Abstract
In Escherichia coli, rRNAs are transcribed as precursors and require processing at the 3′ and 5′ ends to generate mature RNA molecules. The largest of these RNAs, 23S rRNA, is matured at the 3′ end by a set of exonucleases and at the 5′ end by an unknown RNase. Whether the 3′ and 5′ maturation steps occur independently or are coupled has previously been unclear. By assessing the levels of precursors accumulating at the 3′ and 5′ ends, we provide evidence that these processes may be linked. Thus, each of several conditions that led to precursor accumulation at one end also did so at the other end. We also observed that each end undergoes maturation at similar rates, suggesting that the two processes could be coupled. Finally, we provide evidence that processing at the 3′ end facilitates 5′-end maturation. A model to explain the basis for the observed directionality of the reactions is proposed. This information will aid in the search for the enzyme responsible for final maturation of the 5′ end of 23S rRNA.
Many RNAs are transcribed as precursors and require processing for maturation and optimum functionality. This is especially true for rRNAs, and several model organisms have been studied extensively to define the steps involved in rRNA processing, including Escherichia coli, Bacillus subtilis, and Saccharomyces cerevisiae (5, 6, 9, 20). In E. coli, each of the three rRNAs is generated from a single rRNA operon transcript (6). The transcript is first cleaved at two sites by RNase III, generating precursors for 5S, 16S, and 23S rRNAs (16). For 5S rRNA, further processing steps involve endonucleolytic cleavage by RNase E at both the 5′ and 3′ ends, followed by final maturation of the 3′ end by RNase T and by an unidentified enzyme at the 5′ end (7, 11). 16S maturation proceeds via dual cleavages at the 5′ end by RNase E and RNase G, whereas an unknown enzyme is responsible for 3′-end processing. Finally, 23S rRNA requires a set of 3′-to-5′ exonucleases to carry out 3′-end maturation (12). However, the enzyme responsible for 5′-end maturation of 23S rRNA, presumably a novel endonuclease, has remained elusive despite nearly 3 decades of investigation (5). It has also been unclear whether processing of the two ends of this RNA represents coupled events or whether these events occur independently. Here we provide evidence that processing of the two ends constitutes linked events and that prior processing of the 3′ end facilitates 5′-end maturation.
MATERIALS AND METHODS
Strains.
MG1655* is a derivative of the sequenced strain MG1655 (2), which contains an engineered point mutation that converts a defective rph gene to wild type. Derivatives of MG1655* containing ΔdeaD, ΔsrmB, Δrph, Δrnb, or Δrnt alleles were constructed by transduction of deletion alleles that are interrupted by a kanamycin-resistant (Kanr) marker (1). To construct MG1655* Δrph Δrnb Δrnt, the Δrph, Δrnb, and Δrnt alleles were introduced sequentially, and the Kanr marker was removed from the intermediate strains by recombination of frt sites that flank the kan gene prior to each subsequent transduction (1).
RNA preparation and analysis.
RNA was prepared from cultures growing exponentially in LB medium using the hot-phenol method, with variations, as described previously (10). For experiments involving measurement of rRNA processing kinetics, exponentially growing cells were treated with 200 μg/ml of rifampin, and cell aliquots were harvested at different times for RNA preparation. Analysis of 3′-end processing of 23S rRNA was performed by cleavage of the RNA near its 3′ end using a 2′-O-methyl RNA/DNA chimeric oligonucleotide (5′-UGdCdGdCdTUACACACCCGGCC-3′), in which all ribo residues are methylated (12), and Hybridase thermostable RNase H (Epicentre Biotechnologies). The cleaved RNA was fractionated on a 6% polyacrylamide-8 M urea sequencing gel, transferred to a positively charged nylon membrane (Nytran; Whatman Inc.), and probed with a radiolabeled oligonucleotide (5′-AAGGTTAAGCCTCACGGTTC-3′) complementary to the 3′ cleavage product. Primer extension analysis to assess 5′-end maturation of 23S rRNA was carried out using a labeled specific oligonucleotide primer, as described previously (18).
