Abstract
The polysaccharide capsule is a major antigenic factor in Streptococcus agalactiae (Lancefield group B streptococcus [GBS]). Previous observations suggest that exchange of capsular loci is likely to occur rather frequently in GBS, even though GBS is not known to be naturally transformable. We sought to identify and characterize putative capsular switching events, by means of a combination of phenotypic and genotypic methods, including pulsed-field gel electrophoretic profiling, multilocus sequence typing, and surface protein and pilus gene profiling. We show that capsular switching by horizontal gene transfer is not as frequent as previously suggested. Serotyping errors may be the main reason behind the overestimation of capsule switching, since phenotypic techniques are prone to errors of interpretation. The identified putative capsular transformants involved the acquisition of the entire capsular locus and were not restricted to the serotype-specific central genes, the previously suggested main mechanism underlying capsular switching. Our data, while questioning the frequency of capsular switching, provide clear evidence for in vivo capsular transformation in S. agalactiae, which may be of critical importance in planning future vaccination strategies against this pathogen.
Streptococcus agalactiae (group B streptococcus [GBS]) is primarily a colonizing agent of the genitourinary and gastrointestinal tracts, but it is also a leading cause of bacterial sepsis and meningitis in neonates and is increasingly associated with invasive infections in adults (39). The capsular polysaccharide is a major GBS virulence factor and also the main target of antibody-mediated killing (11). In the last decade, conjugated multivalent vaccines have been developed and proved to be highly immunogenic, raising the possibility of the prevention of perinatal GBS disease through maternal immunization (38).
Nine capsular types are recognized: Ia, Ib and II to VIII, along with a new provisional serotype IX, recently proposed (19). Comparison of the capsular locus genes suggested that the structural diversity of the capsular polysaccharide is associated with the genetic diversity of the capsular locus, possibly driven by horizontal gene transfer (9, 24). Capsular serotyping has been the classical method used in epidemiological studies to differentiate GBS isolates, although further characterization of GBS diversity includes the use of a broad range of DNA-based typing methods, such as restriction fragment length polymorphisms (RFLP), pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST). Both PFGE and MLST have provided new clues about the population structure of S. agalactiae, particularly the recognition of diverse lineages among serotype III that were shown to differ in virulence potential and tropism (16, 25, 26, 31, 41). Although the distinction of lineages within a particular serotype has proved useful, a complete correlation between capsular type and the lineages defined by MLST was not found (4, 21, 22). Moreover, whole-genome comparative analysis of isolates expressing different serotypes showed that they sometimes share more genes than strains of the same serotype, suggesting a serotype-independent clustering of strains (43). These observations support the hypothesis that closely and divergently related clones may share the genes coding for a particular capsular type, suggesting that exchange of capsular genes in vivo may have occurred (16, 21, 22). We refer to these phenomena here as capsular switching in vivo, recognizable by the expression of different serotypes and the presence of different capsular loci in otherwise indistinguishable isolates when sampling a set of 11 loci distributed in the genome.
The changes at the capsular locus were proposed to be driven by the equilibrium between the selective pressure imposed by host immunity and conservation of a particular capsular polysaccharide, as an adaptive advantage of virulent clones (4, 9, 21). Capsular switching by homologous recombination would be facilitated by the organization of the locus encoding the capsular polysaccharide synthesis genes (cps), where the highly variable serotype determining region (cpsG-cpsK) is flanked by conserved genes (9, 24). This led to the suggestion that genetic exchange of the central part of the cps operon could be driving capsular switching (9, 22). According to Luan et al., who specifically addressed this issue, horizontal transfer of capsular genes occurs at a high level within a population without restriction to genetic background. The authors of that study also suggest that since only advantageous combinations of genotype-serotype persist, these altered serotypes, due to capsular switching, are recognized at a lower frequency among stable clones (21).
Capsular switching is well established in other streptococcal species such as Streptococcus pneumoniae, where spontaneous in vivo capsular transformation events were observed and characterized (28, 34). In contrast to GBS, S. pneumoniae is naturally transformable, and this is widely believed to be responsible for the ease with which this species exchanges DNA. Capsular switching may have serious impact in pneumococcal vaccination programs since it may provide the selective pressure for virulent genotypes to switch capsules and escape vaccine coverage (6), and a similar response could be seen with a future introduction of GBS vaccination (38).
The aim of the present study was to evaluate the concordance between serotype and the clusters defined by PFGE and to further characterize any putative transformants to establish unequivocally that capsular switching occurs in GBS. We combined PFGE with the analysis of multiple genes spread across the GBS genome in order to identify capsular transformants and concluded that capsular switching events occur less frequently than previously thought.
MATERIALS AND METHODS
Bacterial isolates.
We studied a collection of 463 GBS isolates isolated between December 1999 and December 2004 in 11 Portuguese hospitals. The isolates were recovered from neonates (invasive infections [defined by the isolation of GBS from a normally sterile fluid]) and adults (invasive and noninvasive infections and asymptomatic colonization), some of which were previously characterized (12, 23, 25). Isolates were identified to the species level by Gram stain, colony morphology, catalase test, and the commercial latex agglutination technique Slidex Strepto B (bioMérieux, Marcy l'Etoile, France).
Serotyping.
Capsular serotyping was done by slide agglutination using sera for types Ia, Ib, and II to VIII (hemolytic Streptococcus typing antisera for group B; Seiken, Japan), as previously described (12). Further confirmation of serotyping results was carried out by a latex agglutination assay with a GBS serotyping kit (Essum, Umeå, Sweden) according to the manufacturer's instructions or by the capillary precipitation method (40) with type II, III, and V sera (Statens Serum Institute, Copenhagen, Denmark).
Pulsed-field gel profiling and MLST.
Total bacterial DNA of the strains was isolated, digested with SmaI, and separated by PFGE as previously described (25). PFGE patterns were compared by using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) to create dendrograms by the unweighted pair-group method with arithmetic averages (UPGMA). The Dice similarity coefficient was used with optimization and position tolerance settings of 1.0 and 1.5, respectively. PFGE-based clusters were defined as isolates with ≥80% relatedness on the dendrogram (25).
