Abstract
Transmembrane chemoreceptors are central components in bacterial chemotaxis. Receptors couple ligand binding and adaptational modification to receptor conformation in processes that create transmembrane signaling. Homodimers, the fundamental receptor structural units, associate in trimers and localize in patches of thousands. To what degree do conformational coupling and transmembrane signaling require higher-order interactions among dimers? To what degree are they altered by such interactions? To what degree are they inherent features of homodimers? We addressed these questions using nanodiscs to create membrane environments in which receptor dimers had few or no potential interaction partners. Receptors with many, few, or no interaction partners were tested for conformational changes and transmembrane signaling in response to ligand occupancy and adaptational modification. Conformation was assayed by measuring initial rates of receptor methylation, a parameter independent of receptor-receptor interactions. Coupling of ligand occupancy and adaptational modification to receptor conformation and thus to transmembrane signaling occurred with essentially the same sensitivity and magnitude in isolated dimers as for dimers with many neighbors. Thus, we conclude that the chemoreceptor dimer is the fundamental unit of conformational coupling and transmembrane signaling. This implies that in signaling complexes, coupling and transmembrane signaling occur through individual dimers and that changes between dimers in a receptor trimer or among trimer-based signaling complexes are subsequent steps in signaling.
In motile bacterial cells, thousands of transmembrane chemoreceptor proteins cluster in polar patches (8, 13, 14, 30, 42). The fundamental structural unit of these receptors is a homodimer (18, 32). Dimers interact at their membrane-distal tips to create trimers (18, 38, 39). Interactions among receptor homodimers in trimers and in higher-order associations (Fig. 1A) are thought to be important for function (36, 37), particularly for the high-performance features of the chemotaxis sensory system (15). Understanding the role of receptor-receptor interactions in chemoreceptor function will require definition of the extent to which each receptor activity is an inherent property of individual receptor dimers and the extent to which activities require or are influenced by interactions with neighboring receptors. These issues had not been addressed experimentally because the receptor-receptor interactions could not be easily controlled in vivo or in vitro. However, we found that nanodiscs (2, 5) could be utilized to manipulate the potential for interactions among membrane-embedded chemoreceptors and thus to investigate the influence of receptor-receptor interactions upon chemoreceptor activities (4).
FIG. 1.
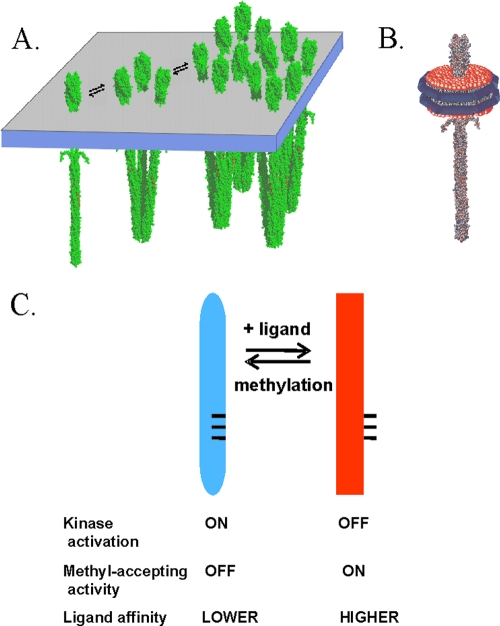
Chemoreceptors. (A) Cartoon of interactions of membrane-embedded chemoreceptors showing a homodimer, a trimer of dimers, and a patch of chemoreceptors. (B) Cartoon of a nanodisc with a single receptor dimer inserted in the plug of the lipid bilayer. (C) Diagram of the chemoreceptor conformational equilibrium.
Nanodiscs.
Nanodiscs are soluble, nanoscale (∼10-nm-diameter) particles of lipid bilayer surrounded by an annulus of amphipathic protein into which transmembrane proteins can be incorporated (2, 5). Activity assays of chemoreceptor Tar from Escherichia coli incorporated into nanodiscs (Fig. 1B) (4) revealed that effective receptor-mediated activation of the signaling histidine kinase occurred only with receptor-bearing nanodiscs containing more than one chemoreceptor dimer. However, homodimers isolated from interaction with other dimers as a result of individual insertion in a nanodisc were still substrates for the enzymes of adaptational modification. Furthermore, the effect of saturating ligand on adaptational modification indicated that transmembrane signaling occurred in individual homodimers. However, the efficiency of that signaling was not investigated.
Chemoreceptors, their conformational equilibrium, and transmembrane signaling.
Bacterial chemoreceptors are elongated, helical coiled-coil homodimers (Fig. 1) (15). Most chemoreceptors, including all receptors in Escherichia coli, span the cytoplasmic membrane. Receptors form noncovalent signaling complexes with the chemotaxis histidine kinase CheA and the coupling protein CheW. In signaling complexes, the inherently low activity of CheA is substantially enhanced and placed under the control of chemoreceptors. An increase in occupancy by an attractant molecule of a specific binding site at one end of the receptor dramatically reduces the activity of the kinase associated with the other end of the elongated receptors, a process that can be viewed as a shift in the conformational equilibrium of the receptor and the signaling complex or as transmembrane signaling (see the following paragraph). The ligand-induced reduction in kinase activity is transient because a feedback loop of sensory adaptation restores the kinase to its initial level of activity. This feedback adaptation is mediated by covalent modification of the receptor cytoplasmic domain. The modifications are formation of methyl esters at specific glutamyl residues, catalyzed by a chemotaxis-specific methyltransferase, and demethylation of those methyl esters, catalyzed by a chemotaxis-specific methylesterase. Methylation counteracts the effects of attractant binding and thus restores the null signaling state of the receptor and the signaling complex, a process that can be viewed both as shifting the conformational equilibrium of the receptor and the signaling complex in the opposite direction from the shift induced by attractant binding and as generating a transmembrane signal that opposes the signal generated by attractant occupancy (20).