RESULTS
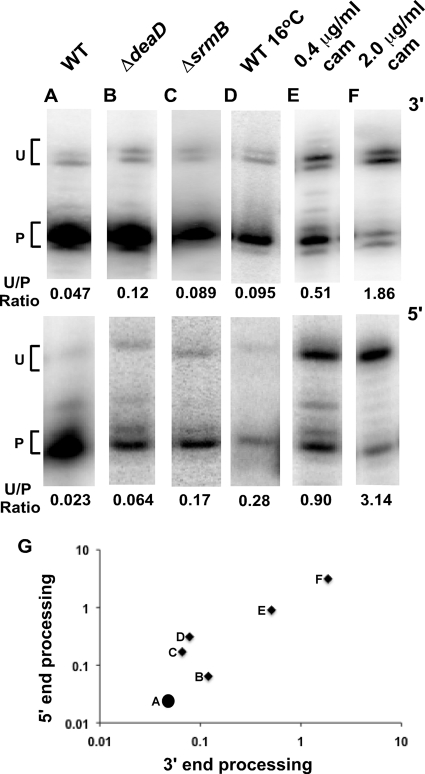
While analyzing 23S rRNA processing in mutant strains lacking the DeaD or SrmB RNA helicases, we observed processing defects at both the 3′ and 5′ ends when these strains were grown in LB medium at 37°C. Similar processing defects have been observed under conditions of low-temperature growth (3, 4). Based on a hypothesis that processing of the two ends could be coordinated, we measured the proportion of unprocessed RNA at either end under several different conditions that cause inefficient processing. These included the aforementioned strains lacking DeaD or SrmB or growth of a wild-type strain in LB at 16°C or at 37°C in LB supplemented with sublethal concentrations of chloramphenicol (16). As shown in Fig. 1, each of the conditions tested led to increased amounts of 23S rRNA precursors at both ends compared to the untreated wild-type strain grown at 37°C. The precursors that were most abundant under these conditions contained 8 or 9 unprocessed nucleotides (nt) at the 3′ end and 7 nt at the 5′ end. These precursors are derived from RNase III cleavage of the rRNA operon transcript (8). We did not observe any condition under which precursors were increased at one end but not the other. These observations suggested that the processing reactions at the two ends could be coordinated and merited further investigation.
FIG. 1.
Processing of the 3′ and 5′ ends of 23S rRNA. (A to F) The extent of 3′- and 5′-end processing of 23S rRNA under different conditions was evaluated by Northern blot analysis or primer extension analysis following the harvest of cultures and RNA isolation, as described in Materials and Methods. 3′-end (top) and 5′-end (bottom) analysis of 23S rRNA is shown. The positions of the unprocessed (U) and processed (P) ends are marked, and the U/P ratio is indicated in each case. MG1655* (A), MG1655* ΔdeaD (B), or MG1655* ΔsrmB (C) was grown in LB at 37°C. (D) MG1655* was grown in LB at 16°C. MG1655* was grown in LB at 37°C supplemented with 0.4 μg/ml (E) or 2.0 μg/ml (F) of chloramphenicol. (G) Relationship between 3′- and 5′-end processing. The ratios of unprocessed to processed RNA at the 3′ and 5′ ends (A to F) are plotted on a logarithmic scale. The data points correspond to panels A to F and are labeled accordingly.