MLST was performed by sequencing seven housekeeping genes as described previously (16), and sequence type (ST) identification was done by using the S. agalactiae MLST database (http://pubmlst.org/sagalactiae). Alleles and sequence types not previously described were deposited at the S. agalactiae MLST database. Clonal complex analysis was done by using the entire GBS MLST database and goeBURST (13).
Surface protein gene profile.
Total bacterial DNA was isolated by treatment of the cells with mutanolysin and boiling. A multiplex PCR assay was performed for direct identification of GBS alpha-protein-like genes, as described elsewhere (10). This assay allowed the determination of the following GBS surface protein genes directly by the analysis of the amplicon size: the alpha-C protein gene (bca); the epsilon protein gene (eps); and the rib, alp2/3, and alp4 genes. Later, we used the sequences of the surface protein antigen genes alp2 and alp3 deposited at GenBank to set up a similar multiplex PCR assay to differentiate these two genes by direct evaluation of the amplicon size. Oligonucleotide primers, their target sites and sequences are listed in Table 1. For detection of the β-antigen gene (bac), a PCR assay was performed as described by previously (27).
TABLE 1.
Oligonucleotide primers designed in this study
| Primer | Target |
Sequence (5′→3′) | Amplicon size (bp)b | |
|---|---|---|---|---|
| Gene | Accession no. (position)a | |||
| alp2/3-D | alp2/alp3 | AF208158 (1070)/AF245663 (1311) | CATTAACCGTCACTCCAGAGCAAC | |
| alp2-R | alp2 | AF208158 (1582) | CTTCATCTGTTGACTTATCTGGATAG | 513* |
| alp3-R | alp3 | AF245663 (1637) | CTTTTGGTTCGTTGCTATCCTTAAG | 327* |
| PI1-UP | gbs80 | AE009948 (632375) | GGTCGTCGATGCTCTGGATTC | 881 |
| PI1-DN | gbs80 | AE009948 (633255) | GTTGCCCAGTAACAGCTTCTCC | 881 |
| PI2a-UP | gbs67 | AE009948 (1417645) | CTATGACACTAATGGTAGAAC | 575 |
| PI2a-DN | gbs67 | AE009948 (1417071) | CACCTGCAATAGACATCATAG | 575 |
| PI2b-UP | san_1519 | AAJR01000022 (10160) | ACACGACTATGCCTCCTCATG | 721 |
| PI2b-DN | san_1519 | AAJR01000022 (10880) | TCTCCTACTGGAATAATGACAG | 721 |
| PI1_all-UP | sal_0710 | NZ_AAJP01000027 (23718) | ACCTATGTTGCTGATTCGGCTGAAAATG | 684† |
| PI1_all-DN | sal_0709 | NZ_AAJP01000027 (23035) | TACGGACACTTTCTAGTGCCTTTGGATC | 684† |
| neuB1-UP | neuB | EF990364 (10162) | CAAGCGGTGAATATTTTACG | 2,797 |
| neuA-DN | neuA | EF990364 (12958) | CATTGCTTCCTTTATATGCCATG | |
| neuB2-UP | neuB | EF990364 (10653) | CTTGGGACAAGAAGCGCAAG | |
| neuC1-UP | neuC | EF990364 (11153) | GCGTTGATTTATAATGTCCCAG | |
| neuC2-UP | neuC | EF990364 (11672) | CTGATTGGTAATTCGTCTTCTGG | |
| neuD-UP | neuD | EF990364 (12133) | GAAGATGGCTCAATAGATGCAG | |
That is, the position of primer from the 5′ end.
*, together with the primer alp2/3-D; †, size if the isolate lacks the PI-1 islet (if this locus is present, a 16.7-kb fragment would be expected).
Pilus-associated gene profile.
The genes encoding pili in GBS are located within two distinct loci in different regions of the genome, designated pilus islands 1 and 2 (PI-1 and PI-2), the later presenting two distinct variants, PI-2a and PI-2b (35). We designed a multiplex PCR targeting the sortase genes to identify the pilus islands present in each isolate by direct evaluation of the amplification product size. Oligonucleotide primers, their target sites, and sequences are listed in Table 1. Briefly, 1 μl of DNA template lysate prepared as described above was added to the PCR mixture containing 1× PCR buffer (Promega, Madison, WI), 200 μM deoxynucleoside triphosphates (MBI Fermentas, Vilnius, Lithuania), 0.5 μM primers, 1.5 mM MgCl2, and 1 U of GoTaq DNA polymerase (Promega) in a final volume of 50 μl. PCR conditions for amplification were as follows: 30 cycles of 95°C for 1 min, 52°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min in a Biometra (Goettingen, Germany) T-gradient thermocycler. The PCR products were analyzed by electrophoresis in a 2% (wt/vol) agarose gel.
In order to confirm that the PI-1-negative isolates did not carry the pilus pathogenicity islet or parts of it, a set of primers that flanked the PI-1 locus were designed with the aim of amplifying the entire pilus islet. Oligonucleotide primers, their target sites, and sequences are listed in Table 1. All isolates that were negative for the presence of the PI-1 sortase gene were subjected to PCR with primers PI1_all-UP and PI1_all-DN. If the pilus locus was absent, the expected PCR product using these primers would be 684 bp. The PCR mixture and PCR conditions were similar to those described above except for the annealing temperature that was 60°C.
Capsule gene cluster RFLP analysis.
Sequence variability within a region of the cps gene cluster (cpsG-neuA) results in different patterns between serotypes upon enzyme digestion, allowing the unambiguous identification of the cps gene clusters. We performed a previously described PCR-based restriction fragment length polymorphism (RFLP) method suitable for genetic serotyping of S. agalactiae (24) to identify which cps cluster was present in each isolate.
Southern blot hybridization.
Chromosomal DNA fragments digested by SmaI and separated by PFGE were transferred to nylon membranes (Hybond N+; Amersham, Uppsala, Sweden) by using the vacuum gene system (Pharmacia LKB Biotech, Uppsala, Sweden), and membranes were hybridized to specific DNA probes labeled with an ECL Direct Labeling System (Amersham, Uppsala, Sweden). Both hybridization and labeling were performed according to the manufacturer's instructions.
Ia, Ib, and II to V serotype-specific DNA probes were generated by PCR with template DNA from reference strains, as previously described (3), and hybridized against all isolates tested. The PCR products were purified by using a High-Pure PCR product purification kit (Roche, Manheim, Germany) and labeled according to the manufacturer's instructions.
