Abstract
A thioredoxin reductase and a thioredoxin were purified to homogeneity from a cell extract of Thermotoga maritima. The thioredoxin reductase was a homodimeric flavin adenine dinucleotide (FAD)-containing protein with a subunit of 37 kDa estimated using SDS-PAGE, which was identified to be TM0869. The amino acid sequence of the enzyme showed high identities and similarities to those of typical bacterial thioredoxin reductases. Although the purified T. maritima thioredoxin reductase could not use thioredoxin from Spirulina as an electron acceptor, it used thioredoxin that was purified from T. maritima by monitoring the dithiothreitol-dependent reduction of bovine insulin. This enzyme also catalyzed the reduction of benzyl viologen using NADH or NADPH as an electron donor with apparent Vmax values of 1,111 ± 35 μmol NADH oxidized min−1mg−1 and 115 ± 2.4 μmol NADPH oxidized min−1mg−1, respectively. The apparent Km values were determined to be 89 ± 1.1 μM, 73 ± 1.6 μM, and 780 ± 20 μM for benzyl viologen, NADH, and NADPH, respectively. Optimal pH values were determined to be 9.5 and 6.5 for NADH and NADPH, respectively. The enzyme activity increased along with the rise of temperature up to 95°C, and more than 60% of the activity remained after incubation for 28 h at 80°C. The purified T. maritima thioredoxin was a monomer with a molecular mass of 31 kDa estimated using SDS-PAGE and identified as TM0868, which exhibited both thioredoxin and thioltransferase activities. T. maritima thioredoxin and thioredoxin reductase together were able to reduce insulin or 5,5′-dithio-bis(2-nitrobenzoic acid) using NAD(P)H as an electron donor. This is the first thioredoxin-thioredoxin reductase system characterized from hyperthermophilic bacteria.
The thioredoxin-thioredoxin reductase system, also called the thioredoxin system, is one of the major players for converting thiol and disulfide bonds in all cells from three domains of life. It is comprised of thioredoxin (Trx), thioredoxin reductase (TR), and NADPH (2). Trxs are a group of small (10- to 12-kDa) ubiquitous proteins which have a conserved CXXC catalytic site that undergoes reversible oxidation/reduction of both cysteine residues. Among many of their functions, Trxs serve as protein disulfide oxidoreductases and interact with a broad range of proteins either for electron transport in substrate reduction or for regulation of activity via thiol-redox control (17). As evidence of their extensive involvement in cellular metabolism, the reduced Trxs can supply reducing equivalents to ribonucleotide reductase, thioredoxin peroxidase, and certain transcription factors (5, 9, 27). TRs are enzymes belonging to the family of pyridine nucleotide-disulfide oxidoreductases that include lipoamide dehydrogenase, mercuric ion reductase, glutathione reductase, and NADH oxidase, and TRs catalyze the NADPH-dependent electron transfer to the active site of oxidized Trx to form dithiol (50). Based on sizes, TRs can be classified into two types: one, with a high molecular mass (∼55 kDa, designated H-TR) and containing selenocysteine, is generally characterized from animals and the protozoan malaria parasite (13, 47); another type, with a low molecular mass (∼35 kDa, designated L-TR), is present in archaea, bacteria, and lower eukarya (16). H-TRs share general characteristics of glutathione reductase, trypanothione reductase, mercuric ion reductase, and lipoamide dehydrogenase, whereas L-TRs have similarities to alkyl hydroperoxide reductases F52A (AhpF) and other NADH:peroxiredoxin oxidoreductases (37).
It was thought that protein disulfide bonds were unlikely to be present in hyperthermophiles because cysteine side chains might be prone to degradation at the high temperatures under which hyperthermophiles thrive (25). However, recent findings of disulfide bond-containing proteins in several hyperthermophiles suggest that there is a positive correlation between the abundance of disulfide bonds and the maximum growth temperature (24, 25). Based on structural analysis, there is an indication that the disulfide bonds may contribute significantly to the thermostability of these proteins by compensating for an insufficient level of electrostatic charge optimization. Therefore, disulfide bonds could be a means of adaptation to high temperatures (45). Despite the growing evidence of the importance of disulfide bonds in hyperthermophiles, the study of the enzymes and mechanisms involved in disulfide bond formation and the maintenance of the thiol-disulfide balance is limited. TRs have been reported from hyperthermophilic archaea such as Pyrococcus horikoshii (22), Aeropyrum pernix K1 (21), and Sulfolobus solfataricus (42). Compared to TRs characterized from mesophilic microorganisms, hyperthermophilic archaeal TRs exhibit different substrate spectra. TRs from A. pernix K1 and S. solfataricus are able to catalyze the reduction of 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) directly (21, 42), which is a characteristic of H-TRs (eukaryotic TRs) and has not been found in other L-TRs yet. Unlike Trx from mesophiles, the purified thermophilic archaeal Trxs are much larger, ranging from 24.8 to 37 kDa, except that isolated from Methanococcus jannaschii, which has a size similar to those of Trxs from mesophiles, 10 kDa. There is no report on the properties of a thioredoxin system in hyperthermophilic bacteria. For a better understanding of the function of the hyperthermophilic thioredoxin system, that in Thermotoga maritima (19) was investigated. Here, we report on the first thioredoxin system from hyperthermophilic bacteria.
MATERIALS AND METHODS
Organism and chemicals.
T. maritima (DSM 3109) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany. All chemicals were commercially available products (Sigma, Oakville, Ontario, Canada) except that dihydrolipoamide was prepared from the reduction of dl-lipoamide with sodium borohydride (38).
Growth of T. maritima.
T. maritima was cultured at 80°C anaerobically in a 15-liter carboy in a medium modified from that of Huber et al. (19). The medium contained (per liter) 20 g of NaCl, 1.14 g of (NH4)2CO3, 2.0 g of KCl, 1.72 g of MgSO4·6H2O, 1.42 g of MgCl2·6H2O, 0.05 g of CaCl2·2H2O, 2.5 g of yeast extract, 4.0 g of glucose, 0.5 g of KH2PO4, 0.05 mg of resazurin, and 10 ml of trace mineral element solution (6). The pH was adjusted to pH 6.8. The growth was monitored by direct cell count using a Petroff-Hausser counting chamber (1/400 mm2, 0.02 mm deep; Hausser Scientific, Horsham, PA) and a Nikon Eclipse E600 phase-contrast light microscope (Nikon Canada, Toronto, Ontario, Canada). The cells were harvested by centrifugation at 13,000 × g, and the cell pellet obtained was frozen at −80°C until use.
Enzyme assays and protein determination.