Many observations about the chemotactic sensory system of E. coli can be understood in the context of the notion that attractant occupancy and adaptational modification alter activities of chemoreceptors and signaling complexes by shifting, in opposite directions, an equilibrium between two conformations (Fig. 1C). The “kinase-on” conformation, favored by methylation and disfavored by attractant occupancy, is thought to activate the histidine kinase to be a substrate for demethylation, not to be a substrate for methylation and have lowered affinity for attractant ligand. The “kinase-off” conformation, favored by attractant occupancy and disfavored by methylation, has the inverse pattern: it does not activate kinase or serve as a substrate for demethylation but serves as a substrate for methylation and has higher affinity for attractant. To what extent are the coupling of the receptor conformational equilibrium to ligand occupancy and adaptational modification and thus transmembrane signaling dependent on higher-order interactions beyond the fundamental receptor structural unit, the homodimer? We addressed this question by utilizing nanodiscs to isolate individual receptor dimers from possible interactions with neighboring dimers inserted in the same membrane or to limit the number of potentially interacting neighbors.
MATERIALS AND METHODS
Strains and plasmids.
Escherichia coli K-12 strain RP3098 (33) carries a chromosomal deletion from flhA to flhD, eliminating expression of genes for chemoreceptors and for che proteins. pAL67 carries a form of tar coding for Tar with a 6-histidine, carboxyl-terminal extension (Tar-6H) and a QEQE arrangement at the four methyl-accepting sites (21). Versions of pAL67 coding for Tar with 0, 1, or 3 glutamines were created by PCR mutagenesis, and the constructs were verified by DNA sequencing. These were pAL529 (0Q; EEEE), pAL531 (1Q; QEEE), pAL535 (3Q; EQQQ), pAL532 (3Q; QEQQ), pAL534 (3Q; QQEQ), and pAL528 (3Q; QQQE).
Membrane vesicle-borne and purified Tar-6H.
Cytoplasmic membrane vesicles containing a form of Tar-6H carrying the desired extent of adaptational modification were prepared by osmotic lysis and sucrose gradient centrifugation (5) from E. coli RP3098 harboring one of the plasmids described in the previous paragraph. Membranes were suspended in 50 mM Tris HCl (pH 7.5) and 10% (wt/vol) glycerol, quick-frozen in liquid N2, and stored at −70°C. Protein and Tar concentrations were determined by using the BCA kit (Pierce, Inc., Rockford, IL) and quantitative immunoblotting (28), respectively. Membranes were solubilized with β-octyl glucoside at 5 mg/mg total membrane protein, and Tar-6H was purified using a nitrilotriacetic acid (NTA)-Ni+2 column (5). Receptor content was quantified by densitometry of Coomassie blue-stained bands on SDS-polyacrylamide gels using Tar-6H standards.
Nanodisc preparation.
Nanodiscs containing Tar-6H were prepared essentially as described previously (5). To make preparations with ∼1 dimer per disc, we used MSP1D1(−) (9) and MSP/lipid and Tar-6H/MSP molar ratios of 1:60 and 1:10, respectively. To make preparations with ∼3 dimers/disc, we used the larger MSP1D1E3(−) (9) and ratios of 1:120 and 1:1.
Methylation assay.
Initial rates of receptor methylation were determined as described previously (1) with the modifications noted in the following description. Tar-6H, in membrane vesicles or nanodiscs, was incubated with or without aspartate in 50 mM Tris HCl (pH 7.5), 0.5 mM EDTA, 2 mM dithiothreitol (DTT), and 10% glycerol for 15 min at room temperature. Reactions were initiated by the addition of a CheR-containing cell extract to which had been added S-adenosyl-l-[methyl-3H]methionine (Amersham Pharmacia) with the specific activity adjusted to ∼1.1 Ci/mmol by the addition of unlabeled S-adenosylmethionine (AdoMet). The final concentrations in the reaction mixture were 3.3 to 5 μM available methyl-accepting sites, 0.125 μM CheR, 50 μM AdoMet, and 0 to 1 mM aspartate. The concentrations of available methyl-accepting sites were determined by incubating Tar-containing membrane vesicle or nanodiscs under conditions that resulted in a maximal extent of receptor methylation (5 μM CheR in a cell extract for 2 h at room temperature), determining the extent of methylation by quantifying the proportion of Tar with an electrophoretic mobility in an SDS-polyacrylamide gel corresponding to the mobility of the fully methylated form relative to the total amount of Tar (routinely 60 to 80%) and using this percentage in conjunction with immunoblot quantification of total Tar. At 10, 20, 30, and 40 s, samples of the reaction mixture were placed in SDS gel electrophoresis sample buffer to stop the reaction. Samples were analyzed by SDS gel electrophoresis, excision of the region containing Tar, alkaline hydrolysis of radiolabeled glutamyl methyl esters to yield radiolabeled methanol, vapor-phase diffusion, and scintillation counting.