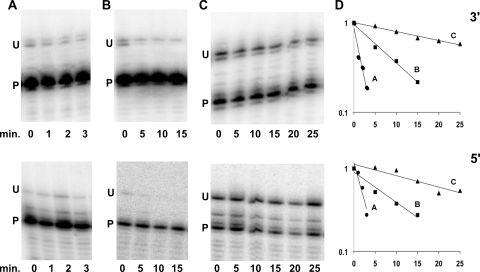
One explanation for the observed accumulation of rRNA precursors could be reduced rates of processing. If processing is coordinated at the two ends, the expectations are that (i) processing of each end in an untreated wild-type strain should occur at similar rates and (ii) any condition that slows processing at one end should similarly affect processing at the other end. To test these hypotheses, processing rates were evaluated at each end for a subset of conditions (Fig. 1). Experiments to determine the processing kinetics were performed by measuring the amount of unprocessed (precursor) or processed (mature) rRNA present either under conditions of steady-state growth (time zero) or at different times after transcription was blocked by the addition of rifampin (Fig. 2A to C). By quantifying the fraction of precursor rRNA remaining at different times after rifampin addition, the half-life for conversion of unprocessed RNA to the mature form could be calculated (Fig. 2D). The processing half-lives from three individual experiments were averaged for each strain, and standard errors were calculated. For a wild-type strain growing in LB at 37°C, average processing half-lives of 1.8 ± 0.3 min at the 3′ end and 1.7 ± 0.2 min at the 5′ end were determined. In contrast, a ΔdeaD strain exhibited processing half-lives of 8.2 ± 1.7 min at the 3′ end and 9.2 ± 1.2 min at the 5′ end. Finally, for a wild-type strain grown in the presence of chloramphenicol, the processing half-lives were 30.0 ± 4.6 and 20.3 ± 2.4 min at the 3′ end and 5′ end, respectively. For each of the situations tested, the processing rates at the two ends were comparable. These results suggested that processing of the 3′ and 5′ ends could be coupled, possibly because initiation of processing at one end of 23S rRNA allows processing at the other end to occur rapidly.
FIG. 2.
Kinetics of 23S rRNA maturation. Cultures were grown to mid-log phase, and aliquots were harvested for RNA preparation at various times, after the addition of rifampin to halt transcription, as indicated. (A to C) The extent of 23S rRNA maturation was assayed at the 3′ end (top) or the 5′ end (bottom) from the following strains: MG1655* grown in LB at 37°C (A), MG1655* ΔdeaD grown in LB at 37°C (B), and MG1655* grown in LB supplemented with 0.4 μg/ml of chloramphenicol at 37°C (C). The positions of the unprocessed (U) and processed (P) RNAs are indicated. The time, in minutes, at which cultures were harvested after rifampin addition is denoted below each panel. (D) The rates of 23S rRNA maturation shown in panels A to C were quantified by plotting the normalized ratio of unprocessed to processed rRNA remaining at different times after transcription inhibition. Individual data points for each graph are averaged over three independent experiments. The data were plotted on a semilogarithmic graph, and straight lines were obtained by linear regression. Maturation kinetics of the 3′ end (top); maturation kinetics of the 5′ end (bottom).
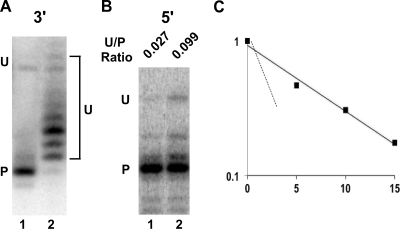
If maturation is coordinated and directional, at which end does processing occur first? Following the cleavage of the ribosomal operon RNA by RNase III, the pre-23S rRNA segment contains 7 to 9 extra nt at the 3′ end (8). The maturation of this end appears to be a two-step exonucleolytic process: first, a set of yet-to-be-identified RNases remove several of the terminal nucleotides, and a second processing step, mediated primarily by RNase T follows, which results in the removal of the remaining unprocessed residues (12). To determine whether defects in 3′-end processing can influence 5′-end maturation, we first analyzed 23S rRNA 3′ ends in a strain lacking three 3′-to-5′ exoribonucleases: RNase II, RNase PH, and RNase T (Fig. 3A). No significant accumulation of the RNase III cleaved product was observed, indicating that the combined loss of the three enzymes is insufficient to block the first step of processing. However, a ladder of unprocessed products containing primarily 1 to 4 unprocessed nt, present at low levels in a wild-type strain, was observed. Such products have also been observed with the singly mutated RNase T strain and are diagnostic of defects in the final maturation step (12). Strikingly, when maturation was evaluated at the 5′ end, increased levels of rRNA precursors were observed at this end as well (Fig. 3B). These results suggested that 3′-end processing might be a prerequisite for efficient and complete 5′-end maturation to occur. To investigate whether impaired processing at the 3′ end leads to delays in 5′-end maturation, the kinetics of 5′-end formation was measured in the triply mutated strain. The half-life for 5′-end maturation in this strain was found to be 6.1 min, nearly four times slower than that in a wild-type strain (Fig. 3C). Although indirect effects cannot be excluded, these results suggest that processing defects at the 3′ end of 23S rRNA cause an accumulation of 5′-end precursors and reduce the rate of maturation at this end directly.