Benzyl viologen oxidoreductase (BVOR) activity was determined in an anaerobic glass cuvette by monitoring the NADH-dependent benzyl viologen (BV) reduction at 580 nm (ɛ580 = 8.8 mM−1cm−1) at 80°C (29). The assay mixture (2 ml) contained 1 mM BV and 0.2 mM NADH in 100 mM glycine-NaOH buffer, pH 9.5. One unit of BVOR activity was defined as 2 μmol BV reduced per min. Ferredoxin:NAD+ oxidoreductase (FNOR) was determined with a method modified from Ma et al. (30). The assay mixture contained 25 μg T. maritima ferredoxin purified as described previously (7), 4 μg T. maritima pyruvate:ferredoxin oxidoreductase purified as described previously (7), 0.5 mM NAD+, 0.4 mM coenzyme A (CoA), 10 mM sodium pyruvate, 2 mM MgCl2, 100 mM N-(2-hydroxyethyl) piperazine-N'-(3-propanesulfonic acid) (EPPS) buffer (pH 8.4), and various amounts of T. maritima TR. The increase of absorbance at 340 nm (ɛ340 = 6.22 mM−1cm−1) was monitored at 80°C. One unit of FNOR activity was defined as 1 μmol NAD+ reduced per min. The substrate specificity of T. maritima TR was surveyed with oxidized glutathione (GSSG) for glutathione reductase activity (3, 34), lipoic acid and lipoamide for lipoamide dehydrogenase activity (3), Na2SeO3 for H-TR activity (23), and molecular oxygen for oxidase activity (42) at 50°C by monitoring the substrate-dependent absorbance change of NADH at 340 nm. One unit of the above activities was defined as 1 μmol NADH oxidized per min. Reduction of DTNB by TR purified from T. maritima was monitored by the increase of absorbance at 412 nm (ɛ412 = 13.6 mM−1 cm−1) using both NADPH and NADH as electron donors. One unit of DTNB reduction activity was defined as 2 μmol NTB produced per min. Utilization of Spirulina Trx (Sigma, Oakville, Ontario, Canada) as a substrate for T. maritima TR was tested with the DTNB reduction method as described previously (22).
Trx activity was monitored by following the disulfide reductase activity according to the method of Holmgren (18). The standard Trx assay mixture contained 100 mM sodium phosphate buffer, pH 7.0, 0.13 mM bovine insulin, 1 mM dithiothreitol, and aliquots of fractions from purification steps or purified T. maritima Trx. The increase of absorbance at 650 nm was monitored at 30°C. The aliquots of purification fractions containing reducing reagents sodium dithionite and dithiothreitol or ammonia sulfate were dialyzed (cutoff, 3 kDa) aerobically against 50 mM Tris-HCl buffer (pH 7.8) containing 5% (vol/vol) glycerol at 4°C overnight. Glutaredoxin (Grx) activity was tested using a thioltransferase assay (11). The assay mixture contained various amounts of T. maritima Trx, 0.35 mM NADPH, 1 mM EDTA, 0.5 mM reduced glutathione, 1 U glutathione reductase from yeast (Sigma, Oakville, Ontario, Canada), and 2.5 mM 2-hydroxyethyl disulfide or 2.5 mM l-cystine in 100 mM sodium phosphate buffer (pH 8.0). The absorbance change at 340 nm was monitored at 30°C. The control without T. maritima Trx was subtracted. One unit of Grx activity was defined as 1 μmol 2-hydroxyethyl disulfide or l-cystine reduced per min.
Both insulin reduction and DTNB reduction methods were used to test if Trx could serve as a substrate for TR purified from T. maritima and thereby to confirm the Trx-TR system. For the reduction of insulin (4), the purified T. maritima TR (50 nM) was added to the assay mixture (0.5 ml) containing 1 mM EDTA, 0.2 mM NAD(P)H, 0.13 mM insulin, and 0 to 1.4 μM T. maritima Trx in 100 mM sodium phosphate buffer (pH 7.0). The increase of absorbance at 650 nm was monitored at 30°C. The control without TR or Trx was performed. In the DTNB reduction method (22), purified T. maritima TR (50 nM) was added to the assay mixture (0.5 ml) containing 1 mM EDTA, 0.2 mM NAD(P)H, 0.1 mM DTNB, and 0 to 1.4 μM T. maritima Trx in 100 mM sodium phosphate buffer (pH 7.0). The increase of absorbance at 412 nm was monitored at 30°C. The control procedure without TR or Trx was performed. One unit of DTNB reduction activity was defined as 2 μmol NTB produced per min. The protein concentration was determined using the Bradford method (8), and bovine serum albumin was used as the standard protein for the assay.
Enzyme purification.
T. maritima cell extracts were prepared anaerobically using procedures similar to those described previously (54). All the purification steps of Trx and TR were carried out anaerobically because the purification of both enzymes was carried out along with other anaerobic enzymes even though Trx and TR are not oxygen sensitive. TR was monitored by following its BVOR activity using sodium phosphate buffer (100 mM, pH 7.0). The cell extracts were applied at a flow rate of 3 ml/min to a DEAE-Sepharose fast flow column (5 × 10 cm; Amersham Biotech, Baie d'Urfe, Quebec, Canada) preequilibrated with buffer A (50 mM Tris-HCl, pH 7.8, 5% [vol/vol] glycerol, 2 mM sodium dithionite, 2 mM dithiothreitol). The column was eluted with a linear gradient of 0 to 0.3 M NaCl in buffer A at a flow rate of 3 ml/min. The fractions containing high levels of activity of TR or Trx were pooled and applied to a hydroxyapatite column (2.6 × 10 cm; Bio-Rad Laboratories, Ontario, Canada) equilibrated with buffer A. The column was eluted with a linear gradient of KH2PO4 (0 to 0.15 M) in buffer A at a flow rate of 2 ml/min. Activity-exhibiting fractions for TR or Trx were pooled separately and applied to a phenyl-Sepharose HP column (2.6 × 8 cm; Amersham Biotech) equilibrated with buffer A containing 0.8 M (NH4)2SO4 (for Trx, sodium dithionite and dithiothreitol were omitted in all buffers for phenyl-Sepharose and columns used later on). The column was eluted with a (NH4)2SO4 gradient (0.8 to 0 M) at a flow rate of 2 ml/min. Fractions exhibiting high levels of activity were pooled and concentrated by ultrafiltration separately (YM 10 membrane; Amicon). The concentrated fraction (3.0 ml) was applied to a preequilibrated Superdex 200 column (2.6 × 60 cm; Amersham Biotech), and the column was eluted with buffer A containing 100 mM KCl at a flow rate of 2 ml/min. Fractions exhibiting high enzyme activity levels were combined and applied to a Q-Sepharose HP column (1.6 × 10 cm; Amersham Biotech) equilibrated with buffer A. The column was eluted with a linear gradient of NaCl (0 to 0.5 M) at a flow rate of 1.0 ml/min. Fractions containing pure TR and Trx, as revealed by a single band on SDS-PAGE (26), were stored at −20°C until use.
