteriovorus cells were harvested by centrifugation at 51,000 rpm (5,525 × g) for 20 min in a Sigma 4K15 centrifuge and resuspended in 10 ml of CaHEPES. The protein concentration of each B. bacteriovorus preparation was measured by using a Lowry assay (25). B. bacteriovorus inocula with matched protein concentrations were used to inoculate 50-ml predatory cultures containing 50 μg of kanamycin/ml and kanamycin-resistant E. coli S17-1pZMR prey at matched optical densities at 600 nm (OD600) of 0.8. These predatory cultures were incubated at 29°C in a shaking incubator and monitored for the prey OD600 as predation proceeded. (B. bacteriovorus attack-phase cells do not give an appreciable absorbance at 600 nm due to their small size.)
B. bacteriovorus synchronous prey infection.
To establish near synchronicity of infection, predatory cocultures were subcultured in 50 ml of CaHEPES buffer for 3 days at 24-h intervals before the resultant B. bacteriovorus cells were used to inoculate a 100-ml predatory lysate. B. bacteriovorus attack-phase cells were harvested after 24 h by centrifugation at 5,525 × g in a Sigma 4K15 centrifuge at 25°C for 20 min. The resulting pellet was resuspended in 10 ml of fresh CaHEPES, and B. bacteriovorus cells were allowed to recover for 3 h in a shaking 29°C incubator. Fifty milliliters of overnight E. coli S17-1:pZMR100 culture was centrifuged at 5,525 × g in a Sigma 4K15 at 25°C for 10 min, and the pellet was diluted in CaHEPES to an OD600 of 1.5. The synchronous predatory culture was set up using 4 ml of B. bacteriovorus preparation, 3 ml of E. coli S17-1:pZMR100 preparation, and 5 ml of fresh CaHEPES preincubated at 29°C. Typically, enumerations revealed that these synchronous cultures contained a B. bacteriovorus/E. coli ratio in excess of 3:1. This ratio was sufficiently high so that the predatory culture was nearly synchronous with >95% of E. coli prey cells becoming bdelloplasts at 30 min after wild-type B. bacteriovorus infection (1, 26).
RESULTS
B. bacteriovorus has two mreB genes that are essential for viability.
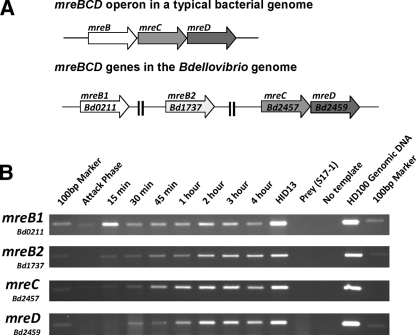
The B. bacteriovorus genome contains two genes encoding proteins with high homology to E. coli MreB, Bd0211 encoding MreB1 (45% identity at the protein level) and Bd1737 encoding MreB2 (58% identity at the protein level). Both of the mreB genes in B. bacteriovorus are not located upstream of mreCD and thus lack an operon organization that is conserved among rod-shaped bacteria (Fig. 2A). Immediately downstream of the mreCD genes in the B. bacteriovorus genome are a peptidoglycan biosynthetic gene pbp2 and rodA, whose protein products are known to form a complex with MreCD in E. coli (21). B. bacteriovorus belongs to the deltaproteobacteria family of Gram-negative bacteria and, although the number of mreB gene homologues in this family is variable, the gene organization of mreC, mreD, pbp2, and rodA is conserved.
FIG. 2.
mreBCD gene organization and expression in B. bacteriovorus. (A) Schematic drawing of the mreBCD locus in a typical bacterial genome versus that of the B. bacteriovorus genome strain HD100. (B) Semiquantitative RT-PCR study of B. bacteriovorus mreBCD expression across a predatory infection cycle. RT-PCR reactions were carried out on RNA extracted at the time points across a synchronous lysate of B. bacteriovorus on the E. coli strain S17-1. Time points at which the RNA sample was taken are labeled on the figure. The HID13 strain used to assay HI gene expression. E. coli S17-1 RNA and no-template reactions provide negative controls, and HD100 genomic DNA was used as a template for a positive control. Primers for gene detection were designed to amplify an internal region of each mreBCD gene. The 100-bp marker from the NEB 100-bp ladder is visible in each marker lane.
Deletion of both mreB genes in B. bacteriovorus was attempted by kanamycin cartridge insertion by methods described previously (24) and using both host dependently growing and host independently growing strains to rescue strains that failed at any point in the HD life cycle (10). Deletions of the mreB genes were also attempted in medium supplemented with 25 mM MgCl2, which is known to support the growth of mreB deletion strains in B. subtilis (12). Despite these provisions, no mreB deletion strains were isolated, although 328 candidates were screened for a mreB1 knockout and 572 candidates were screened for a mreB2 knockout, which was manyfold more than those screened to produce a pilA knockout in the same strain of B. bacteriovorus (2, 10).
B. bacteriovorus mreB1/mreB2 mreCD expression peaks in the growth phase of the predatory cycle.
RT-PCR analyses carried out on RNA samples taken at intervals from synchronous B. bacteriovorus infections were used to monitor the expression of mreB1/mreB2 and mreCD. The RT-PCR had the number of amplification steps limited, so as not proceed to saturation and can therefore be used as a semiquantitative measure of gene expression across a B. bacteriovorus infection cycle, as confirmed previously in comparison to quantitative RT-PCR (10, 34). Multiple sets of reactions on a minimum of two independently prepared RNA sample sets are summarized in Fig. 2B. Despite being located in different regions of the chromosome, expression of mreB, mreC, and mreD genes of B. bacteriovorus showed a peak of transcription at the 2- to 3-h postinfection stage, with almost undetectable expression at the 0-min attack-phase time point (Fig. 2B). This is consistent with the expectation that B. bacteriovorus cell elongation in the growth phase (Fig. 1) involves the MreBCD proteins, as is the case for cell elongation in nonpredatory bacteria. The mreB1 gene showed a reproducible (three repeats) additional peak of expression at 15 min (Fig. 2B) at around the time of periplasmic invasion, when initial establishment of the growth-phase filamentous cell is occurring (Fig. 1). RT-PCR analysis on RNA from both the 2-h synchronous predatory infection time point and on RNA from HI-grown cells showed that the mreCD genes are cotranscribed on the same mRNA molecule but that the pbp2 and rodA genes were transcribed separately to each other and to mreCD (data not shown).