Sequencing of the cps locus.
Sequence analysis of the partial GBS cps gene cluster, namely, the conserved regions cpsD-cpsG and neuB-neuA, reveal polymorphisms that allow the identification of the various cps loci. A previously published method described a GBS serotype identification method by PCR and sequencing of the cpsD-cpsG region. In this region 56 variable sites suitable for molecular serotyping were identified (18). For the neuB-neuA region, we used a similar PCR and sequencing analysis approach. The oligonucleotide primers designed for the amplification and sequencing of this region are listed in Table 1. We aligned the sequences obtained from at least three isolates expressing serotypes Ia, Ib, and II to V in order to identify the variable sites characteristic of each serotype. Later, we analyzed the data obtained by sequencing of the neuB-neuA region of the isolates being tested for concordance with these polymorphisms. Both of these methods allow the confirmation of the conventional capsular serotyping method and also the evaluation of the possibility of partial cps gene cluster transfer and recombination among isolates of different serotypes.
Typing concordance and DNA sequence analysis.
Wallace coefficient (W) provides a quantitative measure of the clustering concordance between different typing methods (7, 30). We assessed the Wallace coefficient in our collection to determine the concordance between PFGE-based clustering and serotyping. Simpson's index of diversity (SID) was calculated to evaluate the diversity found among the isolates studied (8). These calculations were performed at the comparing partitions website (www.comparingpartitions.info).
Analysis of DNA sequences was done by using Vector NTI 10 (Invitrogen, Carlsbad, CA) and GATA (29) software.
Nucleotide sequence accession number.
The sequence of the cpsG-cpsH region of strain 312754 was deposited in GenBank under accession GQ457335.
RESULTS
Serotyping and PFGE.
Among the 463 isolates we found 16 different PFGE clusters (≥5 isolates), of which the major five accounted for nearly 60% of the isolates (Fig. 1). The remaining isolates (n = 67) were included in minor PFGE groups (containing four or fewer isolates) or had unique profiles and were not considered for further analysis. The SID for the classification of the isolates in PFGE clusters was 0.901 (95% confidence interval [CI] = 0.887 to 0.916), indicating that the collection analyzed is very diverse.
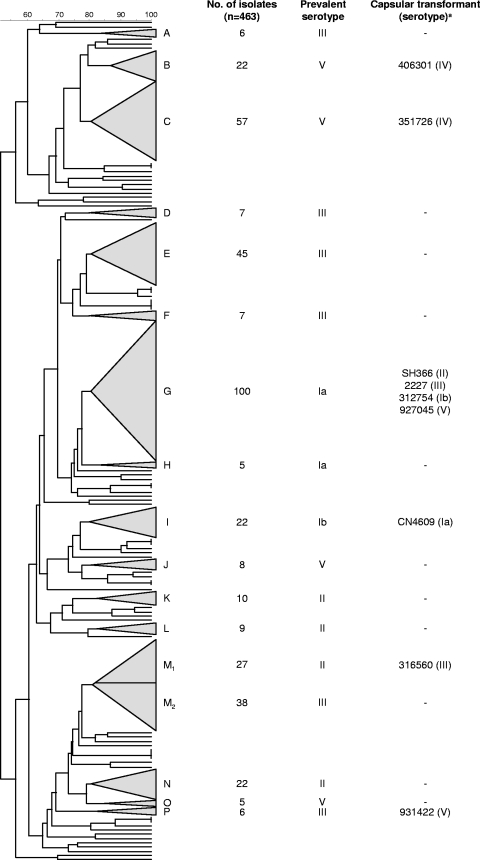
FIG. 1.
Dendrogram analysis of the PFGE profiles of 463 GBS isolates. UPGMA and the Dice coefficient (indicated as percentages in the scale above the dendrogram) were used to construct the dendrogram. Each PFGE cluster (defined as a group of ≥5 isolates with a Dice coefficient of ≥80% in the dendrogram) is represented by a triangle with size proportional to the number of isolates included in the cluster. The clusters are identified by capital letters outside each triangle. Putative capsular transformants were selected based on exhibiting a serotype different from the dominant serotype of the PFGE cluster. M1 and M2 are two sublineages of the same cluster, defined at >80% similarity in the dendrogram.
A few isolates were initially classified as nontypeable by serotyping because of positive agglutination with both types II and III sera when using sera from Denka Seiken. Serotyping of these isolates by the capillary precipitation method (using Statens Serum Institute sera) and by a latex agglutination assay using Essum Probiotics sera, revealed that in all cases the isolates only presented a positive reaction with serotype II specific serum. This phenotype was genetically confirmed by RFLP and Southern hybridization (the latter excluding the possibility of the strains carrying two capsular loci), further confirming that the correct identification was serotype II. With this in mind, all isolates previously classified as either serotype II or III using Denka Seiken sera were independently confirmed by using either of the other sera and conventional serotyping methodologies to exclude any possibility of error. We believe these observations are the result of the cross-reaction between type II and III sera, a possibility admitted in the latest literature provided by the manufacturer. This can lead to errors in serotyping if not further confirmed by other sera or typing methods.
In general, serotype was associated with the overall genotype defined by PFGE. The Wallace coefficient (W) relating the PFGE clusters with the serotype (W = 0.720, 95% CI = 0.666 to 0.775) was high, indicating that isolates grouped in the same PFGE cluster frequently also share the same serotype. However, some clusters were found to contain isolates with serotypes that were different from the most frequently found serotype in the cluster, suggesting that capsular switching could have occurred (n = 27/463 [5.8%]). Confirmation of the serotype of these isolates using conventional serotyping methods revealed in 18 cases that the original serotype was in error. However, we did confirm the serotype of nine isolates (n = 9/463 [1.9%]) that differed from the most prevalent serotype in the same PFGE cluster, and these cases were further studied (Fig. 1).
Every putative transformant was compared to two other isolates in the same PFGE cluster having the prevalent serotype of the cluster and PFGE profiles as similar as possible (maximum of three different bands). Each set of three isolates (the putative transformant and the two isolates used for comparison) was characterized for genes distributed across the GBS genome (Fig. 2) and for confirmation of the genetic determinants of the capsular polysaccharide.