FIG. 3.
Defects in 23S rRNA maturation caused by a lack of 3′-to-5′ exonucleases. MG1655* or MG1655* Δrph Δrnb Δrnt was grown in LB at 37°C, and RNA was extracted from exponentially growing cultures. 23S rRNA processing was evaluated at the 3′ end (A) or at the 5′ end (B). Lane 1, MG1655*; lane 2, MG1655* Δrph Δrnd Δrnt. The positions of the unprocessed (U) and processed (P) RNAs are indicated. The ratio of unprocessed and processed RNA (U/P) at the 5′ end is also shown in panel B. (C) Kinetics of 5′-end processing. Cultures of MG1655* Δrph Δrnd Δrnt were harvested at the indicated times after rifampin addition, and RNA preparations derived from the cultures were assayed by primer extension. The fraction of unprocessed rRNA versus time after the addition of rifampin is plotted on a semilogarithmic graph. Each point represents the average from three independent experiments. A straight line obtained by linear regression is shown. For comparison purposes, a dashed line that corresponds to 5′-end maturation in a wild-type strain (see Fig. 2D) is also shown.
DISCUSSION
rRNAs are the most abundant class of RNA in the cell under a variety of growth conditions, and in prokaryotic organisms, such as E. coli, rRNAs can account for as much as 80% of the total weight of all cellular RNAs (14). Despite their high abundance and obvious importance, the pathway of rRNA processing has yet to be completely elucidated. Specifically, the enzymes responsible for 3′-end maturation of 16S rRNA or for 5′-end maturation of 5S and 23S rRNAs remain to be identified (6). We observed processing defects at both ends of 23S rRNA in mutants that lack RNA helicases, suggesting a possibility that processing of the two ends could be linked events. To investigate this possibility further, additional conditions that cause 23S rRNA processing defects were tested. In each case, the amount of unprocessed RNA was found to increase at both ends (Fig. 1). Moreover, we observed that the 3′ and 5′ ends were processed with similar kinetics either in wild-type cells or under conditions of impaired processing (Fig. 2). These results suggest that the processing events at both ends could be coupled and may be coregulated. Furthermore, we found that defects that impaired the formation of a mature 3′ end led to a reduced rate of 5′-end maturation and to precursor accumulation (Fig. 3). These observations indicate that 3′-end processing in some way facilitates 5′-end maturation.
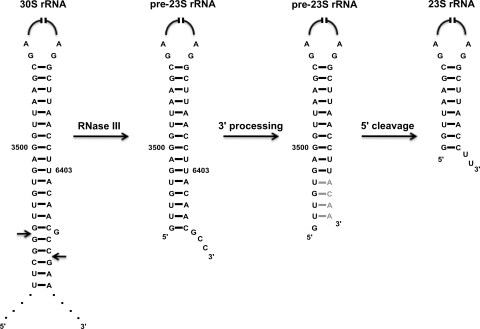
How can the directionality of rRNA processing be explained? Our findings can be rationalized in terms of the interactions between the processing enzymes and the sequences of the 23S rRNA precursor (Fig. 4). After RNase III cleavage, the pre-23S rRNA contains 3 to 7 nt at the 5′ end and 7 to 9 nt at the 3′ end. In the precursor, the 5′ sequences are base paired, forming an extended stem. We hypothesize that maturation at the 5′ end requires an endonuclease that is sensitive to secondary structures and cleavage cannot occur as long as extensive base-pairing is present. For maturation to occur effectively at the 5′ end, it would require the removal of the base-pairing 3′ precursor sequences. Once they were removed, 5′-end maturation would occur very rapidly thereafter, as suggested by the very similar kinetics of processing observed at the two ends.