The initial rates were determined by linear fits of the time course of methylation and adjusted as necessary to the rate at 3.3 μM methyl-accepting sites using the Km values for the methylation reaction (M. Li and G. L. Hazelbauer, unpublished data). For each combination of extent of modification and number of neighboring receptors in the same membrane, the means of at least three determinations of initial rate were plotted as a function of aspartate concentration and the data fit to the relationship v = vu + (vs[Asp]n)/([Asp]1/2n + [Asp]n) where vu is the “unstimulated” initial rate (i.e., the rate in the absence of aspartate), vs is the “maximally stimulated” initial rate (i.e., the rate at saturating aspartate), [Asp]1/2 is the aspartate concentration at which stimulation of the initial rate is half maximal, and n is the Hill coefficient.
RESULTS
Assessing chemoreceptor conformation and signaling using initial rates of methylation.
Investigation of the influence of receptor-receptor interactions on chemoreceptor conformational coupling and transmembrane signaling required an assay of receptor conformation that did not itself depend on those interactions. An attractive candidate was the initial rate of chemoreceptor methylation. The methylation rate reflects the proportion of the receptor population in the “methylation-competent, kinase-off” state versus the “methylation-incompetent, kinase-on” state. Many observations have shown that the shift in the conformational equilibrium generated by a change from ligand-free to ligand-saturated chemoreceptor results in an approximately 2-fold increase in the in vitro initial rate of chemoreceptor methylation (4, 12, 19, 22, 23, 26, 27, 34, 40). Importantly for our goal of investigating the influence of receptor-receptor interaction, this increase in methylation rate occurs even for receptor dimers isolated from interaction with other receptor molecules by placement in individual nanodiscs (4). However, the modest 2-fold difference between no ligand and saturating ligand meant the assay would have a narrow dynamic range. Thus, we investigated whether it could be sufficiently sensitive for our purposes by determining the detailed dose-response relationship between ligand concentration and initial rate of methylation of the E. coli chemoreceptor Tar.
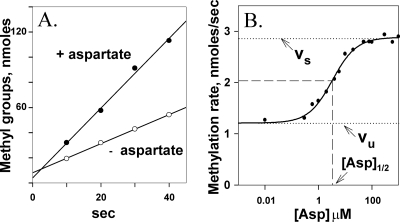
We measured initial rates of methylation for Tar(QEQE), which has the modification pattern encoded by wild-type tar, contained in native membrane vesicles as a function of the concentration of its attractant ligand aspartate over a range from 0.01 to 1,000 μM. Figure 2A shows an example of time course data used to determine initial rates. The plot illustrates the modest difference between rates of methylation of ligand-free and ligand-saturated Tar. Figure 2B displays data from a representative titration experiment and the fit of that data to a dose-response relationship from which can be derived values for vu, the unstimulated initial rate of methylation; vs, the maximally stimulated rate; [Asp]1/2, the ligand concentration at which the initial rate is halfway between vu and vs; and n, the Hill coefficient. The ability of the experimental measurements to track a gradual transition between the unstimulated and maximally stimulated rate and the fit of the data to a simple dose-response relationship indicated that initial rates of methylation could effectively monitor the gradual shift in the receptor conformational equilibrium as a function of ligand concentration.
FIG. 2.
Initial rates of Tar methylation enhanced by aspartate. (A) Time courses of methylation in the absence and presence of a saturating concentration (1 mM) of the Tar ligand aspartate. (B) Initial rate of methylation as a function of aspartate concentration. Data like the data shown in panel A were collected for Tar(QEQE) embedded in native membrane vesicles at the indicated concentrations of aspartate. The curve is a fit of the data to a simple dose-response relationship (see Materials and Methods). Dotted lines labeled vu and vs indicate the initial rates of methylation in the absence of aspartate and in the presence of a saturating concentration of the ligand, respectively. Dashed lines indicate the aspartate concentration, [Asp]1/2, at which enhancement of the initial rate of methylation is half-maximal.
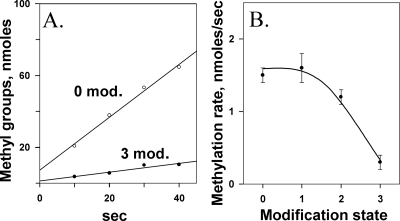
We also investigated whether assays of initial rates of methylation could monitor the influence of adaptational modification on receptor conformation by testing representative forms of Tar with 0 to 3 of its 4 methyl-accepting sites modified by introduction of a glutamine in place of glutamates, a substitution that mimics the functional effects of a glutamyl methylester (20). Tar with four modified sites could not be probed by this assay, since such a protein would have no sites available for methylation. Because our principal goal was to investigate effects of dimer-dimer interactions by comparing activities of receptor dimers with different potentials for interaction with neighbors, it was not necessary to assay all possible combinations of one, two, or three modifications among the four sites. Instead we chose representative combinations: Tar(QEQE), encoded by the wild-type gene, for the two-modification receptor, and Tar(QEEE) and Tar(QEQQ), each related to the two-modification receptor by a single change, for the one- and three-modification receptors. Figure 3A shows an example of primary data for Tar with zero or three modifications. Characterization of methylation rates for Tar with 0, 1, 2, or 3 modifications illustrated that the assay could monitor the shift in receptor conformational equilibrium generated by adaptational modification (Fig. 3B). Thus, the effects on receptor conformation of ligand occupancy (Fig. 2) and adaptational modification (Fig. 3) could be assessed by measuring initial rates of methylation.
FIG. 3.