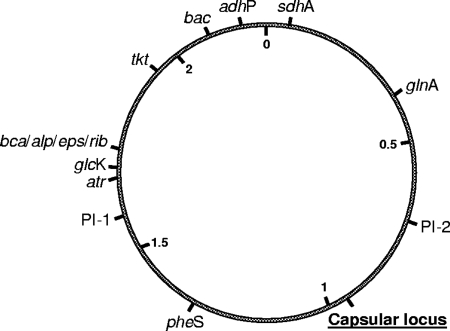
FIG. 2.
Genome map of S. agalactiae indicating loci of interest. The relative positions of housekeeping genes characterized in the MLST scheme (pheS, atr, tkt, glnA, sdhA, glcK, and adhP), genes identified for surface protein gene profiling (same relative positions for bca, alp2, alp3, alp4, rib, or eps), bac, regions characterized for pilus island profiling PI-1 and PI-2 (same relative positions for PI-2a or PI-2b), and the capsular locus region are depicted. Numbers inside the circle represent the scale in megabases.
MLST, surface protein gene profiling, and pilus-associated gene profiling.
In the nine sets of isolates studied the STs found (Table 2) were associated with serotypes in agreement with previous observations (17, 21, 25). With no exceptions, all isolates were positive for only one alpha or alpha-like protein gene and occasionally the isolates were also positive for the β-antigen gene (Table 2). None of the isolates tested were positive for the alp2 or alp4 genes. The isolates were also tested for the presence of the genetic determinants of the pilus-like structures, and all were found to contain at least one of the identified islands. The PI-2a variant was present in all isolates, whereas PI-1 was found together with PI-2a in 16 isolates, and none were positive for the PI-2b variant (Table 2).
TABLE 2.
Detailed characterization of the putative transformants and other isolates from the same PFGE cluster
| Set | PFGE profile (cluster) | Isolate | Serotype | MLST (ST) | Surface protein gene profile | Sequencinga |
RFLP (cpsG-neuA) | Hybridization-positive fragment size (kb)b | Hybridization probeb | Pilus island gene profile | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cpsD-cpsG | neuB-neuA | ||||||||||
| S1 | Identical (G) | SH366 | II | 24 | bca | II | II | II | 450 | II | PI-2a |
| 707618 | Ia | 24 | bca | Ia | Ia | Ia | 450 | Ia | PI-2a | ||
| 12631 | Ia | 24 | bca | Ia | Ia | Ia | 450 | Ia | PI-2a | ||
| S2 | Identical (I) | CN4609 | Ia | 10 | bca + bac | Ia | Ia | Ia | 582 | Ia | PI-1 + PI-2a |
| 275586 | Ib | 10 | bca + bac | Ib | Ib | Ib | 582 | Ib | PI-1 + PI-2a | ||
| 321927 | Ib | 10 | bca + bac | Ib | Ib | Ib | 582 | Ib | PI-1 + PI-2a | ||
| S3 | Identical (G) | 312754 | Ib | 24 | bca | Ia | Ia | Iac | 436.5 | Ia | PI-2a |
| 333425 | Ia | 24 | bca | Ia | Ia | Ia | 436.5 | Ia | PI-2a | ||
| 401936 | Ia | 24 | bca | Ia | Ia | Ia | 436.5 | Ia | PI-2a | ||
| S4 | Identical (M1) | 316560 | III | 295 | rib + bac | III | III | III | 500 | III | PI-1 + PI-2a |
| 768348 | II | 28 | rib + bac | II | II | II | 500 | II | PI-1 + PI-2a | ||
| 850409 | II | 28 | rib + bac | II | II | II | 500 | II | PI-1 + PI-2a | ||
| S5 | One-band difference (B) | 406301 | IV | 2 | eps | IV | IV | IV | 300 | IV | PI-1 + PI-2a |
| 424176 | V | 1 | alp3 | V | V | V | 300 | V | PI-1 + PI-2a | ||
| 345352 | V | 1 | alp3 | V | V | V | 300 | V | PI-1 + PI-2a | ||
| S6 | Three-band difference (C) | 351726 | IV | 2 | eps | IV | IV | IV | 300 | IV | PI-1 + PI-2a |
| 314460 | V | 2 | eps | V | V | V | 339.5 | V | PI-1 + PI-2a | ||
| 371990 | V | 2 | eps | V | V | V | 339.5 | V | PI-1 + PI-2a | ||
| S7 | Three-band difference (G) | 927045 | V | 23 | eps | V | V | V | 582 | V | PI-2a |
| 401207 | Ia | 23 | eps | Ia | Ia | Ia | 582 | Ia | PI-2a | ||
| 314927 | Ia | 23 | eps | Ia | Ia | Ia | 582 | Ia | PI-2a | ||
| S8 | Three-band difference (P) | 931422 | V | 19 | eps + bac | V | V | V | 582 | V | PI-1 + PI-2a |
| 950541 | III | 19 | rib | III | III | III | 582 | III | PI-1 + PI-2a | ||
| 880372 | III | 19 | rib | III | III | III | 582 | III | PI-1 + PI-2a | ||
| S9 | Three-band difference (G) | 2227 | III | 396 | rib | III | III | III | 535.5 | III | PI-1 + PI-2a |
| SH436 | Ia | 24 | bca | Ia | Ia | Ia | 500 | Ia | PI-2a | ||
| 342399 | Ia | 24 | bca | Ia | Ia | Ia | 500 | Ia | PI-2a | ||
The capsular type compatible with the single nucleotide polymorphisms and insertions found using the methodology described in the text is indicated.
That is, the size of the fragment in the PFGE profile that presented a positive hybridization signal and the corresponding serotype identified by the probe (see the text).
One fragment different from the characteristic serotype Ia RFLP profile.
Sequencing of the conserved regions of the cps locus.
All 55 single nucleotide polymorphisms and one repetitive sequence suitable for molecular serotyping in the cpsD-cpsG region described previously (18) were analyzed, and a molecular capsular type was assigned to each isolate according to this methodology. A similar analysis was done for the neuB-neuA region. The single nucleotide polymorphism patterns found were characteristic of the serotype identified by using conventional immunological techniques for all isolates except strain 312754 in set S3 (Table 2).