FIG. 4.
Model for 23S rRNA maturation. The first processing step involves cleavage of the 30S ribosomal operon RNA by RNase III. The resulting pre-23S rRNA contains unprocessed sequences at the 3′ and 5′ ends that are partially base paired (21). We propose that this RNA is first processed at the 3′ end by 3′-to-5′ exonucleases. Exonucleolytic digestion may be facilitated by RNA helicases that open up the duplex region. The removal of 3′-end sequences that base pair with the 5′-end precursor is proposed to allow rapid cleavage by an unknown enzyme(s) that leads to 5′-end maturation. The removal of all of the 3′ unprocessed nucleotides, however, may not be a prerequisite for 5′-end maturation to occur. In strain MG1655* Δrph Δrnb Δrnt, most of the 3′ ends retain 1 to 4 nt of unprocessed RNA (shown in gray); nonetheless, most of the 5′ ends undergo maturation, albeit with a reduced kinetics of processing compared to that of the wild type (Fig. 3).
In the context of this model, it is interesting to note that in a strain lacking RNase II, RNase PH, and RNase T, there was a significant lack of complete 3′-end processing, yet despite an increased accumulation of 5′-end precursors, the majority of the 5′ ends were fully mature (Fig. 3A and B). We believe that the observed maturation of the 5′ end is possible because several unprocessed nucleotides at the 3′ end have already been removed, which is apparently sufficient to permit 5′-end maturation, albeit with reduced kinetics (Fig. 3C). A prediction of the model is that only when the first step of processing is blocked will it cause a substantial defect in 5′-end maturation. This prediction is not yet directly testable, because the set of RNases that mediate this process still remain to be identified. It is interesting to note, however, that some of the conditions that cause the first step of maturation at the 3′ end to become impeded, albeit indirectly, result in the accumulation of substantial levels of immature 5′ ends (Fig. 1).
Some degree of coupling has also been observed for the maturation of the 3′ and 5′ ends of 16S rRNA in E. coli. Thus, mutations in the endonucleases RNase E and RNase G, which are involved in formation of the mature 5′ end, cause delayed maturation of the 3′ end (13). However, the requirement for 5′ processing is not absolute, as 3′-end maturation proceeds even when 5′-end processing is completely blocked, albeit at reduced rates. In contrast, we observe a high degree of coupling between the processing events at the two ends of 23S rRNA, since maturation of the 5′ end occurs at virtually the same rate as that of the 3′ end under a wide range of conditions (Fig. 2 and data not shown). Maturation of 23S rRNA in Bacillus subtilis is also a coupled process, but in this case it is due to the action of a double-strand-specific endonuclease, Mini-III, which simultaneously generates both the mature 3′ and 5′ ends (15).
The notion that maturation of the two ends of E. coli 23S rRNA could be linked processes was mooted several years ago, but it was speculated that 5′-end maturation would precede 3′-end maturation (17). The rationale for the proposed directionality was that the 5′ precursor sequences would need to be removed in order for exonucleases, which could be sensitive to secondary structures, to process the 3′ ends. However, that proposal did not take into consideration the presence of RNA helicases in the cell, which could have a role in disrupting terminal base-pairing and allow RNA digestion by exonucleases. That helicases could play such a role is consistent with the processing defects observed in the ΔsrmB and ΔdeaD strains (Fig. 1B and C). Further support for 3′-end processing being the first step is provided by an examination of published data: quantitative maturation of the 3′ end has been reproduced in vitro using cell extracts or purified exonucleases, whereas partial maturation of the 5′ end has been accomplished only under conditions of protein synthesis (12, 17).