Determination of molecular mass and protein identification.
The native molecular masses of both T. maritima TR and Trx were estimated by gel filtration (Superdex 200, 2.6 × 60 cm; Amersham Biotech). The column was calibrated with protein standard (Amersham Biosciences, Piscataway, NJ). The molecular weights of subunits were determined using SDS-PAGE (26) with a standard curve obtained using the low-molecular-weight standard (Bio-Rad Laboratories). The single band of TR or Trx on SDS-PAGE was cut in a flow hood and digested in-gel with trypsin. The resulting peptides were extracted and cleaned as described previously (44). The cleaned peptide samples were subjected to mass spectrometry for protein identification, which was carried out on a Waters Micromass Q-TOF Ultima using nanospray injection as the sample delivery method (Mass Spectrometry Facility, University of Waterloo, Waterloo, Ontario, Canada). MS/MS profiling was carried out using PEAK software (BSI, University of Waterloo). The sequence fragments of T. maritima TR or Trx were BLASTed over a protein database (1). Deduced protein sequences of T. maritima TR or Trx and their corresponding homologues were aligned and compared with ClustalW (48).
Analysis of flavin cofactor of TR.
The oxidized and NADH-reduced TR samples were scanned with a Cary 50 Bio UV-visible spectrophotometer from 190 to 600 nm. FAD was released from TR by boiling in methanol (water: methanol = 1:9) for 10 min (46). The amount of FAD was estimated using the absorbance value at 450 nm (ɛ450 = 11.3 mM−1cm−1 [49]). The sample was concentrated by flushing with nitrogen gas before it was spotted on a thin-layer silica gel plate (5 × 10 cm, 200 μm; Selecto Scientific, Suwanee, GA) together with commercially available flavin standards, including riboflavin, FMN, and flavin adenine dinucleotide (FAD). Samples were chromatographed in a dark room with n-butanol-acetic acid-H2O (12:3:5) as a solvent. Samples on the plate were visualized using FluorChem (FluorChem 8000 chemiluminescence and visible imaging system; Alpha Innotech Corporation, CA) under UV light after the silica plate was dried with a hair dryer.
RESULTS
Purification of T. maritima TR and Trx.
T. maritima TR was purified from five chromatographic columns by monitoring its BVOR activity. BVOR activity appeared to be in two peaks after elution from the DEAE-Sepharose column. The major peak (∼70% of the total BVOR activity) started to elute when 0.08 M NaCl was applied to the column. The second peak (∼30% of the total BVOR activity), corresponding to a highly active NADH oxidase (53), started to elute when 0.1 M NaCl was applied to the column. For this study, only the fractions of the first peak were pooled and applied to the hydroxyapatite column, and BVOR activity eluted as a predominant single peak for all the columns thereafter. The enzyme was purified 378-fold after the last Q-Sepharose column (Table 1). The purity of the enzyme was confirmed by SDS-PAGE showing a single band with a molecular mass of about 37 kDa (Fig. 1). The native molecular mass of the purified enzyme was estimated to be about 67 kDa from the gel filtration chromatography, indicating that the purified BVOR was a homodimer.
TABLE 1.
Purification of T. maritima TR
| Steps | Total protein (mg) | Total activity (U)a | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 2,004 | 1,680 | 0.7 | 1 | 100 |
| DEAE-Sepharose | 570 | 995 | 1.74 | 2.5 | 59 |
| Hydroxyapatite | 176 | 779 | 4.42 | 6.3 | 46 |
| Phenyl-Sepharose | 25.2 | 529 | 21 | 30 | 31 |
| Gel filtration | 4.16 | 458 | 110 | 157 | 27 |
| Q-Sepharose | 0.78 | 206 | 265 | 378 | 12 |
TR activity was measured using the BVOR assay at 80°C as described in Materials and Methods.
FIG. 1.
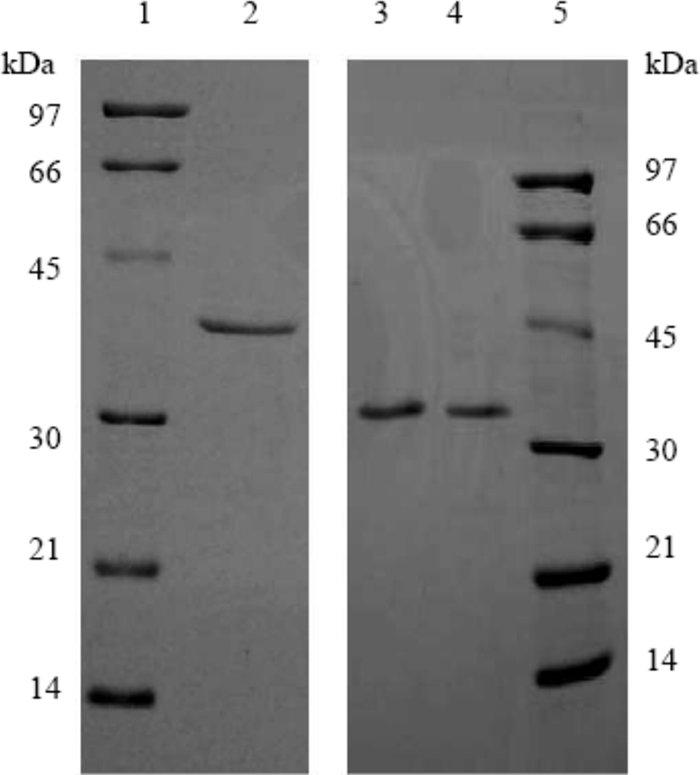
SDS-PAGE (12.5%) of the purified thioredoxin reductase (TR) and thioredoxin (Trx) from Thermotoga maritima. Lanes 1 and 5, low-molecular-mass standards along with their corresponding molecular masses; lane 2, purified T. maritima TR (1 μg); lanes 3 and 4, purified T. maritima Trx (0.4 and 0.6 μg, respectively).
T. maritima Trx was purified from the same batch of cell extract by following dithiothreitol-dependent reduction of bovine insulin. It eluted as a single peak after all the five columns. Trx eluted from the phenyl-Sepharose column with water but not 100% buffer A, indicating that it is more hydrophobic than most soluble proteins. After the elution from the Q-Sepharose column, the purified enzyme resolved as a single band on SDS-PAGE with a molecular mass of about 31 kDa (Fig. 1). The native molecular mass of T. maritima Trx was estimated to be 23 kDa from the gel filtration chromatography, indicating that the purified T. maritima Trx is a monomer.
Protein identification using mass spectrometry and sequence analysis.