Southern hybridization and capsule gene cluster RFLP analysis.
The Southern blot hybridization methodology used to probe the capsular locus aimed at determining if a serotype-specific gene of the cps locus was present in each isolate and, if so, its location in the chromosome. This information was essential to exclude the possibility of “binary encapsulated” strains (1) or the generation of hybrid cps loci that could cross-react with two existing sera. We observed that all isolates tested presented only one capsular locus, as hybridization occurred with a single probe and in a single chromosomal fragment. We also observed that the positive hybridization fragment was of the same size in sets S1 to S5 and S7 to S8 but not for sets S6 and S9 (Table 2). In the latter cases one of the three different bands in the PFGE profiles of the isolates included the fragment with the capsular locus, suggesting a possible link between capsular transformation and the differences in PFGE profile.
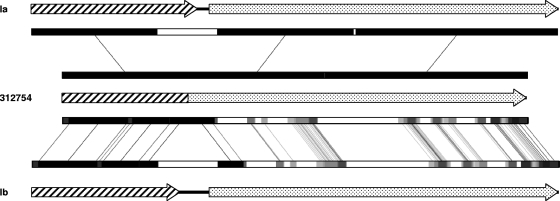
RFLP of the cps region was concordant with the phenotypically determined serotype in all isolates with the exception of isolate 312754 (phenotypically serotype Ib), whose pattern was different from the predicted pattern for serotype Ib isolates. In fact, the cps RFLP pattern was very similar to the one expected for serotype Ia isolates, with the exception of a single fragment (1,135 bp), which was smaller in strain 312754. An in silico analysis suggested that this fragment corresponded to the cpsG-cpsH region. Sequencing of this region showed two deletions (one of 197 bp and a smaller one of 6 bp) in isolate 312754 that generated a smaller fragment (932 bp instead of 1,135 bp) in the restriction profile. The deletion resulted in the fusion of the cpsG and cpsH genes without compromising the reading frame, and leading to the possible production of a fusion protein lacking 23 amino acids of the C-terminal region of cpsG, 9 amino acids of the N-terminal region of cpsH, and 2 amino acids close to the C-terminal region of cpsH (Fig. 3).
FIG. 3.
Diagram representing the DNA sequence of the cpsG-cpsH region of the capsular locus of S. agalactiae serotypes Ia and Ib compared to that of strain 312754. The cpsG and cpsH genes are represented by dashed and dotted arrows, respectively. The cpsG-cpsH fusion in strain 312754 is predicted to generate a fusion protein since the reading frame is maintained (see the text). The fragments with high DNA identity are represented by black boxes, whereas lower identity fragments are represented by lighter shades of gray. Similar fragments are connected by lines.
DISCUSSION
Clonal structure of the S. agalactiae population.
PFGE analysis showed a diverse population of GBS isolates with 16 different PFGE clusters identified (Fig. 1). Such diversity is in agreement with the isolates being collected from multiple sources, in different years and age groups and associated with different severity of infections and colonization. Nevertheless, we observed that most isolates were grouped in clusters sharing the same serotype, implying an association between the serotype and the PFGE-based genotypes reflected in a high value of the Wallace coefficient and this coefficient would be even higher if we excluded nontypeable isolates (W = 0.838, 95% CI = 0.804 to 0.872). Nontypeable isolates frequently produce small amounts of capsule polysaccharide that easily revert to fully encapsulated variants (36) or result from mutations or from the insertion of mobile genetic elements in the cps locus, leading to absent or reduced expression of the capsule polysaccharide due to loss of function of the proteins involved in its biosynthesis (33). Thus, nontypeable isolates probably do not represent capsular switching events but carry cps loci very similar to those of other isolates in the same PFGE cluster. Therefore, their inclusion in the calculation of the Wallace coefficients leads to an artificial underestimation of the concordance between the classification in PFGE clusters and serotype.
Still, in certain PFGE clusters, the presence of isolates whose serotype is different from the prevalent serotype in the cluster, but having similar PFGE profiles, suggested that capsular switching by horizontal gene transfer of the capsular genes could have recently occurred. The number of putative capsular transformant isolates in the present study was much larger when we first analyzed the PFGE profiles (5.8%), and this value was in line with previous studies that addressed this issue (21). However, only nine isolates (2% of the total isolates) could be confirmed by a second round of traditional serotyping using different methods and sera. This significant decrease in putative capsule switching events can be explained by the fact that serotyping using phenotypic methods is particularly prone to errors due to user-dependent evaluation of an agglutination reaction that relies heavily on the experience of the user. On the other hand, the specificity of the polyclonal sera used may also be an issue in particular situations, as discussed previously. In fact, a validation study to evaluate the quality of serotyping of S. pneumoniae, another species of the Streptococcus genus, in European reference laboratories identified a 5% error rate (20). Similarly, our data argue that serotyping errors in S. agalactiae may be frequent and contribute significantly to overestimate capsular switching events in this species. However, studies using capsular genotyping, and therefore not subjected to these errors, have also found evidence of capsular switching (32).
We used the classification in PFGE clusters as the main screening method to identify putative transformation events. To further identify recent capsular switching events, we characterized isolates with no more than three different bands in their PFGE profiles, which could result from a single genetic event meaning that the isolates are very closely related (42). For capsular switching to have occurred recently, the isolate with an unusual serotype must have been identical to other isolates of the same bacterial clone, all sharing the same genotype as defined by MLST, surface protein, and pili island gene profile.
Capsular switching.
Table 2 summarizes the characterization of the nine isolates identified as potentially resulting from capsular switching. Whereas some of the isolates had exactly the same genotype as the isolates they were being compared to (sets S1 and S2), other capsular transformants were found having different genotypic properties compared to additional isolates in the same PFGE cluster (sets S3 to S9). Sets S1 and S2 provide strong evidence for recent capsular switching since in these cases the potential transformants share the same genotype as defined by PFGE, ST, surface protein gene, and pilus island gene profile as the comparator isolates, with the only difference residing in the capsular locus.