Initial rates of Tar methylation reduced by adaptational modification. (A) Time courses of methylation for Tar with 0 or 3 (QEQQ) methyl-accepting sites modified by the introduction of glutamine, a functional analog of a methyl ester. (B) Initial rate of methylation as a function of adaptational modification. Data like the data shown in panel A were collected for Tar embedded in native membrane vesicles carrying 0, 1 (QEEE), 2 (QEQE), or 3 (QEQQ) glutamines at the 4 methyl-accepting sites of that chemoreceptor. The curve is drawn to guide the eye.
Opposing influences of ligand occupancy and adaptational modification.
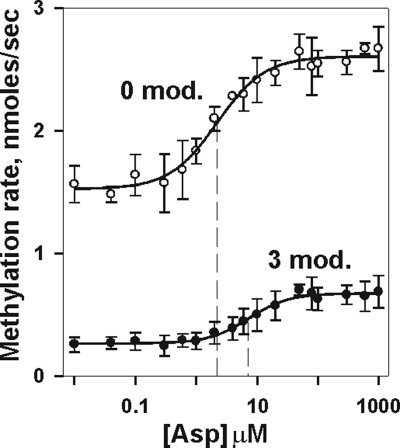
The chemotaxis system functions by balancing the opposing influences of ligand occupancy and adaptational modification (15). We used initial rates of receptor methylation to investigate the combined effects of these two inputs on chemoreceptor conformation in the absence of other chemotaxis components (Fig. 4). Ligand enhanced the initial rate of Tar methylation, whether the receptor was modified, and modification reduced methylation rate, no matter what the concentration of ligand. Thus, the two parameters were in large part independent influences on the receptor conformational equilibrium. However, extensive adaptational modification shifted the dose-response curve to a higher aspartate concentration, indicating that modification reduced functional affinity of receptor for ligand, an effect observed more dramatically in signaling complexes (6, 7, 24, 25, 36, 37). Interestingly, the ∼6-fold effect on initial rate of methylation generated by increasing adaptational modification from zero to three was larger than the ∼2-fold shift in initial rate generated by the transition from ligand free to ligand saturated.
FIG. 4.
Initial rates of methylation as a function of adaptational modification and ligand concentration. Rates were determined for Tar embedded in native membrane vesicles carrying 0 or 3 (QEQQ) glutamines at the indicated aspartate concentrations. The graph shows the means ± standard deviations (error bars) for ≥3 independent experiments. The curves are fits of the data to a simple dose-response relationship (see Materials and Methods). The vertical dashed line shows the respective values for [Asp]1/2.
Influence of interaction partners on the receptor conformation.
We investigated the influence of receptor interaction partners on conformational coupling and transmembrane signaling using membrane-embedded chemoreceptor Tar with three different potentials for dimer-dimer interactions: (i) no neighboring dimers and thus no interaction partners, a condition achieved by making nanodisc preparations containing ∼1 receptor dimer per disc; (ii) a few neighbors/interaction partners, a condition achieved by preparing nanodiscs containing ∼3 receptor dimers per disc, in which ∼80% of receptor dimers would have at least one parallel interaction partner but few would have more than two; and (iii) many neighbors and interaction partners, a condition provided by native membrane vesicles isolated from cells producing high levels of chemoreceptors. Initial rates of methylation were determined as a function of ligand concentration for Tar carrying 0, 1, 2, or 3 adaptational modifications, the mean values were plotted, and each aspartate dose-response curve was fitted to the relationship described for Fig. 2B. Table 1 lists the parameters derived from fitting dose-response data. Figure 5 compares receptors with no interaction partners to those with many partners, and Fig. 6 compares receptors with no or few potential partners.
TABLE 1.
Parameters derived from fitting dose-response dataa
| Condition | No. of modifications (modification) | Parameterb |
|||
|---|---|---|---|---|---|
| [Asp]1/2 (μM) | vu (nM/s) | vs (nM/s) | n | ||
| Vesicles | 0 (EEEE) | 2.2 ± 0.4 | 1.5 ± 0.1 | 2.6 ± 0.1 | 1.1 ± 0.3 |
| 1 (QEEE) | 3.0 ± 0.4 | 1.6 ± 0.2 | 3.7 ± 0.4 | 1.3 ± 0.2 | |
| 2 (QEQE) | 3.2 ± 1.3 | 1.2 ± 0.1 | 2.9 ± 0.1 | 1.2 ± 0.1 | |
| 3 (QEQQ) | 7.3 ± 1.6 | 0.3 ± 0.1 | 0.7 ± 0.1 | 1.3 ± 0.05 | |
| 3 (QQEQ) | 8.6 ± 0.8 | 0.5 ± 0.05 | 3.0 ± 0.4 | 1.2 ± 0.2 | |
| 1 dimer/disc | 0 (EEEE) | 1.8 ± 0.5 | 2.8 ± 0.5 | 5.1 ± 0.3 | 1.2 0± 0.1 |
| 1 (QEEE) | 1.9 ± 0.6 | 3.0 ± 0.1 | 6.1 ± 0.05 | 1.1 ± 0.2 | |
| 2 (QEQE) | 2.5 ± 0.7 | 3.2 ± 0.5 | 5.2 ± 0.9 | 1.2 ± 0.1 | |
| 3 (QEQQ) | 3.2 ± 0.4 | 1.1 ± 0.05 | 3.1 ± 0.05 | 1.3 ± 0.1 | |
| 3 (QQEQ) | 5.7 ± 1.7 | 1.9 ± 0.8 | 4.1 ± 1.3 | 1.2 ± 0.2 | |
| ∼3 dimers/disc | 0 (QEEE) | 0.8 ± 0.3 | 2.3 ± 0.2 | 4.8 ± 0.5 | 1.0 ± 0.2 |
| 1 (QEEE) | 1.5 ± 0.7 | 2.8 ± 0.4 | 5.2 ± 0.8 | 1.1 ± 0.1 | |
| 2 (QEQE) | 2.5 ± 0.4 | 2.7 ± 0.1 | 5.2 ± 0.1 | 0.9 ± 0.05 | |
| 3 (QEQQ) | 7.1 ± 1.4 | 0.9 ± 0.05 | 3.0 ± 0.1 | 1.1 ± 0.3 | |
| 3 (QQEQ) | 3.6 ± 0.7 | 1.0 ± 0.1 | 3.4 ± 0.3 | 1.0 ± 0.2 | |
Data shown in Fig. 5 and 6 were fit to the relationship v = vu + (vs[Asp]n)/([Asp]1/2n + [Asp]n) where vu is the “unstimulated” initial rate (i.e., the rate in the absence of aspartate), vs is the “maximally stimulated” initial rate (i.e., the rate at saturating aspartate), [Asp]1/2 is the aspartate concentration at which stimulation of the initial rate is half maximal, and n is the Hill coefficient.