C-terminal mTFP tagging of each MreB caused developmental changes in predatory B. bacteriovorus.
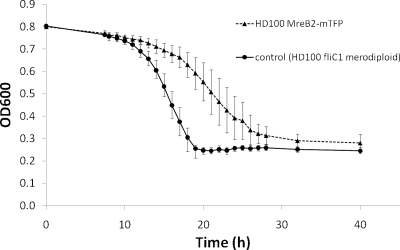
Previous studies with other bacteria showed that C-terminal GFP tagging altered the turnover dynamics of MreBs; since we could not inactivate mreB genes, we used this approach (18, 33). To determine the involvement of mreB1 or mreB2 in the predatory cycle of B. bacteriovorus, we constructed two mreB-mtfp-tagged strains in which each MreB protein was individually C-terminally labeled with a bright monomeric teal-fluorescent protein (mTFP) (2). A 3′ fragment of each mreB coding region lacking the stop codon was PCR amplified and cloned in frame with the mtfp ORF. This construct was integrated into the B. bacteriovorus genome, giving a strain in which each mreB-mtfp fusion was expressed from its native promoter (verified by sequencing), along with a promoterless corresponding mreB 3′ORF fragment in the presence of a wild-type copy of the other mreB gene on the genome (see Materials and Methods). Progression through the predatory cycle was monitored for both strains using fluorescence and is shown also in terms of prey killing efficiency for MreB2-mTFP (Fig. 3) and discussed below for MreB1-mTFP. As shown in Fig. 3, the MreB2-mTFP strain was effectively predatory but killed E. coli at a slightly slower rate than the control.
FIG. 3.
Predatory kill curves of HD100 mreB2-mTFP versus control. OD600 values for 50-ml B. bacteriovorus cultures containing E. coli S17-1:pZMR100 prey matched to a starting OD600 of 0.8. Matched protein concentrations of B. bacteriovorus lysates were used to start infections. Values fall as prey are lysed given that B. bacteriovorus attack-phase cells do not register at OD600. The B. bacteriovorus fliC1 merodiploid strain was used as a control, since it has no predatory phenotype but has an integrated kanamycin cassette of the same type as the MreB2-mTFP strain. Points on the graph represent three biological repeats, with each experiment set up in duplicate.
The MreB1-mTFP strain perturbed development only within the bdelloplast.
TFP activity within the MreB1-mTFP strain was low, and detectable fluorescence was constrained to the growth-phase cell, within the bdelloplast, which is partly consistent with the RT-PCR results (Fig. 2B and 4D). The MreB1-mTFP strain showed a phenotype in which the development of the growth-phase cell, within the bdelloplast, had been compromised instead of proceeding rapidly through the cycle like the wild-type B. bacteriovorus. These MreB1-mTFP B. bacteriovorus failed to plaque bacterial lawns and in prey-killing experiments in 50-ml cultures the MreB1-mTFP strain took between five to seven times longer than the wild-type strain (which took 24 h to lyse all of the prey cells present and clear the culture) to partially but never totally clear the prey from the culture due to the presence of many stable although abortive bdelloplasts, as seen by phase-contrast microscopy (Fig. 4Bg, h, and i).
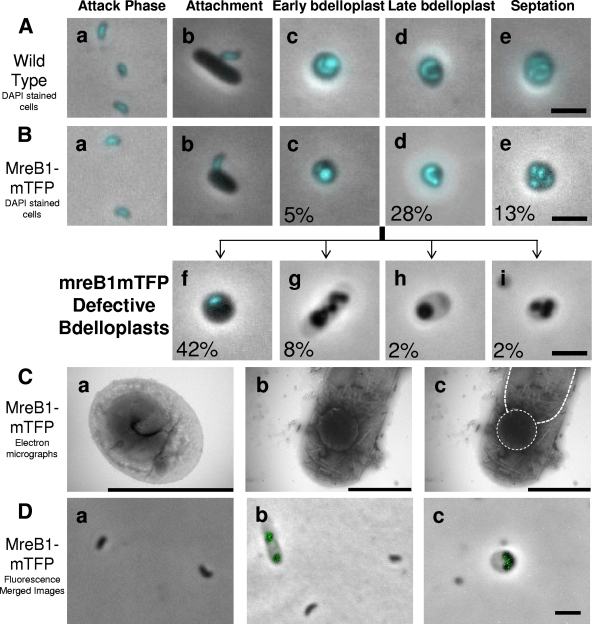
FIG. 4.