Initially, we considered the possibility that SH366 had acquired only the type II-specific genes cpsG-cpsK, as proposed in the literature (9, 22), and so the capsular locus of the putative transformant would be expected to be a hybrid between the recipient and the donor cps loci. For instance, strain SH366 was expected to exhibit features of both serotype II (recently acquired central genes cpsG-cpsK) and Ia (conserved regions of the operon cpsA-cpsF and cpsL-neuA). If that were so, we should find the type Ia-specific polymorphisms when sequencing the conserved regions of SH366 capsular locus. This was not the case, and the entire capsular locus of SH366 was consistent with the type II capsule. These observations led us to conclude that SH366 had acquired the entire capsular locus coding for the type II capsule and that the recombination breakpoints are located outside of the capsular locus. An in silico analysis of the published GBS genomes showed that the regions flanking the capsular locus are extremely conserved among S. agalactiae isolates (data not shown); therefore, homologous recombination in these regions is possible independently of the overall genetic background of the isolates. Moreover, recent data describe the region surrounding the capsular locus as showing evidence of a high density of recombination events corroborating our data, while identifying the transfer of DNA fragments as large as 334 kb (5). The sequence type of SH366 (ST24) is almost exclusively found among serotype Ia isolates (16, 21), with a high prevalence in Portugal (25), and accounts for one of two main lineages of serotype Ia (unpublished data). A similar situation was found in set S2, with strain CN4609 possessing a cps locus entirely consistent with serotype Ia but found in a PFGE cluster of serotype Ib isolates, indicating the acquisition of a large chromosomal fragment encompassing the entire cps locus (Table 2). CN4609 is a representative of ST10, frequently found among serotype Ib isolates but previously not described in serotype Ia isolates (16, 21, 25), further supporting its identification as a capsular transformant. Taken together, these findings provide evidence of recent capsular switching events in SH366 and CN4609.
In set S3, we found that strain 312754 (phenotypically identified as serotype Ib) presented two deletions in a cps locus that was closely related to that of serotype Ia (Fig. 3). These deletions result in the loss of 23 amino acids in the cpsG gene product, a β-1,4-galactosyltransferase, and of 11 amino acids in the cpsH gene product, the capsular polymerase, with the consequent production of a 505-amino-acid fusion protein. Nuclear magnetic resonance studies showed that the types Ia and Ib polysaccharides have an identical composition and differ only in the glycosidic bond of the galactose residue to the N-acetylglucosamine in the lateral chain—type Ia polysaccharide has a β(1→4) bond and type Ib polysaccharide a β(1→3)—and these bonds are critical for the immune specificity of both polysaccharides (15). The characterization of the genes that compose the capsular locus of type Ia attributed to each gene a specific role in assembling the capsular polysaccharide (44). The two β-1,4-galactosyltransferases identified were encoded in the cpsG and cpsJ genes, responsible for the glucose-galactose bond in the polysaccharide backbone and the galactose-N-acetylglucosamine bond of the lateral chain, respectively (44). Since this last bond is the only difference between type Ia and Ib capsules, and considering that the cpsJ gene is not altered in 312754, it would be expectable that, in this isolate, the merging of cpsG and cpsH genes would result in the production of an Ia or Ia-like capsular polysaccharide. However, the data presented here seems to argue that cpsG would be responsible for the β-(1→4)-galactose-N-acetylglucosamine bond. The cps locus of strain 312754 is therefore closely related to that of serotype Ia isolates (Fig. 3) not being clear, in the light of previous knowledge, how the deletions described above justify the expression of a type Ib capsule. Discrepant results between phenotypic serotyping and the capsular locus were described previously for a GBS isolate (19), but no detailed study of its capsular locus was performed. It is also possible that the capsular polysaccharide expressed by strain 312754 is biochemically different from both type Ia and Ib capsules but is somehow recognizable by the Ib polyclonal serum used in conventional serotyping; however, a detailed characterization of this polysaccharide is outside the scope of the present study.
In sets S4 to S9 the isolates express a serotype that is unique in the cluster and show different genetic features compared to other isolates of the same PFGE cluster. These differences are shown in Table 2 and include MLST sequence type (sets S4, S5, and S9), PFGE profile (sets S5 to S9), surface protein gene profile (sets S5, S8, and S9), and the presence of the pilus island-associated genes (set S9). All of the markers used to evaluate the genetic background of the isolates are distributed in an arrangement conserved across serotypes and spread throughout the GBS genome (Fig. 2), assuring that if horizontal gene transfer occurs these should be independently transferred among isolates providing a good measure of the genetic background against which to detect capsular transformation.
In set S4 the isolates have different STs and share the remaining features tested. ST295 and ST28 belong to the same clonal complex (CC19) and are double-locus variants, presenting different alleles for the genes adhP and glnA. Since the distance between these genes is more than 400 kb (Fig. 2) and each is even further distant from the capsular locus, it is unlikely that the event driving capsule switching and the acquisition of the two alleles occurred simultaneously. This implies that at least three independent genetic events occurred between the putative transformant and its ancestors.
In set S5 the putative capsular transformant and the representatives of the cluster show differences in the PFGE profile, the MLST profile, and the surface protein gene profile. ST1 and ST2 are single locus variants in the same clonal complex (CC19) and differ only in the atr allele. The eps and alp3 genes code for epsilon and alpha-like 3 proteins, respectively; both are members of the alpha-like surface protein family and mutually exclusive that have been mapped to the same genomic location. Considering that the surface protein and the atr gene are ∼64 kb apart, both of these genes could be transferred among isolates in the same DNA fragment. In this case, a minimum of two independent genetic events would have to be invoked to explain the differences observed between the putative transformant and its ancestors.
Sets S6 and S7 include isolates that share all of the characteristics tested with the exception of the PFGE profile, which differs in three bands between the putative capsular transformants and the representatives of the PFGE cluster where they were found. In set S6 some of the detected differences in the PFGE profile involve the fragment where the cps locus is found, as demonstrated by Southern hybridization, suggesting that the capsular switching itself could be linked to the changes in the PFGE profile. A single genetic event, involving recombination of a large DNA fragment, could result in both capsular switching and a different PFGE profile. This hypothesis is supported by the successful incorporation of DNA fragments up to 334 kb in S. agalactiae (5). In set S7 the changes in the PFGE profile of the putative transformant do not involve the fragment containing the capsular locus, not suggesting a direct link between the changes in PFGE profile and cps locus, although this cannot also be formally excluded.