The mean values and standard deviations for these parameters derived from fitting dose-response data from at least three independent experiments determining initial rates of methylation for Tar with 0 to 3 adaptational modifications and in conditions with many potentially interacting neighboring receptor dimers (vesicles), no potentially interacting neighboring receptor dimers (1 dimer/disc), or few potentially interacting neighboring receptor dimers (∼3 dimers/disc) are shown.
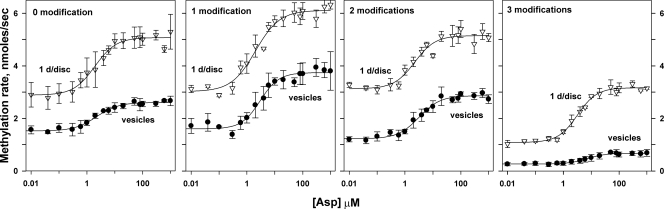
FIG. 5.
Effects of ligand and adaptational modification on initial rate of methylation persist in chemoreceptor dimers isolated from interacting neighbors. Experiments, data, and curves are as described in the legend to Fig. 4, using the Tar with patterns of modification described in the legend to Fig. 3. The figures compare Tar inserted in native membrane vesicles (vesicles) in which each receptor dimer had many potentially interacting neighbors and inserted in nanodiscs at 1 dimer/disc (1 d/disc) a condition with no potentially interacting neighbors.
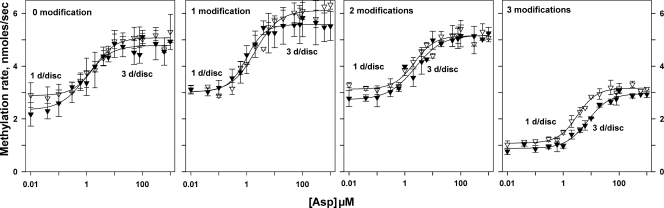
FIG. 6.
Effects of ligand and adaptational modification on initial rate of methylation for chemoreceptors with no or few interacting neighbors. Dose-response curves of initial rates of methylation as a function of aspartate concentration for chemoreceptor Tar with no (1 dimer/disc) or few (∼3 dimers/disc) potentially interacting neighboring receptors were determined as described in the legend to Fig. 4.
Comparison of nanodisc-inserted receptor dimers with no interaction partners to dimers with many potential interaction partners in native membranes (Fig. 5 and Table 1) shows that absence of partners did not fundamentally alter the effects of ligand occupancy or adaptational modification on the receptor conformational equilibrium. Dimers with no interaction partners or few potential interaction partners were essentially indistinguishable in terms of the effects of ligand occupancy or adaptational modification on the receptor conformational equilibrium (Fig. 6 and Table 1). Independent of the presence of interaction partners, aspartate enhanced the initial rate of methylation, and adaptational modification reduced that rate. Dose-response relationships for methylation rate as a function of ligand concentration were similar for receptors with or without partners in terms of fold enhancement, concentration of ligand for half-maximal enhancement, and a lack of significant cooperativity, i.e., Hill coefficients near one (Table 1). Thus, transmembrane signaling was fully functional in isolated chemoreceptor dimers as well as in receptors with a few interaction partners. However, dimers with few or no interaction partners exhibited a systematic shift of initial rates of methylation to higher values (Fig. 5 and 6 and Table 1). This shift could reflect greater accessibility of methyl-accepting sites on isolated receptor dimers in comparison to sites on receptors in dense arrays (3) or an influence of dimer-dimer interactions on receptor conformation.