Defective bdelloplast development in the HD100 MreB1-mTFP strain. (A) Merged fluorescent images of a DAPI-stained wild-type HD100 B. bacteriovorus infection. The images show labeled key stages of the B. bacteriovorus life cycle: small vibrioid attack-phase cells (a), cells attaching to and invading intact prey (b), early-stage bdelloplasts where B. bacteriovorus and prey DNA are visible as the prey cell wall has been partially degraded making the infected cell more transparent (c), late-stage bdelloplasts where the B. bacteriovorus has grown to a long filament and the prey DNA and other cytoplasmic contents are digested (d), and mature bdelloplasts in which septation of B. bacteriovorus has occurred prior to prey lysis (e). (B) Merged fluorescent images of DAPI-stained HD100 MreB1-mTFP strain infection. The images were obtained after 24 h of incubation. (a to e) B. bacteriovorus life cycle stages in common with the wild type; (f to i) defective bdelloplasts, including stalled bdelloplasts where prey DNA has been degraded and yet there is still no growth of B. bacteriovorus to form a filament, as is seen in wild-type late bdelloplasts (f), elongated but spheroplasting B. bacteriovorus that is not producing intact filamentous growth (g), and spheroplasting B. bacteriovorus within bdelloplasts (h and i). These are morphologies novel to this MreB1-mTFP strain. Percentage values indicate the prevalence of the various morphologies within the 24-h prey lysate for this strain in which the synchrony of infection was breaking down. (C) Electron micrographs of MreB1-mTFP bdelloplasts. A wild-type bdelloplast in which the growth-phase cell has formed correctly (a) and an example of the MreB1-mTFP strain, stalled bdelloplast morphotype (Fig. 4Bh), (b), are shown with the outlines of the rounded Bdellovibrio and the prey cell cytoplasm outlined (c). The stain used was 0.5% uranyl acetate. (D) Merged fluorescence micrographs showing the very low level of TFP fluorescence in the MreB1-mTFP cells and that this is not present in attack-phase cells (a) but is seen in cells within a bdelloplast (b and c). Scale bars, 2 μm.
For analysis of the bdelloplast stalling phenotype of the MreB1-mTFP, a starting B. bacteriovorus MreB1-mTFP “attack-phase only” inoculum was generated by filtering five independent aliquots of 2-ml replicate prey lysates, raised individually from frozen stocks and incubated for 5 days, through a 0.45-μm-pore-size filter, to remove all previously stalled bdelloplasts but allow harvesting of pure attack-phase MreB1-mTFP B. bacteriovorus. These attack-phase cells were used in infections with fresh prey, so that the approximate age of bdelloplasts in the resulting prey lysate could be determined. These infections were surveyed microscopically at 24-h intervals, and a survey of stalled bdelloplasts taken at day 1 (24 h) is displayed as percentages in Fig. 4B. For a comparison, the stages in a wild-type B. bacteriovorus HD100 infection are shown in Fig. 4A. Since each prey cell was lysed completely by HD100 within 4 h (Fig. 4Ae, for example, was obtained after 3.5 h) and since no stalled bdelloplast stages were ever seen for HD100, the images in Fig. 4Ac, d, and e could not be time matched to the MreB1-mTFP culture but are shown for morphological and staining comparisons. It was clear that some of the B. bacteriovorus cells in the stalled bdelloplasts were spheroplasting (Fig. 4Bg, h, and i and Fig. 4Cb and c compared to nonspheroplasting Fig. 4Ca), whereas others remained as attack-phase-sized cells within the bdelloplasts, even after the prey cytoplasmic material (including DNA, which stains bright with DAPI [4′,6′-diamidino-2-phenylindole] and Hoechst 33372 stain used in this study) had been degraded (compare Fig. 4Bf to Fig. 4Ac; the wild-type B. bacteriovorus is elongating even while the prey DNA remains; but in the stalled MreB1-mTFP strain the B. bacteriovorus is not elongated even though the prey DNA is not detected by staining with DAPI).
To better understand the morphologies and behaviors of MreB1-mTFP stalled growth-phase cells in bdelloplasts, DAPI staining (with Hoechst 33372) was used to label the B. bacteriovorus chromosomes and prey DNA (where still present) within the usually phase-dark, stalled bdelloplasts and in the minority of the MreB1-mTFP B. bacteriovorus that did escape the block and proceed through the predatory cycle (Fig. 4Bc, d, and e). DAPI staining of B. bacteriovorus lysates had not been reported until now; it was readily taken up by both attack-phase cells and growing cells within bdelloplasts (Fig. 4A and B). DAPI staining of B. bacteriovorus gave a higher fluorescence intensity than for the E. coli prey in the attachment images (Fig. 4Ab and Bb) due to the relatively high concentration of DNA per unit volume within the predator cells, with B. bacteriovorus being ca. 1/5 the volume of a E. coli and yet containing a genome of comparable size (27). In early bdelloplasts, DAPI staining revealed both the B. bacteriovorus and E. coli prey chromosomes (Fig. 4Ac and Bc). In these bdelloplasts the B. bacteriovorus chromosome appeared diffuse throughout the growth-phase cell where the chromosome of the prey was concentrated within a rounded prey cytoplasm. (Fig. 4Ac and Bc). Where the predatory infection continued normally and the prey chromosome was digested in bdelloplasts, produced by wild-type B. bacteriovorus, or mutants managing to complete the predatory cycle; DAPI staining could be used to observe the morphology of the B. bacteriovorus growth-phase cells within the bdelloplast (Fig. 4Ad and Bd) and their ultimate septation (Fig. 4Ae and Be).
Strikingly, 42% of MreB1-mTFP bdelloplasts contained B. bacteriovorus that had not yet formed a filamentous growth-phase cell at 24 h of infection (Fig. 4Bf) and appeared morphologically similar to wild-type cells in the early stage of development (Fig. 4Ac). However, the lack of DAPI stain fluorescence within the prey cytoplasm in these bdelloplasts, compared to true early-stage bdelloplasts (Fig. 4Bf, in contrast to Fig. 4Ac and Bc), suggests that prey chromosome digestion had been completed in those MreB1-mTFPderived bdelloplasts due to the action of B. bacteriovorus nucleases (26). Thus, the lack of B. bacteriovorus filament elongation was due to the MreB1-mTFP-tagging and not to a lack of growth resources. The decrease in the presence of stalled MreB1-mTFP-derived bdelloplasts over time suggests that a fraction of these mutants must eventually find escape and give at least some progeny cells (which we were able to harvest by filtration after 5 days and use as the B. bacteriovorus inoculum for this experiment). Only 28% of MreB1-mTFP bdelloplasts had successfully formed an elongated growth-phase filament (Fig. 4Bd) after 24 h, which should allow attack-phase cells to septate from it and escape the stalled bdelloplast.