The purified T. maritima TR had FNOR activity detected with reduced T. maritima ferredoxin (2.1 U/mg). The only FNOR reported from hyperthermophiles is that from Pyrococcus furiosus, which is a multifunctional enzyme showing BVOR, FNOR, sulfide dehydrogenase, and NADPH oxidase activities (29). It was speculated that the purified TR might have other functions in addition to its FNOR and BVOR activities. Since the genome of T. maritima is available (33), the purified BVOR from T. maritima was identified to be TM0869 by peptide mass fingerprints. The gene was annotated to encode thioredoxin reductase in the T. maritima genome with a predicted mass of 34,347 Da. The amino acid sequence of T. maritima TR showed 94 to 98% identity to those of its homologues in Thermotoga neapolitana, Thermotoga sp. (strain RQ2), and Thermotoga petrophila RKU-1. It was found that T. maritima TR sequence had both NAD and FAD-binding regions and the signatures of a class II pyridine nucleotide-disulfide oxidoreductase active site when analyzed for secondary structures using InterProScan (55). Similar to other L-TRs, T. maritima TR showed two active cysteine residues (CXXC), an NAD(P)H binding motif, and FAD-binding motifs when its amino acid sequence was aligned with those of other identified TRs in hyperthermophilic archaea and well-characterized TR in Escherichia coli (data not shown).
The purified T. maritima Trx was identified to be an annotated glutaredoxin (Grx)-related protein encoded by TM0868, which is adjacent to that encoding TR, TM0869. The predicted molecular mass of T. maritima Trx is 25,158 Da (33). The protein sequence of T. maritima Trx showed two Trx folds and two redox active sites, CQYC at the N terminus and CPYC at the C terminus. The T. maritima Trx amino acid sequence had identities ranging from 54 to 99% to those of annotated Grx-related proteins in other Thermotogales species and an identity of 41% to the characterized Grx-like protein from P. furiosus (39).
Identification of a flavin cofactor of T. maritima TR.
The purified TR was yellowish, which was an indication of the presence of flavin. The oxidized enzyme solution showed characteristic flavin absorbance maxima at 380 nm and 450 nm. The peaks disappeared upon the addition of 0.1 mM NADH into the enzyme solution. A yellowish cofactor was released after the enzyme was mixed with methanol and boiled for 10 min in the dark. This flavin cofactor was further identified as FAD using thin-layer chromatography (data not shown). The T. maritima TR contained 1.88 ± 0.10 mol of FAD per mol native enzyme based on the absorbance value at 450 nm and the protein amount from which the FAD was extracted. Therefore, each subunit contained one noncovalently bound FAD moiety.
Catalytic properties of the purified TR and Trx from T. maritima.
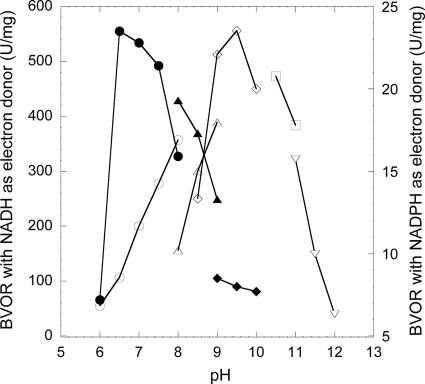
The highest BVOR activity of the purified T. maritima TR was observed at pH 9.5 in 100 mM glycine-NaOH buffer, when NADH was used as an electron donor (Fig. 2). However, the enzyme exhibited maximum activity at pH 6.5, in 100 mM sodium phosphate buffer, when NADPH was used as an electron donor. BVOR activity increased as temperatures increased from 30 (64 U mg−1) to 95°C (868 U mg−1). The activity doubled every 10°C from 30 to 50°C, and the increase of activities was slowed down gradually at temperatures above 50°C, similar to the activity of TR characterized from A. pernix K1 (21), but no decrease of activity for the former was observed from 90 to 95°C. The enzyme was stable at elevated temperatures. Following an initial drop of activity in the first 2 h of incubation, over 60% of the BVOR activity remained after incubation for 28 h at 80°C and 50% remained after 9 h at 95°C. The loss of activity upon heat at 80 and 95°C did not follow the first-order kinetics. BVOR activity was dependent on concentrations of both NAD(P)H and BV. The kinetic parameters of BVOR were determined under specified assay conditions by measuring the initial rate at different starting concentrations of NAD(P)H or BV. Apparent Km and Vmax values were obtained using a computer-aided direct fit to the Michaelis-Menten equation (Sigmaplot 10, Enzyme module; Systat Software, San Jose, CA; data not shown). The apparent Km value for NADH and the apparent Vmax value were determined to be 73 ± 1.6 μM and 1,111 ± 35 U/mg, respectively. The apparent Km value for BV was determined to be 89 ± 1.1 μM. The TR enzyme showed a preference for NADH over NADPH in the BV reduction. The activities with NADPH were only 11% (pH 7.0, 100 mM sodium phosphate buffer) and 1.4% (pH 9.5, 100 mM glycine-NaOH) of that with NADH. The apparent Km for NADPH and the apparent Vmax were determined to be 780 ± 20 μM and 115 ± 2.4 U/mg, respectively, which were about 11 times higher and lower, respectively, than those values when NADH was used.
FIG. 2.
pH dependency of the purified thioredoxin reductase (TR) from Thermotoga maritima. The activity was assayed at 80°C with NADH (open symbols) or NADPH (filled symbols) as an electron donor. Circles, 100 mM sodium phosphate buffer, pH 6.0 to 8.0; triangles, 100 mM glycylglycine-NaOH buffer, pH 8.0 to 9.0; diamonds, 100 mM glycine-NaOH buffer, pH 8.5 to 10.0; squares, 100 mM CAPS buffer, pH 10.0 to 11.0; inverted triangles, 100 mM sodium phosphate buffer, pH 11.0 to 12.0.
The substrate specificity of T. maritima TR was also surveyed with Spirulina Trx, oxidized glutathione (GSSG), lipoic acid, lipoamide, Na2SeO3, DTNB, and molecular oxygen. Spirulina Trx, GSSG, lipoic acid, lipoamide, and Na2SeO3 could not be used as substrates for T. maritima TR. When molecular oxygen was used as an electron acceptor, the enzyme showed NADH oxidase activity of 22.3 U/mg, which was only 16% of BVOR activity (139 U/mg) at 50°C. Interestingly, T. maritima TR could also catalyze the reduction of DTNB directly using either NADH or NADPH as an electron donor (see thioredoxin system in T. maritima section), which is unusual for a L-TR. T. maritima TR showed a preference for NADH over NADPH for all the substrates tested.