In set S8 the isolates differ in their PFGE profiles by three bands and in the surface protein gene profiles. Again, since the genomic locations for the eps or rib genes and the bac gene are more than 350 kb apart and even more distant from the capsular locus, all of these genes should be independently transferred among the isolates. Any of these genetic events could also give rise to the differences observed in the PFGE profiles, but a minimum of three independent recombination events would still be needed to explain the differences between the isolates.
Finally, set S9 comprises isolates showing very different genetic properties and thus unlikely to be closely related. These differences include the PFGE profile, all seven MLST alleles, surface protein, and the pilus island gene profiles. The grouping of these isolates by PFGE seems to have been spurious, and all other evidence argues against capsular transformation.
Capsular switching is thought to contribute to the rise of new serotype-genotype combinations, allowing evasion of immune pressure. However, the success of these new variants is possibly restrained by interactions between the new capsule and the original properties of the isolate. If immune pressure were an overwhelming selective force, we would expect to see the expansion of these capsule switching sublineages. We did detect in our sample a PFGE cluster that contains two sublineages, identified as such by PFGE (i.e., defined as two independent groups of isolates related at >80% similarity in the dendrogram), presenting different serotypes, STs, and surface proteins, suggesting the existence of stable related lineages (Fig. 1 and 3, PFGE cluster M). However, after confirmation of the initial serotyping results, we found only nine isolates that met our initial criteria and, upon more detailed analysis, only a fraction of these could result from recent capsular switching events. This could be due to either a small rate of capsular transformation, the clearing of these variants from the population or a combination of both. The stability of several GBS clones, such as the hypervirulent ST17 clone found among serotype III isolates that is characterized by a specific combination of genetic markers, including mobile genetic elements and virulence genes, that are part of the variable genome and thus not shared by all GBS (2, 4, 21), suggests that the rate of gene exchange in this species may be lower than that of other streptococci. In fact, the PFGE analysis of collections of S. agalactiae isolates shows fewer clones than in other streptococcal species, such as Streptococcus pyogenes (14) and S. pneumoniae (37), a finding consistent with a less diverse clonal structure of the population. On the other hand, genomic analysis of eight fully sequenced strains of S. agalactiae provided evidence for the exchange of large chromosomal fragments, hinting at a high rate of gene exchange through an unusual mechanism (5).
The analysis presented here used several loci and took into account both the core and the variable portions of the genome when analyzing GBS isolates. This approach is expected to allow us to identify changes in the genomic background of the transformants with high resolution, enabling the identification of recent capsular switching events. Our data suggest that serotyping errors may account for a significant proportion of putative capsular transformation events described previously, reinforcing the importance of different methods and sera or a genetic approach in confirming all putative transformation events. Our analysis also provided unambiguous evidence for the existence of capsular transformation in GBS and indicated that the exchange of the entire capsular locus is more widespread than switching restricted to capsule-specific genes. Taken together, our data support the existence of capsular switching in GBS but at a lower frequency than previously suggested.
Acknowledgments
This study was partly supported by a grant from Fundação Calouste Gulbenkian, by Fundação para a Ciência e a Tecnologia (POCI/SAU-ESP/57646/2004) and an unrestricted grant from Glaxo Smithkline Portugal. E.R.M. was supported by a grant from Fundação para a Ciência e a Tecnologia (SFRH/BD/41761/2007).
We thank Francisco Pinto for help with data analysis.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Austrian, R., H. P. Bernheimer, E. E. B. Smith, and G. T. Mills. 1959. Simultaneous production of two capsular polysaccharides by pneumococcus. II. The genetic and biochemical bases of binary capsulation. J. Exp. Med. 110:585-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidet, P., N. Brahimi, C. Chalas, Y. Aujard, and E. Bingen. 2003. Molecular characterization of serotype III group B-streptococcus isolates causing neonatal meningitis. J. Infect. Dis. 188:1132-1137. [DOI] [PubMed] [Google Scholar]
- 3.Borchardt, S. M., B. Foxman, D. O. Chaffin, C. E. Rubens, P. A. Tallman, S. D. Manning, C. J. Baker, and C. F. Marrs. 2004. Comparison of DNA dot blot hybridization and Lancefield capillary precipitin methods for group B streptococcal capsular typing. J. Clin. Microbiol. 42:146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brochet, M., E. Couvé, M. Zouine, T. Vallaeys, C. Rusniok, M. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 8:1227-1243. [DOI] [PubMed] [Google Scholar]
- 5.Brochet, M., C. Rusniok, E. Couvé, S. Dramsi, C. Poyart, P. Trieu-Cuot, F. Kunst, and P. Glaser. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 105:15961-15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brueggemann, A. B., R. Pai, D. W. Crook, and B. Beall. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carriço J. A., F. R. Pinto, C. Simas, S. Nunes, N. G. Sousa, N. Frazão, H. de Lencastre, and J. S. Almeida. 2005. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:5483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carriço, J. A., C. Silva-Costa, J. Melo-Cristino, F. R. Pinto, H. de Lencastre, J. S. Almeida, and M. Ramirez. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cieslewicz, M. J., D. Chaffin, G. Glusman, D. Kasper, A. Madan, S. Rodrigues, J. Fahey, M. R. Wessels, and C. E. Rubens. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creti, R., F. Fabretti, G. Orefici, and C. von Hunolstein. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42:1326-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, H. D., N. Jones, T. S. Whittam, S. Elsayed, N. Bisharat, and C. J. Baker. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J. Infect. Dis. 189:1097-1102. [DOI] [PubMed] [Google Scholar]