For MreB1-mTFP B. bacteriovorus within bdelloplasts, 12% (Fig. 4Bg, h, and i) had failed to develop correctly at 24 h, and the bdelloplasts had become transparent as the prey contents were completely digested. These B. bacteriovorus cells were either totally spherical (Fig. 4Bh), spherical and septated (Fig. 4Bi), or at an intermediate phase (Fig. 4Bg). These morphologies are similar to those reported for MreB depletion strains in other nonpredatory bacteria (8). Spherical MreB1-mTFP B. bacteriovorus cells within bdelloplasts persisted in these cultures and fail to lyse prey, suggesting that the expression of genes required for prey lysis was not fully activated in these abortive structures. Electron microscopy confirmed that these bdelloplasts contained a spherical B. bacteriovorus (see Fig. 4Cb and c, which shows a spherical B. bacteriovorus within the periplasmic space of an E. coli prey cell).
Curiously, all attack-phase cells of the MreB1-mTFP strain that did complete predation had wild-type morphology, and all contained a centrally located copy of the genome, as revealed by DAPI staining (compare Fig. 4Aa to Ba). Infections with high prey/Bdellovibrio ratios, using MreB1-mTFP attack-phase cells filtered out from previous predatory cultures also containing stalled bdelloplasts, revealed that all attack-phase cells of this strain were capable of prey attachment and entry (Fig. 4Bb), but again the same levels of stalled bdelloplasts were produced; thus, the main impact of the MreB-mTFP tagging appeared to take place after prey entry.
MreB2-mTFP causes unusual attack-phase morphologies.
In contrast to the MreB1-mTFP strain, which had a developmental defect within the bdelloplast, the MreB2-mTFP strain completed the predatory cycle and lysed prey cells at almost wild-type rates in synchronous prey lysate cultures with high Bdellovibrio/prey ratios (3:1) (data not shown), whereas at low Bdellovibrio/prey ratios (1:6) the predation rates seemed significantly slower than those of the control strain (Fig. 3).
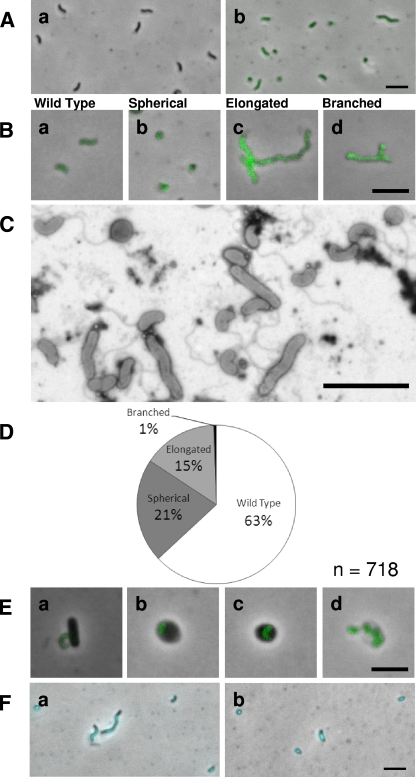
The MreB2-mTFP strain, probably due to a change in the dynamics of the MreB2 protein caused by the addition of the mTFP, yielded fluorescent, attack-phase B. bacteriovorus cells with differing morphologies (Fig. 5A and B). In contrast to the MreB1-mTFP strain, which could most readily be visualized by DAPI staining in the fluorescence microscope due to faint TFP fluorescence in the bdelloplast (Fig. 4D), all of the MreB2-mTFP strain cells showed mTFP-fluorescence from pole to pole. TFP activity was detected at all points of the B. bacteriovorus cycle from prey attachment to prey lysis (Fig. 5E). The distribution of morphotypes in the MreB2-mTFP strain was observed by using fluorescence microscopy (Fig. 5B) and scored in five separate experiments summarized in the pie chart (Fig. 5D). Observations made by using Electron microscopy confirmed this distribution (Fig. 5C).
FIG. 5.
Analysis of the attack-phase morphologies of HD100 MreB2-mTFP strain. (A) Fluorescence images of the H100ABC, nonfluorescent “wild-type” control strain (a) and HD100 MreB2-mTFP attack-phase cells (b). (B) Attack-phase morphologies of MreB2-mTFP strain wild-type (a), spherical (b), elongated (c), and branched (d) cells. (C) Representative electron micrograph showing MreB2-mTFP attack-phase cells. The cells were stained with 0.5% uranyl acetate. (D) Pie chart representing a survey of 718 HD100 MreB2-mTFP attack-phase cell morphologies. (E) Fluorescence images of MreB2-mTFP strain at the attachment (a), early (b), and late (c) bdelloplast and lysis (d) stages of the HD predatory life cycle. (F) DAPI-stained fluorescence images of MreB2-mTFP attack-phase cells. Scale bar, 3 μm.
Wild-type B. bacteriovorus attack-phase cells are usually very uniform in size and shape, having a length, measured by cryoelectron microscopy, of 1.02 ± 0.15 μm and a width of 0.30 ± 0.02 μm (6). Although the majority of MreB2-mTFP attack-phase cells were wild type in shape (63%; Fig. 5D), a significant number of cells fell into three additional categories; elongated, spherical, and branched (Fig. 5B). Elongated cells of the MreB2-mTFP strain showed significant length variation and were classified as cells that were over double the length of a single wild-type cell (Fig. 5Ab, Bc, and C). Since these cells varied from 2 to 10 times the length of a wild-type cell and were scored as one cell, the score in Fig. 5D is an under-representation of the relative biomass of this category. Branched MreB2-mTFP cells were in all cases also elongated and only ever branched near one pole (Fig. 5Bd). Cells of the MreB2-mTFP spherical morphotype were motile and thus alive; electron microscopy confirmed that these cells had a single flagellum (Fig. 5C). DAPI staining revealed that all MreB2-mTFP cells contained a similar intensity of DNA staining, removing the possibility that the spherical cells of this strain were mini-cells (Fig. 5Fb). For MreB2-mTFP elongated cells, the chromosomal material was never partitioned along the filament but appeared as a single elongated focus always located to the midpoint of the cells (Fig. 5F).