Trxs are known as disulfide reductases that reduce insulin with dithiothreitol as an electron donor. The reduced ß-chain of insulin precipitates out and causes an increase of turbidity at 650 nm. The insulin reduction was dependent on the amount of Trx in the assay system (Fig. 3). Since T. maritima Trx has Grx-like structure at the C terminus (based on amino acid sequence information), Grx activity was tested using a thioltransferase assay, and the Grx activity was observed with both l-cystine (4.9 U/mg) and 2-hydroxyethyl disulfide (4.5 U/mg) as substrates.
FIG. 3.
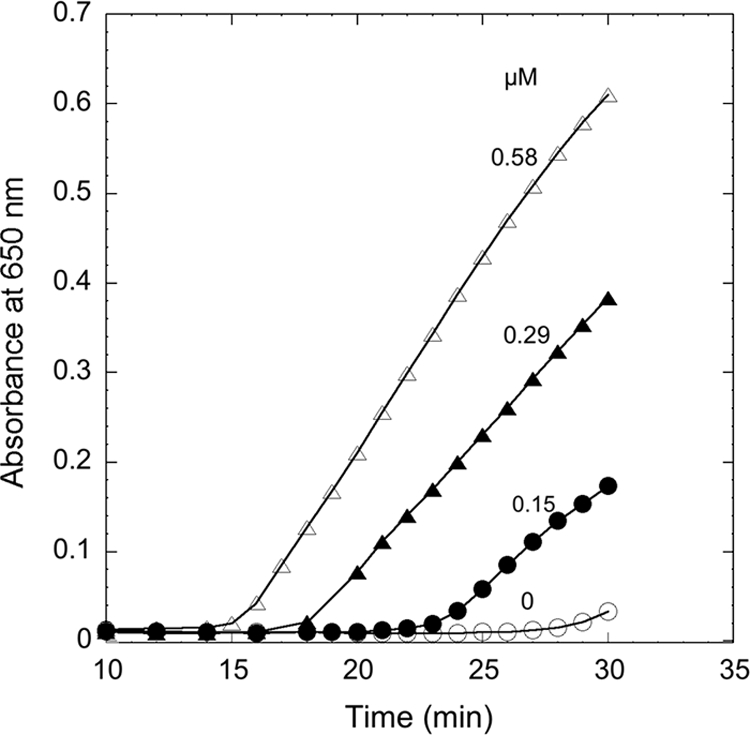
Reduction of insulin by Thermotoga maritima thioredoxin (Trx). The assay mixture contained 1 mM dithiothreitol and 1 mg/ml insulin in 100 mM sodium phosphate buffer, pH 7.0. The reaction was carried out at 30°C by monitoring the increase of absorbance at 650 nm in the absence (open circles) or presence of 0.15 μM (filled circles), 0.29 μM (filled triangles), or 0.58 μM (open triangles) T. maritima Trx.
The thioredoxin system in T. maritima.
Trx-mediated reduction of DTNB and insulin was carried out in order to examine whether the purified Trx and TR from T. maritima could function as a thioredoxin system. The results showed that the insulin disulfide bonds were reduced in the presence of T. maritima TR and Trx when either NADH or NADPH was used as an electron donor (Fig. 4). The rate of insulin disulfide bond reduction was higher when NADH was used as an electron donor. Apparently, T. maritima TR together with T. maritima Trx formed a thioredoxin system, and the DTNB reduction activity of TR was dependent on the concentration of Trx (Fig. 5). There was a very low level of activity of DTNB reduction in the absence of Trx. These results clearly indicate that T. maritima Trx, which shares homology with the P. furiosus Grx-like protein (41), has the capacity to form a redox system with TR in T. maritima cells. The system formed by T. maritima TR and Trx was capable of rapidly reducing both small-molecule (DTNB)- and protein (bovine insulin) disulfide-containing substrates in the presence of NADPH or NADH.
FIG. 4.
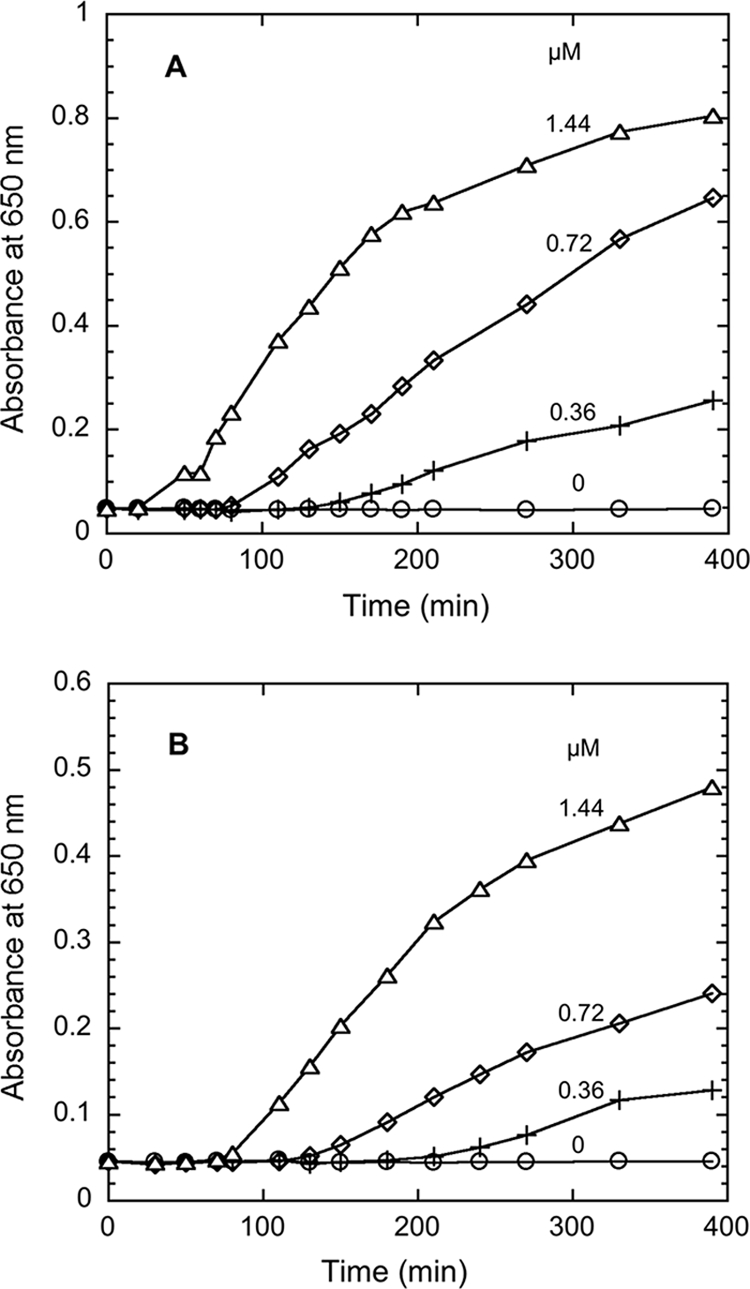
Reduction of insulin by the Thermotoga maritima thioredoxin system. The assay mixture contained 50 nM T. maritima thioredoxin reductase, 0.2 mM NADH (A) or NADPH (B), 0.13 mM insulin, 1 mM EDTA, and 0 μM (open circles), 0.36 μM (crosses), 0.72 μM (diamonds), or 1.44 μM (triangles) T. maritima thioredoxin in 100 mM sodium phosphate buffer, pH 7.0. The increase of absorbance at 650 nm was monitored at 30°C.