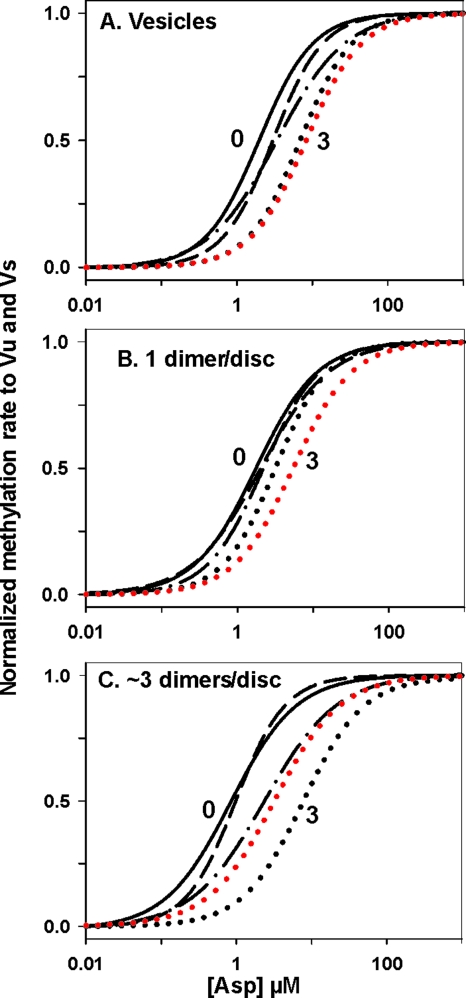
Coupling of adaptational modification to receptor conformation had the same general features independent of the presence of interaction partners. However, the extent to which the dose-response curves were spread by modification appeared less for dimers with no interaction partners (Fig. 7B) than for dimers with some (Fig. 7C) or many (Fig. 7A) partners. The differences might reflect effects of dimer-dimer interactions, but for all three conditions, the spread was less than 10-fold in Asp1/2 values and much of the difference in the spread of the curves was the result of shifts relative to the other curves of the dose-response curve for the three-modification form we had characterized, Tar(QEQQ). Thus, we examined the other three-modification forms. Tar(EQQQ) and Tar(QQQE) had initial rates of methylation too low to allow collection of reliable data, but we could determine dose-response curves for Tar(QQEQ). The parameters derived from these data are shown in Table 1, and the normalized dose-response curves for this alternative three-modification receptor are shown in red in Fig. 7. The data for the two three-modification forms of Tar are in large part within the variation in the assay. However, the difference in spread of the Tar(QQEQ) curves is less than the difference in spread of the Tar(QEQQ) curves. Considering the data for both three-modification forms of Tar, differences in the effects of modification on operational ligand affinity between isolated dimers and those with interaction partners appear marginal at best. Thus, we conclude that the reverse transmembrane signaling of adaptational modification to the ligand-binding site is essentially the same for isolated dimers as for dimers with few or many interacting partners.
FIG. 7.
Effects of chemoreceptor adaptational modification on functional affinity for ligand as influenced by the presence of potentially interacting neighboring receptors. Tar with many potentially interacting neighbors (A), no potentially interacting neighbors (B), or few potentially interacting neighbors (C) were compared for effects of adaptational modification on the dose-response relationship of the Tar ligand aspartate and initial rate of methylation. To facilitate comparison of the dose-response relationships for receptors with 0 (solid black line), 1 (QEEE; dashed line), 2 (QEQE; dot-dash line), and 3 (QEQQ [black dotted line] and QQEQ [red dotted line]) modifications, the curves shown in Fig. 5 and 6 were normalized to vu and to vs and displayed without the data points from which they had been derived.
DISCUSSION
Transmembrane signaling and conformational coupling.
We found that the essential chemoreceptor functions of conformational coupling and transmembrane signaling were properties of the receptor homodimer, the fundamental receptor structural unit. Our data showed that ligand occupancy was coupled to receptor conformation through transmembrane signaling with essentially the same efficiency and sensitivity in isolated individual receptor dimers as for dimers with many neighbors. Furthermore, for isolated receptor dimers, the compensatory parameter of adaptational modification was coupled to receptor conformation in a pattern very similar to that observed for receptors with neighbors. Finally, we found that the crucial interplay of ligand occupancy and adaptational modification occurred at the level of the homodimer. Identification of the receptor dimer as the fundamental unit of conformational coupling and transmembrane signaling implies that in signaling complexes, as well as in isolated dimers, the initial step in these processes is through individual receptor dimers. Our data demonstrated that effects of ligand occupancy, transmembrane signaling, and adaptational modification on the receptor conformational equilibrium are not dependent on higher-order interactions among chemoreceptor dimers or between receptors and other components of the signaling complex.
Our conclusion implies that mutations that block trimer formation without otherwise disrupting receptor structure should not block transmembrane signaling. Our laboratory has observed that this is the case. Tar carrying a single amino acid substitution that does not form trimers in vivo (C. Studdert and J. S. Parkinson, personal communication) or activate kinase in vitro does exhibit an increased initial rate of methylation in the presence of aspartate (M. Li and G. L. Hazelbauer, unpublished data).
Nanodiscs.
Nanodiscs provided a way to isolate receptor dimers from interaction with neighboring dimers or to limit the number of interacting neighbors. This allowed us to characterize the functional properties of the chemoreceptor homodimer and determine the central role of this structural unit in conformational coupling and transmembrane signaling. There are few, if any, alternative approaches to address such issues.
However, it is conceivable that receptor dimers isolated in individual nanodiscs might associate at their membrane-distal tips to form trimers and that only dimers in trimers exhibit the signaling we observed experimentally. This seems unlikely, since one-dimer-per-disc preparations do not exhibit the trimer property of effective activation of kinase (3), and electron microscopy shows no indication of trimer formation by nanodisc-isolated dimers (M. Li, C. Khursigara, S. Subramaniam, and G. L. Hazelbauer, unpublished data). In addition, 10-fold dilution of nanodisc preparations containing ∼1 Tar dimer/disc, which would be expected to drastically reduce the probability of the collision of three isolated dimers necessary for trimer formation, did not significantly reduce transmembrane signaling as assayed by an aspartate-induced increase in initial rate of methylation (data not shown).