Predatory cultures set up with a low Bdellovibrio/prey ratio of 1:8 were observed frequently before and after B. bacteriovorus prey entry to determine whether any MreB2-mTFP cell morphotypes were defective for prey entry. This showed that the spherical cells were no longer able to enter prey, remaining outside prey cells even after 3 h of incubation. We conclude that, although motile and alive, these spherical MreB2-mTFP cells are no longer viable in the longer term, since they cannot enter prey and so replicate via the predatory growth cycle.
Surprisingly, the MreB2-mTFP elongated cells were still able to invade the host periplasm despite their increased size. The rate of development of MreB2-mTFP B. bacteriovorus within bdelloplasts was assayed (by the microscopic monitoring of the first appearance of septated attack-phase Bdellovibrio escaping from lysed bdelloplasts [data not shown]) as the wild type, in synchronous infections, at a high Bdellovibrio/prey ratio (3:1). In the nonsynchronous, low-Bdellovibrio/prey-ratio (1:6) prey-killing assay, the spherical MreB2-mTFP cells, which were liberated as a some fraction of the attack-phase cells after previous predation, played no part in the infection since they failed to enter the prey (Fig. 3).
Addition of MreB inhibitor A22 caused MreB2-mTFP attack-phase cells, but not wild-type B. bacteriovorus, to alter shape.
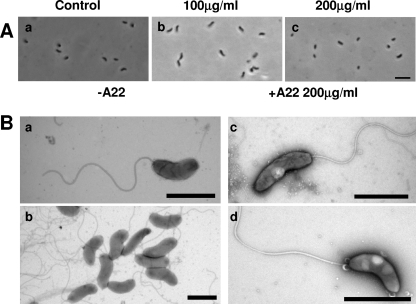
MreB inhibitor A22 has revealed details of the protein function previously in other bacteria (8, 14) causing shape deformations due to the loss of directionally specific peptidoglycan incorporation into the cell wall (36). The addition of A22 at a final concentration 100 μg/ml to wild-type B. bacteriovorus attack-phase cells caused no change in shape after 24 h of incubation (Fig. 6) or even after 48 h; this was an A22 concentration 10 times that reported to induce spherical and/or lemon-shaped cells in E. coli and C. crescentus within 6 h (14, 16). Attack-phase B. bacteriovorus cells, incubated at final A22 concentrations of 200 μg/ml for 24 h, lost motility, formed inclusion bodies, and failed to plaque bacterial lawns but maintained their cell shape (Fig. 6), whereas E. coli control cells show rounding with A22 concentrations as low as 5 μg/ml. Thus, at 200 μg/ml, A22 is toxic to the B. bacteriovorus and, at lower concentrations, the wild-type B. bacteriovorus attack-phase cell lacks the capacity to alter its shape.
FIG. 6.
Effect of the MreB inhibitor A22 on wild-type B. bacteriovorus HD100 attack-phase cells. (A) HD100 attack-phase cells after 24 h of incubation with 0 μg (a), 100 μg (b), or 200 μg (c) of A22/ml. Scale bar, 3 μm. (B) Representative electron micrographs of HD100 treated with 200 μg of A22/ml (c and d) versus untreated cells (a and b). Inclusion bodies were only observed in the A22-treated cells at this concentration. The cells were stained with 0.5% uranyl acetate. Scale bars, 1 μm.
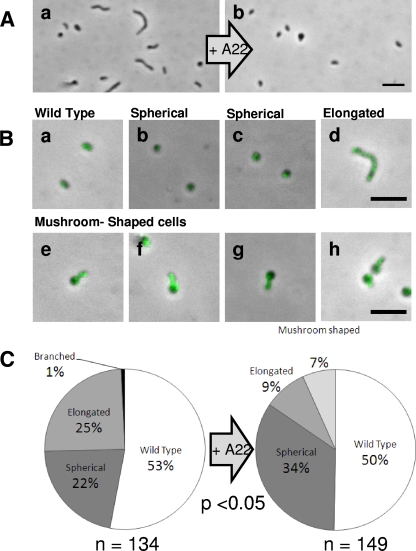
The attack-phase cells of the MreB2-mTFP strain gained the capacity to change shape with the addition of nontoxic levels of A22 (Fig. 7A). Overnight incubation of MreB2-mTFP attack-phase B. bacteriovorus with A22 at 100 μg/ml caused statistically significant changes in morphology compared to control MreB2-mTFP cultures of the same age, as summarized in Fig. 7. TFP fluorescence of the MreB2-mTFP A22-treated cells was maintained throughout (Fig. 7A and B). Overnight treatments at 100 μg of A22/ml reduced 10-fold, but did not abolish, predatory plaque counts on E. coli prey for the MreB2-mTFP strain compared to when it had no A22 added. Surveys of morphologies of MreB2-mTFP B. bacteriovorus, after A22 treatment, showed a reduction in the number of elongated cells an increase in the number of spherical MreB2-mTFP attack-phase cells (Fig. 7C) and a new intermediate form of B. bacteriovorus that was mushroom-shaped (Fig. 7Be and f) and which was not present in the controls.
FIG. 7.