FIG. 5.
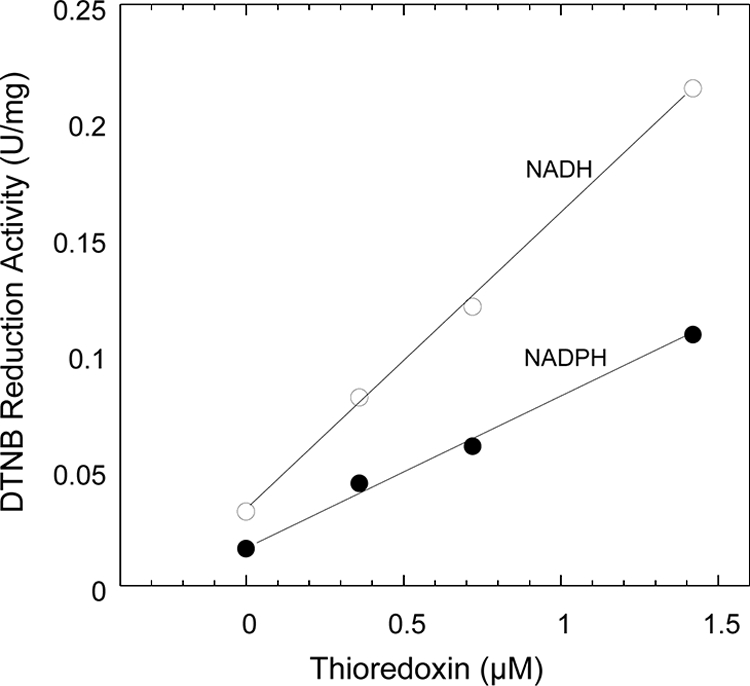
Reduction of DTNB by the Thermotoga maritima thioredoxin system. The assay mixture contained 50 nM T. maritima thioredoxin reductase, 0.2 mM NADH (open circles) or NADPH (filled circles), 0.1 mM DTNB, and 0 to 1.4 μM T. maritima thioreodxin. The increase of absorbance at 412 nm was monitored at 30°C.
DISCUSSION
The thioredoxin system plays several key roles in maintaining the redox balance inside the cell and responding to oxidative stress in all three domains of life (16, 51). The TR purified from T. maritima is a dimeric FAD-containing protein encoded by the gene TM0869, with a deduced amino acid sequence 98% identical to that from T. petrophila and 33% identical to the E. coli TR. T. maritima TR has typical FAD-binding motifs, one near the N terminus (GXGXXG) and another at the C terminus (GXYAVAGD), an NAD(P)H-binding motif close to the middle of the protein (GGGXXA), and an active redox center (CXXC) that is common in all enzymes showing TR activity (16). The active redox center is within a class II pyridine nucleotide-disulfide oxidoreductase motif, CATCDGYLFAGKDVIVVGGGD, that overlaps with the NAD(P)H-binding region in the primary structure. The proximity of the NAD(P)H-binding region and the pyridine nucleotide-disulfide oxidoreductase active site is a feature present in all L-TRs, while in H-TRs they are spatially separated (42). This further confirms T. maritima TR to be one of the L-TRs. The nicotinamide nucleotide-binding motif of T. maritima TR, GXGXXA, is common to many NADPH-dependent enzymes (43). However, it has also been found in NADH-utilizing enzymes, such as TR-like protein from Clostridium pasteurianum (40) and both NADH and NADPH-utilizing TR from S. solfataricus (32, 42). Unlike the values for most of the TRs that use NADPH as their reducing power, the apparent Km value of T. maritima TR for NADH is 11 times lower than that for NADPH. The preference for NADH could have resulted from a lack of the conserved arginine/lysine residues that are present in NADPH-binding flavoproteins, required for binding of the 2′-phosphate group of NADPH (43), and located approximately 20 amino acids downstream of the NAD(P)H binding region in the primary structure of T. maritima TR.
Active cysteine residues are critical for the enzymatic catalysis of the thioredoxin family proteins such as Trx, Grx, protein disulfide isomerase, and DsbA, which have CXXC as their active-site motif (where X is any amino acid other than cysteine). The residues between the two cysteines at the active sites are distinct in conventional Trxs (CGPC) and Grxs (CPYC), although they are highly conserved within each family (31). Gene annotation lists a protein as a Trx-like protein if it has a CGPC active site or a Grx-like protein if it has a CPYC active site. T. maritima Trx has two redox-active motifs, CPYC and CQYC, which resulted in its annotation as a Grx-like protein (33). In contrast to the lack of homology to conventional Trxs, the primary structure of T. maritima Trx has high levels of identities to some protein disulfide oxidoreductases (PDO) that have molecular masses of around 25 kDa or higher and two redox-active motifs (CXXC and CXXC), which are exclusively isolated from hyperthermophiles, such as P. horikoshii (22), P. furiosus (14), Aquifex aeolicus (35), S. solfataricus (15), and A. pernix (21). T. maritima Trx exhibited both Trx and Grx (thioltransferase) activities, similar to the archaeal PDO from S. solfataricus (36). The unusually large sizes of T. maritima Trx and S. solfataricus PDO as substrates for their respective TRs may indicate catalytic mechanisms that are different from those of their mesophilic counterparts. PDOs from P. furiosus and A. aeolicus have not been reconstituted in any TR system, while both proteins have Grx activity. From structural and direct mutagenesis studies, it seems that the two active CXXC centers in P. furiosus PDO have very different accessibilities and redox properties (39) and it is likely that the S. solfataricus CXXC at the C terminus is involved in catalytic activities (28). It is not clear if both centers present in T. maritima Trx or only one of them would be involved in catalytic activities.