Cooperativity and reverse transmembrane signaling.
The relationship between ligand concentration and receptor conformation, assayed by initial rate of methylation, could be fit by a simple binding isotherm exhibiting no significant cooperativity (Fig. 4 and Table 1). This indicates a direct link between ligand occupancy and receptor conformation and thus provides further support for the role of individual chemoreceptor dimers as the basic signaling units. The lack of cooperativity was not altered by the extent of adaptational modification and was independent of the availability of other receptor dimers as interaction partners (Fig. 5 to 7 and Table 1). Thus, the direct link between ligand occupancy and receptor conformation was not altered by shifts in the receptor conformational equilibrium or by interactions among receptors, again supporting the central role of individual dimers in transmembrane signaling and conformational coupling.
Changing the extent of receptor adaptational modification from 0 to 3 shifted the position of dose-response curve for initial rate of methylation to higher ligand concentrations (Fig. 7 and Table 1). The shift was less than an order of magnitude, corresponding to similarly modest effects of adaptational modification on affinity for ligand (6, 10, 16, 24, 29). This reverse transmembrane signaling, which involves the sliding of a single helix in the periplasmic/transmembrane signaling module of chemoreceptors in the opposite direction (toward the periplasm) from the sliding induced by ligand occupancy (20), occurred in isolated receptor dimers.
Investigating central tenants of models for bacterial chemotaxis.
The bacterial chemotaxis system, particularly the well-characterized system of E. coli, has become a popular and productive focus of mathematical modeling (for instance, see references 11, 31, and 35). Such models generally include assumptions that there is an equilibrium between two conformations of signaling complexes, one kinase on, methylation incompetent and the other kinase off, methylation competent, and that adaptational modification shifts the equilibrium toward the former and ligand occupancy toward the latter. There is extensive experimental evidence for the graded effects of receptor modification and ligand occupancy on CheA kinase activity in signaling complexes, but no published evidence of graded effects of these two parameters on the propensity of receptors for methylation. Data summarized in Fig. 4 to 7 provide such evidence. The use of initial rate of methylation as a probe of chemoreceptor conformation allowed us to investigate effects of ligand and adaptational modification on receptor dimers independent of the presence of other receptor dimers or other components of the chemotaxis system. We found that receptors alone, independent of any interactions with partners, are influenced in a graded manner by ligand occupancy and adaptational modification. This provides experimental documentation of a central tenant in models of the chemotaxis system.
Steps in receptor signaling.
At what stage in signaling are ligand-induced, noncooperative changes in individual receptor dimers or the modest, modification-induced shifts in the aspartate dose-response curve transformed into the cooperative effect of ligand binding and the several orders of magnitude changes in operational kinase activity upon adaptational modification observed in signaling complexes (6, 7, 24, 25, 36, 37)? A first putative signaling step downstream from the conformational change in individual receptor dimers is a change in the relative orientation of receptor dimers in trimers of dimers (17, 41). However, where characterized, these changes are not cooperative as a function of ligand concentration, and increasing the extent of adaptational modification of Tar from 0 to 3 shifted the concentration of aspartate for half-maximal response less than 10-fold (41), much like our observations of effects on the conformation of individual dimers. This implies that cooperativity and increased effects of adaptational modification are created at the level of signaling complexes. This implication will be the subject of subsequent investigations.
Acknowledgments
We thank Angela Lilly for creating the strains and plasmids used and an anonymous reviewer for comments that improved the manuscript.
This work was supported in part by grant GM29963 to G.L.H. from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 8 January 2010.
REFERENCES
- 1.Barnakov, A., L. Barnakova, and G. Hazelbauer. 1998. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J. Bacteriol. 180:6713-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayburt, T. H., and S. G. Sligar. 2003. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 12:2476-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besschetnova, T. Y., D. J. Montefusco, A. E. Asinas, A. L. Shrout, F. M. Antommattei, and R. M. Weis. 2008. Receptor density balances signal stimulation and attenuation in membrane-assembled complexes of bacterial chemotaxis signaling proteins. Proc. Natl. Acad. Sci. U. S. A. 105:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldog, T., S. Grimme, M. Li, S. G. Sligar, and G. L. Hazelbauer. 2006. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. U. S. A. 103:11509-11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boldog, T., M. Li, and G. L. Hazelbauer. 2007. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 423:317-335. [DOI] [PubMed] [Google Scholar]
- 6.Borkovich, K. A., L. A. Alex, and M. I. Simon. 1992. Attenuation of sensory receptor signaling by covalent modification. Proc. Natl. Acad. Sci. U. S. A. 89:6756-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornhorst, J. A., and J. J. Falke. 2001. Evidence that both ligand binding and covalent adaptation drive a two-state equilibrium in the aspartate receptor signaling complex. J. Gen. Physiol. 118:693-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briegel, A., D. R. Ortega, E. I. Tocheva, K. Wuichet, Z. Li, S. Chen, A. Müller, C. V. Iancu, G. E. Murphy, M. J. Dobro, I. B. Zhulin, and G. J. Jensen. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U. S. A. 106:17181-17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denisov, I. G., Y. V. Grinkova, A. A. Lazarides, and S. G. Sligar. 2004. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126:3477-3487. [DOI] [PubMed] [Google Scholar]