Addition of A22 to the MreB2-mTFP strain alters attack-phase cell morphology. (A) Untreated attack-phase MreB2-mTFP cells (a) and MreB2-mTFP cells treated with 24 h of incubation with 100 μg of A22/ml (b). (B) Attack-phase morphologies of the MreB2-mTFP A22-treated wild type (a), spherical (b and c), elongated (d), novel mushroom-shaped (e to h) cells with fluorescently bright “stalk” shapes indicative of high MreB2-mTFP content. (C) Pie charts summarizing the survey of 134 MreB2-mTFP untreated attack-phase cells and 149 cells treated with 100 μg of A22/ml, both incubated for 24 h. Chi-square analysis of morphologies shared between both surveys gave a chi-square values of 26.75, giving a P value of <0.05 at 2 degrees of freedom. Scale bars, 3 μm.
The addition of A22 at low concentrations (0.01 μg/ml) seemed to support the predatory growth of and reduce bdelloplast persistence in theMreB1-mTFP strain, since large numbers of attack-phase B. bacteriovorus were seen microscopically, after 24 h, in predatory infections set up in the presence of that concentration of A22 but not without A22 (data not shown). In these cultures, the prey E. coli cells not morphologically affected by the A22 concentration (data not shown). Additional control prey killing experiments by the B. bacteriovorus filC1 merodiploid kanamycin-resistant “wild-type” control strain showed that A22 concentrations of 0.01 μg/ml had no effect on the predatory growth rate (data not shown).
DISCUSSION
Both mreB genes of Bdellovibrio were found to be essential for B. bacteriovorus viability and could not be deleted. This is consistent with the data for mreB from both E. coli and B. subtilis (18, 21). Despite their essentiality, the function of the B. bacteriovorus MreBs in the bdelloplast could be examined using C-terminal GFP fusions, with mTFP, to both gene products. In each case this fusion gave an MreB-mTFP strain with a phenotype very different from that of the wild type, a finding consistent with work on a B. subtilis MreB homologue, Mbl, wherein C-terminal GFP tagging resulted in a MreB protein with altered dynamics (18).
We also treated the wild-type B. bacteriovorus with the classical MreB-modifying agent A22, which produces shape changes in other bacteria due to MreB filament modifications at concentrations from 3 to 100 μg/ml (16) and found that A22 at 200 μg/ml was toxic to the B. bacteriovorus, and it formed inclusion bodies, but at lower concentrations (those active in shape-changing other bacteria) the wild-type B. bacteriovorus attack-phase cell never altered shape upon incubation with A22, although an E. coli control in our hands did round up at 5 μg/ml.
RT-PCR (Fig. 2) analyses suggested that the two mreB gene products in B. bacteriovorus are expressed during developmental stages of the B. bacteriovorus growth phase within the bdelloplast and at undetectable levels in the attack-phase cells. This is consistent with the expectation that B. bacteriovorus cell elongation in the growth phase (Fig. 1) involves MreBs. The pattern of increasing expression in the bdelloplast up to a peak at the 2- to 3-h point is consistent with the increasing size of the elongating growth-phase cell (Fig. 1). At 3 to 4 h the B. bacteriovorus growth-phase cells are septating, forming progeny, and repressing growth, yielding a slight reduction in mreB expression. The expression pattern of both mreB genes was similar to that of the mreCD operon despite being expressed from different promoters (Fig. 2). In addition, the mreB1 gene has a second peak of expression at the 15-min attachment phase, providing some circumstantial evidence for the involvement of MreB1 early in prey invasion. The mreB1 gene showed a reproducible additional peak of expression at 15 min (Fig. 2B), around the time of periplasmic invasion, when initial establishment of the growth-phase filamentous cell is occurring (Fig. 1). This may suggest that MreB1 may has a role in early-growth-phase cell development. These roles were further investigated using the mTFP fusion strains.
The MreB1-mTFP strain had a major defect in predatory growth, with normal development arresting in the early-growth-phase B. bacteriovorus inside the bdelloplast (Fig. 4Bf). The fluorescence activity within the MreB1-mTFP strain was low, and detectable fluorescence was constrained to the growth-phase cell, within the bdelloplast (Fig. 4D).
Growth-phase arrest of the MreB1-mTFP strain in the bdelloplast seemed not to be caused by an absence of growth resources, since DAPI-stained stalled bdelloplasts showed no prey DNA labeling, suggesting that the prey chromosome had been digested by B. bacteriovorus nucleases. Thus, there were at least nucleic acid resources available for B. bacteriovorus growth, but the B. bacteriovorus were not elongating for other reasons. B. bacteriovorus cells, with abortive development within bdelloplasts, failed to lyse prey, and the stalled bdelloplasts accumulated within the population (Fig. 4Bg to i). The partial rescue of this phenotype by the addition of low levels of A22, a molecule known to stabilize ADP-bound MreB filaments at subinhibitory concentrations in vitro using MreB purified from Thermotoga maritima (3), suggests that the MreB1-mTFP had a reduced capacity to form filaments within the early bdelloplast or that its filaments were less stable and that this impaired early predatory growth. Thus, the stabilization of the MreB1-mTFP filaments by A22 partly aided rescue and produced more B. bacteriovorus attack-phase cells, liberated from bdelloplasts, by the completion of predation.
A proportion of the MreB1-mTFP cells escaped the arrested state, without A22 addition, and did continue to the late bdelloplast, filamentous growth-phase septating to yield attack-phase cells that were wild type in shape, contained DNA, and were capable of HD growth (Fig. 4Ba and e). These attack-phase cells were clearly seen after 24 h of incubation with prey, and 13% of the bdelloplast population in Figure 4Be contained sepatated B. bacteriovorus at 24 h that looked indistinguishable from the wild type (Fig. 4Ae). We do not know what genetic events caused these few MreB1-mTFP cells to proceed slowly to lyse bdelloplasts at 24 h when so many others arrested (Fig. 4Bf) or spheroplasted and died within bdelloplasts (Fig. 4Bg to i). It must be remembered that a wild-type copy of mreB2 was present in the MreB1-mTFP strains but, because of the asynchrony of the predatory process for the MreB1-mTFP strain, due to the stalling bdelloplasts, it was not possible to meaningfully monitor changes in MreB2 expression in the MreB1-mTFP strains during predation.