Unlike TRs from mammals that can catalyze the reduction of lipoic acid efficiently (3), T. maritima TR did not show any activity of lipoamide dehydrogenase and glutathione reductase. It could catalyze the reduction of T. maritima Trx with NADH or NADPH as an electron donor, which was shown by the reduction of DTNB and insulin, respectively. Interestingly, the purified T. maritima TR could also catalyze the direct reduction of DTNB, which is a common feature of H-TRs (16) and not normally found in L-TRs. Recently, it has been reported that the L-TRs from the hyperthermophilic archaea S. solfataricus (32, 42), which was characterized as an NADH oxidase previously, and A. pernix K1 (21) showed the capability of catalyzing the reduction of DTNB directly. However, there has been no bacterial TR showing this property. This indicates that T. maritima TR resembles catalytic properties closer to those of some archaeal and eukaryotic types of TRs and may represent a novel type of TRs that combine catalytic properties of both L-TRs and H-TRs. The broader substrate spectrum of H-TR may have resulted from the presence of selenocysteine at the C terminus (Gly-Cys-Sec-Gly) (12, 13, 47). However, there is no selenocysteine present in the TRs from T. maritima, S. solfataricus, and A. pernix based on the deduced amino acid sequences. The reason for the direct use of DTNB as the electron acceptor for these enzymes remains unclear.
Glutathione metabolism, closely associated with oxygenic photosynthesis and aerobic respiration, is absent in many prokaryotes, including archaea and anaerobes (10). Some low-molecular-weight thiols other than glutathione, such as coenzyme M in methanogens (52) and coenzyme A in the hyperthermophilic archaea P. furiosus, Thermococcus litoralis, and S. solfataricus (20), are present at substantial concentrations, and they may function as the equivalent of glutathione in their respective hosts. T. maritima Trx was able to accept electrons from reduced glutathione. There are no reports on the presence of glutathione nor are there predicted homologues of glutathione reductase and the enzymes for glutathione biosynthesis found in the genome of T. maritima (33). There is also no homologue of classical Trx genes that encode smaller Trxs (∼12 kDa) with a single CXXC redox-active motif present in the T. maritima genome. Therefore, the characterized thioredoxin system might be the sole protein disulfide oxidoreductase system functioning in T. maritima.
It has been reported that in the mesophilic anaerobic bacterium Clostridium pasteurianum, a TR (Cp34), a Grx (Cp9) and a peroxiredoxin (Cp20) form an electron transfer system that is proposed to be involved in the oxidative response of this anaerobe (40). Previous studies showed that T. maritima could tolerate micromolar levels of oxygen in the growth media and that it possesses a highly active H2O2-forming NADH oxidase (53). An NADH-dependent peroxidase activity in cell extracts of T. maritima has also been demonstrated, and a gene predicted to encode peroxiredoxin, TM0807, is present in the genome of T. maritima. It warrants further investigation to see whether the T. maritima thioredoxin system provides electrons to peroxiredoxin, which would catalyze the reduction of hydrogen peroxide to water, thereby forming a complete oxygen defense pathway.
Acknowledgments
This work was supported by research grants from Natural Sciences and Engineering Research Council (Canada) and Canada Foundation for Innovation to K.M.
Footnotes
Published ahead of print on 8 January 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnér, E. S. J., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. [DOI] [PubMed] [Google Scholar]
- 3.Arnér, E. S. J., J. Nordberg, and A. Holmgren. 1996. Efficient reduction of lipoamide and lipoic acid by mammalian thioredoxin reductase. Biochem. Biophys. Res. Commun. 225:268-274. [DOI] [PubMed] [Google Scholar]
- 4.Arnér, E. S. J., L. Zhong, A. Holmgren, and P. Lester. 1999. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 300:226-239. [DOI] [PubMed] [Google Scholar]
- 5.Baker, A., C. M. Payne, M. M. Briehl, and G. Powis. 1997. Thioredoxin, a gene found overexpressed in human cancer, inhibits apoptosis in vitro and in vivo. Cancer Res. 57:5162-5167. [PubMed] [Google Scholar]
- 6.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Mol. Biol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blamey, J. M., and M. W. W. Adams. 1994. Characterization of an ancestral type of pyruvate ferredoxin oxidoreductase from the hyperthermophilic bacterium, Thermotoga maritima. Biochemistry 33:1000-1007. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Chae, H. Z., S. J. Chung, and S. G. Rhee. 1994. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269:27670-27678. [PubMed] [Google Scholar]
- 10.Fahey, R. C. 2001. Novel thiols of prokaryotes. Annu. Rev. Microbiol. 55:333-356. [DOI] [PubMed] [Google Scholar]
- 11.Gan, Z. R., and W. W. Wells. 1986. Purification and properties of thioltransferase. J. Biol. Chem. 261:996-1001. [PubMed] [Google Scholar]
- 12.Gasdaska, P. Y., J. R. Gasdaska, S. Cochran, and G. Powis. 1995. Cloning and sequencing of a human thioredoxin reductase. FEBS Lett. 373:5-9. [DOI] [PubMed] [Google Scholar]
- 13.Gladyshev, V. N., K. T. Jeang, and T. C. Stadtman. 1996. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc. Natl. Acad. Sci. U. S. A. 93:6146-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guagliardi, A., D. de Pascale, R. Cannio, V. Nobile, S. Bartolucci, and M. Rossi. 1995. The purification, cloning, and high level expression of a glutaredoxin-like protein from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270:5748-5755. [DOI] [PubMed] [Google Scholar]
- 15.Guagliardi, A., V. Nobile, S. Bartolucci, and M. Rossi. 1994. A thioredoxin from the extreme thermophilic Archaeon Sulfolobus solfataricus. Int. J. Biochem. 26:375-380. [Google Scholar]
- 16.Hirt, R. P., S. Müller, T. Martin Embley, and G. H. Coombs. 2002. The diversity and evolution of thioredoxin reductase: new perspectives. Trends Parasitol. 18:302-308. [DOI] [PubMed] [Google Scholar]
- 17.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54:237-271. [DOI] [PubMed] [Google Scholar]
- 18.Holmgren, A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254:9627-9632. [PubMed] [Google Scholar]
- 19.Huber, R., T. A. Langworthy, H. König, M. Thomm, C. R. Woese, U. B. Sleytr, and K. O. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 20.Hummel, C. S., K. M. Lancaster, and E. J. Crane III. 2005. Determination of coenzyme A levels in Pyrococcus furiosus and other Archaea: implications for a general role for coenzyme A in thermophiles. FEMS Microbiol. Lett. 252:229-234. [DOI] [PubMed] [Google Scholar]