It now remains to identify the enzyme that mediates formation of the 5′ mature end. Despite several decades of research, the enzyme responsible for this process has proven to be notoriously difficult to identify. It seems unlikely that 5′ maturation of 23S rRNA is mediated by any of the known E. coli RNases, and the current evidence suggests the presence of a novel activity. Interestingly, some studies of this topic have indicated that 5′-end maturation occurs within the context of translating ribosomes, rather than on naked RNA or within the 50S large ribosomal subunit (19), resulting in the speculation that this process is either mediated by a factor tightly associated with translating ribosomes or caused by the ribosome itself.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM-081735).
We thank Murray Deutscher for comments on the manuscript.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32:2751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charollais, J., D. Pflieger, J. Vinh, M. Dreyfus, and I. Iost. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48:1253-1265. [DOI] [PubMed] [Google Scholar]
- 5.Condon, C. 2007. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 10:271-278. [DOI] [PubMed] [Google Scholar]
- 6.Deutscher, M. P. 2009. Maturation and degradation of ribosomal RNA in bacteria. Prog. Mol. Biol. Transl. Sci. 85:369-391. [DOI] [PubMed] [Google Scholar]
- 7.Ghora, B. K., and D. Apirion. 1978. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell 15:1055-1066. [DOI] [PubMed] [Google Scholar]
- 8.Ginsburg, D., and J. A. Steitz. 1975. The 30 S ribosomal precursor RNA from Escherichia coli. A primary transcript containing 23 S, 16 S, and 5 S sequences. J. Biol. Chem. 250:5647-5654. [PubMed] [Google Scholar]
- 9.Henras, A. K., J. Soudet, M. Gerus, S. Lebaron, M. Caizergues-Ferrer, A. Mougin, and Y. Henry. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 65:2334-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain, C., A. Deana, and J. G. Belasco. 2002. Consequences of RNase E scarcity in Escherichia coli. Mol. Microbiol. 43:1053-1064. [DOI] [PubMed] [Google Scholar]
- 11.Li, Z., and M. P. Deutscher. 1995. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc. Natl. Acad. Sci. U. S. A. 92:6883-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, Z., S. Pandit, and M. P. Deutscher. 1999. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA 5:139-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Z., S. Pandit, and M. P. Deutscher. 1999. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18:2878-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nomura, M., R. Gourse, and G. Baughman. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 53:75-117. [DOI] [PubMed] [Google Scholar]
- 15.Redko, Y., D. H. Bechhofer, and C. Condon. 2008. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol. Microbiol. 68:1096-1106. [DOI] [PubMed] [Google Scholar]
- 16.Schlessinger, D., M. Ono, N. Nikolaev, and L. Silengo. 1974. Accumulation of 30S preribosomal ribonucleic acid in an Escherichia coli mutant treated with chloramphenicol. Biochemistry 13:4268-4271. [DOI] [PubMed] [Google Scholar]
- 17.Sirdeshmukh, R., and D. Schlessinger. 1985. Ordered processing of Escherichia coli 23S rRNA in vitro. Nucleic Acids Res. 13:5041-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slagter-Jager, J. G., L. Puzis, N. S. Gutgsell, M. Belfort, and C. Jain. 2007. Functional defects in transfer RNAs lead to the accumulation of ribosomal RNA precursors. RNA 13:597-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srivastava, A. K., and D. Schlessinger. 1988. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. U. S. A. 85:7144-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivastava, A. K., and D. Schlessinger. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44:105-129. [DOI] [PubMed] [Google Scholar]
- 21.Young, R. A., and J. A. Steitz. 1978. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc. Natl. Acad. Sci. U. S. A. 75:3593-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]