What was very clear was that the low numbers of MreB1-mTFP attack-phase cells that completed the predatory cycle were not revertants to a wild-type (mreB1+) state. When they were separated by filtration from the stalled bdelloplasts and applied again to a fresh culture of E. coli prey, the same bdelloplast stalling effect was produced again, with only a few cells making it through to prey lysis, producing attack-phase B. bacteriovorus again.
The MreB2-mTFP strain gave a phenotypic change that occurred later in B. bacteriovorus predatory development in the bdelloplast. This strain had an almost wild-type growth rate within the bdelloplast (Fig. 3), the slightly lowered predation rate being due to the generation of a subpopulation of nonpredatory, spherical cells (Fig. 5A to C). However, upon completion of the predatory cycle, bdelloplast lysis gave MreB2-mTFP progeny with altered morphologies, including branched and elongated, in addition to spherical and wild-type vibrioid cells (Fig. 5B). We were not able to assess how many of each morphotype was produced from each bdelloplast since the cells are quite constricted within the bdelloplast before release. However, we presume that a spherical cell or branched cell was produced, along with some attack-phase cells from at least some of the bdelloplasts, rather than pure morphotypes being liberated from single bdelloplasts. For MreB2-mTFP elongated cells the chromosomal material was never partitioned along the filament but appeared as a single elongated focus always located to the midpoint of the cells (Fig. 5F). This is similar to the chromosomal distribution reported in E. coli for elongated cells produced by cephalexin treatment expressing MreB with point changes at aspartic acid 165, although this was not mutated in our MreB1-mTFP fusion (22). We found that, although the spherical cells contained DNA (Fig. 5Fb) and were motile, with a single flagellum (Fig. 5C), they were not able to predatorily enter prey. Thus, the spherical cells were a dead-end side product of the predatory cycle of MreB2-mTFP cells, which also produced the other morphotypes. Interestingly, the elongated morphotypes did enter prey. Thus, the generation of nonpredatory spherical cells effectively reduced the predatory B. bacteriovorus yield per bdelloplast of the MreB2-mTFP strain and was responsible for the slight reduction in the rate of E. coli prey killing that we observed (Fig. 3).
A strain that significantly alters attack-phase morphology in B. bacteriovorus has not been described previously. It is worth noting that although cell length and shape were affected in this strain, no significant defects in the widths of viable cells of this strain were observed; this is also the case for C-terminally GFP tagged Mbl in B. subtilis (18). The division-inhibitory properties giving rise to an elongated morphology of the MreB2-mTFP strain are consistent with published observations for the Mbl mutant of B. subtilis, but in that bacterium the block in septation involved a change in cell width (18).
Interestingly, although the cell shape of wild-type, attack-phase B. bacteriovorus was not affected by A22 addition, shape changes were seen in attack-phase cells of the MreB2-mTFP strain after the addition of A22 at 100 μg/ml, suggesting that this strain may have been turning over its cell wall/cytoskeleton in the attack phase, whereas the wild type did not seem to do so to the same extent (Fig. 6 and 7).
Incubation with A22 led to a loss of elongated cells and an increase in the number of spherical cells, suggesting that as in other model organisms, B. bacteriovorus MreB2-mTFP cells (but not the wild type) became spherical upon addition of A22 (Fig. 6 and 7). The novel mushroom-shaped morphology present (Fig. 7Be to h) in a subset of the MreB2-mTFP A22-treated population, could represent an intermediate in MreB2-mTFP attack-phase cells becoming spherical. Since B. bacteriovorus attack-phase cells rely on a finite reserve of stored material from the last infection as a nutrition source (and do not take up organic nutrients), there were few resources available to turn over cell wall and allow full conversion of the vibrioid cell-shaped attack-phase cells to spheres. This is likely why adding A22 gave rise to the mushroom-shaped cells, which are a known intermediate in the process of rod-shaped bacteria becoming spherical (8). Fluorescence activity within these A22-treated, mushroom-shaped, MreB2-mTFP cells was predominantly in the rod-shaped “mushroom stalk” part of the cell and less strongly in the hemispherical “cap” shape, suggesting that turnover of the MreB2-mTFP protein at this point was beginning to cause a shape change from vibroid-shaped to spherical. The fully spherical MreB2-mTFP cells produced without A22 did show some fluorescence, suggesting that MreB2-mTFP protein remained within them, although it was no longer functional in shape determination.
This mushroom-shaped cell production in the MreB2-mTFP strain with A22 matches that seen in Vibrio parahaemolyticus cells entering a crisis, where MreB filament breakdown at the pole of the cell caused that pole to round up, ultimately spreading over the whole cell in that case (8). We suggest that the altered dynamics of the MreB2 caused by the mTFP addition gave a distribution of B. bacteriovorus with altered MreB filament states either stabilizing the MreB2 filament directly (suggested by the 15% elongated cells, Fig. 5D) or increasing MreB2 filament turnover (suggested by the 21% spherical; Fig. 5D), the continued presence of MreB2-mTFP giving the cells the capacity to change shape by interaction with MreCD and cell wall synthetic machinery. This would suggest that such additional factors required for MreB2 to direct cell wall biosynthesis were present and active in the MreB2-mTFP strain.
Our study represents the first investigation into cytoskeletal activities during predatory B. bacteriovorus development. We show how mreB gene duplication and specialization has facilitated a developmental process that yields septated vibrioid attack-phase cells, from an intermediate elongated, filamentously growing cell growing within the confines, and finite nutritional resources, of a dead prey bacterium.