- 21.Jeon, S.-J., and K. Ishikawa. 2002. Identification and characterization of thioredoxin and thioredoxin reductase from Aeropyrum pernix K1. Eur. J. Biochem. 269:5423-5430. [DOI] [PubMed] [Google Scholar]
- 22.Kashima, Y., and K. Ishikawa. 2003. A hyperthermostable novel protein-disulfide oxidoreductase is reduced by thioredoxin reductase from hyperthermophilic archaeon Pyrococcus horikoshii. Arch. Biochem. Biophys. 418:179-185. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, S., M. Bjornstedt, and A. Holmgren. 1992. Selenite is a substrate for calf thymus thioredoxin reductase and thioredoxin and elicits a large non-stoichiometric oxidation of NADPH in the presence of oxygen. Eur. J. Biochem. 207:435-439. [DOI] [PubMed] [Google Scholar]
- 24.Ladenstein, R., and B. Ren. 2006. Protein disulfides and protein disulfide oxidoreductases in hyperthermophiles. FEBS J. 273:4170-4185. [DOI] [PubMed] [Google Scholar]
- 25.Ladenstein, R., and B. Ren. 2008. Reconsideration of an early dogma, saying “there is no evidence for disulfide bonds in proteins from archaea”. Extremophiles 12:29-38. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Laurent, T. C., E. C. Moore, and P. Reichard. 1964. Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 239:3436-3444. [PubMed] [Google Scholar]
- 28.Limauro, D., M. Saviano, I. Galdi, M. Rossi, S. Bartolucci, and E. Pedone. 2009. Sulfolobus solfataricus protein disulphide oxidoreductase: insight into the roles of its redox sites. Protein Eng. Des. Sel. 22:19-26. [DOI] [PubMed] [Google Scholar]
- 29.Ma, K., and M. W. Adams. 1994. Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus: a new multifunctional enzyme involved in the reduction of elemental sulfur. J. Bacteriol. 176:6509-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, K., M. W. W. Adams, W. W. A. Michael, and M. K. Robert. 2001. Ferredoxin:NADP oxidoreductase from Pyrococcus furiosus. Methods Enzymol. 334:40-45. [DOI] [PubMed] [Google Scholar]
- 31.Martin, J. L. 1995. Thioredoxin—a fold for all reasons. Structure 3:245-250. [DOI] [PubMed] [Google Scholar]
- 32.Masullo, M., G. Raimo, A. Dello Russo, V. Bocchini, and J. V. Bannister. 1996. Purification and characterization of NADH oxidase from the archaea Sulfolobus acidocaldarius and Sulfolobus solfataricus. Biotechnol. Appl. Biochem. 23:47-54. [PubMed] [Google Scholar]
- 33.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 34.Patel, M. P., J. Marcinkeviciene, and J. S. Blanchard. 1998. Enterococcus faecalis glutathione reductase: purification, characterization and expression under normal and hyperbaric O2 conditions. FEMS Microbiol. Lett. 166:155-163. [DOI] [PubMed] [Google Scholar]
- 35.Pedone, E., K. D'Ambrosio, G. De Simone, M. Rossi, C. Pedone, and S. Bartolucci. 2006. Insights on a new PDI-like family: structural and functional analysis of a protein disulfide oxidoreductase from the bacterium Aquifex aeolicus. J. Mol. Biol. 356:155-164. [DOI] [PubMed] [Google Scholar]
- 36.Pedone, E., D. Limauro, R. D'Alterio, M. Rossi, and S. Bartolucci. 2006. Characterization of a multifunctional protein disulfide oxidoreductase from Sulfolobus solfataricus. FEBS J. 273:5407-5420. [DOI] [PubMed] [Google Scholar]
- 37.Poole, L. B., C. M. Reynolds, Z. A. Wood, P. A. Karplus, H. R. Ellis, and M. L. Calzi. 2000. AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur. J. Biochem. 267:6126-6133. [DOI] [PubMed] [Google Scholar]
- 38.Reed, L. J., F. R. Leach, and M. Koike. 1958. Studies on a lipoic acid-activating system. J. Biol. Chem. 232:123-142. [PubMed] [Google Scholar]
- 39.Ren, B., G. Tibbelin, D. de Pascale, M. Rossi, S. Bartolucci, and R. Ladenstein. 1998. A protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus contains two thioredoxin fold units. Nat. Struct. Mol. Biol. 5:602-611. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds, C. M., J. Meyer, and L. B. Poole. 2002. An NADH-dependent bacterial thioredoxin reductase-like protein in conjunction with a glutaredoxin homologue form a unique peroxiredoxin (AhpC) reducing system in Clostridium pasteurianum. Biochemistry 41:1990-2001. [DOI] [PubMed] [Google Scholar]
- 41.Robb, F. T., D. L. Maeder, J. R. Brown, J. DiRuggiero, M. D. Stump, R. K. Yeh, R. B. Weiss, D. M. Dunn, W. W. A. Michael, and M. K. Robert. 2001. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134-157. [DOI] [PubMed] [Google Scholar]
- 42.Ruocco, M. R., A. Ruggiero, L. Masullo, P. Arcari, and M. Masullo. 2004. A 35 kDa NAD(P)H oxidase previously isolated from the archaeon Sulfolobus solfataricus is instead a thioredoxin reductase. Biochimie 86:883-892. [DOI] [PubMed] [Google Scholar]
- 43.Scrutton, N. S., A. Berry, and R. N. Perham. 1990. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature 343:38-43. [DOI] [PubMed] [Google Scholar]
- 44.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 45.Spassov, V. Z., A. D. Karshikoff, and R. Ladenstein. 1994. Optimization of the electrostatic interactions in proteins of different functional and folding type. Protein Sci. 3:1556-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanton, T. B., and N. S. Jensen. 1993. Purification and characterization of NADH oxidase from Serpulina (Treponema) hyodysenteriae. J. Bacteriol. 175:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura, T., and T. C. Stadtman. 1996. A new selenoprotein from human lung adenocarcinoma cells: purification, properties, and thioredoxin reductase activity. Proc. Natl. Acad. Sci. U. S. A. 93:1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitby, L. G. 1953. A new method for preparing flavin-adenine dinucleotide. Biochem. J. 54:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, C. H., Jr. 1992. Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and mercuric ion reductase: a family of flavoenzyme transhydrogenases, p. 121-211. In F. Müller (ed.), Chemistry and biochemistry of flavoenzymes, vol. 3. CRC Press, Boca Raton, FL. [Google Scholar]
- 51.Williams, C. H., Jr., L. D. Arscott, S. Müller, B. W. Lennon, M. L. Ludwig, P.-F. Wang, D. M. Veine, K. Becker, and R. H. Schirmer. 2000. Thioredoxin reductase. Eur. J. Biochem. 267:6110-6117. [DOI] [PubMed] [Google Scholar]
- 52.Wolfe, R. S., and B. C. McBride. 1971. New coenzyme of methyl transfer, coenzyme M. Biochemistry 10:2317-2324. [DOI] [PubMed] [Google Scholar]
- 53.Yang, X., and K. Ma. 2007. Characterization of an exceedingly active NADH oxidase from the anaerobic hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 189:3312-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, X., and K. Ma. 2005. Purification and characterization of an NADH oxidase from extremely thermophilic anaerobic bacterium Thermotoga hypogea. Arch. Microbiol. 183:331-337. [DOI] [PubMed] [Google Scholar]
- 55.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]