- 12.Figueira-Coelho, J., M. Ramirez, M. J. Salgado, and J. Melo-Cristino. 2004. Streptococcus agalactiae in a large Portuguese teaching hospital: antimicrobial susceptibility, serotype distribution, and clonal analysis of macrolide-resistant isolates. Microb. Drug Resist. 10:31-36. [DOI] [PubMed] [Google Scholar]
- 13.Francisco, A. P., M. Bugalho, M. Ramirez, and J. A. Carriço. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friães, A., M. Ramirez, and J. Melo-Cristino. 2007. Nonoutbreak surveillance of group A streptococci causing invasive disease in Portugal identified internationally disseminated clones among members of a genetically heterogeneous population. J. Clin. Microbiol. 45:2044-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennings, H. J., E. Katzenellenbogen, C. Lugowski, and D. L. Kasper. 1983. Structure of native polysaccharide antigens of type Ia and type Ib group B Streptococcus. Biochemistry 22:1258-1264. [DOI] [PubMed] [Google Scholar]
- 16.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, N., K. A. Oliver, J. Barry, R. M. Harding, N. Bisharat, B. G. Spratt, T. Peto, and D. W. Crook. 2006. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B streptococcus is independent of capsular serotype. Clin. Infect. Dis. 42:915-924. [DOI] [PubMed] [Google Scholar]
- 18.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong, F., L. M. Lambertsen, H. Slotved, D. Ko, H. Wang, and G. L. Gilbert. 2008. Use of phenotypic and molecular serotype identification methods to characterize previously nonserotypeable group B streptococci. J. Clin. Microbiol. 46:2745-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konradsen H. B., and Pneumococcus Reference laboratories in Europe. 2005. Validation of serotyping of Streptococcus pneumoniae in Europe. Vaccine 23:1368-1373. [DOI] [PubMed] [Google Scholar]
- 21.Luan, S., M. Granlund, M. Sellin, T. Lagergård, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manning, S. D., M. A. Lewis, A. C. Springman, E. Lehotzky, T. S. Whittam, and H. D. Davies. 2008. Genotypic diversity and serotype distribution of group B streptococcus isolated from women before and after delivery. Clin. Infect. Dis. 46:1829-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins, E. R., C. Florindo, F. Martins, I. Aldir, M. J. Borrego, L. Brum, M. Ramirez, and J. Melo-Cristino. 2007. Streptococcus agalactiae serotype Ib as an agent of meningitis in two adult non-pregnant women. J. Clin. Microbiol. 45:3850-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martins, E. R., J. Melo-Cristino, and M. Ramirez. 2007. Reevaluating the serotype II capsular locus of Streptococcus agalactiae. J. Clin. Microbiol. 45:3384-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins E. R., M. A. Pessanha, M. Ramirez, J. Melo-Cristino, and The Portuguese Group for the Study of Streptococcal Infections. 2007. Analysis of group B streptococcal isolates from infants and pregnant women in Portugal revealing two lineages with enhanced invasiveness. J. Clin. Microbiol. 45:3224-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. U. S. A. 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagano, N., Y. Nagano, and F. Taguchi. 2002. High expression of a C protein beta antigen gene among invasive strains from certain clonally related groups of type Ia and Ib group B streptococci. Infect. Immun. 70:4643-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nesin, M., M. Ramirez, and A. Tomasz. 1998. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J. Infect. Dis. 177:707-713. [DOI] [PubMed] [Google Scholar]
- 29.Nix, D. A., and M. B. Eisen. 2005. GATA: a graphic alignment tool for comparative sequence analysis. BMC Bioinform. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinto, F. R., J. Melo-Cristino, and M. Ramirez. 2008. A confidence interval for the Wallace coefficient of concordance and its application to microbial typing methods. PLoS One 3:e3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quentin, R., H. Huet, F. S. Wang, P. Geslin, A. Goudeau, and R. K. Selander. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 33:2576-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramaswamy, S. V., P. Ferrieri, A. E. Flores, and L. C. Paoletti. 2006. Molecular characterization of nontypeable group B streptococcus. J. Clin. Microbiol. 44:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramaswamy, S. V., P. Ferrieri, L. C. Madoff, A. E. Flores, N. Kumar, H. Tettelin, and L. C. Paoletti. 2006. Identification of novel cps locus polymorphisms in nontypeable group B Streptococcus. J. Med. Microbiol. 55:775-783. [DOI] [PubMed] [Google Scholar]
- 34.Ramirez, M., and A. Tomasz. 1999. Acquisition of new capsular genes among clinical isolates of antibiotic-resistant Streptococcus pneumoniae. Microb. Drug Resist. 5:241-246. [DOI] [PubMed] [Google Scholar]
- 35.Rosini, R., C. D. Rinaudo, M. Soriani, P. Lauer, M. Mora, D. Maione, A. Taddei, I. Santi, C. Ghezzo, C. Brettoni, S. Buccato, I. Margarit, G. Grandi, and J. L. Telford. 2006. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 61:126-141. [DOI] [PubMed] [Google Scholar]
- 36.Sellin, M., C. Olofsson, S. Håkansson, and M. Norgren. 2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serrano, I., J. Melo-Cristino, J. A. Carriço, and M. Ramirez. 2005. Characterization of the genetic lineages responsible for pneumococcal invasive disease in Portugal. J. Clin. Microbiol. 43:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinha, A., T. A. Lieu, L. C. Paoletti, M. C. Weinstein, and R. Platt. 2005. The projected health benefits of maternal group B streptococcal vaccination in the era of chemoprophylaxis. Vaccine 23:3187-3195. [DOI] [PubMed] [Google Scholar]
- 39.Skoff, T. H., M. M. Farley, S. Petit, A. S. Craig, W. Schaffner, K. Gershman, L. H. Harrison, R. Lynfield, J. Mohle-Boetani, S. Zansky, B. A. Albanese, K. Stefonek, E. R. Zell, D. Jackson, T. Thompson, and S. J. Schrag. 2009. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin. Infect. Dis. 49:85-92. [DOI] [PubMed] [Google Scholar]
- 40.Swift, H. F., A. T. Wilson, and R. C. Lancefield. 1943. Typing group a hemolytic streptococci by m precipitin reactions in capillary pipettes. J. Exp. Med. 78:127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi, S., E. E. Adderson, Y. Nagano, N. Nagano, M. R. Briesacher, and J. F. Bohnsack. 1998. Identification of a highly encapsulated, genetically related group of invasive type III group B streptococci. J. Infect. Dis. 177:1116-1119. [DOI] [PubMed] [Google Scholar]
- 42.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto, S., K. Miyake, Y. Koike, M. Watanabe, Y. Machida, M. Ohta, and S. Iijima. 1999. Molecular characterization of type-specific capsular polysaccharide biosynthesis genes of Streptococcus agalactiae type Ia. J. Bacteriol. 181:5176-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]