Acknowledgments
We thank Jon McMaster and Ashley Board for the synthesis of A22, Laura Hobley for the gift of strain HD100Bd2345::Kmr, and Marilyn Whitworth for technical assistance.
This study was funded by a BBSRC Ph.D. Studentship for A.K.F. to R.E.S. and by Wellcome grant AL077459 to R.E.S. for C.L. P.C.W. was an undergraduate summer student supported by a bursary from the Society for General Microbiology UK.
A.K.F. carried out the majority of the experiments, designed parts of the experimental program including the mTFP fusions and coauthored the paper; C.L. critically read and contributed to the manuscript and carried out mreB transcription and some of the cloning experiments with P.W.; and R.E.S. designed the experimental program, supervised the research, and coauthored the paper.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Abram, D., J. Castro e Melo, and D. Chou. 1974. Penetration of Bdellovibrio bacteriovorus into host cells. J. Bacteriol. 118:663-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ai, H. W., J. N. Henderson, S. J. Remington, and R. E. Campbell. 2006. Directed evolution of a monomeric, bright, and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem. J. 400:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bean, G. J., S. T. Flickinger, W. M. Westler, M. E. McCully, D. Sept, D. B. Weibel, and K. J. Amann. 2009. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry 48:4852-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendezu, F. O., and P. A. de Boer. 2008. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J. Bacteriol. 190:1792-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Borgnia, M. J., S. Subramaniam, and J. L. Milne. 2008. Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J. Bacteriol. 190:2588-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnham, J. C., T. Hashimoto, and S. F. Conti. 1968. Electron microscopic observations on the penetration of Bdellovibrio bacteriovorus into gram-negative bacterial hosts. J. Bacteriol. 96:1366-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, S. W., S. Y. Chen, and H. C. Wong. 2008. Localization and expression of MreB in Vibrio parahaemolyticus under different stresses. Appl. Environ. Microbiol. 74:7016-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 10.Evans, K. J., C. Lambert, and R. E. Sockett. 2007. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J. Bacteriol. 189:4850-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 12.Formstone, A., and J. Errington. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol. Microbiol. 55:1646-1657. [DOI] [PubMed] [Google Scholar]
- 13.Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 16.Iwai, N., T. Ebata, H. Nagura, T. Kitazume, K. Nagai, and M. Wachi. 2004. Structure-activity relationship of S-benzylisothiourea derivatives to induce spherical cells in Escherichia coli. Biosci. Biotechnol. Biochem. 68:2265-2269. [DOI] [PubMed] [Google Scholar]
- 17.Iwai, N., K. Nagai, and M. Wachi. 2002. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci. Biotechnol. Biochem. 66:2658-2662. [DOI] [PubMed] [Google Scholar]
- 18.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, Y., R. A. Daniel, and J. Errington. 2009. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol. Microbiol. 71:1131-1144. [DOI] [PubMed] [Google Scholar]
- 20.Kessel, M., and M. Shilo. 1976. Relationship of Bdellovibrio elongation and fission to host cell size. J. Bacteriol. 128:477-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78-89. [DOI] [PubMed] [Google Scholar]
- 22.Kruse, T., J. Moller-Jensen, A. Lobner-Olesen, and K. Gerdes. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert, C., K. J. Evans, R. Till, L. Hobley, M. Capeness, S. Rendulic, S. C. Schuster, S. Aizawa, and R. E. Sockett. 2006. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol. Microbiol. 60:274-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert, C., M. C. Smith, and R. E. Sockett. 2003. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J. reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 5:127-132. [DOI] [PubMed] [Google Scholar]
- 25.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 26.Matin, A., and S. C. Rittenberg. 1972. Kinetics of deoxyribonucleic acid destruction and synthesis during growth of Bdellovibrio bacteriovorus strain 109D on Pseudomonas putida and Escherichia coli. J. Bacteriol. 111:664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rendulic, S., P. Jagtap, A. Rosinus, M. Eppinger, C. Baar, C. Lanz, H. Keller, C. Lambert, K. J. Evans, A. Goesmann, F. Meyer, R. E. Sockett, and S. C. Schuster. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689-692. [DOI] [PubMed] [Google Scholar]
- 28.Rogers, M., N. Ekaterinaki, E. Nimmo, and D. Sherratt. 1986. Analysis of Tn7 transposition. Mol. Gen. Genet. 205:550-556. [DOI] [PubMed] [Google Scholar]
- 29.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 30.Seidler, R. J., and M. P. Starr. 1969. Isolation and characterization of host-independent bdellovibrios. J. Bacteriol. 100:769-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih, Y. L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. U. S. A. 100:7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon, R., U. Preifer, and A. Puhler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (NY) 9:184-191. [Google Scholar]
- 33.Slovak, P. M., G. H. Wadhams, and J. P. Armitage. 2005. Localization of MreB in Rhodobacter sphaeroides under conditions causing changes in cell shape and membrane structure. J. Bacteriol. 187:54-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steyert, S. R., S. A. Messing, L. M. Amzel, S. B. Gabelli, and S. A. Pineiro. 2008. Identification of Bdellovibrio bacteriovorus HD100 Bd0714 as a Nudix dGTPase. J. Bacteriol. 190:8215-8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomashow, M. F., and S. C. Rittenberg. 1978. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: solubilization of Escherichia coli peptidoglycan. J. Bacteriol. 135:998-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varma, A., and K. D. Young. 2009. In Escherichia coli, MreB and FtsZ direct the synthesis of lateral cell wall via independent pathways that require PBP 2. J. Bacteriol. 191:3526-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vats, P., Y. L. Shih, and L. Rothfield. 2009. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol. Microbiol. 72:170-182. [DOI] [PubMed] [Google Scholar]
- 38.Vats, P., J. Yu, and L. Rothfield. 2009. The dynamic nature of the bacterial cytoskeleton. Cell Mol. Life Sci. 66:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]