- 10.Dunten, P., and D. E. Koshland, Jr. 1991. Tuning the responsiveness of a sensory receptor via covalent modification. J. Biol. Chem. 266:1491-1496. [PubMed] [Google Scholar]
- 11.Endres, R. G., and N. S. Wingreen. 2006. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods.” Proc. Natl. Acad. Sci. U. S. A. 103:13040-13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falke, J. J., A. F. Dernburg, D. A. Sternberg, N. Zalkin, D. L. Milligan, and D. E. Koshland, Jr. 1988. Structure of a bacterial sensory receptor. A site-directed sulfhydryl study. J. Biol. Chem. 263:14850-14858. [PubMed] [Google Scholar]
- 13.Gestwicki, J. E., A. C. Lamanna, R. M. Harshey, L. L. McCarter, L. L. Kiessling, and J. Adler. 2000. Evolutionary conservation of methyl-accepting chemotaxis protein location in bacteria and archaea. J. Bacteriol. 182:6499-6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenfield, D., A. L. McEvoy, H. Shroff, G. E. Crooks, N. S. Wingreen, E. Betzig, and J. Liphardt. 2009. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 7:e1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazelbauer, G. L., J. J. Falke, and J. S. Parkinson. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwama, T., M. Homma, and I. Kawagishi. 1997. Uncoupling of ligand-binding affinity of the bacterial serine chemoreceptor from methylation- and temperature-modulated signaling states. J. Biol. Chem. 272:13810-13815. [DOI] [PubMed] [Google Scholar]
- 17.Khursigara, C. M., X. Wu, P. Zhang, J. Lefman, and S. Subramaniam. 2008. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 105:16555-16560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, K. K., H. Yokota, and S.-H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 19.Kleene, S. J., A. C. Hobson, and J. Adler. 1979. Attractants and repellents influence methylation and demethylation of methyl-accepting chemotaxis proteins in an extract of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 76:6309-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, W.-C., B. D. Beel, and G. L. Hazelbauer. 2006. Adaptational modification and ligand occupancy have opposite effects on positioning of the transmembrane signalling helix of a chemoreceptor. Mol. Microbiol. 61:1081-1090. [DOI] [PubMed] [Google Scholar]
- 21.Lai, W.-C., and G. L. Hazelbauer. 2005. Carboxyl-terminal extensions beyond the conserved pentapeptide reduce rates of chemoreceptor adaptational modification. J. Bacteriol. 187:5115-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Moual, H., T. Quang, and D. E. Koshland, Jr. 1998. Conformational changes in the cytoplasmic domain of the Escherichia coli aspartate receptor upon adaptive methylation. Biochemistry 37:14852-14859. [DOI] [PubMed] [Google Scholar]
- 23.Le Moual, H., T. Quang, and D. E. Koshland, Jr. 1997. Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry 36:13441-13448. [DOI] [PubMed] [Google Scholar]
- 24.Levit, M. N., and J. B. Stock. 2002. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J. Biol. Chem. 277:36760-36765. [DOI] [PubMed] [Google Scholar]
- 25.Li, G., and R. M. Weis. 2000. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100:357-365. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., G. Li, and R. M. Weis. 1997. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry 36:11851-11857. [DOI] [PubMed] [Google Scholar]
- 27.Li, M., and G. L. Hazelbauer. 2006. The carboxyl-terminal linker is important for chemoreceptor function. Mol. Microbiol. 60:469-479. [DOI] [PubMed] [Google Scholar]
- 28.Li, M., and G. L. Hazelbauer. 2004. Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 186:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, L. N., J. Li, J. F. Brandts, and R. M. Weis. 1994. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry 33:6564-6570. [DOI] [PubMed] [Google Scholar]
- 30.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 31.Mello, B. A., and Y. Tu. 2007. Effects of adaptation in maintaining high sensitivity over a wide range of backgrounds for Escherichia coli chemotaxis. Biophys. J. 92:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milburn, M. V., G. G. Prive, D. L. Milligan, W. G. Scott, J. Yeh, J. Jancarik, D. E. Koshland, Jr., and S. H. Kim. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342-1347. [DOI] [PubMed] [Google Scholar]
- 33.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro, M. J., I. Chakrabarti, and D. E. Koshland, Jr. 1995. Contributions made by individual methylation sites of the Escherichia coli aspartate receptor to chemotactic behavior. Proc. Natl. Acad. Sci. U. S. A. 92:1053-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu, T. S., S. V. Aksenov, and D. Bray. 2003. A spatially extended stochastic model of the bacterial chemotaxis signalling pathway. J. Mol. Biol. 329:291-309. [DOI] [PubMed] [Google Scholar]
- 36.Sourjik, V., and H. C. Berg. 2004. Functional interactions between receptors in bacterial chemotaxis. Nature 428:437-441. [DOI] [PubMed] [Google Scholar]
- 37.Sourjik, V., and H. C. Berg. 2002. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 99:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studdert, C. A., and J. S. Parkinson. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U. S. A. 101:2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studdert, C. A., and J. S. Parkinson. 2005. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. U. S. A. 102:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swain, K. E., and J. J. Falke. 2007. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry 46:13684-13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaknin, A., and H. C. Berg. 2007. Physical responses of bacterial chemoreceptors. J. Mol. Biol. 366:1416-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, P., C. M. Khursigara, L. M. Hartnell, and S. Subramaniam. 2007. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 104:3777-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]